Abstract
In this study, we describe our results on the evaluation of the ability of different permissive mammalian cell lines to support the biological enrichment of mycoplasma species known to be bacterial contaminants of cell substrates. The study showed that this approach is able to significantly improve the efficiency of mycoplasma detection based on nucleic acid testing or biochemical technologies (e.g., MycoAlert mycoplasma detection). Of 10 different cell lines (Vero, MDBK, HEK-293, Hep-G2, CV-1, EBTr, WI-38, R9ab, MDCK, and High Five) used in the study, only MDCK cell culture was found to support the efficient growth of all the tested mycoplasmas (Mycoplasma arginini, M. bovis, M. fermentans, M. gallinaceum, M. gallisepticum, M. synoviae, M. hominis, M. hyorhinis, M. orale, M. salivarium, and Acholeplasma laidlawii) known to be most frequently associated with contamination of cell substrates and cell lines in research laboratories or manufacturing facilities. The infection of MDCK cells with serial dilutions of each mycoplasma species demonstrated that these common cell line contaminants can be detected reliably after 7-day enrichment in MDCK cell culture at contamination levels of 0.05 to 0.25 CFU/ml. The High Five insect cell line was also found to be able to support the efficient growth of most mycoplasma species tested, except for M. hyorhinis strain DBS1050. However, mycoplasma growth in insect cell culture was demonstrated to be temperature dependent, and the most efficient growth was observed when the incubation temperature was increased from 28°C to between 35 and 37°C. We believe that this type of mycoplasma enrichment is one of the most promising approaches for improving the purity and safety testing of cell substrates and other cell-derived biologics and pharmaceuticals.
Mycoplasmas are known to be broadly distributed cell wall-less prokaryotes with one of the smallest known genomes among the species of the Bacteria (16, 17, 49). Most mycoplasma species are naturally harmless commensals that colonize skin and mucosal surfaces of their natural hosts (8, 45, 48, 49). However, some mycoplasmas have evolved to become pathogenic to either poikilothermic or homeothermic organisms (8, 48, 49). From the regulatory aspects of biosafety, the species of the genera Mycoplasma and Acholeplasma are of major concern as a group of agents known frequently to contaminate primary and continuous cell lines (14, 33, 48, 49, 58, 59). Generally, mycoplasma contamination represents a serious problem for biomedical research laboratories and facilities involved in development and manufacturing of cell-derived biological and pharmaceutical products. Potentially, administration of mycoplasma-containing products could cause an iatrogenic or nosocomial bacterial infection, especially in pediatric, geriatric, or immunocompromised patients (19, 42, 64).
Despite precautionary measures and systematic mycoplasma monitoring, mycoplasma contamination is periodically detected in veterinary and human live virus vaccines or viral stocks produced by multiple manufacturers worldwide (4, 5, 10, 11, 28, 30, 34, 46, 55, 66). Other biologics (nonvaccine products, e.g., commercial diagnostic antigens) were also reported to be contaminated with mycoplasmas (37, 60). Mycoplasma testing of cell substrates and cell-derived biological products in the United States is regulated by Title 21 of the Code of Federal Regulations (21 CFR 610.30) and additionally described in “Points to Consider (PTC) in the Characterization of Cell Lines Used to Produce Biologicals (1993)” (39). The procedure, which includes broth/agar and indicator cell line tests, was developed to detect all mycoplasma species previously isolated from contaminated cell substrates, viral vaccines, and virus stocks. Despite the reported ability of this mycoplasma testing procedure to efficiently detect all possible cell culture mycoplasmal contaminants, the overall testing procedure is time-consuming (a minimum of 28 days) and tedious and might not be suitable for biologics with shelf-lives that are shorter than the turnaround time for testing. In order to reduce the time required for mycoplasma testing, various approaches based on nucleic acid testing (NAT) technologies targeting different genetic markers of Mollicutes have been developed and proposed as potential alternatives to the current methods (15, 31, 33, 54, 56, 57, 63). However, although PCR methods have definite advantages over conventional microbiological methods in terms of analytical sensitivity, simplicity, and turnaround time required for testing, it continues to be unclear whether PCR-based methods can provide a limit of detection comparable or superior to those of conventional methods. Furthermore, NAT methods do not allow for accurate discrimination between viable and nonviable mycoplasma contaminants that might lead to false-positive results in mycoplasma testing (35). From this standpoint, the development of additional pretesting sample preparation procedures able to improve the sensitivity of the NAT-based assay as well as to determine whether mycoplasmas are viable is of high importance to ensure the safety and purity of cell substrates and cell-derived biologics. Biological enrichment is believed to be one of the more promising approaches due to its ability to significantly increase the number of viable mycoplasma cells to levels readily and reliably detected by routine PCR or other molecular methods. Alternatively, mycoplasma species known to be common cell line contaminants can be amplified using either cultivation in broth media (1, 3, 11, 29, 40, 50) or cocultivation with permissive mammalian cell lines (6, 11, 21, 29, 40). However, the use of broth media for mycoplasma enrichment has three inherent shortcomings. First, there are no universal axenic medium and environmental conditions that support equally efficient growth of all known mycoplasma species (isolates). Second, some mycoplasma species or individual strains are fastidious or even uncultivable in broth media (e.g., some strains of Mycoplasma hyorhinis, M. fermentans, M. pneumoniae, M. genitalium, etc.) Finally, the growth of fastidious mycoplasmas in broth media is relatively slow, with culture turnaround times of up to 30 days. In contrast, the use of permissive mammalian cell cultures can avoid the above problems while expediting and simplifying mycoplasma testing procedures in general. The use of cell cultures could efficiently enrich any mycoplasma contaminant to titers of 102 to 103 CFU/ml and above (32, 36), which allows for reliable detection of mycoplasmas by routine single-round NAT-based assays. Usually, these titers of mycoplasmas can be reached in less than 1 week. The feasibility of cell culture enrichment for improving the efficiency of NAT-based mycoplasma testing methods was recently addressed in the latest version of the European Pharmacopeia (monograph 2.6.7).
Recently, the successful use of Vero cell culture for the recovery, detection, and antibiotic susceptibility testing of M. genitalium from clinical specimens was demonstrated (25, 26, 32). At the same time, different cell cultures may exhibit a difference in the ability to support growth of different mycoplasma species. Thus, the ability of Vero cells to support rapid and uniform growth of common mycoplasma cell line contaminants was evaluated (36). Surprisingly, significant differences in the growth rates of different mycoplasma species in Vero cell culture were observed, questioning the utility of Vero cells as a universal cell culture for mycoplasma enrichment (36).
Herein we describe our results on evaluation of the feasibility of different permissive mammalian cell cultures to support a uniform enrichment of low levels of mycoplasma agents in cell substrates and biologics, using infections with different mycoplasma species known to be common cell line contaminants. We believe that the use of this approach can significantly enhance the sensitivity and efficiency of mycoplasma detection in cell substrates and biologics by either NAT-based or biochemical methods.
MATERIALS AND METHODS
Mollicutes strains and culture.
Eleven Mollicutes species, namely, M. arginini ATCC 23838, M. bovis ATCC 25523, M. fermentans ATCC 19989, M. gallinaceum ATCC 33550, M. gallisepticum ATCC 19610, M. hominis ATCC 27545, M. hyorhinis ATCC 29052, M. orale ATCC 23714, M. salivarium ATCC 23064, M. synoviae ATCC 25204, and Acholeplasma laidlawii ATCC 14089, were obtained from the American Type Culture Collection (ATCC [Manassas, VA]). The growth of selected species was carried out using media and culture conditions recommended by the supplier (http://www.atcc.org). The authenticity of all mycoplasma species used in the study was confirmed by analyzing multiple mycoplasmal genetic markers as described elsewhere (61, 62).
Mammalian and insect cell cultures.
The mammalian cell cultures (Table 1) obtained from ATCC were propagated in Corning T75 flasks (Corning Life Sciences, Acton, MA), using Dulbecco's modified Eagle medium (DMEM) supplemented with glucose, GlutaMAX-I, sodium pyruvate, and 5% heat-inactivated fetal bovine serum (FBS; Invitrogen, Carlsbad, CA). All cell cultures were grown without antibiotics at 37°C ± 1°C under 5% CO2. High Five insect cells (Invitrogen) were grown at 30°C ± 1°C under an air atmosphere, using T75 flasks and Grace's insect cell culture medium (Grace's medium; Invitrogen) supplemented with 3.33 mg/liter lactalbumin hydrolysate, 3.33 mg/liter yeast hydrolysate, and 10% heat-inactivated FBS (Invitrogen). During the study, all working cell culture stocks were periodically tested for the absence of mycoplasmal, bacterial, and fungal contaminations by using light microscope examination of monolayers, fluorescence microscopy of Hoechst-stained cells, mycoplasma testing with a broth/agar culture method, and PCR amplification of the 16S rRNA gene (15) and the 16S-23S internal transcribed spacer (ITS) region (61). Using these methods, no mycoplasmal or any other bacterial contamination was ever detected in the working cell culture stocks.
TABLE 1.
ATCC cell lines used in this study
| ATCC no. | Designation | Organism | Organ | Cell type |
|---|---|---|---|---|
| CCL-22 | MDBK (NBL-1) | Cow | Kidney | Epithelial |
| CRL-1573 | 293 (HEK-293) | Human | Kidney | Epithelial |
| HB-8065 | Hep-G2 | Human | Liver/hepatocellular carcinoma | Epithelial |
| CCL-34 | MDCK (NBL-2) | Dog | Kidney | Epithelial |
| CCL-81 | Vero | Green monkey | Kidney | Epithelial |
| CCL-193 | R9ab | Rabbit | Lung | Fibroblast |
| CCL-44 | EBTr (NBL-4) | Cow | Trachea | Fibroblast |
| CCL-75 | WI-38 | Human | Lung | Fibroblast |
| CCL-70 | CV-1 | Green monkey | Kidney | Fibroblast |
Cocultivation of mycoplasmas with cell cultures.
Approximately 90% confluent monolayers of different cell cultures grown in Corning T75 flasks were used for infection with the mycoplasma species used in the study. On the day of infection, old medium was removed and replaced with 10 ml of fresh DMEM or Grace's medium (depending on the type of cells) supplemented with 2% FBS and containing dilutions of viable mycoplasma with expected concentrations of 0.025, 0.05, 0.1, 0.25, 0.5, and 1.0 CFU/ml. On the third day after infection, 10 ml of fresh medium containing 2% FBS was added to each flask, and incubation was continued. The flasks with infected cells were incubated at 37°C for a maximum of 7 days postinfection (dpi). One flask per infection dose was frozen (−80°C) on the day of infection as well as at 3 and 7 dpi. Insect cell cultures infected with mycoplasmas were incubated at temperatures of 30°C ± 1°C and 35°C ± 1°C under air and at 37°C ± 1°C under a 5% CO2 atmosphere. All mycoplasma infection cell culture experiments were repeated on two separate occasions.
The mycoplasma titers (CFU/ml) in working stocks were determined by plating 0.2-ml dilutions with expected mycoplasma concentrations of 10, 100, and 1,000 CFU/ml on agar plates of ATCC medium 243 (for all mycoplasmas except M. synoviae) or ATCC medium 486 for M. synoviae. All media were supplemented with heat-inactivated horse or swine serum (ATCC, Manassas, VA) and 10% yeast extract solution (Invitrogen, Carlsbad, CA). The plates were incubated for a maximum of 14 days at 37°C ± 1°C under anaerobic (GasPak EZ anaerobe pouch system; BD Biosciences, Franklin Lakes, NJ) or aerobic (5% CO2) conditions, depending on the species, and CFU numbers were counted at the end of incubation. Bacteroides fragilis (ATCC 25285; BD Biosciences) was used as the indicator culture to confirm anaerobic conditions when the GasPak EZ anaerobe pouch system was used.
Genomic DNA isolation and PCR amplification.
While thawing at room temperature, frozen cells were vigorously shaken to obtain well-homogenized cell debris. Four milliliters of this mixture was centrifuged at 16,110 × g for 20 min. Total DNA from the cell pellet was extracted using a DNeasy blood and tissue kit (Qiagen, Chatsworth, CA) according to the manufacturer's protocol. The 16S-23S rRNA ITS regions of all Mollicutes species used in the study were amplified using the forward PCR primer 16S-F-MYC (GGTGAATACGTTCTCGGGTCTTGTACACAC) and the reverse PCR primer 23S-R1-MYC (TNCTTTTCACCTTTCCCTCACGGTAC) (61). Depending on the species, the sizes of ITS-derived amplicons varied from 600 bp to 1,000 bp (61).
Amplification of the rpoB gene of the Mollicutes strains used in this study was carried out using the forward PCR primer rpoB-F1-MYC (ATGGGTGCVAACATGCAACGTCAAGC) and a mixture of two reverse primers, rpoB-R-MYC (GCTCAHACTTCCATTTCHCCAAA) and rpoB-R1-MYC (CGTTTTGWGCTTTACCACCCATTGGTTGTTG) (36). The sizes of rpoB-specific amplicons varied from 1,250 to 1,600 bp, depending on the mycoplasma species.
Briefly, the standard PCR mixture (50 μl) contained 1.5 U of HotStar Taq DNA polymerase, 1× reaction buffer supplemented with 2.5 mM MgCl2 (Qiagen, Chatsworth, CA), 500 nM of each forward and reverse primer, a 200 μM concentration of each deoxyribonucleoside triphosphate (dATP, dGTP, dCTP, and dTTP), and 5 μl of DNA template (ca. 0.1 to 0.2 μg of total DNA). The PCR was performed using a GeneAmp PCR system model 9600 thermocycler (PE Applied Biosystems, Foster City, CA), with the following cycle conditions: initial activation at 95°C for 15 min; 40 cycles of 94°C for 1 min, 60°C (for ITS PCR) or 55°C (for rpoB PCR) for 1 min, and 72°C (extension) for 1 min; and a final extension at 72°C for 7 min. The synthesis of PCR products with the expected molecular weights was confirmed by electrophoresis using a 1% SeaKem Gold TAE-agarose gel with ethidium bromide (Lonza, Allendale, NJ), followed by UV visualization. In order to identify mycoplasma species, the rpoB amplicons were additionally sequenced, and the sequences obtained were compared with those available in the GenBank database. The absence of PCR inhibitors in isolated DNAs was confirmed by PCR amplification of cell housekeeping genes. Thus, primers F1-Animal (CCWAYCGAGCYKGGTGATAGCTGGTT) and R1-Animal (TCCGGTCTGAACTCAGATCACGTAGGA) were used for amplification of the mitochondrial 16S rRNA of mammalian cells, while primers Trichoplusia_HSP70_f (GCTCAGCGTCAAGCCACCAAGGAC) and Trichoplusia_HSP70_r (TGACACCTCCCACAGTCTCGATAC) were used to amplify the heat shock protein gene (hsp70) from insect High Five or Sf9 cells. The synthesis of amplicons with approximate sizes of 1,085 bp and 763 bp was observed for mammalian and insect cells, respectively. The PCR conditions used for amplification of these housekeeping genes were the same as those described above for mycoplasmal ITS-specific PCR.
Detection of mycoplasma in cell cultures by use of a MycoAlert kit.
During the mycoplasma enrichment study, samples of supernatants from infected cultures were collected on days 0, 3, and 7 postinfection. The supernatant was cleared by low-speed centrifugation and stored at −80°C until analysis using a MycoAlert mycoplasma detection kit (Lonza, Frederick, MD) according to the manufacturer's instructions. The luminescent signals in analyzed samples were read using an FB12 luminometer (Berthold Detection Systems, Oak Ridge, TN). Samples with ratios of reading B to reading A of >1 were considered to be mycoplasma positive. In cases where borderline ratios (e.g., 0.9 to 1.3) were observed, the samples were retested after concentration of mycoplasmas by centrifugation of 2 ml of cell culture supernatant at 16,000 × g for 20 min. If the ratio remained unchanged between readings, the sample was considered to be mycoplasma negative in the MycoAlert assay.
RESULTS AND DISCUSSION
Mycoplasma enrichment using different mammalian cell cultures.
The main goal of this study was to evaluate the feasibility of the use of mammalian cell cultures for biological enrichment for improving the efficiency of subsequent mycoplasma detection by using NAT or other suitable molecular methods. Biological enrichment via cocultivation with cell cultures is believed to be one of the more promising approaches to rapidly increase the concentration of mycoplasmal agents to levels that can reliably be detected by suitable molecular methods. The enrichment potential of different mammalian and insect cell cultures was evaluated.
The study included the screening of several mammalian cell cultures that had different growth characteristics and were derived from two different tissue types, i.e., fibroblasts or epithelium (Table 1). Four of them, Vero, MDCK, MDBK, and HEK-293 cells, were derived from kidney epithelial cells of different mammals, with average doubling times of 18 to 22 h (9, 44). In contrast to epithelial cells, fibroblast cells derived from respiratory tract tissues (WI-38, EBTr, and R9ab cells) or kidneys (CV-1) of different mammals grow more slowly, with average doubling times ranging from 24 to 72 h (7). The initial screening of cell cultures was carried out using infection of 80 to 90% confluent cell monolayers with low infection doses (0.1 to 1.0 CFU/ml) of M. salivarium strain PG-20. The use of M. salivarium for initial screening relied on the previously observed less efficient growth of this species in Vero cell culture than that of other tested mycoplasma species (36). It is necessary to note that M. salivarium is known as an infrequent cause of cell line contamination (43). Nevertheless, if the nutrition and environmental conditions are favorable, it can be grown in cell cultures to high titers. For example, the titer of M. salivarium in human lymphocyte cell cultures infected with M. salivarium was reported to be as high as 108 CFU/ml (43). Thus, M. salivarium was certainly a useful reference species with which to carry out a preliminary screening of cell cultures for the ability to support the growth of fastidious mycoplasma agents. Based on the results of the screening, three cell lines, WI-38, EBTr, and CV-1, were excluded from further study because, even at 7 dpi, M. salivarium could not be detected in these cell cultures by in-house single-round PCR assays developed for detection of mycoplasmal DNA.
To assess the relative capabilities of six other cell lines (Vero, MDBK, HEK-293, Hep-G2, R9ab, and MDCK) to support the enrichment of potential mycoplasma contaminants, we used several mycoplasma species currently recommended by the European Pharmacopoeia as positive controls in mycoplasma testing procedures (M. arginini, M. fermentans, M. gallisepticum, M. hyorhinis, M. orale, M. synoviae, and Acholeplasma laidlawii), as well as a few more species (M. gallinaceum, M. bovis, M. hominis, and M. salivarium) known to cause contamination of vaccine cell substrates. The cell cultures were infected with serial dilutions of each analyzed mycoplasma species, and the mycoplasmal growth was tested using PCR analysis of total DNA isolated from infected cells on days 0, 3, and 7 postinfection. The results of this study are summarized in Table 2, which shows that the lowest mycoplasma infection doses resulted in positive PCR-based detection of mycoplasma in infected cell cultures at 3 and/or 7 dpi. The data clearly demonstrated a significant variability in the growth efficiencies of mycoplasma species in the tested cell lines. Of all analyzed cell cultures, only MDCK cells were found to demonstrate the unique ability to support efficient growth of all mycoplasma species at low infection doses ranging from 0.05 to 0.25 CFU/ml.
TABLE 2.
Results of biological enrichment of Mycoplasma species by use of ATCC cell cultures
| Species | Lowest concn (CFU/ml) or serial dilution (M. hyorhinis only) detectable by PCRa
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Vero cells
|
MDCK (NBL-2) cells
|
MDBK (NBL-1) cells
|
R9ab cells
|
293 (HEK-293) cells
|
Hep-G2 cells
|
|||||||
| 3 dpi | 7 dpi | 3 dpi | 7 dpi | 3 dpi | 7 dpi | 3 dpi | 7 dpi | 3 dpi | 7 dpi | 3 dpi | 7 dpi | |
| A. laidlawii | 0.5 | 0.025 | 0.25 | 0.05 | 0.25 | 0.1 | 0.25 | 0.1 | 10 | 0.25 | 0.25 | 0.2 |
| M. arginini | 0.5 | 0.25 | 0.25 | 0.1 | 0.5 | 0.25 | 0.5 | 0.25 | ND | ND | 0.25 | 0.25 |
| M. hyorhinis | 10−4 | 10−5 | 10−4 | 10−6 | 10−3 | 10−6 | 10−3 | 10−3 | 10−3 | 10−4 | 10−4 | 10−5 |
| M. fermentans | ND | 0.25 | ND | 0.025 | ND | 0.25 | ND | ND | ND | 2.5 | ND | ND |
| M. salivarium | NG | 1.0 | 1.0 | 0.25 | 0.25 | 0.5 | NG | 0.5 | NG | NG | NG | NG |
| M. orale | 0.25 | 0.25 | 0.25 | 0.05 | NG | 0.1 | 1.0 | 0.05 | NG | 0.05 | 1.0 | 0.05 |
| M. hominis | 0.25 | 0.25 | 0.1 | 0.1 | 1.0 | 0.25 | 0.5 | 0.01 | NG | 0.5 | 0.1 | 0.1 |
| M. gallisepticum | 0.5 | 0.05 | 0.25 | 0.05 | 1.0 | 0.25 | NG | 0.5 | NG | 0.1 | NG | 0.1 |
| M gallinaceum | 0.25 | 0.25 | 0.25 | 0.25 | 0.25 | 0.25 | 0.25 | 0.25 | 1.0 | 0.5 | 0.25 | 0.25 |
| M. bovis | 0.05 | 0.1 | 0.05 | 0.05 | 0.25 | 0.1 | 0.1 | 0.1 | NG | 0.05 | 0.1 | 0.05 |
| M. synoviae | NG | NG | 0.5 | 0.05 | NG | NG | NG | NG | NG | NG | NG | NG |
NG, no growth; ND, not done.
The results of the study also revealed that some mycoplasma species demonstrated specific growth in infected cell cultures, which we describe and discuss in detail below.
Growth features of M. synoviae in mammalian cell cultures.
M. synoviae was included in the study because it is a well-known avian mycoplasma agent with significant poultry importance. Moreover, M. synoviae is a fastidious species which does not grow in media commonly used for cultivation of other mycoplasmas and requires specially formulated media (Frey's or Chalquist's) supplemented with NAD coenzyme (67). Despite its fastidious growth character, M. synoviae was found to be able to infect and efficiently grow in chicken embryo cell cultures (2). It was shown that the growth of M. synoviae depended on the presence of avian cells, because no growth of the mycoplasma was detected in the axenic cell culture medium (2). The results of our study using the infection of different cell cultures with low infection doses of M. synoviae strain WVU 1853, ranging from 0.05 to 1.0 CFU/ml, revealed a dramatic difference in the ability of this mycoplasma species to replicate in different mammalian cells (Table 2). Of all tested mammalian cells, only MDCK cells were found to enable efficient M. synoviae growth when the aforementioned mycoplasmal infection doses were used. Another striking observation was that Vero cells, recommended by European and Japanese pharmacopoeias and the PTC and widely used as the indicator cell culture for mycoplasma testing, were unable to support the enrichment of M. synoviae at the infection doses used in the screening study.
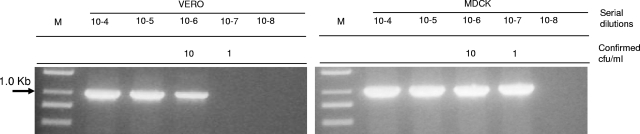
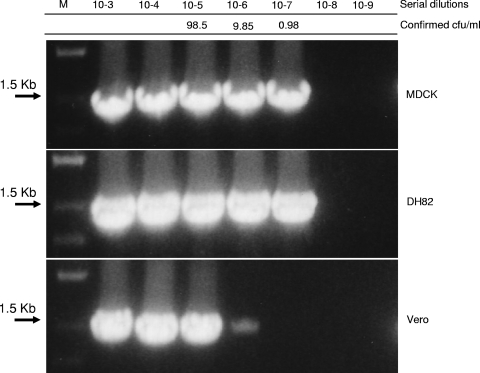
To confirm the ability of M. synoviae to grow in Vero cell culture and to accurately assess the difference in susceptibility between Vero and MDCK cell cultures, we infected these two cultures with serial 10-fold dilutions of an M. synoviae stock. The mycoplasmal growth was tested at 7 dpi, using PCR amplification of the mycoplasmal ITS genetic marker (Fig. 1). The results of this study showed that enrichment in MDCK cells allowed for at least a 1-log increase in sensitivity of M. synoviae detection in comparison with Vero cells. It is noteworthy that comparison of the growth of M. synoviae in two different types of canine-derived cells, i.e., DH82 (ATCC CRL-10389), a macrophage-monocyte cell line derived from neoplastic progenitor cells of canine malignant histiocytosis (65), and kidney-derived epithelial MDCK cells, showed that both cell lines provide equivalent sensitivities of M. synoviae detection at 7 dpi (Fig. 2). The reason for the equivalency between the two canine cell lines as well as their superior efficiency in comparison with other tested mammalian cell lines continues to be unclear. We may only speculate that both epithelial and macrophage-monocyte canine cells provide an optimal spectrum of nutritional factors required for the efficient growth of M. synoviae in mammalian cell cultures.
FIG. 1.
Growth of M. synoviae WVU 1853 in MDCK and Vero cell cultures, detected by PCR on the 16S-23S ITS region. Both cultures were infected with serial dilutions of the strain stock and incubated for 7 days at 37°C and 5% CO2. The mycoplasma titers (CFU/ml) in the dilutions were determined using ATCC 486 medium.
FIG. 2.
Growth of M. synoviae WVU 1853 in MDCK, Vero, and DH82 cell cultures, detected by PCR on the rpoB gene. The cultures were infected with serial dilutions of the strain stock and incubated for 7 days at 37°C and 5% CO2. The mycoplasma titers (CFU/ml) in the dilutions were determined using ATCC 486 medium.
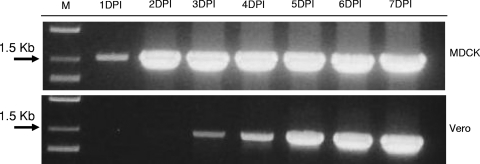
The study of the dynamics of M. synoviae growth in MDCK and Vero cells also revealed a significant difference in mycoplasma replication rates between those cell cultures (Fig. 3). Even though both MDCK and Vero cells were simultaneously infected with the same dose (25 CFU/ml), the reliable detection of M. synoviae in MDCK cells was possible at 2 dpi, 2 days earlier than that in Vero cells. The difference in lag phase times observed for M. synoviae grown in MDCK and Vero cell cultures is likely to be attributed to the different efficiencies and/or spectra of extracellular metabolites and nutritional components produced by these two cell lines. Potentially, identification and isolation of the cellular factors which are essential for efficient growth of M. synoviae can be used to improve the growth of fastidious mycoplasma species and for the development of universal media for rapid mycoplasma testing of cell substrates and cell-derived biological products.
FIG. 3.
Daily dynamics of growth of M. synoviae WVU 1853 in MDCK and Vero cell cultures, detected by PCR on the rpoB gene. Both cultures were infected with the same infectious dose (25 CFU/ml), and flasks were taken daily until 7 dpi (at 37°C and 5% CO2) and frozen at −80°C until DNA extraction.
Growth features of M. fermentans in mammalian cell cultures.
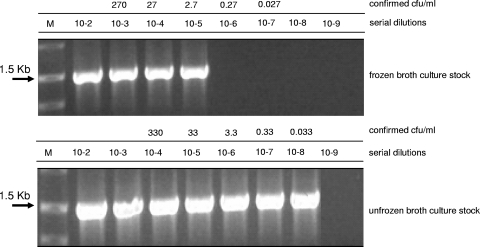
In our previous study, where Vero cells were tested for the ability to support the growth of different mycoplasma species, we demonstrated that the infection dose of M. fermentans strain PG18 required for reliable mycoplasma PCR-based detection was at least 2 logarithms higher (100 CFU/ml) than that observed for all other mycoplasma species used in the study (36). This was surprising because all stocks used in the study, including the stock of M. fermentans, were freshly prepared and stored at −80°C until use. The unique behavior of M. fermentans allowed us to assume that the difference observed in infection dose could be caused by the effect of stress conditions during the freeze-thawing procedure. This assumption was also supported by data previously obtained on the effect of freeze-thawing on the lag phase of different microorganisms (38, 41, 47). To prove the hypothesis and to demonstrate whether freezing affects the following mycoplasma growth in cell cultures, we conducted an experiment which included the infection of MDCK cells with both frozen and not frozen M. fermentans cells. The titers of both types of cells were determined using an agar plating procedure on the day of infection of MDCK monolayers. The results of the study showed that although we did not detect a significant reduction of the viability of the stock (measured using agar plating) caused by freezing and storage of cells at −80°C (data not shown), nevertheless the freezing procedure resulted in an approximately 2-logarithm reduction of the infectivity of M. fermentans required for mycoplasma detection at 7 dpi (Fig. 4). It seems very plausible that a significant loss of the cell viability occurred when mycoplasma cells were inoculated into DMEM instead of ATCC medium 243. The viability loss of M. fermentans may have resulted from the significant difference in the compositions of DMEM and ATCC medium 243 and might additionally be aggravated by the use of different atmospheric conditions, i.e., aerobic and anaerobic, respectively.
FIG. 4.
Growth of M. fermentans PG18 in MDCK cell cultures, detected by PCR on the rpoB gene. Serial dilutions of frozen and unfrozen stocks of the strain were used for infection of MDCK cell cultures, and infected flasks were incubated until 7 dpi at 37°C and 5% CO2. The mycoplasma titers (CFU/ml) in the dilutions were determined using ATCC 243 medium under anaerobic conditions.
Although M. fermentans is known to be a facultative anaerobe, it grows more efficiently under anaerobic conditions (27, 48). We may hypothesize that with all of these changes, only approximately 1% of injured M. fermentans cells were able to survive in MDCK culture.
Analysis of the ability of High Five insect cell culture to support enrichment of mycoplasmal agents.
In parallel with the testing of mammalian cell lines, we also conducted experiments to assess the feasibility of insect cell cultures to carry out enrichment of low levels of mycoplasma contamination prior to the application of NAT detection methods. Similar to mammalian cell lines, insect cell lines are also known to be susceptible to mycoplasma infection (51, 52). However, in contrast to mammalian cells, insect cells demonstrate natural resistance to infection with a variety of mammalian viruses or support very limited virus replication without visible cytopathic effects (68). From this standpoint, the use of insect cells may offer a certain advantage in mycoplasma testing of live viral vaccines. Commonly, mycoplasma testing using an indicator cell culture assay requires a neutralization of viral infectivity by specific antibodies. The use of antibodies always raises the question of the potential inhibition of mycoplasmal growth. The replacement of mammalian cells with insect cells in some cases avoids the use of neutralizing antibodies when cells are resistant to the tested virus. However, the application of insect cells for mycoplasma testing in indicator cell culture tests is restricted by the limited number of mycoplasma species able to replicate in insect cell cultures at 25 to 28°C (permissive temperature range for insect cells). An increase of the temperature above 28°C allows a broader range of mycoplasma species to efficiently replicate in insect cell cultures (52). Thus, the use of insect cell cultures promises to significantly simplify the testing of both mycoplasmal and spiroplasmal contaminations by use of biological enrichment (22-24, 52, 53). However, until now, it has been uncertain if insect cells were able to support the growth of the vast majority of mycoplasma target species able to cause contamination of mammalian cell cultures (51, 52). To address this issue, we carried out a study aimed at assessing the enrichment efficiencies of common mycoplasmal cell line contaminants, using two insect cell lines, Sf9 (Spodoptera frugiperda) and High Five (Trichoplusia ni). The results of the study showed that in comparison to Sf9 cells, High Five cells demonstrated more efficient support of growth of mycoplasmas (data not shown). We also observed that the use of a permissive temperature for propagation of insect cells (from 25 to 30°C) and mycoplasma enrichment resulted in selective growth of some, but not all, target mycoplasma species (data not shown).
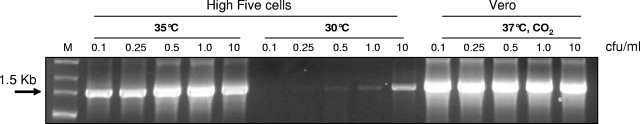
Our results for infection of High Five cell cultures at nonpermissive temperatures showed that increases of the incubation temperature from 30°C ± 1°C to 35°C ± 1°C and, further, to 37°C ± 1°C resulted in an additional increase (up to 2 log) of mycoplasma growth in High Five cell cultures and finally allowed for mycoplasma detection at contamination levels as low as 0.05 or 0.1 CFU/ml. This level of sensitivity is equivalent to that observed previously when mammalian cell cultures were used for mycoplasma enrichment. The positive effect of temperature increase on mycoplasma growth was observed for several mycoplasma species, including M. gallisepticum, M. bovis, and M. orale (Fig. 5).
FIG. 5.
M. gallisepticum growth in H5 insect cell culture at different temperatures, detected by PCR on the rpoB gene. H5 cells were infected with different infectious doses of mycoplasma and incubated for 7 days at 30°C and 35°C. Vero cells were used as a growth control.
However, it continued to be unclear if insect cells were able to survive and function properly above permissive temperatures. The visual inspection (by light microscopy) of High Five cells during incubation at 37°C ± 1°C revealed significant morphological changes of cells (i.e., cytoplasmic and perinuclear vacuolization, which indicates cell dystrophy) (data not shown). Nevertheless, despite all these serious morphological changes, the trypan blue exclusion test showed that approximately 60 to 80% of cells sustained their viability after 7 days of incubation at 37°C ± 1°C. However, the attempt to passage the cells after incubation at nonpermissive temperatures (37°C ± 1°C) was unsuccessful. Thus, the use of insect cells for enrichment requires the selection of cell clones able to grow at 35 to 37°C. The principle for the possibility of the establishment of such cells (derived from Sf21 and Sf9 insect cell lines) able to grow at 37°C ± 1°C has previously been demonstrated (20).
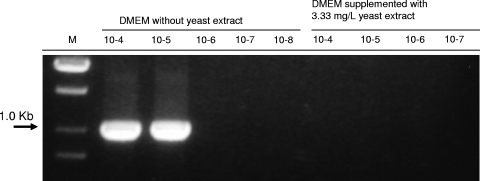
Although we successfully demonstrated the enrichment of several mycoplasma species by use of High Five insect cell culture, M. hyorhinis strain DBS1050 was unable to grow in these cells to levels readily detected by PCR. Thus, all our attempts to optimize growth conditions (medium, temperature, supplements, etc.) to achieve efficient growth of M. hyorhinis strain DBS1050 in High Five insect cell culture did not display any positive results. M. hyorhinis strain DBS1050 is known to have atypical cultivation features, and in contrast to the type strains (BTS-7 and GDL) of M. hyorhinis, this strain does not grow in defined mycoplasma broth media. Unsuccessful attempts to cultivate M. hyorhinis strain DBS1050 in defined microbiological media were previously attributed to the sensitivity of this strain to compounds in peptones/hydrolysates and yeast extracts used for medium formulation (12, 13, 18). We also showed that addition of yeast extract to DMEM at a concentration of 3.33 mg/liter, which is equivalent to that in Grace's medium, and the use of this modified DMEM for growth of MDCK cells and cocultivation with M. hyorhinis DBS1050 resulted in dramatic suppression of the growth of this mycoplasma strain (Fig. 6). Thus, the failure to enrich M. hyorhinis DBS1050 in insect cells was most likely caused by the use of Grace's medium containing peptones and yeast extract. We propose that the use of other artificial media not containing any peptones and yeast extracts but suitable for growth of insect cells may enable efficient enrichment of inhibitor-sensitive M. hyorhinis strains.
FIG. 6.
Growth of M. hyorhinis DBS1050 in MDCK cell culture grown in DMEM with and without yeast extract at 37°C and 5% CO2. MDCK culture was infected with serial dilutions of the strain stock and incubated for 7 days. Growth of M. hyorhinis was detected by PCR on the 16S-23S ITS region.
The use of MDCK cells for biological enrichment of mycoplasmal agents prior to their detection using NAT-based or other molecular methods seems to be able to eliminate a major technical problem associated with the demonstration of equivalency (in terms of detection limits) of novel and compendial methods. It is necessary to note that although PCR-based methods have definite advantages over conventional microbiological methods in terms of intrinsic analytical sensitivity, simplicity, and turnaround time required for testing, it still continues to be unclear whether PCR-based methods could become equivalent alternatives to the microbiological methods currently used for mycoplasma testing. The ambiguity stems from the difference in the nature of the biological characteristics measured by NAT and microbiological methods. NAT methods detect the presence of mycoplasmal DNAs from viable and nonviable cells. In contrast, compendial microbiological methods detect the presence of viable mycoplasma via the enumeration of CFU on agar media or the color-changing units in broth media. All PCR-based methods have some shortcomings, which considerably limit the potential of these methods for detection of mycoplasma contamination in cell substrates and cell-derived biologics. Thus, the very limited sample volume of cell substrates that can be analyzed by PCR methods restricts the overall sensitivity of assay, even as the intrinsic sensitivity of PCR reaches the theoretical maximum of a single DNA molecule per sample. In contrast, compendial microbiological methods can test sample volumes of 10 ml, and thus their theoretical maximum limits of detection could be higher than those of PCR methods. The sample volume limitation as well as the susceptibility of PCR to the presence of inhibitors is the main cause of false-negative results in PCR-based mycoplasma testing. In addition, PCR methods based on the amplification of genomic DNA of the target pathogen generally do not permit discrimination between viable and nonviable microorganisms in analyzed biological samples.
Our results demonstrate that the application of biological enrichment prior to application of NAT-based detection methods, as well as other suitable molecular methods, shortens the time required for mycoplasma testing from between 28 and 30 days to 1 week. Moreover, the ability of MDCK cell culture to support the efficient growth of different mycoplasmal agents opens the real opportunity to simplify the mycoplasma testing procedure by using one universal cell culture for enrichment of the vast majority of mycoplasmas, including all known common cell line contaminants.
Acknowledgments
The findings and conclusions in this article have not been formally disseminated by the Food and Drug Administration and should not be construed to represent any agency determination or policy.
Published ahead of print on 7 July 2008.
REFERENCES
- 1.Abele-Horn, M., U. Busch, H. Nitschko, E. Jacobs, R. Bax, F. Pfaff, B. Schaffer, and J. Heesemann. 1998. Molecular approaches to diagnosis of pulmonary diseases due to Mycoplasma pneumoniae. J. Clin. Microbiol. 36:548-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aldridge, K. E. 1975. Growth and cytopathology of Mycoplasma synoviae in chicken embryo cell cultures. Infect. Immun. 12:198-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baird, S. C., J. Carman, R. P. Dinsmore, R. L. Walker, and J. K. Collins. 1999. Detection and identification of Mycoplasma from bovine mastitis infections using a nested polymerase chain reaction. J. Vet. Diagn. Investig. 11:432-435. [DOI] [PubMed] [Google Scholar]
- 4.Benisheva, T., V. Sovova, I. Ivanov, and G. Opalchenova. 1993. Comparison of methods used for detection of mycoplasma contamination in cell cultures, sera, and live-virus vaccines. Folia Biol. (Praha) 39:270-276. [PubMed] [Google Scholar]
- 5.Benton, W. J., M. S. Cover, and F. W. Melchior. 1967. Mycoplasma gallisepticum in a commercial laryngotracheitis vaccine. Avian Dis. 11:426-429. [PubMed] [Google Scholar]
- 6.Boatman, E. S., and G. E. Kenny. 1971. Morphology and ultrastructure of Mycoplasma pneumoniae spherules. J. Bacteriol. 106:1005-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bota, D. A., J. K. Ngo, and K. J. Davies. 2005. Downregulation of the human Lon protease impairs mitochondrial structure and function and causes cell death. Free Radic. Biol. Med. 38:665-677. [DOI] [PubMed] [Google Scholar]
- 8.Brown, D. R. 2002. Mycoplasmosis and immunity of fish and reptiles. Front. Biosci. 7:d1338-d1346. [DOI] [PubMed] [Google Scholar]
- 9.Cho, M. J., D. P. Thompson, C. T. Cramer, T. J. Vidmar, and J. F. Scieszka. 1989. The Madin Darby canine kidney (MDCK) epithelial cell monolayer as a model cellular transport barrier. Pharm. Res. 6:71-77. [DOI] [PubMed] [Google Scholar]
- 10.Darai, G., R. M. Flugel, L. Zoller, B. Matz, A. Kreig, H. Gelderblom, H. Delius, and R. H. Leach. 1981. The plaque-forming factor for mink lung cells present in cytomegalovirus and herpes-zoster virus stocks identified as Mycoplasma hyorhinis. J. Gen. Virol. 55:201-205. [DOI] [PubMed] [Google Scholar]
- 11.Darai, G., L. Zoller, R. M. Flugel, and H. Gelderblom. 1983. Mink lung cells as a tool for detection of Mycoplasma hyorhinis contamination in cell cultures and virus stocks. In Vitro 19:7-15. [DOI] [PubMed] [Google Scholar]
- 12.Del Giudice, R. A. 1998. M-CMRL, a new axenic medium to replace indicator cell cultures for the isolation of all strains of Mycoplasma hyorhinis. In Vitro Cell Dev. Biol. Anim. 34:88-89. [DOI] [PubMed] [Google Scholar]
- 13.Del Giudice, R. A., R. S. Gardella, and H. E. Hopps. 1980. Cultivation of formerly noncultivable strains of Mycoplasma hyorhinis. Curr. Microbiol. 4:75-80. [Google Scholar]
- 14.Drexler, H. G., C. C. Uphoff, W. G. Dirks, and R. A. MacLeod. 2002. Mix-ups and mycoplasma: the enemies within. Leukoc. Res. 26:329-333. [DOI] [PubMed] [Google Scholar]
- 15.Eldering, J. A., C. Felten, C. A. Veilleux, and B. J. Potts. 2004. Development of a PCR method for mycoplasma testing of Chinese hamster ovary cell cultures used in the manufacture of recombinant therapeutic proteins. Biologicals 32:183-193. [DOI] [PubMed] [Google Scholar]
- 16.Fadiel, A., K. D. Eichenbaum, N. El Semary, and B. Epperson. 2007. Mycoplasma genomics: tailoring the genome for minimal life requirements through reductive evolution. Front. Biosci. 12:2020-2028. [DOI] [PubMed] [Google Scholar]
- 17.Fraser, C. M., J. D. Gocayne, O. White, M. D. Adams, R. A. Clayton, R. D. Fleischmann, C. J. Bult, A. R. Kerlavage, G. Sutton, J. M. Kelley, R. D. Fritchman, J. F. Weidman, K. V. Small, M. Sandusky, J. Fuhrmann, D. Nguyen, T. R. Utterback, D. M. Saudek, C. A. Phillips, J. M. Merrick, J. F. Tomb, B. A. Dougherty, K. F. Bott, P. C. Hu, T. S. Lucier, S. N. Peterson, H. O. Smith, C. A. Hutchison III, and J. C. Venter. 1995. The minimal gene complement of Mycoplasma genitalium. Science 270:397-403. [DOI] [PubMed] [Google Scholar]
- 18.Gardella, R. S., and R. A. Del Giudice. 1995. Growth of Mycoplasma hyorhinis cultivar alpha on semisynthetic medium. Appl. Environ. Microbiol. 61:1976-1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geissdorfer, W., G. Sandner, S. John, A. Gessner, C. Schoerner, and K. Schroppel. 2008. Ureaplasma urealyticum meningitis in an adult patient. J. Clin. Microbiol. 46:1141-1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gerbal, M., P. Fournier, P. Barry, M. Mariller, F. Odier, G. Devauchelle, and M. Duonor-Cerutti. 2000. Adaptation of an insect cell line of Spodoptera frugiperda to grow at 37 degrees C: characterization of an endodiploid clone. In Vitro Cell Dev. Biol. Anim. 36:117-124. [DOI] [PubMed] [Google Scholar]
- 21.Gouriet, F., F. Fenollar, J. Y. Patrice, M. Drancourt, and D. Raoult. 2005. Use of shell-vial cell culture assay for isolation of bacteria from clinical specimens: 13 years of experience. J. Clin. Microbiol. 43:4993-5002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hackett, K. J., and D. E. Lynn. 1985. Cell-assisted growth of a fastidious Spiroplasma. Science 230:825-827. [DOI] [PubMed] [Google Scholar]
- 23.Hackett, K. J., D. E. Lynn, A. S. Ginsberg, S. Rottem, R. B. Henegar, J. Adams, D. L. Williamson, and R. F. Whitcomb. 1987. Cell-assisted culture of fastidious spiroplasmas: initial analysis of growth factors. Isr. J. Med. Sci. 23:667-670. [PubMed] [Google Scholar]
- 24.Hackett, K. J., D. E. Lynn, D. L. Williamson, A. S. Ginsberg, and R. F. Whitcomb. 1986. Cultivation of the Drosophila sex-ratio spiroplasma. Science 232:1253-1255. [DOI] [PubMed] [Google Scholar]
- 25.Hamasuna, R., Y. Osada, and J. S. Jensen. 2005. Antibiotic susceptibility testing of Mycoplasma genitalium by TaqMan 5′ nuclease real-time PCR. Antimicrob. Agents Chemother. 49:4993-4998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hamasuna, R., Y. Osada, and J. S. Jensen. 2007. Isolation of Mycoplasma genitalium from first-void urine specimens by coculture with Vero cells. J. Clin. Microbiol. 45:847-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hayes, M. M., H. H. Foo, H. Kotani, D. J. Wear, and S. C. Lo. 1993. In vitro antibiotic susceptibility testing of different strains of Mycoplasma fermentans isolated from a variety of sources. Antimicrob. Agents Chemother. 37:2500-2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hlinak, P., J. Friedemann, and G. Starke. 1969. Problems of the state examination of viral vaccines in Mycoplasma contaminations. Pharmazie 24:189-192. [PubMed] [Google Scholar]
- 29.Hopps, H. E., and R. A. Del Giudice. 1984. Cell culture models as ancillary tools in the isolation and characterization of mycoplasmas. Isr. J. Med. Sci. 20:927-930. [PubMed] [Google Scholar]
- 30.Ikoev, V. N., G. M. Gonskii, and S. G. Dzagurov. 1973. A method of ultracentrifugation used for control of live viral vaccines with regard to removal of mycoplasma. Vopr. Virusol. 18:625-628. [PubMed] [Google Scholar]
- 31.Ishikawa, Y., T. Kozakai, H. Morita, K. Saida, S. Oka, and Y. Masuo. 2006. Rapid detection of mycoplasma contamination in cell cultures using SYBR green-based real-time polymerase chain reaction. In Vitro Cell Dev. Biol. Anim. 42:63-69. [DOI] [PubMed] [Google Scholar]
- 32.Jensen, J. S., H. T. Hansen, and K. Lind. 1996. Isolation of Mycoplasma genitalium strains from the male urethra. J. Clin. Microbiol. 34:286-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jules Mattes, M. 2004. Control of the mycoplasma epidemic. In Vitro Cell Dev. Biol. Anim. 40:253-254. [DOI] [PubMed] [Google Scholar]
- 34.Kojima, A., T. Takahashi, M. Kijima, Y. Ogikubo, M. Nishimura, S. Nishimura, R. Harasawa, and Y. Tamura. 1997. Detection of Mycoplasma in avian live virus vaccines by polymerase chain reaction. Biologicals 25:365-371. [DOI] [PubMed] [Google Scholar]
- 35.Kojima, A., T. Takahashi, M. Kijima, Y. Ogikubo, Y. Tamura, and R. Harasawa. 1996. Detection of mycoplasma DNA in veterinary live virus vaccines by the polymerase chain reaction. J. Vet. Med. Sci. 58:1045-1048. [DOI] [PubMed] [Google Scholar]
- 36.Kong, H., D. V. Volokhov, J. George, P. Ikonomi, D. Chandler, C. Anderson, and V. Chizhikov. 2007. Application of cell culture enrichment for improving the sensitivity of mycoplasma detection methods based on nucleic acid amplification technology (NAT). Appl. Microbiol. Biotechnol. 77:223-232. [DOI] [PubMed] [Google Scholar]
- 37.Krausse-Opatz, B., P. Dollmann, H. Zeidler, J. G. Kuipers, and L. Kohler. 2000. Frequent contamination of Chlamydia trachomatis and Chlamydia pneumoniae strains with mycoplasma. Biological relevance and selective eradication of mycoplasma from chlamydial cultures with mupirocin. Med. Microbiol. Immunol. (Berlin) 189:19-26. [DOI] [PubMed] [Google Scholar]
- 38.Kuo, C. C., and J. T. Grayston. 1988. Factors affecting viability and growth in HeLa 229 cells of Chlamydia sp. strain TWAR. J. Clin. Microbiol. 26:812-815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee, C. K. 1996. Issues of biological assays for viral vaccines. Dev. Biol. Stand. 88:41-47. [PubMed] [Google Scholar]
- 40.Levy, J. A., P. E. Sumner, and L. E. Hooser. 1982. Rapid tissue culture method for detection of Mycoplasma hyorhinis. J. Gen. Microbiol. 128:2817-2820. [DOI] [PubMed] [Google Scholar]
- 41.Mackey, B. M., and C. M. Derrick. 1984. Conductance measurements of the lag phase of injured Salmonella typhimurium. J. Appl. Bacteriol. 57:299-308. [DOI] [PubMed] [Google Scholar]
- 42.Madoff, S., B. Q. Pixley, R. A. DelGiudice, and R. C. Moellering, Jr. 1979. Isolation of Mycoplasma bovis from a patient with systemic illness. J. Clin. Microbiol. 9:709-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McGarrity, G. J., D. M. Phillips, and A. B. Vaidya. 1980. Mycoplasmal infection of lymphocyte cell cultures: infection with M. salivarium. In Vitro 16:346-356. [DOI] [PubMed] [Google Scholar]
- 44.Nahapetian, A. T., J. N. Thomas, and W. G. Thilly. 1986. Optimization of environment for high density Vero cell culture: effect of dissolved oxygen and nutrient supply on cell growth and changes in metabolites. J. Cell Sci. 81:65-103. [DOI] [PubMed] [Google Scholar]
- 45.Pitcher, D. G., and R. A. Nicholas. 2005. Mycoplasma host specificity: fact or fiction? Vet. J. 170:300-306. [DOI] [PubMed] [Google Scholar]
- 46.Polster, U. 1986. Demonstration of Mycoplasma bovis as a contaminant in live virus vaccine. Arch. Exp. Veterinarmed. 40:147-150. [PubMed] [Google Scholar]
- 47.Prentice, M. J., and J. Farrant. 1977. Survival of chlamydiae after cooling to −196°C. J. Clin. Microbiol. 6:4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Razin, S., and E. A. Freundt. 1984. The mycoplasmas, p. 740-775. In N. R. Krieg and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 1. Williams & Wilkins, Baltimore, MD. [Google Scholar]
- 49.Razin, S., D. Yogev, and Y. Naot. 1998. Molecular biology and pathogenicity of mycoplasmas. Microbiol. Mol. Biol. Rev. 62:1094-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stakenborg, T., J. Vicca, P. Butaye, H. Imberechts, J. Peeters, A. De Kruif, F. Haesebrouck, and D. Maes. 2006. A multiplex PCR to identify porcine mycoplasmas present in broth cultures. Vet. Res. Commun. 30:239-247. [DOI] [PubMed] [Google Scholar]
- 51.Steiner, T., and G. McGarrity. 1983. Mycoplasmal infection of insect cell cultures. In Vitro 19:672-682. [DOI] [PubMed] [Google Scholar]
- 52.Steiner, T., G. J. McGarrity, J. M. Bove, D. M. Phillips, and M. Garnier. 1984. Insect cell cultures in the study of attachment and pathogenicity of spiroplasmas and mycoplasmas. Ann. Microbiol. (Paris) 135A:47-53. [DOI] [PubMed] [Google Scholar]
- 53.Steiner, T., G. J. McGarrity, and D. M. Phillips. 1982. Cultivation and partial characterization of spiroplasmas in cell cultures. Infect. Immun. 35:296-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sung, H., S. H. Kang, Y. J. Bae, J. T. Hong, Y. B. Chung, C. K. Lee, and S. Song. 2006. PCR-based detection of Mycoplasma species. J. Microbiol. 44:42-49. [PubMed] [Google Scholar]
- 55.Thornton, D. H. 1986. A survey of mycoplasma detection in veterinary vaccines. Vaccine 4:237-240. [DOI] [PubMed] [Google Scholar]
- 56.Timenetsky, J., L. M. Santos, M. Buzinhani, and E. Mettifogo. 2006. Detection of multiple mycoplasma infection in cell cultures by PCR. Braz. J. Med. Biol. Res. 39:907-914. [DOI] [PubMed] [Google Scholar]
- 57.Uphoff, C. C., and H. G. Drexler. 2005. Detection of mycoplasma contaminations. Methods Mol. Biol. 290:13-23. [DOI] [PubMed] [Google Scholar]
- 58.Uphoff, C. C., and H. G. Drexler. 1999. Detection of mycoplasma contaminations in cell cultures by PCR analysis. Hum. Cell 12:229-236. [PubMed] [Google Scholar]
- 59.Uphoff, C. C., and H. G. Drexler. 2002. Detection of mycoplasma in leukemia-lymphoma cell lines using polymerase chain reaction. Leukemia 16:289-293. [DOI] [PubMed] [Google Scholar]
- 60.Verkooyen, R. P., M. Sijmons, E. Fries, A. Van Belkum, and H. A. Verbrugh. 1997. Widely used, commercially available Chlamydia pneumoniae antigen contaminated with mycoplasma. J. Med. Microbiol. 46:419-424. [DOI] [PubMed] [Google Scholar]
- 61.Volokhov, D. V., J. George, S. X. Liu, P. Ikonomi, C. Anderson, and V. Chizhikov. 2006. Sequencing of the intergenic 16S-23S rRNA spacer (ITS) region of Mollicutes species and their identification using microarray-based assay and DNA sequencing. Appl. Microbiol. Biotechnol. 71:680-698. [DOI] [PubMed] [Google Scholar]
- 62.Volokhov, D. V., A. A. Neverov, J. George, H. Kong, S. X. Liu, C. Anderson, M. K. Davidson, and V. Chizhikov. 2007. Genetic analysis of housekeeping genes of members of the genus Acholeplasma: phylogeny and complementary molecular markers to the 16S rRNA gene. Mol. Phylogenet. Evol. 44:699-710. [DOI] [PubMed] [Google Scholar]
- 63.Wang, H., F. Kong, P. Jelfs, G. James, and G. L. Gilbert. 2004. Simultaneous detection and identification of common cell culture contaminant and pathogenic Mollicutes strains by reverse line blot hybridization. Appl. Environ. Microbiol. 70:1483-1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Webster, D., H. Windsor, C. Ling, D. Windsor, and D. Pitcher. 2003. Chronic bronchitis in immunocompromised patients: association with a novel Mycoplasma species. Eur. J. Clin. Microbiol. Infect. Dis. 22:530-534. [DOI] [PubMed] [Google Scholar]
- 65.Wellman, M. L., S. Krakowka, R. M. Jacobs, and G. J. Kociba. 1988. A macrophage-monocyte cell line from a dog with malignant histiocytosis. In Vitro Cell Dev. Biol. 24:223-229. [DOI] [PubMed] [Google Scholar]
- 66.Woods, S. B. 1983. The isolation of a ‘non-cultivable’ strain of Mycoplasma hyorhinis from a mammalian live virus vaccine. J. Biol. Stand. 11:247-250. [DOI] [PubMed] [Google Scholar]
- 67.Yagihashi, T., and K. Kato. 1984. Diversity in nicotinamide-adenine dinuscleotide requirement for the growth of different strains of Mycoplasma synoviae. Res. Vet. Sci. 37:353-354. [PubMed] [Google Scholar]
- 68.Zhang, P. F., M. Klutch, J. Muller, and C. J. Marcus-Sekura. 1994. Susceptibility of the Sf9 insect cell line to infection with adventitious viruses. Biologicals 22:205-213. [DOI] [PubMed] [Google Scholar]