Abstract
Yersinia enterocolitica biovar 1B is one of a number of strains pathogenic to humans in the genus Yersinia. It has three different type III secretion systems, Ysc, Ysa, and the flagella. In this study, the effect of flagella on biofilm formation was evaluated. In a panel of 31 mutant Y. enterocolitica strains, we observed that mutations that abolish the structure or rotation of the flagella greatly reduce biofilm formation when the bacteria are grown under static conditions. These results were further evaluated by assessing biofilm formation under continuous culture using a flow cell chamber. The results confirmed the important contribution of flagella to the initiation of biofilm production but indicated that there are differences in the progression of biofilm development between static growth and flow conditions. Our results suggest that flagella play a critical role in biofilm formation in Y. enterocolitica.
Yersinia enterocolitica is a food-borne, pathogenic, gram-negative bacterium that causes gastroenteritis. In some cases, septicemia can occur, depending on the health of the host. The most serious illnesses are developed by young children under the age of 1 year (1). Y. enterocolitica is one of three human-pathogenic species in the genus Yersinia that, along with Yersinia pseudotuberculosis, causes gastroenteritis. The third species, Yersinia pestis, is the causative agent of plague. Historically, the formation of biofilms by Y. pestis has been better studied than biofilms of the other two. It has been suggested that Y. pestis biofilms induce starvation of fleas by blocking their digestive tracts. These biofilms seem to affect the fleas but play no role in mammalian pathogenesis, as Y. pestis strains unable to form biofilms are still able to be transmitted by fleas and cause disease (23-25, 53).
There are many studies showing the importance of flagella in biofilm formation in other bacteria (32, 61, 67). In Escherichia coli, a screen showed that 34 of 72 adhesion-deficient strains had changes in motility characteristics (31). Another screen showed that about half the strains that were biofilm defective also exhibited changes in motility function (67), and while motility was a critical factor under certain biofilm formation conditions, chemotaxis was not required (67). Flagella are involved in the adherence of enteropathogenic E. coli to epithelial cells (32), and flagella are also important for biofilm development in Pseudomonas aeruginosa and Aeromonas spp. (61). However, there are other studies that suggest that flagella are not essential for biofilm formation in E. coli (68), P. aeruginosa (47), and Pseudomonas fluorescens (74). These studies suggest that the critical contribution of flagella to biofilm formation is conditional.
Among the three pathogenic Yersinia strains, only Y. enterocolitica and Y. pseudotuberculosis are motile. Y. pestis has a frameshift in flhD, the flagellar-biosynthesis regulator protein, and is therefore nonmotile. The goal of this study was to determine whether flagella and motility were important for biofilm formation in Y. enterocolitica. To address these questions, we used selected conditions for biofilm growth, along with flagellar and motility mutants from our strain collection, and assessed their abilities to form biofilms. Interestingly, we found that mutations that either abolish the structure of the flagella or paralyze the flagella greatly reduce the ability of Y. enterocolitica to establish biofilms on abiotic surfaces.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Y. enterocolitica biovar 1B was used in these experiments. The media used for growth conditions were as follows: M63GM medium is minimal M63 medium supplemented with 0.02% MgSO4 (final concentration) and 0.2% glucose (final concentration) (57, 92); Luria-Bertani broth (LB) was purchased from Becton Dickinson and Company (91, 93); tryptone-yeast extract (TYE) medium is 1% tryptone and 0.5% yeast extract (93). Bacteria were routinely grown on LB plates at 26°C for 2 days prior to the initiation of experiments. Cells were grown overnight in TYE broth at 26°C and then subcultured to an optical density at 600 nm (OD600) of 0.05 in 100 μl of M63GM broth in 96-well polyvinyl chloride (PVC) microplates (this is equivalent to approximately 108 CFU ml−1) (see Fig. 1 and 2). The cells were cultured for 6 h or 24 h at 26°C. The cell growth was measured at an absorbance of 595 nm before the quantification of biofilm formation using an ELx808 microplate reader equipped with KC4 software (Bio-Tek Instruments, Inc., Winooski, VT). The absorbance was measured at 595 nm, rather than 600 nm, because of the filter set present on the plate reader.
FIG. 1.
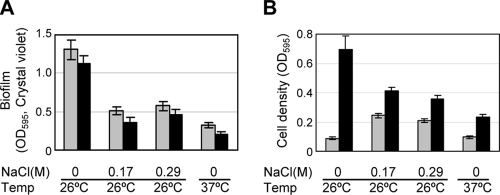
Flagellar expression is important for biofilm formation. Different overnight culture conditions that induced or repressed flagellar expression were evaluated for their effects on biofilm formation. Cultures were grown overnight in 0 M NaCl, 0.17 M NaCl, or 0.29 M NaCl at 26°C or 37°C and then subcultured into M63MG medium and grown for 6 or 24 h at 26°C. (A) Biofilms were assayed by the crystal violet method. (B) Cell density was determined by measuring the OD595 before biofilm production was assessed. The gray bars represent 6-h cultures, and the black bars represent 24-h cultures. The error bars indicate standard deviations.
FIG. 2.
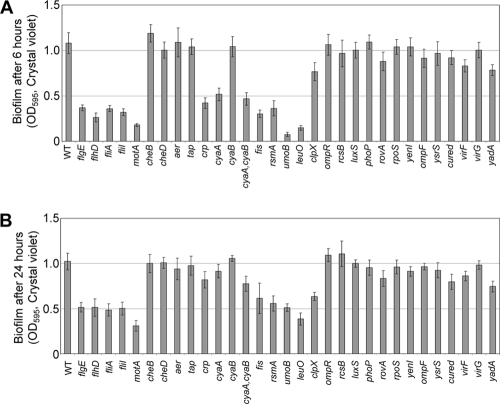
Biofilm formation under static conditions. Selected mutants were grown overnight in TYE medium, subcultured into M63GM medium in 96-well PVC plates, and incubated at 26°C for 6 h (A) or 24 h (B). Biofilms were assayed by the crystal violet method. The error bars indicate the standard deviations. WT, wild type.
Crystal violet assay for quantification of biofilm formation.
Quantification of biofilms was performed by the crystal violet assay, with minor modifications of a previously reported method (62). After cell culture in 96-well PVC microplates, excess cells were removed by aspiration. After three rinses with double-distilled water, 100 μl of 1% crystal violet was added to each well, and the plates were incubated for 15 min at room temperature. The plates were rinsed three times with double-distilled water, after which 100 μl of 95% ethanol was added to each well to solubilize the crystal violet, and the plates were incubated for an additional 15 min. An ELx808 microplate reader equipped with KC4 software (Bio-Tek Instruments, Inc., Winooski, VT) was used to measure the absorbance at 595 nm. Uninoculated wells were used as the blank and were subtracted from each value to calculate the final A595 value.
Motility tests.
Motility tests were preformed as previously described (93). Y. enterocolitica strains were grown for 2 days on TYE plates at 26°C. Colonies of approximately equal sizes were picked from these plates and were stabbed into the centers of fresh TYE plates containing 0.3% agar and incubated at 26°C for 24 h. The plates were photographed to document motility.
Flagellum staining.
Flagella were stained using a slight modification of a previously reported method (42, 55). The stain was made by mixing 10 parts mordant (2 g tannic acid, 10 ml 5% phenol, 10 ml saturated aqueous AlKO8S2·12H2O) with 1 part dye (12% crystal violet in ethanol). The cultures were grown overnight in TYE broth at 26°C. Three microliters of culture in TYE broth, corresponding to an OD600 of 0.5, was applied to a microscope slide and covered with a cover glass (50 mm by 24 mm), after which 10 μl of stain was applied to the edge of the cover glass and drawn into the sample by capillary action. The flagella were observed at ×2,500 magnification using a Zeiss Axioplan microscope and Axiophot software.
Biofilm formation in continuous-culture flow cells.
Flow cells and bubble traps were purchased from the Center for Biomedical Microbiology, Technical University of Denmark, Lyngby, Denmark. The chamber in the flow cell was 1 mm deep by 4 mm wide by 40 mm long. The chamber was sealed with a PVC coverslip. M63GM was used for continuous feeding of medium by a peristaltic pump (model 2058; Watson Marlow, Wilmington, MA). The cells were cultured overnight in TYE broth at 26°C and were then diluted to an OD600 of 0.15 in TYE broth. A 350-μl aliquot of the subculture was inoculated into the flow cell and allowed to attach for 1 hour, after which the flow was initiated at 0.5 ml/minute. At the indicated time points, pictures were taken using bright-field microscopy to document the formation of the biofilms using an Axioplan microscope equipped with an AxioCam camera and AxioVision4 software (Zeiss, Germany).
RESULTS
The expression of flagella in preculture is important for biofilm formation.
Initially, experiments were conducted to establish the conditions for robust biofilm formation by wild-type Y. enterocolitica. We found that cultures grown overnight in TYE broth at 26°C that were then subcultured into M63GM medium exhibited greater biofilm density, as assayed by the crystal violet method, than cultures that had been pregrown in higher-salt media or at 37°C. It has been established that culturing in TYE broth at 26°C leads to flagellar expression in Y. enterocolitica (91, 93). Therefore, based on the well-documented contribution of flagella to biofilm development in other bacteria and the preculture conditions tested, we began to assess the contribution of motility to biofilm formation by Y. enterocolitica. For the experiments shown in Fig. 1, cultures were grown overnight under different flagellum-inducing or -repressing conditions and then subcultured into M63GM medium. Specifically, bacteria were precultured in TYE medium containing sodium chloride at concentrations of 0 M, 0.17 M, and 0.29 M, which reflect the sodium chloride contents of TYE and LB and that which is required to induce the Ysa type III secretion system (TTSS), respectively (91). In addition to salt, we also evaluated the contribution of increased temperature to the repression of flagellar expression and the subsequent effect on biofilm formation by growing the bacteria overnight in TYE broth at 37°C before subculturing them in M63GM medium. We previously established that these growth conditions affect the expression of flagella by monitoring the β-galactosidase activities of lacZYA fusions to flhD, flhB, fliA, and motA (66; B. M. Young, unpublished results). The growth of cells was determined by measuring the absorbance at OD595 (Fig. 1B) before samples were processed by the crystal violet assay to determine levels of biofilm formation (Fig. 1A). The results indicated that conditions that have been shown to repress flagellar expression in Y. enterocolitica also lead to a decrease in biofilm formation (Fig. 1A). This occurred despite the fact that cell growth was more robust under some of the culturing conditions (no salt and 26°C) (Fig. 1B). These observations led us to further examine, by genetic means, whether motility or the presence of flagella itself was required for biofilm production by Y. enterocolitica.
Biofilm formation under static growth conditions.
Y. enterocolitica has three different TTSSs. One of these, the flagellar TTSS, has been extensively studied by our laboratory, and thus, we have a large collection of mutant strains whose flagellar regulation, structure, and motility are affected. From this collection, 31 strains were selected based on motility defects and regulatory function (Tables 1 and 2). Additional mutant strains studied included those with defects in global regulators of Y. enterocolitica virulence and in the two other TTSSs of Y. enterocolitica, the Ysa and Ysc TTSSs. As shown in Table 2, many of these are insertion mutations, and there may be polar effects on the downstream genes in the operon. Biofilm formation (Fig. 2) and motility (Fig. 3) were measured, and the existence of flagella (Fig. 4) was assessed for these selected strains. The results are summarized in Table 3.
TABLE 1.
Y. enterocolitica strains used in this study
| Strain name | Relevant genotype | Method for mutations | Source or reference |
|---|---|---|---|
| JB580v | Serogroup O:8; Nalr ΔyenR (R− M+) | None | 45 |
| GY274 | rovA::mTn5Kn2 inv′-′phoA inv+ | Transposon insertion | 73 |
| GY294 | clpX::pEP185.2 | Plasmid insertion | Laboratory collection |
| GY316 | phoP::pEP185.2 | Plasmid insertion | Laboratory collection |
| GY448 | rsmA::TnMod-R-Kn′ | Transposon insertion | Laboratory collection |
| GY452 | umoB::TnMod-R-Kn′ | Transposon insertion | Laboratory collection |
| GY453 | flhD::TnMod-R-Kn′ | Transposon insertion | Laboratory collection |
| GY512 | ompF::mTn5lacZYA′ | Transposon insertion | —b |
| GY572 | cyaA::TnMod-R-Kn′ | Transposon insertion | 66 |
| GY574 | fliA::TnMod-R-Kn′ | Transposon insertion | —a |
| GY656 | fis::TnMod-R-Kn′ | Transposon insertion | —a |
| GY663 | flgE::TnMod-R-Kn′ | Transposon insertion | —a |
| GY689 | leuO::TnMod-R-Kn′; 60-bp upstream of leuO | Transposon insertion | —a |
| GY744 | fliI::mTn5lacZYA′ | Transposon insertion | —b |
| GY751 | motA::mTn5lacZYA′ | Transposon insertion | —b |
| GY752 | cheD(tsr)::mTn5lacZYA′ | Transposon insertion | —b |
| GY762 | tap(tar)::mTn5lacZYA′ | Transposon insertion | —b |
| GY818 | cheB::mTn5lacZYA′ | Transposon insertion | —b |
| GY854 | aer::mTn5lacZYA′ | Transposon insertion | —b |
| GY939 | ompR::pEP185.2 | Plasmid insertion | —b |
| GY4319 | rpoS::Kn | Drug marker insertion | Laboratory collection |
| GY4419 | ysrS::mTn5lacZYA′ Pcat-yplAB in pTM100 | Transposon insertion | Laboratory collection |
| GY4478 | pYV8081− | Plasmid elimination | 91 |
| GY4493 | yenI::Kn | Drug marker insertion | Laboratory collection |
| GY4531 | crp::Sm′ | Drug marker insertion | 2 |
| GY4548 | virF::TnMod-lacZYA | Transposon insertion | Laboratory collection |
| GY4575 | rcsB::Tn5Mod-lacZYA Pcat-yplAB in pTM100 | Transposon insertion | —b |
| GY4629 | virG::mTn5Kn | Transposon insertion | Laboratory collection |
| GY4647 | luxS::pEP185.2 | Plasmid insertion | Laboratory collection |
| GY5254 | cyaB::pEP185.2 | Plasmid insertion | This study |
| GY5255 | cyaA::TnMod-R-Kn′ cyaB::pEP185.2 | Transposon insertion | This study |
| GY5375 | yadA::pEP185.2 | Plasmid insertion | This study |
S. Petersen and G. M. Young, unpublished data.
K. Venecia and G. M. Young, unpublished data.
TABLE 2.
Functions of mutated genes used in this study
| Strain | Genotype | Function | Reference for function |
|---|---|---|---|
| JB580v | Wild type | 45a | |
| GY663 | flgE-flgF | Flagellar hook protein | 93a |
| GY453 | flhD-flhC | Flagellar biosynthesis regulator protein (class I) | 93a |
| GY574 | fliA-fliZ | Sigma-28; flagellar sigma factor | 41a |
| GY744 | fliI-fliJ | Flagellar assembly protein | 27b, 20e |
| GY751 | motA-motB-cheA | Protein for flagellar rotation energy | 18, 27b |
| GY818 | cheB | Protein methylesterase for chemotaxis | 16, 64, 90b |
| GY752 | cheD | Methyl-accepting chemotaxis protein | 43b |
| GY854 | aer | Putative methyl-accepting chemotaxis protein | 69, 70b |
| GY762 | tap-YE4140 | Methyl-accepting chemotaxis protein IV | 77b |
| GY4531 | crp-YE3956 | Cyclic AMP receptor protein | 66a |
| GY572 | cyaA | Adenylate cyclase A | 66a |
| GY5254 | cyaB | Adenylate cyclase B | 54c |
| GY5255 | cyaA, cyaB | Adenylate cyclases A and B | 66a, 54c |
| GY656 | fis | DNA binding protein | 6b, 44d |
| GY448 | rsmA | RNA binding global posttranscriptional regulator | 34a |
| GY452 | umoB | Putative membrane protein | 22f |
| GY689 | leuO | LysR family transcriptional regulator | 46b |
| GY294 | clpX | ATP-dependent Clp protease ATP binding subunit | 65a |
| GY939 | ompR-envZ | Osmolarity response regulator | 19a |
| GY4575 | rcsB-rcsA-rcsC | Response regulator of two-component system | 85a |
| GY4647 | luxS | Synthase of quorum-sensing autoinducer 2 protein | 75g |
| GY316 | phoP-phoQ-YE1716 | PhoP-PhoQ two-component system | 88a |
| GY274 | rovA | Global regulator of virulence | 21, 73a |
| GY4319 | rpoS | Alternative sigma factor | 4, 35a |
| GY4493 | yenI | Methyltransferase endonuclease | 81a |
| GY512 | ompF | Outer membrane protein F (porin) | 59b, 60e |
| GY4419 | ysrS-YE3561A | Sensor of two-component system | 85, 86a |
| GY4478 | pYV8081− | Virulence plasmid-cured strain | 13a |
| GY4548 | virF-yscA | Transcriptional activator for TTSS | 12a |
| GY4629 | virG | Pilot protein for yscC | 2a |
| GY5375 | yadA-ymoA | Adhesin | 10, 11, 26, 33, 63, 79a |
Y. enterocolitica.
E. coli.
P. aeruginosa.
S. enterica.
S. enterica serovar Typhimurium.
P. mirabilis.
Vibrio harveyi.
FIG. 3.
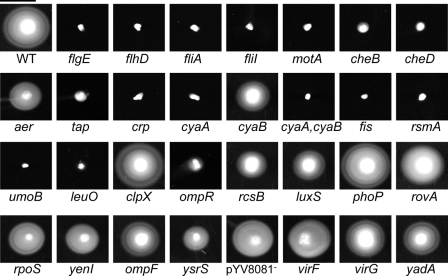
Motility phenotypes in soft agar. Motility in TYE soft agar was assessed by point inoculating bacterial strains and incubating them for 24 h at 26°C. The large white circles indicate bacterial movement from the point of inoculation. Bar (upper left), 1 cm. WT, wild type.
FIG. 4.
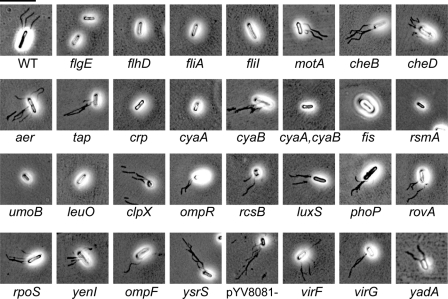
Analysis of flagellar production by microscopy. Flagella were stained as described in Materials and Methods and observed by bright-field microscopy. Flagella appear as black lines attached to the cells. More than 10 cells were observed for each mutant, with representative pictures shown here. Magnification, ×2,500. Bar (upper left), 10 μm.
TABLE 3.
Summary of results
| Genotype | Biofilma
|
Motilityb | Flagellac | |
|---|---|---|---|---|
| 6 h | 24 h | |||
| WTd | +++++ | +++++ | + | + |
| flgE | ++ | +++ | − | − |
| flhD | + | +++ | − | − |
| fliA | ++ | +++ | − | − |
| fliI | ++ | +++ | − | − |
| motA | + | ++ | − | + |
| cheB | +++++ | +++++ | − | + |
| cheD | +++++ | +++++ | − | + |
| aer | +++++ | +++++ | + | + |
| tap | +++++ | +++++ | − | + |
| crp | +++ | ++++ | − | − |
| cyaA | +++ | +++++ | − | − |
| cyaB | +++++ | +++++ | + | + |
| cyaA, cyaB | ++ | ++++ | − | − |
| fis | ++ | +++ | − | − |
| rsmA | ++ | +++ | − | − |
| umoB | + | +++ | − | − |
| leuO | + | ++ | − | − |
| clpX | ++++ | +++ | + | + |
| ompR | +++++ | +++++ | − | + |
| rcsB | +++++ | +++++ | + | + |
| luxS | +++++ | +++++ | + | + |
| phoP | +++++ | +++++ | + | + |
| rovA | ++++ | ++++ | + | + |
| rpoS | +++++ | +++++ | + | + |
| yenI | +++++ | +++++ | + | + |
| ompF | ++++ | +++++ | + | + |
| ysrS | +++++ | +++++ | + | + |
| Cured | +++++ | ++++ | + | + |
| virF | ++++ | ++++ | + | + |
| virG | +++++ | +++++ | + | + |
| yadA | ++++ | ++++ | + | + |
Biofilm production in PVC plates. +, less than 20% of WT; ++, 20 to 40% of WT; +++, 40 to 60% of WT; ++++, 60 to 80% of WT; +++++, more than 80% of WT.
Motility assay. +, motile; −, nonmotile.
Flagellar staining. +, flagella were observed; −, flagella were not observed.
WT, wild type.
We found that strains with mutations in structural genes and in positive regulators of the flagella showed defects in biofilm production. flgE and fliI are genes that encode structural components of the flagella. Mutations in either of these genes resulted in a lack of flagella. The master regulator gene flhD and fliA, a sigma factor gene, are both required for expression of flagella. Mutations in any of these four genes resulted in defective or absent flagella, and each showed a reduction in the amounts of biofilm made after 6 and 24 h (Table 3 and Fig. 2, 3, and 4) under static growth conditions. Additional positive regulators of flagellar expression in Y. enterocolitica include Crp, a cyclic AMP receptor protein, and CyaA, a catabolic regulatory adenylate cyclase (class I). In an earlier study, we established that mutations in crp and cyaA abolish flagellar expression in Y. enterocolitica, as well as affecting Ysa and Ysc TTSS expression (66). Subsequently, a hypothetical adenylate cyclase (class II) gene, cyaB, was discovered, which we included in this study. The crp and cyaA mutants and the cyaA cyaB double mutant do not produce flagella and are not motile; however, the cyaB mutant is unaffected and produces motile flagella (Fig. 3 and 4). When the crp, cyaA, and cyaAcyaB mutants were assessed for biofilm formation, it was found that there was a decrease in biofilms at 6 h. By 24 h, the biofilms made by the mutants approached the levels seen for wild-type Y. enterocolitica. The cyaB mutant displayed wild-type levels of biofilm production at both time points.
Other possible positive regulators of flagellar expression in Y. enterocolitica are Fis, RsmA, and UmoB. Fis, an H-NS protein, is a positive regulator of full flagellar expression in Salmonella enterica serovar Typhimurium (44). Additionally, H-NS proteins have been shown to regulate flagellar expression in E. coli (6). CsrA, a homologue of RsmA in E. coli, (28), and UmoB in Proteus mirabilis (22) both have positive regulatory functions in flagellar expression. Y. enterocolitica with mutations in fis, csrA, or umoB made no flagella, was not motile, and exhibited decreased biofilm formation in the PVC plate assay.
In contrast, mutations in negative regulators of flagellar expression do not affect motility. RcsB is a two-component response regulator of the RcsCBD system and has been shown to be a negative regulator of flagellar expression in E. coli (29). A mutation in Y. enterocolitica rcsB resulted in cells that exhibited wild-type flagellar expression and motility (Fig. 3 and 4). Additionally, biofilm production was unaffected for an rcsB mutant (Fig. 2). These results further establish for Y. enterocolitica that mutations in genes that are positive regulators of flagellar expression reduce the production of biofilms while mutations in genes that encode negative regulators of flagellar expression exhibit wild-type biofilm initiation and development (Table 3).
In addition to regulatory and structural genes, we also looked at genes that control the rotation of the flagella. MotA provides the energy for flagellar rotation, and mutations in motA result in intact, but paralyzed, flagella. Biofilms made by the motA strain were greatly reduced compared to those of wild-type Y. enterocolitica (Table 3 and Fig. 2, 3, and 4). These results begin to suggest that not only the presence of flagella, but motility itself, is a key to biofilm production.
A set of chemotaxis mutants was examined to further elucidate the contribution of flagella versus flagellar rotation to the formation of biofilms. These mutants make flagella that rotate (Fig. 2) but cannot control the direction of movement, and as a result, cheB, cheD, and tap mutants appear to be nonmotile in soft agar (Fig. 3). Another gene involved in chemotaxis, aer, had a motile phenotype in soft agar (Fig. 3). Each of these four chemotaxis mutants was able to produce biofilms comparable to those seen for wild-type Y. enterocolitica at 6 and 24 h. The ompR gene encodes an osmolarity response regulator that negatively regulates flagellar expression through flhDC in E. coli (76). Additionally, OmpR of E. coli is homologous to CheY of S. enterica serovar Typhimurium (78), suggesting it may play a role in chemotaxis. An ompR mutant in Y. enterocolitica made flagella (Fig. 4) but had a phenotype similar to those of cheB, cheD, and tap mutants in soft agar (Fig. 3), suggesting that it may be affected for chemotaxis but not for flagellar expression in general. The ompR mutant was unaffected for biofilm production (Fig. 2). Further experiments will be required to determine whether OmpR plays a role in chemotaxis in Y. enterocolitica. However, taken together, these results suggest that random movement of the flagella is sufficient to allow initiation of the biofilm phenomenon, while absence or paralysis of the flagella inhibits the production of biofilms.
Included in our study were several genes known to affect biofilm production in other bacteria. LeuO is a LysR-like transcriptional regulator that has been shown to exhibit decreased biofilm formation in Vibrio cholerae (58). In Y. enterocolitica, a leuO mutant did not have flagella and was not motile, and it subsequently produced biofilms in decreased amounts (Fig. 2, 3, and 4). ClpX is an ATP-dependent specificity component of the ClpP serine protease (87), and in P. fluorescens, ClpP mutants were shown to have a defect in biofilm formation (62). In Y. enterocolitica, we found that clpX mutants possessed flagella and were motile but exhibited reduced biofilm formation at 24 h compared with wild-type bacteria (Table 3 and Fig. 2, 3, and 4). It has been shown that ClpX and ClpP affect the expression of many cell surface proteins in Y. enterocolitica, including Ail (65) and the Ysc TTSS (37). This may suggest that motility is not the only significant factor for biofilm formation in Y. enterocolitica.
Another set of mutants used in this study were predicted to be affected for flagellar expression and/or biofilm formation based on their functions in other bacteria. LuxS is involved in quorum sensing and is required for motility in Vibrio alginolyticus (82). RpoS, an alternate sigma factor, is required for biofilm formation in V. cholerae (89). Interestingly, while both of these mutants exhibited a slight motility defect (Fig. 3), both were able to form biofilms comparable to those of wild-type Y. enterocolitica at 6 and 24 h (Fig. 2). This may suggest that the genetic cascade leading to the process of initiation and development of biofilms by Y. enterocolitica is distinct from those of other bacteria. These results also support our initial findings, which suggested that rotating flagella, rather than chemotaxis, are required for the formation of biofilms by Y. enterocolitica.
Finally, we screened a number of mutants predicted to be unaffected for flagellar expression. These mutants served as internal controls for the biofilm assay (Fig. 2). The mutant strains studied included those with genes that encode proteins for regulators (ysrS, virF, phoP, rovA, and yenI), outer membrane proteins (ompF), an adhesin involved in virulence (yadA), and a pilot protein for a structural component of the Ysc system (virG) and a virulence plasmid-cured strain (defective for the Ysc TTSS apparatus). Each of these mutants was unaffected for biofilm formation, flagellar expression, and motility under the tested conditions (Fig. 2, 3, and 4), suggesting that mutations made by insertion of plasmids or transposons per se do not affect the biofilm process (Table 3). Additionally, these results suggest that the other TTSSs (Ysa and Ysc) do not play a role in biofilm formation under the tested conditions. It should be noted that YadA would not be expressed under the conditions tested. It is interesting that mutants exhibiting small defects in motility (Fig. 3, aer, cyaB, rcsB, luxS, rpoS, yenI, and ysrS) were still able to make robust biofilms. This indicates that minimal motility is sufficient to establish contact with abiotic surfaces, leading to subsequent development of biofilms.
Biofilm formation in continuous medium flow.
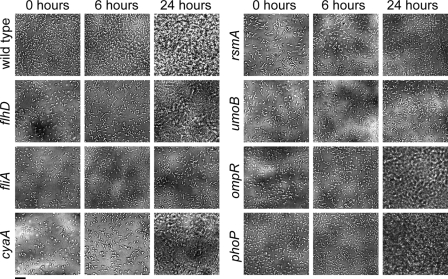
Flow cell chambers were used to investigate the effects of selected mutations on biofilm formation (Fig. 5). Based on phenotypes from the PVC plate assay (Fig. 2), eight Y. enterocolitica mutant strains were selected to demonstrate negative or positive effects on biofilm production under a continuous flow of culture medium. Normally, Y. enterocolitica attaches to the surface of the flow cell and begins to form microcolonies by 6 h. At 24 h, well-developed biofilms can be seen (Fig. 5). In contrast, strains with mutations in fliA, rsmA, and umoB exhibited reduced attachment to PVC coverslips at 0 h, and even after 24 h, there was no apparent microcolony or biofilm formation. flhD and cyaA mutants, which completely lacked flagella, showed delays in attachment at 0 h and microcolony formation at 6 h but, surprisingly, had robust biofilms at 24 h (Fig. 5). This result for the flhD mutation is in contrast to the results seen in the PVC plate assays (Fig. 2). In agreement with the PVC plate assay, an ompR mutant exhibited wild-type biofilm production. Also in agreement with results from the static biofilm assay, phoP mutants made thick biofilms that were indistinguishable from those seen for wild-type Y. enterocolitica. These results confirmed the importance of the rotation of flagella for the initiation of biofilm production by Y. enterocolitica but suggested that there are other compensatory factors which may allow the formation of biofilms under flow conditions.
FIG. 5.
Biofilm formation in flow cell chambers. Selected strains were inoculated into flow cell chambers to assess biofilm formation under continuous medium flow. Pictures were taken at 0-, 6-, and 24-h time points. Normal development proceeds through the following stages: (i) attachment of individual bacteria at 0 h, (ii) microcolony formation by 6 h, and (iii) mature biofilm structures present at 24 h. Magnification, ×1,000. Bar (lower left), 10 μm.
DISCUSSION
The ability of cells to form biofilms is significant in that many physiological changes occur (49) that can serve to protect the bacteria from various stresses (30). During the course of this study, we evaluated many different mutant strains of Y. enterocolitica for the ability to form biofilms under both static and continuous-flow culture conditions. The importance of flagella in biofilm formation was demonstrated previously (51, 56, 67), and it has been shown that flagella contribute not only to early attachment, but also to the maturation of biofilms (51, 72). Our results extend these observations for Y. enterocolitica and indicate that mutants lacking flagella, or possessing paralyzed flagella, exhibit a defect in biofilm formation under static conditions, while chemotaxis mutants that possess flagella with unorganized motility make wild-type biofilms. We suggest that the presence of rotating flagella is required for the bacteria to gain access to microniches on abiotic surfaces, where attachment is favored. Additionally, the ability of Y. enterocolitica to establish biofilms may be a means of environmental survival, rather than a mechanism of virulence. This hypothesis is suggested because the higher temperature encountered in an infected host is predicted to repress the expression of flagella.
The regulation of biofilm development in Y. enterocolitica seems to be different from that which occurs in Y. pestis. Y. pestis is nonmotile and requires hmsHFRS, hmsT, and hmsP to form biofilms in the flea midgut (38, 39). hmsHFRS and hmsP were identified in the complete genomic sequence of Y. enterocolitica, but hmsT was absent (80). hmsT encodes a diguanylate cyclase, which positively regulates exopolysaccharide production and is required for biofilm formation by Y. pestis (39). The absence of hmsT in Y. enterocolitica and the nonmotile phenotype of Y. pestis suggest that the mechanisms of biofilm development used by these two species are distinctly different.
Biofilm development by Y. enterocolitica does not require either LuxS or RpoS. This is in contrast to many other bacterial species. For example, Lactobacillus rhamnosus, Campylobacter jejuni, Y. pestis, and Vibrio sp. with mutations in luxS all exhibit significant defects in biofilm formation (7, 50, 71). However, E. coli strain 536 (5) and the Y. enterocolitica strain tested in this study were not dependent on LuxS for biofilm formation. RpoS also plays a complex role in the regulation of biofilms. In S. enterica and Vibrio sp., mutations in rpoS repressed the development of biofilms (8), and in E. coli MG1655, the rpoS mutation repressed the maturation of biofilms (36). In contrast, the rpoS mutation enhanced biofilm formation in E. coli K-12 and Pseudomonas sp. strain KL28 (14, 94). In this study, the rpoS mutant did not exhibit any apparent change in biofilm formation, motility, or flagellar expression, suggesting Y. enterocolitica may use a different genetic cascade for biofilm development.
Quorum sensing is central to biofilm development for many bacteria (9, 17, 48, 52), but it appears that Y. enterocolitica does not share this requirement. We suggest this because mutations in genes known to play a role in quorum sensing, such as luxS, rpoS, and yenI, do not show a reduced biofilm phenotype under the conditions tested. This is striking, because it has been documented that a mutation in yenI delays motility in Y. enterocolitica and results in reduced expression of FleB, one of the flagellin subunits (3). It is possible that this delay in motility can eventually be overcome and that the presence of shortened, but rotating, flagella is sufficient to establish what appears to be wild-type biofilm development by 6 h. Experiments specifically addressing the role of quorum sensing in biofilm formation by Y. enterocolitica will be needed to address these issues.
The flhD mutant under continuous-flow conditions represents an exception to the hypothesis that motility is required for the formation of biofilms by Y. enterocolitica. It would be predicted that a nonmotile strain would be completely deficient in the ability to form biofilms. However, we observed a low initial attachment of the flhD mutant to the abiotic surface, little or no microcolony formation by 6 h, and then, surprisingly, distinct biofilm structures at 24 h. This discrepancy between static and flow conditions is not fully understood but may result from altered genetic signaling (unlike in the plate assay, flow cells do not become nutrient depleted) or altered expression of surface-exposed proteins on the bacteria, including adhesins (15) and exopolysaccharides (67).
The results obtained for the cyaA and crp mutants suggest that flagella are not the only determining factor for the development of biofilms by Y. enterocolitica. Because these mutants are completely defective for flagellar expression, it would be expected that no biofilms would be formed under static or flow conditions. In fact, what was observed was that there was very little biofilm present at 6 h (under both static and flow conditions), but by 24 h, the mutants exhibited wild-type levels of biofilms (Fig. 2). It should be noted that the crp and cyaA mutants exhibited a growth defect in complex media, such as LB and TYE. However, when the mutants were grown in M63GM medium for the biofilm experiments, the cell densities reached levels that were 30% higher than that of wild-type Y. enterocolitica (data not shown). It is possible that the biofilm produced by these mutants at 24 h simply reflects a higher cell density. However, a recent study indicated that mutations in crp and cyaA may indeed affect biofilm formation. In Serratia marcescens, cyaA and crp regulate biofilm production by responding to glucose (40). Normally, Crp and CyaA upregulate flagella as glucose is depleted, and an increase in biofilm formation occurs. crp and cyaA mutants are no longer able to respond to glucose, and these strains show upregulated expression of the type I fimbriae encoded by fimABCD, which then results in a large increase in biofilm formation (40). A search of the Y. enterocolitica genome revealed the presence of fimA. In Y. enterocolitica cyaA and crp mutant strains, in which no flagella are expressed, we hypothesize that fimbriae may eventually lead to the initiation of attachment to the abiotic surface and to development of robust biofilms. To date, no studies have been performed to determine whether type I fimbriae encoded by fimABCD are expressed in Y. enterocolitica stain 8081. It will be interesting to pursue experiments to elucidate the contribution of flagella versus fimbriae to biofilm development by Y. enterocolitica under different growth conditions.
Y. enterocolitica has a wide tolerance for temperature and growth conditions, suggesting it can survive in soil, water, and other environmental sites. Besides the common presence of Y. enterocolitica in the tonsils of swine (83, 84), environmental reservoirs for the bacteria have not yet been determined. The conditions that induce expression of the flagella (ambient temperature and low salt concentrations) suggest that biofilms may be a means for Y. enterocolitica to persist in environmental niches.
Acknowledgments
We thank members of the Young laboratory for helpful discussions relating to this project.
This work was supported by University of California, Davis, start-up funds awarded to G.M.Y.
Footnotes
Published ahead of print on 7 July 2008.
REFERENCES
- 1.Abdel-Haq, N., B. Asmar, W. Abuhammour, and W. Brown. 2000. Yersinia enterocolitica infection in children. Pediatr. Infect. Dis. J. 19:954-958. [DOI] [PubMed] [Google Scholar]
- 2.Allaoui, A., R. Scheen, C. Lambert de Rouvroit, and G. R. Cornelis. 1995. VirG, a Yersinia enterocolitica lipoprotein involved in Ca2+ dependency, is related to exsB of Pseudomonas aeruginosa. J. Bacteriol. 177:4230-4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atkinson, S., C.-Y. Chang, R. E. Sockett, M. Camara, and P. Williams. 2006. Quorum sensing in Yersinia enterocolitica controls swimming and swarming motility. J. Bacteriol. 188:1451-1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Badger, J. L., and V. L. Miller. 1995. Role of RpoS in survival of Yersinia enterocolitica to a variety of environmental stresses. J. Bacteriol. 177:5370-5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beloin, C., K. Michaelis, K. Lindner, P. Landini, J. Hacker, J.-M. Ghigo, and U. Dobrindt. 2006. The transcriptional antiterminator RfaH represses biofilm formation in Escherichia coli. J. Bacteriol. 188:1316-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bertin, P., E. Terao, E. H. Lee, P. Lejeune, C. Colson, A. Danchin, and E. Collatz. 1994. The H-NS protein is involved in the biogenesis of flagella in Escherichia coli. J. Bacteriol. 176:5537-5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bobrov, A. G., S. W. Bearden, J. D. Fetherston, A. A. Khweek, K. D. Parrish, and R. D. Perry. 2007. Functional quorum sensing systems affect biofilm formation and protein expression in Yersinia pestis. Adv. Exp. Med. Biol. 603:178-197. [DOI] [PubMed] [Google Scholar]
- 8.Cabeza, M. L., A. Aguirre, F. C. Soncini, and E. G. Vescovi. 2007. Induction of RpoS degradation by the two-component system regulator RstA in Salmonella enterica. J. Bacteriol. 189:7335-7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carniol, K., and M. S. Gilmore. 2004. Signal transduction, quorum-sensing, and extracellular protease activity in Enterococcus faecalis biofilm formation. J. Bacteriol. 186:8161-8163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.China, B., B. T. N′Guyen, M. de Bruyere, and G. R. Cornelis. 1994. Role of YadA in resistance of Yersinia enterocolitica to phagocytosis by human polymorphonuclear leukocytes. Infect. Immun. 62:1275-1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.China, B., M. P. Sory, B. T. N′Guyen, M. De Bruyere, and G. R. Cornelis. 1993. Role of the YadA protein in prevention of opsonization of Yersinia enterocolitica by C3b molecules. Infect. Immun. 61:3129-3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cornelis, G., C. Sluiters, C. L. de Rouvroit, and T. Michiels. 1989. Homology between virF, the transcriptional activator of the Yersinia virulence regulon, and AraC, the Escherichia coli arabinose operon regulator. J. Bacteriol. 171:254-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cornelis, G. R., A. Boland, A. P. Boyd, C. Geuijen, M. Iriarte, C. Neyt, M.-P. Sory, and I. Stainier. 1998. The virulence plasmid of Yersinia, an antihost genome. Microbiol. Mol. Biol. Rev. 62:1315-1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Corona-Izquierdo, F. P., and J. Membrillo-Hernández. 2002. A mutation in rpoS enhances biofilm formation in Escherichia coli during exponential phase of growth. FEMS Microbiol. Lett. 211:105-110. [DOI] [PubMed] [Google Scholar]
- 15.Danese, P. N., L. A. Pratt, and R. Kolter. 2000. Exopolysaccharide production is required for development of Escherichia coli K-12 biofilm architecture. J. Bacteriol. 182:3593-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dang, C. V., M. Niwano, J. Ryu, and B. L. Taylor. 1986. Inversion of aerotactic response in Escherichia coli deficient in cheB protein methylesterase. J. Bacteriol. 166:275-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davies, D. G., M. R. Parsek, J. P. Pearson, B. H. Iglewski, J. W. Costerton, and E. P. Greenberg. 1998. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280:295-298. [DOI] [PubMed] [Google Scholar]
- 18.Dean, G. E., R. M. Macnab, J. Stader, P. Matsumura, and C. Burks. 1984. Gene sequence and predicted amino acid sequence of the motA protein, a membrane-associated protein required for flagellar rotation in Escherichia coli. J. Bacteriol. 159:991-999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dorrell, N., S.-R. Li, P. H. Everest, G. Dougan, and B. W. Wren. 1998. Construction and characterisation of a Yersinia enterocolitica O:8 ompR mutant. FEMS Microbiol. Lett. 165:145-151. [DOI] [PubMed] [Google Scholar]
- 20.Dreyfus, G., A. W. Williams, I. Kawagishi, and R. M. Macnab. 1993. Genetic and biochemical analysis of Salmonella typhimurium FliI, a flagellar protein related to the catalytic subunit of the F0F1 ATPase and to virulence proteins of mammalian and plant pathogens. J. Bacteriol. 175:3131-3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dube, P. H., S. A. Handley, P. A. Revell, and V. L. Miller. 2003. The rovA mutant of Yersinia enterocolitica displays differential degrees of virulence depending on the route of infection. Infect. Immun. 71:3512-3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dufour, A., R. B. Furness, and C. Hughes. 1998. Novel genes that upregulate the Proteus mirabilis flhDC master operon controlling flagellar biogenesis and swarming. Mol. Microbiol. 29:741-751. [DOI] [PubMed] [Google Scholar]
- 23.Eisen, R. J., S. W. Bearden, A. P. Wilder, J. A. Montenieri, M. F. Antolin, and K. L. Gage. 2006. Early-phase transmission of Yersinia pestis by unblocked fleas as a mechanism explaining rapidly spreading plague epizootics. Proc. Natl. Acad. Sci. USA 103:15380-15385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eisen, R. J., J. L. Lowell, J. A. Montenieri, S. W. Bearden, and K. L. Gage. 2007. Temporal dynamics of early-phase transmission of Yersinia pestis by unblocked fleas: secondary infectious feeds prolong efficient transmission by Oropsylla montana (Siphonaptera: Ceratophyllidae). J. Med. Entomol. 44:672-677. [DOI] [PubMed] [Google Scholar]
- 25.Eisen, R. J., A. P. Wilder, S. W. Bearden, J. A. Montenieri, and K. L. Gage. 2007. Early-phase transmission of Yersinia pestis by unblocked Xenopsylla cheopis (Siphonaptera: Pulicidae) is as efficient as transmission by blocked fleas. J. Med. Entomol. 44:678-682. [DOI] [PubMed] [Google Scholar]
- 26.El Tahir, Y., and M. Skurnik. 2001. YadA, the multifaceted Yersinia adhesin. Int. J. Med. Microbiol. 291:209-218. [DOI] [PubMed] [Google Scholar]
- 27.Fernandez, L. A., and J. Berenguer. 2000. Secretion and assembly of regular surface structures in Gram-negative bacteria. FEMS Microbiol. Rev. 24:21-44. [DOI] [PubMed] [Google Scholar]
- 28.Fettes, P. S., V. Forsbach-Birk, D. Lynch, and R. Marre. 2001. Overexpresssion of a Legionella pneumophila homologue of the E. coli regulator csrA affects cell size, flagellation, and pigmentation. Int. J. Med. Microbiol. 291:353-360. [DOI] [PubMed] [Google Scholar]
- 29.Francez-Charlot, A., B. Laugel, A. Van Gemert, N. Dubarry, F. Wiorowski, M. Castanié-Cornet, C. Gutierrez, and K. Cam. 2003. RcsCDB His-Asp phosphorelay system negatively regulates the flhDC operon in Escherichia coli. Mol. Microbiol. 49:823-832. [DOI] [PubMed] [Google Scholar]
- 30.Fux, C. A., J. W. Costerton, P. S. Stewart, and P. Stoodley. 2005. Survival strategies of infectious biofilms. Trends Microbiol. 13:34-40. [DOI] [PubMed] [Google Scholar]
- 31.Genevaux, P., S. Muller, and P. Bauda. 1996. A rapid screening procedure to identify mini-Tn10 insertion mutants of Escherichia coli K-12 with altered adhesion properties. FEMS Microbiol. Lett. 142:27-30. [DOI] [PubMed] [Google Scholar]
- 32.Giron, J. A., A. G. Torres, E. Freer, and J. B. Kaper. 2002. The flagella of enteropathogenic Escherichia coli mediate adherence to epithelial cells. Mol. Microbiol. 44:361-379. [DOI] [PubMed] [Google Scholar]
- 33.Gripenberg-Lerche, C., M. Skurnik, L. Zhang, K. O. Soderstrom, and P. Toivanen. 1994. Role of YadA in arthritogenicity of Yersinia enterocolitica serotype O:8: experimental studies with rats. Infect. Immun. 62:5568-5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heeb, S., S. A. Kuehne, M. Bycroft, S. Crivii, M. D. Allen, D. Haas, M. Camara, and P. Williams. 2006. Functional analysis of the post-transcriptional regulator RsmA reveals a novel RNA-binding site. J. Mol. Biol. 355:1026-1036. [DOI] [PubMed] [Google Scholar]
- 35.Iriarte, M., I. Stainier, and G. R. Cornelis. 1995. The rpoS gene from Yersinia enterocolitica and its influence on expression of virulence factors. Infect. Immun. 63:1840-1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ito, A., T. May, K. Kawata, and S. Okabe. 2008. Significance of rpoS during maturation of Escherichia coli biofilms. Biotechnol. Bioeng. 99:1462-1471. [DOI] [PubMed] [Google Scholar]
- 37.Jackson, M. W., E. Silva-Herzog, and G. V. Plano. 2004. The ATP-dependent ClpXP and Lon proteases regulate expression of the Yersinia pestis type III secretion system via regulated proteolysis of YmoA, a small histone-like protein. Mol. Microbiol. 54:1364-1378. [DOI] [PubMed] [Google Scholar]
- 38.Jarrett, C. O., E. Deak, K. E. Isherwood, P. C. Oyston, E. R. Fischer, A. R. Whitney, S. D. Kobayashi, F. R. DeLeo, and B. J. Hinnebusch. 2004. Transmission of Yersinia pestis from an infectious biofilm in the flea vector. J. Infect. Dis. 190:783-792. [DOI] [PubMed] [Google Scholar]
- 39.Jones, H. A., J. W. Lillard, and R. D. Perry. 1999. HmsT, a protein essential for expression of the haemin storage (Hms+) phenotype of Yersinia pestis. Microbiology 145:2117-2128. [DOI] [PubMed] [Google Scholar]
- 40.Kalivoda, E. J., N. A. Stella, D. M. O'Dee, G. J. Nau, and R. M. Q. Shanks. 2008. The cyclic AMP-dependent catabolite repression system of Serratia marcescens mediates biofilm formation through regulation of type 1 fimbriae. Appl. Environ. Microbiol. 74:3461-3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kapatral, V., J. W. Olson, J. C. Pepe, V. L. Miller, and S. A. Minnich. 1996. Temperature-dependent regulation of Yersinia enterocolitica class III flagellar genes. Mol. Microbiol. 19:1061-1071. [DOI] [PubMed] [Google Scholar]
- 42.Kearns, D. B., and R. Losick. 2003. Swarming motility in undomesticated Bacillus subtilis. Mol. Microbiol. 49:581-590. [DOI] [PubMed] [Google Scholar]
- 43.Kehry, M. R., T. G. Doak, and F. W. Dahlquist. 1985. Aberrant regulation of methylesterase activity in cheD chemotaxis mutants of Escherichia coli. J. Bacteriol. 161:105-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kelly, A., M. D. Goldberg, R. K. Carroll, V. Danino, J. C. D. Hinton, and C. J. Dorman. 2004. A global role for Fis in the transcriptional control of metabolism and type III secretion in Salmonella enterica serovar Typhimurium. Microbiology 150:2037-2053. [DOI] [PubMed] [Google Scholar]
- 45.Kinder, S. A., J. L. Badger, G. O. Bryant, J. C. Pepe, and V. L. Miller. 1993. Cloning of the YenI restriction endonuclease and methyltransferase from Yersinia enterocolitica serotype O8 and construction of a transformable R-M+ mutant. Gene 136:271-275. [DOI] [PubMed] [Google Scholar]
- 46.Klauck, E., J. Böhringer, and R. Hengge-Aronis. 1997. The LysR-like regulator LeuO in Escherichia coli is involved in the translational regulation of rpoS by affecting the expression of the small regulatory DsrA-RNA. Mol. Microbiol. 25:559-569. [DOI] [PubMed] [Google Scholar]
- 47.Klausen, M., A. Heydorn, P. Ragas, L. Lambertsen, A. Aaes-Jorgensen, S. Molin, and T. Tolker-Nielsen. 2003. Biofilm formation by Pseudomonas aeruginosa wild type, flagella and type IV pili mutants. Mol. Microbiol. 48:1511-1524. [DOI] [PubMed] [Google Scholar]
- 48.Kong, K.-F., C. Vuong, and M. Otto. 2006. Staphylococcus quorum sensing in biofilm formation and infection. Int. J. Med. Microbiol. 296:133-139. [DOI] [PubMed] [Google Scholar]
- 49.Lazazzera, B. A. 2005. Lessons from DNA microarray analysis: the gene expression profile of biofilms. Curr. Opin. Microbiol. 8:222-227. [DOI] [PubMed] [Google Scholar]
- 50.Lebeer, S., T. L. A. Verhoeven, M. Perea Velez, J. Vanderleyden, and S. C. J. De Keersmaecker. 2007. Impact of environmental and genetic factors on biofilm formation by the probiotic strain Lactobacillus rhamnosus GG. Appl. Environ. Microbiol. 73:6768-6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lemon, K. P., D. E. Higgins, and R. Kolter. 2007. Flagellar motility is critical for Listeria monocytogenes biofilm formation. J. Bacteriol. 189:4418-4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li, Y.-H., N. Tang, M. B. Aspiras, P. C. Y. Lau, J. H. Lee, R. P. Ellen, and D. G. Cvitkovitch. 2002. A quorum-sensing signaling system essential for genetic competence in Streptococcus mutans is involved in biofilm formation. J. Bacteriol. 184:2699-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lillard, J. W., Jr., S. W. Bearden, J. D. Fetherston, and R. D. Perry. 1999. The haemin storage (Hms+) phenotype of Yersinia pestis is not essential for the pathogenesis of bubonic plague in mammals. Microbiology 145:197-209. [DOI] [PubMed] [Google Scholar]
- 54.Lory, S., M. Wolfgang, V. Lee, and R. Smith. 2004. The multi-talented bacterial adenylate cyclases. Int. J. Med. Microbiol. 293:479-482. [DOI] [PubMed] [Google Scholar]
- 55.Mayfield, C. I., and W. E. Inniss. 1977. A rapid, simple method for staining bacterial flagella. Can. J. Microbiol. 23:1311-1313. [DOI] [PubMed] [Google Scholar]
- 56.Merritt, J. H., K. M. Brothers, S. L. Kuchma, and G. A. O'Toole. 2007. SadC reciprocally influences biofilm formation and swarming motility via modulation of exopolysaccharide production and flagellar function. J. Bacteriol. 189:8154-8164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 58.Moorthy, S., and P. I. Watnick. 2005. Identification of novel stage-specific genetic requirements through whole genome transcription profiling of Vibrio cholerae biofilm development. Mol. Microbiol. 57:1623-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nakae, T. 1976. Identification of the outer membrane protein of that produces transmembrane channels in reconstituted vesicle membranes. Biochem. Biophys. Res. Commun. 71:877-884. [DOI] [PubMed] [Google Scholar]
- 60.Nakae, T. 1976. Outer membrane of Salmonella. Isolation of protein complex that produces transmembrane channels. J. Biol. Chem. 251:2176-2178. [PubMed] [Google Scholar]
- 61.O'Toole, G. A., and R. Kolter. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30:295-304. [DOI] [PubMed] [Google Scholar]
- 62.O'Toole, G. A., and R. Kolter. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28:449-461. [DOI] [PubMed] [Google Scholar]
- 63.Paerregaard, A., F. Espersen, and M. Skurnik. 1991. Role of the Yersinia outer membrane protein YadA in adhesion to rabbit intestinal tissue and rabbit intestinal brush border membrane vesicles. APMIS 99:226-232. [DOI] [PubMed] [Google Scholar]
- 64.Parkinson, J. S. 1976. cheA, cheB, and cheC genes of Escherichia coli and their role in chemotaxis. J. Bacteriol. 126:758-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pederson, K., S. Carlson, and D. Pierson. 1997. The ClpP protein, a subunit of the Clp protease, modulates ail gene expression in Yersinia enterocolitica. Mol. Microbiol. 26:99-107. [DOI] [PubMed] [Google Scholar]
- 66.Petersen, S., and G. M. Young. 2002. Essential role for cyclic AMP and its receptor protein in Yersinia enterocolitica virulence. Infect. Immun. 70:3665-3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pratt, L. A., and R. Kolter. 1998. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol. Microbiol. 30:285-293. [DOI] [PubMed] [Google Scholar]
- 68.Prigent-Combaret, C., G. Prensier, T. T. Le Thi, O. Vidal, P. Lejeune, and C. Dorel. 2000. Developmental pathway for biofilm formation in curli-producing Escherichia coli strains: role of flagella, curli and colanic acid. Environ. Microbiol. 2:450-464. [DOI] [PubMed] [Google Scholar]
- 69.Pruss, B. M., J. W. Campbell, T. K. Van Dyk, C. Zhu, Y. Kogan, and P. Matsumura. 2003. FlhD/FlhC is a regulator of anaerobic respiration and the Entner-Doudoroff pathway through induction of the methyl-accepting chemotaxis protein Aer. J. Bacteriol. 185:534-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rebbapragada, A., M. S. Johnson, G. P. Harding, A. J. Zuccarelli, H. M. Fletcher, I. B. Zhulin, and B. L. Taylor. 1997. The Aer protein and the serine chemoreceptor Tsr independently sense intracellular energy levels and transduce oxygen, redox, and energy signals for Escherichia coli behavior. Proc. Natl. Acad. Sci. USA 94:10541-10546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Reeser, R. J., R. T. Medler, S. J. Billington, B. H. Jost, and L. A. Joens. 2007. Characterization of Campylobacter jejuni biofilms under defined growth conditions. Appl. Environ. Microbiol. 73:1908-1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Reisner, A., J. A. J. Haagensen, M. A. Schembri, E. L. Zechner, and S. Molin. 2003. Development and maturation of Escherichia coli K-12 biofilms. Mol. Microbiol. 48:933-946. [DOI] [PubMed] [Google Scholar]
- 73.Revell, P. A., and V. L. Miller. 2000. A chromosomally encoded regulator is required for expression of the Yersinia enterocolitica inv gene and for virulence. Mol. Microbiol. 35:677-685. [DOI] [PubMed] [Google Scholar]
- 74.Robleto, E. A., I. Lopez-Hernandez, M. W. Silby, and S. B. Levy. 2003. Genetic analysis of the AdnA regulon in Pseudomonas fluorescens: nonessential role of flagella in adhesion to sand and biofilm formation. J. Bacteriol. 185:453-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schauder, S., and B. L. Bassler. 2001. The languages of bacteria. Genes Dev. 15:1468-1480. [DOI] [PubMed] [Google Scholar]
- 76.Shin, S., and C. Park. 1995. Modulation of flagellar expression in Escherichia coli by acetyl phosphate and the osmoregulator OmpR. J. Bacteriol. 177:4696-4702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Slocum, M. K., and J. S. Parkinson. 1985. Genetics of methyl-accepting chemotaxis proteins in Escherichia coli: null phenotypes of the tar and tap genes. J. Bacteriol. 163:586-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Stock, A., D. E. Koshland, and J. Stock. 1985. Homologies between the Salmonella typhimurium CheY protein and proteins involved in the regulation of chemotaxis, membrane protein synthesis, and sporulation. Proc. Natl. Acad. Sci. 82:7989-7993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tertti, R., M. Skurnik, T. Vartio, and P. Kuusela. 1992. Adhesion protein YadA of Yersinia species mediates binding of bacteria to fibronectin. Infect. Immun. 60:3021-3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Thomson, N. R., S. Howard, B. W. Wren, M. T. G. Holden, L. Crossman, G. L. Challis, C. Churcher, K. Mungall, K. Brooks, T. Chillingworth, T. Feltwell, Z. Abdellah, H. Hauser, K. Jagels, M. Maddison, S. Moule, M. Sanders, S. Whitehead, M. A. Quail, G. Dougan, J. Parkhill, and M. B. Prentice. 2006. The complete genome sequence and comparative genome analysis of the high pathogenicity Yersinia enterocolitica strain 8081. PLoS Gene 2:e206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Throup, J., M. Camara, G. Briggs, M. Winson, S. Chhabra, B. Bycroft, P. Williams, and G. Stewart. 1995. Characterisation of the yenI/yenR locus from Yersinia enterocolitica mediating the synthesis of two N-acylhomoserine lactone signal molecules. Mol. Microbiol. 17:345-356. [DOI] [PubMed] [Google Scholar]
- 82.Tian, Y., Q. Wang, Q. Liu, Y. Ma, X. Cao, L. Guan, and Y. Zhang. 2008. Involvement of LuxS in the regulation of motility and flagella biogenesis in Vibrio alginolyticus. Biosci. Biotechnol. Biochem. 72:1063-1071. [DOI] [PubMed] [Google Scholar]
- 83.Toma, S., and V. R. Deidrick. 1975. Isolation of Yersinia enterocolitica from swine. J. Clin. Microbiol. 2:478-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tsubokura, M., K. Otsuki, and K. Itagaki. 1973. Studies on Yersinia enterocolitica. I. Isolation of Y. enterocolitica from swine. Nippon Juigaku Zasshi 35:419-424. [DOI] [PubMed] [Google Scholar]
- 85.Venecia, K., and G. M. Young. 2005. Environmental regulation and virulence attributes of the Ysa type III secretion system of Yersinia enterocolitica biovar 1B. Infect. Immun. 73:5961-5977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Walker, K. A., and V. L. Miller. 2004. Regulation of the Ysa type III secretion system of Yersinia enterocolitica by YsaE/SycB and YsrS/YsrR. J. Bacteriol. 186:4056-4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wojtkowiak, D., C. Georgopoulos, and M. Zylicz. 1993. Isolation and characterization of ClpX, a new ATP-dependent specificity component of the Clp protease of Escherichia coli. J. Biol. Chem. 268:22609-22617. [PubMed] [Google Scholar]
- 88.Wren, B., A. Olsen, R. Stabler, and S. Li. 1995. Genetic analysis of phoP and ompR from Yersinia enterocolitica, Yersinia pseudotuberculosis and Yersinia pestis. Contrib. Microbiol. Immunol. 13:318-320. [PubMed] [Google Scholar]
- 89.Yildiz, F. H., X. S. Liu, A. Heydorn, and G. K. Schoolnik. 2004. Molecular analysis of rugosity in a Vibrio cholerae O1 El Tor phase variant. Mol. Microbiol. 53:497-515. [DOI] [PubMed] [Google Scholar]
- 90.Yonekawa, H., H. Hayashi, and J. S. Parkinson. 1983. Requirement of the cheB function for sensory adaptation in Escherichia coli. J. Bacteriol. 156:1228-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Young, B. M., and G. M. Young. 2002. YplA is exported by the Ysc, Ysa, and flagellar type III secretion systems of Yersinia enterocolitica. J. Bacteriol. 184:1324-1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Young, G. M., and V. L. Miller. 1997. Identification of novel chromosomal loci affecting Yersinia enterocolitica pathogenesis. Mol. Microbiol. 25:319-328. [DOI] [PubMed] [Google Scholar]
- 93.Young, G. M., M. J. Smith, S. A. Minnich, and V. L. Miller. 1999. The Yersinia enterocolitica motility master regulatory operon, flhDC, is required for flagellin production, swimming motility, and swarming motility. J. Bacteriol. 181:2823-2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yun, J. I., K. M. Cho, J.-K. Kim, S. O. Lee, K. Cho, and K. Lee. 2007. Mutation of rpoS enhances Pseudomonas sp. KL28 growth at higher concentrations of m-cresol and changes its surface-related phenotypes. FEMS Microbiol. Lett. 269:97-103. [DOI] [PubMed] [Google Scholar]