Abstract
We have developed a fast and accurate method to engineer the Bacillus subtilis genome that involves fusing by PCR two flanking homology regions with an antibiotic resistance gene cassette bordered by two mutant lox sites (lox71 and lox66). The resulting PCR products were used directly to transform B. subtilis, and then transient Cre recombinase expression in the transformants was used to recombine lox71 and lox66 into a double-mutant lox72 site, thereby excising the marker gene. The mutation process could also be accomplished in 2 days by using a strain containing a cre isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible expression cassette in the chromosome as the recipient or using the lox site-flanked cassette containing both the cre IPTG-inducible expression cassette and resistance marker. The in vivo recombination efficiencies of different lox pairs were compared; the lox72 site that remains in the chromosome after Cre recombination had a low affinity for Cre and did not interfere with subsequent rounds of Cre/lox mutagenesis. We used this method to inactivate a specific gene, to delete a long fragment, to realize the in-frame deletion of a target gene, to introduce a gene of interest, and to carry out multiple manipulations in the same background. Furthermore, it should also be applicable to large genome rearrangement.
Bacillus subtilis is the best-characterized gram-positive bacterial organism: its biochemistry, physiology and genetics have been studied intensely for more than 50 years. B. subtilis and the closely related Bacillus species are nonpathogenic and free of endotoxins, and their fermentation technology has been well characterized, making them important cell factories for industrial enzymes, fine biochemicals, antibiotics, and insecticides (8, 31). The completion of the sequencing and annotation of the B. subtilis 168 genome supplies a complete view of the B. subtilis protein machinery, inspiring new approaches for analyzing biochemical pathways (23, 25). Postgenomic studies require simple and highly efficient tools to enable genetic manipulation. Classically, these chromosomal modifications have been achieved by a method that uses a positive selection marker, usually an antibiotic resistance marker, which is generated by insertion of the marker gene into the chromosome. When introducing multiple modifications into the same background, it is better to evict the selection marker gene, usually through a single-crossover event. Selection of the strain that has lost marker gene is tedious without the aid of counterselectable markers that, under appropriate growth conditions, can promote the death of the microorganisms that harbor them. Until now, four counterselectable markers have been described (6, 12, 17, 38) that improve the genetic manipulations of B. subtilis by allowing the subsequent excision of the selection marker, coupled with positive selection. The genetic manipulations described above, however, are mainly based on restriction enzyme and DNA ligase-dependent vector construction, which require about 1 to 2 weeks before a marker-free modification can be achieved.
A simple PCR-based method for gene replacement was used on a genome-wide scale in Saccharomyces cerevisiae and Escherichia coli, depending on a mitotic recombination system and bacteriophage-encoded recombination systems, respectively (4, 5, 11, 29). In contrast to E. coli, B. subtilis naturally develops a physiological state of competence, in which it has the ability to bind, take up, and integrate exogenous linear DNA efficiently (15). Integration of linear DNA, however, requires a minimal length of homology of about 400 to 500 bp (15), compared to just 30 to 50 bp in S. cerevisiae and E. coli (5, 11). In addition, transformation frequency in B. subtilis falls off sharply as the size of transforming DNA decreases (14). These behaviors preclude the use of DNA molecules with short homology extensions in a gene replacement method in B. subtilis. Thus, genetic manipulations were mainly based on the cloning of segments from every target site into an integrative vector. Recently, some researchers have started using joining PCR (17) or long-flanking homology PCR (LFH PCR) techniques (35) to fuse long-flanking homologous regions with a selection marker to delete a given target gene in B. subtilis (16, 27). These PCR-based methods greatly improve mutation efficiency, but selection markers are left behind. Furthermore, the fidelity of PCR amplification should also be considered, as any undesired PCR amplification errors that arise in the flanking homology can be introduced into the chromosome of B. subtilis through a double-crossover event. In 2004, Shevchuk et al. reported a procedure for fast and precise fusion of several fragments into a linear DNA construct, and for shorter projects (<4 kb), the accuracy is expected to be better since high-fidelity PCR can be used (32).
Site-specific recombination systems such as Flp/FRT (7) and Cre/lox (1) have much higher recombination efficiency than the endogenous recombination systems, making them ideal for many genetic manipulation strategies. The Cre/loxP recombination system is a simple two-component system currently recognized as a powerful DNA recombination tool (24). Cre recombinase catalyzes reciprocal site-specific recombination between two loxP sites without requiring any host cofactors or accessory proteins. A DNA sequence that is flanked by loxP sites is excised when the loxP sites are convergently oriented or inverted when the loxP sites are divergently oriented. Notably, the use of native loxP sites for consecutive rounds of manipulations in the same background would lead to the integration into the genome of multiple loxP sites that could still be recognized by Cre. To minimize genetic instability, a pair of mutant lox sites (Fig. 1), a right element mutant lox site (lox66) and a left element mutant lox site (lox71) are usually used (2). Recombination of lox71 and lox66 results in a double-mutant lox72 site that has a strongly reduced binding affinity for Cre, allowing for repeated manipulations in a single genetic background (26, 34).
FIG. 1.
Schematic representation of mutant lox66 and lox71 sites, which, after Cre-mediated recombination, result in a double-mutant lox72 site. Boldfaced sequences are mutated in comparison to the native loxP site. Solid arrows between pairs of lox sites indicate the relative Cre-mediated recombination efficiencies in the forward and reverse directions. “X” indicates recombination between two lox sites. Arrows with dotted lines depict the orientation of the 8-bp spacer.
Here, combining the mutant Cre/lox system and Shevchuk's (32) fusion PCR method, we developed a fast and accurate B. subtilis genome engineering procedure that enabled us to inactivate a specific gene, to delete a long fragment, to realize the in-frame deletion of a target gene, to introduce a gene of interest, and to do multiple manipulations in the same background. The flanking homology could be shortened to approximately 450 bp, and together with the 500-bp zeocin resistance (Zeor) lox71-zeo-lox66 cassette, the resulting PCR-fused fragment was about 1,500 bp. This length remarkably reduced the number of the errors generated by PCR and ensured the accuracy of the method since the undesired PCR amplification errors arising in the flanking homology may be introduced into the chromosome of Bacillus subtilis. Using the strain containing a cre isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible expression cassette in the chromosome as the recipient or using the lox site-flanked cassette containing the cre IPTG-inducible expression cassette and resistance marker, the mutation procedure could be finished in 2 days, allowing it to be used for genome-scale mutagenesis.
MATERIALS AND METHODS
Bacterial strains, plasmids, oligonucleotides, and DNA sequencing.
The bacterial strains and plasmids used in this study are listed in Table 1. All B. subtilis recombinant strains were derived from the Marburg 168 trpC2 strain. The synthesis of oligonucleotides (see Table S1 in the supplemental material) and DNA sequencing were performed by Invitrogen Biotechnology Co., Ltd. Taq DNA polymerase, PrimeSTAR HS DNA polymerase, and TA cloning vectors were purchased from TaKaRa Biotechnology (Dalian) Co., Ltd.
TABLE 1.
Strains and plasmids used in this study
| B. subtilis strain or plasmid | Characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| 168 | trpC2 | BGSC |
| BSPC1 | ΔamyE::lox72 | This work |
| BSPC2 | Δprophage 3::lox72 | This work |
| BSPC3 | ΔhisZ::lox72 | This work |
| BSPC4 | ΔamyE::(lox72-spc-lox72 cassette) | This work |
| BSPC5 | ΔamyE::(lox72-spc-lox71 cassette) | This work |
| BSPC6 | ΔamyE::(lox72-spc-lox66 cassette) | This work |
| BSPC7 | ΔamyE::(lox71-spc-lox66 cassette) | This work |
| BSPC8 | ΔamyE::lox72 Δprophage 3::lox72 ΔhisZ::lox72 | This work |
| BSPC9 | ΔamyE::(mpd expression cassette, lox72) | This work |
| BSPC10 | ΔamyE::(Pspac-cre spc lacI) | This work |
| BSPC11 | ΔamyE::(Pspac-cre spc lacI) ΔhisZ::(lox71-zeo-lox66 cassette) | This work |
| BSPC12 | ΔhisZ::(lox71-zeo-lox66 cassette) | This work |
| BSPC13 | ΔlacA::(Pspac-cre spc lacI) | This work |
| Plasmids | ||
| pMD18-T | Ampr; MCS | Takara |
| p7S6 | pMD18-T ligated with lox71-spc-lox66 cassette | This work |
| p2S6 | pMD18-T ligated with lox72-spc-lox66 cassette | This work |
| p2S1 | pMD18-T ligated with lox72-spc-lox71 cassette | This work |
| p2S2 | pMD18-T ligated with lox72-spc-lox72 cassette | This work |
| pPICZαA | Zeor; P. pastoris expression vector | Invitrogen |
| p7Z6 | pMD18-T containing lox71-zeo-lox66 cassette | This work |
| pTS | Emr Ampr; temperature sensitive in B. subtilis | This work |
| pTSC | pTS containing Pspac-cre expression cassette | This work |
| pDG148 | Ampr Kmr; B.subtilis-E. coli shuttle expression vector | 33 |
| pDGC | pDG148 containing cre fused to Pspac promoter | This work |
| pDG1730 | Ampr Spcr; B. subtilis amyE locus integration vector | 18 |
| pDGI | pDG1730 containing IPTG-inducible expression elements | This work |
| pDGIC | pDGI containing cre under control of Pspac | This work |
| pDGICZ | pDGIC, Sh ble | This work |
| pAX01 | Emr Ampr; B. subtilis lacA locus integration vector | 19 |
| pAXC | Ampr Spcr; pAX01 containing cre IPTG-inducible expression cassette | This work |
| pM7Z6M | pMD18T-simple ligated with PCR-fused fragment containing lox71-spac-lox66 cassette and homology-flanking amyE site | This work |
| pUBC19 | Ampr Kmr; B.subtilis-E. coli shuttle vector | 39 |
| pPhisZ | pUBC19 containing hisZ gene and its promoter region | This work |
| pP43NMK | Ampr Kmr; B. subtilis expression vector, mpd expression cassette | 39 |
Culture, growth conditions, and phenotypic characterization.
All microorganisms were grown at 37°C in Luria-Bertani (LB) broth or on LB agar. Minimal medium (MM) was used for auxotrophy determination (20). To check histidine auxotrophy, MM was supplemented with histidine or histidinol at a concentration of 50 μg/ml. Amylase expression by B. subtilis colonies was detected by growing colonies overnight on LB agar containing 1% starch and then staining the plate with iodine, as described elsewhere (10). Methyl parathion hydrolase (MPH) hydrolyzes methyl parathion to bright yellow p-nitrophenol, which was detected by incubating the strains on LB agar containing methyl parathion (250 μg/ml) (9). E. coli transformation was performed as described by Sambrook et al. (30). B. subtilis transformation was performed by the competent cell method (3). The following antibiotics were used for selections: ampicillin, 100 μg/ml; spectinomycin, 100 μg/ml; erythromycin, 0.3 μg/ml; and zeocin, 20 μg/ml.
DNA manipulation techniques.
The isolation and manipulation of recombinant DNA were performed using standard techniques. All enzymes were commercial preparations and used as specified by the supplier (TaKaRa Biotechnology Co., Ltd.).
Plasmids.
The lox71-spc-lox66 cassette was amplified by PCR using pDG1730 (18) as a template and oligonucleotide pair P1/P2 as primers; lox71 and lox66 sites were introduced by the forward and reverse primers, respectively. The PCR-generated fragment was TA cloned into pMD18-T, yielding plasmid p7S6 (Fig. 2A). With p7S6 as template, the three primer pairs (forward/reverse) P3/P2, P1/P4, and P3/P4 were used to amplify fragments the lox72-spc-lox66, lox71-spc-lox72, and lox72-spc-lox72 fragments, respectively. These fragments were each cloned into pMD18-T to generate p2S6, p1S2, and p2S2. The promoter of the B. subtilis DNA-binding protein HBsu gene hbs (28) and two mutant lox sites (lox71 and lox66) were added to the Zeor gene Sh ble by PCR using pPICZαA (http://www.invitrogen.com) as a template and oligonucleotides P5, P6, and P7 as primers. The PCR-produced lox71-zeo-lox66 cassette was cloned into pMD18-T to create p7Z6 (Fig. 2A).
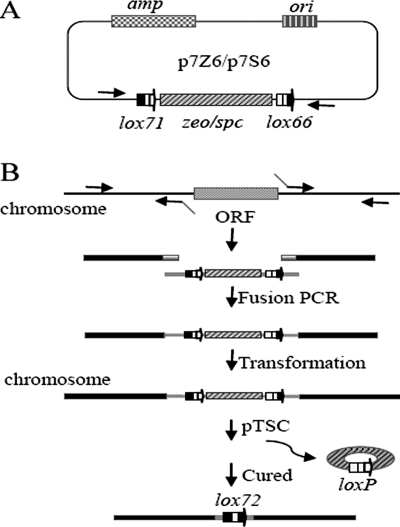
FIG. 2.
Vectors containing antibiotic resistance marker cassettes (A) and scheme for Cre/lox and PCR-based mutation delivery in B. subtilis (B). (A) ori, ColE1 replication origin; amp, Ampr marker; zeo/spc, Zeor or Spcr marker; horizontal solid arrows, primer binding site. (B) (i) lox71-spc/zeo-lox66,lox71-spc-lox66/lox71-zeo-lox66 cassette. The front and back regions flanking the target to be deleted are PCR amplified, gel purified and fused by PCR. (ii) PCR-fused products are directly used to transform B. subtilis, and Spcr or Zeor transformants are selected. (iii) pTSC is introduced into a Spcr or Zeor clone, and the constitutively expressed Cre recombinase mediates the recombination between lox71 and lox66. (iv) pTSC is eliminated to get the target strain by patching a transformant on antibiotic-free LB agar and incubating it at 51°C.
To construct a temperature-sensitive vector for this study, the plasmids pUC19 carrying an ampicillin resistance (Ampr) marker (37) and pE194 (21) carrying an erythromycin resistance (Emr) marker were each digested with NdeI, ligated together, and transformed into E. coli DH5α. Transformants were selected on LB agar containing both ampicillin (100 μg/ml) and erythromycin (300 μg/ml). Transformants were examined by restriction digestion to identify an appropriate shuttle plasmid containing both pUC19 and pE194 fragments. One recombinant, pTS, was confirmed by further restriction enzyme digestion. The Cre recombinase gene cre was amplified using the primer pair P8/P9, with the HindIII site and the Shine-Dalgarno sequence (UAAGGAGG) introduced by the forward primer and the SphI site introduced by the reverse primer. The HindIII/SphI-digested product was then cloned into the corresponding site of pDG148 (33) to yield pDGC, putting cre under the control of the Pspac promoter. Pspac-cre expression cassette (without lacI) was excised from pDGC with EcoRI/SphI and ligated into pTS, also cut with EcoRI/SphI, yielding plasmid pTSC. For the elimination of pTSC from B. subtilis, a transformant was patched on antibiotic-free LB agar and incubated at 51°C (13): the resulting colonies all lost pTSC.
For complementation analysis of the hisZ gene in-frame deletion mutation, the hisZ gene and its promoter region was amplified with the primer pair P32/P33. The PCR product was digested by BamHΙ/PstΙ and ligated into the corresponding sites of the B. subtilis-E. coli shuttle vector pUBC19 (39), yielding pPhisZ.
For integration of the cre IPTG-inducible expression cassette into the B. subtilis chromosome at the amyE site, plasmid pDGIC was constructed as follows. The EcoRΙ- and BamHΙ-flanked fragment containing an IPTG-inducible Pspac promoter and the lac repressor-encoding gene lacI were transferred from pDG148 to the corresponding sites of pDG1730, giving pDGI; then, the cre gene was amplified using the primer pair P8/P9 above, and the HindIII/SphI-digested PCR product was cloned into the corresponding sites of pDGI to put cre under the control of the IPTG-inducible Pspac promoter and to yield pDGIC. For insertion of cre IPTG-inducible expression cassette at the lacA locus of B. subtilis, the cre IPTG-inducible expression cassette and Spcr marker were PCR amplified using pDGIC as template and P34 and P35 as primers. The SmaI-digested PCR product was ligated with pAX01 (19), which was also digested by SmaI, to generate pAXC. To generate a lox-free big cassette containing the cre IPTG-inducible expression cassette and the Zeor marker, the Zeor marker was amplified using primer pair P36/P37, and the BamHΙ-digested PCR product was inserted downstream of the lacI gene in pDGIC, yielding pDGICZ.
PCR-based fusion of antibiotic resistance marker cassette with LFH regions.
Fusion of the antibiotic resistance marker cassette with LFH regions by PCR was done as described by Shevchuk et al. (32). In brief, it was carried out as follows. Marker cassettes were amplified from vector p7Z6 or p7S6 with primer pair P10/P11 or P12/P13. Two primer pairs were used to amplify ∼450-bp or ∼900-bp DNA fragments flanking the region to be mutated at its front and back ends. Extensions of 25 to 40 nucleotides (nt) that were complementary to the 5′ and 3′ ends of the amplified marker cassette were added to the 5′ end of the reverse and forward primers of the front and back flanking regions, respectively. Three fragments were amplified using PrimeSTAR HS DNA polymerase, and all PCR products were gel purified with extraction from agarose using the AxyPrep DNA gel purification and extraction kit (Axygen). The purified marker cassette fragment could be used repeatedly. The steps were as follows. Step A included 12 μl water, 5 μl PrimeSTAR buffer (5×), 2 μl deoxynucleoside triphosphates (dNTP) mix (2.5 mM each), 2.5 μl (25 ng) front flanking fragment, 2.5 μl (25 ng) back flanking fragment, 0.5 μl (5 ng) marker cassette fragment, and 0.5 μl PrimeSTAR HS DNA polymerase. Step B included 32 μl water, 10 μl PrimeSTAR buffer, 4 μl dNTP mix, 1 μl forward primer of front flanking fragment, 1 μl reverse primer of back flanking fragment, 1 μl of unpurified PCR product from step A, and 1 μl PrimeSTAR HS DNA polymerase. The resulting PCR product was analyzed by electrophoresis in 1% agarose.
Nucleotide sequence accession numbers.
The sequences of plasmids p7S6, p7Z6, pTSC, and pDGICZ have been submitted to NCBI under accession no. EU541493, EU541492, EU864234, and EU864235, respectively.
RESULTS
Scheme for Cre/lox system and PCR-based genetic modification system in B. subtilis.
The lox71-spc-lox66 and lox71-zeo-lox66 resistance marker cassettes were first cloned into the pMD18-T vector to yield the marker cassette templates p7S6 and p7Z6 (Fig. 2A). Zeocin is rarely used in B. subtilis for selection, but we found it worked very well at 20 μg/ml. The rationale of this system for genetic modification is shown in Fig. 2B. The front and back regions flanking the target to be deleted were amplified by PCR with the appropriate primers, the lox71-spc-lox-66 and lox71-zeo-lox66 cassettes, and these two region fragments were fused by PCR, the PCR products were directly used to transform B. subtilis competent cells, and Spcr or Zeor transformants were selected. pTSC was introduced into an Spcr or Zeor clone, and most (>90%) of the Emr transformants were spectinomycin and zeocin sensitive (Spcs Zeos), as the constitutively expressed Cre recombinase efficiently promoted the recombination between lox71 and lox66, thereby deleting the resistance marker cassette and leaving a 34-bp double-mutant lox72 site in the chromosome of B. subtilis; then pTSC was eliminated to get the target strain. Note that the lox71 should be designed upstream of lox66, so that, after recombination between them, the double-mutant lox72 site is left in the chromosome and the wild-type loxP site is eliminated with the resistance marker cassette. The whole process can be accomplished in about 4 days.
Knockout of the amyE gene and deletion of the prophage 3 region.
To test the feasibility of this strategy, we first used it to knock out the well-known amyE gene, which is usually used as an integration site (18). The upstream and downstream fragments flanking amyE were PCR amplified using B. subtilis 168 chromosomal DNA as the template and oligonucleotide pairs P14/P15 and P16/P17 as primers, respectively. The lox71-spc-lox66 cassette was PCR amplified using p7S6 as the template and oligonucleotide pair P10/P11 as primers. According to the procedure described above, the 1,166-bp fragment of the amyE gene was deleted, and a double-mutant lox72 site remained. The resulting mutant, BSPC1, was confirmed by PCR amplification with primers P14 and P17 (Fig. 3A), sequencing of homologous regions (data not shown), and detection of α-amylase activity (halo assay). These results suggest that the strategy described above worked efficiently.
FIG. 3.
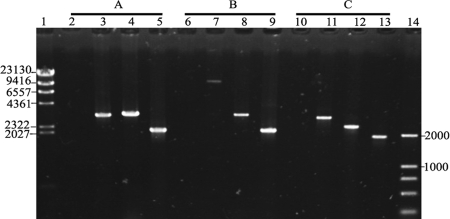
Confirmation of the amyE knockout (A), prophage 3 deletion (B) and hisZ in-frame deletion (C) by PCR. The PCR products were analyzed by agarose gel electrophoresis. Lanes 1 and 14, DNA markers. (A) P14 and P17 were used as primers, and water (lane 2), 168 (lane 3), 168 [ΔamyE::(lox71-spc-lox66)] (lane 4), and BSPC1 (ΔamyE::lox72)(lane 5) were used as templates. (B) P18 and P21 were used as primers, and water (lane 6), 168 (lane 7), BSPC7 [ΔamyE::(lox71-spc-lox66)] (lane 8), and BSPC2 (Δprophage 3::lox72) (lane 9) were used as templates. (C) P21 and P25 were used as primers, and water (lane 10), 168 (lane 11), 168 [ΔhisZ::(lox71-zeo-lox66)] (lane 12), and BSPC3 (ΔhisZ::lox72) (lane 13) were used as templates.
DNA sequencing of the B. subtilis chromosomal rrnE region revealed a 13-kb region with a lower G+C content and lower gene density between the groESL operon and the gut operon (22); this region was called prophage 3, which contains the genes for both BsuM modification (ydiO and ydiP) and BsuM restriction (ydiR, ydiS, and ydjA) (36). Here, we used the method described above to delete the prophage 3 region (SubtiList coordinates bp 652840 to 664137). Two primer pairs, P18/P19 and P20/P21, were used to amplify the upstream and downstream fragments flanking prophage 3 from B. subtilis 168 chromosomal DNA. The lox71-spc-lox66 fragment was already gel purified above. An ∼11-kb fragment was deleted, and lox72 remained, yielding strain BSPC2, which was confirmed by PCR amplification with primers P18 and P21 (Fig. 3B) and sequencing of homologous regions (data not shown).
In-frame deletion of the hisZ gene in the histidine operon.
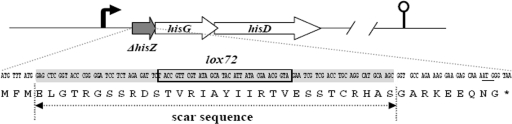
An in-frame deletion can inactivate a protein by removing its central part while preserving the signals for translation regulation, which minimizes the chance of exerting a polar effect on the expression of the downstream genes. hisZ, the first gene of the B. subtilis histidine operon (Fig. 4), was used to generate an in-frame deletion. Primer pairs P22/P23 and P24/P25 were designed to amplify upstream and downstream regions from B. subtilis chromosomal DNA. The lox71-zeo-lox66 cassette was amplified using p7Z6 as template and P12 and P13 as primers. The primers were designed so that, after Cre-mediated excision of the lox-flanked Zeor marker, the remnant open reading frame (ORF) contained a translatable scar sequence in-frame with the hisZ gene initiation codon, its N-terminal 6-nt coding region, and its C-terminal 30-nt coding region (Fig. 4). Translation from the ΔhisZ Shine-Dalgarno sequence and start codon was expected to produce a 43-residue peptide with the HisZ N-terminal MetPheMet, 31 scar-specific residues, and 9 residues of the HisZ C terminus (Fig. 4). The mutation process and the remnant ORF of hisZ were confirmed by PCR amplification (Fig. 3C) and DNA sequencing. The resulting ΔhisZ strain, BSPC3, required histidine for growth in MM. For complementation analysis, pUBC19 (control) and pPhisZ were separately introduced into BSPC3 to generate BSPC3(pUBC19) and BSPC3(pPhisZ). BSPC3(pPhisZ) grew well in MM, but BSPC3(pUBC19) required histidine for growth. The hisZ and hisG genes overlap by 8 nt, and hisG and hisD overlap by 4 nt. The addition of histidinol, which is converted in histidine by histidinol dehydrogenase (HisD), restored the growth of BSPC3 in MM. These results prove that few secondary mutations were caused by hisZ gene in-frame deletion.
FIG. 4.
In-frame deletion of hisZ gene in the histidine operon. The designed structure of the hisZ in-frame deletion. The ORFs are represented by open arrows, the promoter region is represented by a bent arrow, and the transcription terminator is represented by a lollipop. Cre-mediated excision of the lox-flanked zeocin-resistant marker is predicted to create a translatable scar sequence that is in frame with the hisZ gene (N-terminal 9-nt coding region and C-terminal 30-nt coding region). Translation from the ΔhisZ Shine-Dalgarno sequence and start codon is expected to produce a 43-residue peptide with the HisZ N-terminal MetPheMet, 31 scar-specific residues, and 9 residues of the HisZ C terminus. The scar sequence is shaded, and the lox72 site is boxed.
Shortening the length of the PCR-fused fragment.
The disadvantage of the PCR-based allele replacement strategy is as obvious as its advantage. It is more time saving than a cloning-based procedure, but as the amplified fragment lengthens, the number of undesired errors increases. Here, PCR-generated errors in the flanking homology regions were not expected to be introduced into the B. subtilis chromosome through a double-crossover event. On the premise of getting recombination-mediated transformants, we managed to reduce the length of the resulting PCR-fused fragment. Integration of linear DNA into B. subtilis requires a minimal length of homology of about 400 to 500 bp (15), so the recombination-mediated integration efficiencies of the fusion fragments containing ∼450-bp flanking homology regions were investigated here. Four primer pairs were used to generate different lengths of the homology regions (upstream/downstream regions of amyE and hisZ sites) flanking the lox71-zeo-lox66 cassette (500 bp): P14/P15 (986/931 bp), P26/P27 (401/401 bp), P22/P25 (900/916 bp), and P28/P29 (514/512 bp). As shown in Table 2, under our conditions, PCR-generated products with homology of about 900 bp on either side were sufficient to promote effective recombination-mediated transformation (∼103/μg), and products with homology of ∼450 bp gave approximately tens of transformants (<102/μg). This result implies that, with a larger amount of PCR products added, products with homology of ∼500 bp also work well. A short Zeor marker lox71-zeo-lox66 cassette (500 bp) was constructed; this cassette was found to work well in B. subtilis. So the length of the resulting PCR-fused fragment was shortened to about 1,500 bp, thereby decreasing the number of the errors generated by PCR amplification and the possibility of introducing undesired mutations into the regions flanking the target site in the chromosome of B. subtilis. The 1,500-bp PCR-fused products amplified with PrimeSTAR HS DNA polymerase were TA cloned and sequenced. No error was found in two of the three sequenced clones, and one error arose in the primer region of the last clone (data not shown), which may have been a result of the synthesis of the long oligonucleotide.
TABLE 2.
Recombination-mediated transformation efficiencies of the PCR-fused fragments in B. subtilis
| Integration site | Homology length (bp)
|
Transformation efficiencya | |
|---|---|---|---|
| Upstream | Downstream | ||
| amyE | 401 | 401 | 0.9 × 102 |
| 986 | 931 | 1.7 × 103 | |
| hisZ | 514 | 512 | 0.5 × 102 |
| 900 | 916 | 1.5 × 103 | |
The transformation efficiency was calculated as the number of Zeor colonies/μg of PCR-fused products.
Comparing the in vivo Cre-mediated recombination efficiencies of different lox pairs.
Cre-mediated recombination between lox71 and lox66 generates a wild-type loxP site and a double-mutant lox72 site that has low affinity for Cre (2). To investigate the potential effect of the lox72 site remaining in the chromosome on the repeated manipulations in the same background, we compared the in vivo Cre-mediated recombination efficiencies of different lox pairs (lox72/lox72, lox71/lox72, lox72/lox66, and lox71/lox66). The lox72-spc-lox72, lox71-spc-lox72, lox72-spc-lox66, and lox71-spc-lox66 cassettes were each integrated into the B. subtilis chromosome at the amyE site by the method described above, yielding strains BSPC4, BSPC5, BSPC6, and BSPC7, respectively. These four strains were each transformed with plasmids pTSC and pTS (control) and incubated at 37°C for 24 h. Then, 100 transformants of each strain were transferred into LB broth containing 100 μg/ml spectinomycin in 96-well plates and incubated at 37°C with shaking; Spcs transformants (resulting from the loss of the spectinomycin cassette during Cre-mediated recombination between two lox sites) were counted after 24 h. As shown in Table 3, none of the checked transformants harboring pTS was Spcs, indicating that the recombination of different lox pairs was Cre mediated. Among them, lox71/lox66 had the highest Cre-mediated recombination efficiency, and about 92% of the tranformants had undergone a Cre-mediated recombination event between lox71 and lox66, losing the Spcr marker; on the contrary, 90% of the BSPC4 [ΔamyE::(lox72-spc-lox72)] transformants were still Spcr, which validated the low affinity of lox72 to Cre recombinase and also suggested that lox72/lox72 has a low Cre-mediated recombination efficiency in vivo. Compared with the recombination efficiency of lox71/lox66, the relative Cre-mediated recombination efficiencies were approximately 0.41 for lox71/lox72 and about 0.39 for lox72/lox66. So, the main effect of the lox72 site remaining in the chromosome on multiple mutations in the same background is its recombination with the lox71 or lox66 sites subsequently introduced. The lox71/lox66 pair has higher recombination efficiency, however, than lox71/lox72 or lox72/lox66. Furthermore, if the lox72 sites are physically far from lox71 or lox66, lox72 sites left from previous manipulations should have little effect on the next manipulation. To prove this point, we successfully introduced a prophage 3 deletion and a hisZ in-frame deletion mutation into BSPC1 (ΔamyE::lox72), yielding BSPC8 (ΔamyE::lox72 ΔhisZ::lox72 Δprophage 3::lox72).
TABLE 3.
In vivo recombination efficiencies of different lox pairs
| lox pair | Spcs transformants (pTSC/pTS)a | Efficiency (%)b | Relative efficiency vs. lox71/lox66 |
|---|---|---|---|
| lox71/lox66 | 92/0 | 92 | 1 |
| lox72/lox71 | 36/0 | 36 | 0.41 |
| lox72/lox66 | 38/0 | 38 | 0.39 |
| lox72/lox72 | 10/0 | 10 | 0.11 |
The strains containing different lox pairs were each transformed with plasmids pTSC and pTS (control) and incubated at 37°C for 24 h on LB agar containing erythromycin.
The efficiency of Cre-mediated recombination was calculated as the no. of Spcs transformants/no. of all tested transformants (100).
Introduction of the mpd gene into B. subtilis chromosome.
This system can also be used to conveniently introduce a gene of interest into a target site without selection markers. There were two multiple cloning sites (MCS) flanking the lox71-zeo-lox66 cassette in the plasmid p7Z6. The resulting PCR fusion fragment (amyE upstream region, lox71-zeo-lox66 cassette, and amyE downstream region) containing the same MCS between the regions of homology and the lox71-zeo-lox66 cassette was TA cloned into pMD18T-simple, which lacks an MCS, giving pM7Z6M. The mpd (MPH gene from Plesiomonas sp. strain M6) (40) expression cassette was PCR amplified using plasmid pP43NMK (39) as the template and oligonucleotide pair P30/P31 as primers and inserted into the MCS of pM7Z6M at EcoRΙ and BamHΙ sites. The resulting plasmid, pAZP43M (see Fig. S1 in the supplemental material), was linearized and introduced into B. subtilis competent cells. Then, the lox71-zeo-lox66 cassette was evicted through transient Cre recombinase expression, yielding strain BSPC9. A yellow halo (p-nitrophenol) around the BSPC9 colony indicated the functional expression of the mpd gene (see Fig. S2 in the supplemental material).
Integration of the cre IPTG-inducible expression cassette into the B. subtilis chromosome.
To further simplify the mutation procedure, the plasmid pDGIC was linearized and introduced into B. subtilis 168 to generate BSPC10, in which the IPTG-inducible cre expression cassette was integrated at the amyE site. In so doing, we bypassed the introduction and elimination of plasmid pTSC. The lox71-zeo-lox66 cassette was inserted at the hisZ site of BSPC10 and B. subtilis 168, respectively, yielding BSPC11 [ΔamyE::(Pspac-cre spc lacI) ΔhisZ::(lox71-zeo-lox66 cassette)] and BSPC12 [ΔhisZ::(lox71-zeo-lox66 cassette)]. BSPC11 and BSPC12 (control) were each transferred into LB broth or LB broth containing 1 mM IPTG and incubated for 8 h, and then cells were diluted and spread on LB agar. One hundred colonies of each treatment were transferred into LB broth containing zeocin. All IPTG-induced BSPC11 cells were Zeos. Without the addition of IPTG, approximately 10% of BSPC11 cells were Zeos. With or without the addition of IPTG, all BSPC12 (control) cells were Zeor. This result suggests that IPTG-induced cre expression could efficiently promote the recombination of lox71/lox66 and that any leaky expression of cre from the IPTG-inducible Pspac promoter could also work in some cells. Inserting the cre gene at the frequently used amyE site may compromise the use of this site for other insertions, so we transferred the cre IPTG-inducible expression cassette to the lacA locus to produce BSPC13.
As an alternative, we tried to put the cre IPTG-inducible expression cassette inside the lox71-zeo-lox66 cassette so that, after the induction, the Cre recombinase would excise both the marker gene and the cre gene. We could not accomplish this construction, however, even in E. coli strain JM109 (lacIq), probably due to leaky cre expression from the IPTG-inducible Pspac promoter. To solve this problem, we constructed the vector pDGICZ to generate a big lox-free cassette containing the cre IPTG-inducible expression cassette and Zeor marker, and then, in the PCR fusion process, lox71 and lox66 were added to the both sides of the big cassette by the primer pair P38/P39. The amyE gene was successfully deleted using this strategy. (Primer pairs P14/P40 and P41/P17 were used to amplify upstream and downstream regions of amyE, respectively.) In this manner, after the induction, both the marker gene and the cre gene were excised, obviating the need for the introduction and elimination of plasmid pTSC. The length of the resulting PCR fragment increased (4 to 5 kb), however, so the accuracy of this approach was possibly affected.
DISCUSSION
In this study, we developed a Cre/lox system and PCR-based strategy for genetic manipulation of the B. subtilis chromosome that could rapidly introduce mutations and eliminate the selection marker. This method bypasses the traditional time-consuming vector construction procedure and overcomes the drawback of methods that leave selection markers behind. Furthermore, the length of the resulting PCR-fused fragment could be shortened to approximately 1,500 bp, which remarkably reduces the errors generated by PCR amplification and the possibility of introducing undesired mutations into the regions flanking the targeted site in the chromosome of B. subtilis. However, it should be noted that sequencing of the flanking regions is necessary and our approach only reduces the number of clones that have to be sequenced to identify a clone without errors. Introduction of a cre expression temperature-sensitive vector promoted the efficient recombination between two single-mutant lox sites and created a double-mutant lox72 site that did not affect further manipulations; three mutations were successfully introduced into the same background by this method.
To further simplify our scheme, an IPTG-inducible cre expression cassette was integrated at the amyE site of the B. subtilis chromosome. IPTG-induced cre expression could efficiently promote the recombination between lox71 and lox66. Therefore, the whole mutation procedure could be accomplished in 2 days when using BSPC11 as the recipient. As a result, genome-scale mutations could be carried out efficiently in B. subtilis, as has been done for the construction of Escherichia coli K-12 in-frame, single-gene-knockout mutants (4). The IPTG-inducible cre gene was ligated with the Zeor marker to produce a big lox site-free cassette, and lox71 and lox66 were added to both sides of the big cassette by the primers. After induction, the Cre recombinase would excise both the marker gene and the cre gene, eliminating the need for the introduction and elimination of pTSC.
An about-11-kb prophage 3 region was successfully replaced using the lox71-spc-lox66 cassette flanked by upstream and downstream homologies (∼900 bp each). When a much longer region needs to be deleted, however, this procedure may not work. Another strategy should work in this case: first, lox71 and lox66 would be placed at the two sides of the target region, and then transiently expressed Cre recombinase would delete the target region together with the selection markers. Alternatively, when the lox71 and lox66 are divergently oriented, the target region will be inverted, a property that could be exploited to create large genome rearrangements.
In conclusion, this method is simpler and more applicable than existing methods for genome engineering in B. subtilis. The strains BSPC10 and BSPC13, as well as the plasmids p7S6, p7Z6, pTSC, and pDGICZ, will be accessible from the Bacillus Genetic Stock Center (http://www.bgsc.org) (accession no. 1A872, 1A873, ECE202, ECE203, ECE204, and ECE205).
Supplementary Material
Acknowledgments
We are grateful to the Bacillus Genetic Stock Center for providing bacterial strains and vectors. We appreciate Pu-Yan Chen for offering us the cre gene.
This work was supported by grants from the JiangSu Province Innovation Project (CX07B_053z), Chinese National Natural Science Foundation (40471073), National “863” Plan (2006AA10Z402, 2007AA10Z405, and 2007AA061101), and National Support Plan (2006BAD17D04 and 2006D90204007).
Footnotes
Published ahead of print on 18 July 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Abremski, K., R. Hoess, and N. Sternberg. 1983. Studies on the properties of P1 site-specific recombination: evidence for topologically unlinked products following recombination. Cell 32:1301-1311. [DOI] [PubMed] [Google Scholar]
- 2.Albert, H., E. C. Dale, E. Lee, and D. W. Ow. 1995. Site-specific integration of DNA into wild-type and mutant lox sites placed in the plant genome. Plant J. 7:649-659. [DOI] [PubMed] [Google Scholar]
- 3.Anagnostopoulos, C., and J. Spizizen. 1961. Requirements for transformation in Bacillus subtilis. J. Bacteriol. 81:741-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baba, T., T. Ara, M. Hasegawa, Y. Takai, Y. Okumura, M. Baba, K. A. Datsenko, M. Tomita, B. L. Wanner, and H. Mori. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baudin, A., O. Ozier-Kalogeropoulos, A. Denouel, F. Lacroute, and C. Cullin. 1993. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 21:3329-3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brans, A., P. Filée, A. Chevigné, A. Claessens, and B. Joris. 2004. New integrative method to generate Bacillus subtilis recombinant strains free of selection markers. Appl. Environ. Microbiol. 70:7241-7250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Broach, J. R., V. R. Guarascio, and M. Jayaram. 1982. Recombination within the yeast plasmid 2μ circle is site-specific. Cell 29:227-234. [DOI] [PubMed] [Google Scholar]
- 8.Bron, S., R. Meima, J. M. van Dijl, A. Wipat, and C. Harwood. 1999. Molecular biology and genetics of Bacillus spp., p. 392-416. In A. L. Demain and J. E. Davies (ed.), Manual of industrial microbiology and biotechnology, 2nd ed. ASM Press, Washington, DC.
- 9.Cui, Z. L., X. Z. Zhang, Z. H. Zhang, and S. P. Li. 2004. Construction and application of a promoter-trapping vector with methyl parathion hydrolase gene mpd as the reporter. Biotechnol. Lett. 26:1115-1118. [DOI] [PubMed] [Google Scholar]
- 10.Dahl, M. K., and C. G. Meinhof. 1994. A series of integrative plasmids for Bacillus subtilis containing unique cloning sites in all three open reading frames for translational lacZ fusions. Gene 145:151-152. [DOI] [PubMed] [Google Scholar]
- 11.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Defoor, E., M. B. Kryger, and J. Martinussen. 2007. The orotate transporter encoded by oroP from Lactococcus lactis is required for orotate utilization and has utility as a food-grade selectable marker. Microbiology 153:3645-3659. [DOI] [PubMed] [Google Scholar]
- 13.Dempsey, L. A., and D. A. Dubnau. 1989. Localization of the replication origin of plasmid pE194. J. Bacteriol. 171:2866-2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dubnau, D. 1991. Genetic competence in Bacillus subtilus. Microbiol. Rev. 55:395-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dubnau, D. 1993. Genetic exchange and homologous recombination, p. 555-584. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. American Society for Microbiology, Washington, DC.
- 16.Ellermeier, C. D., and R. Losick. 2006. Evidence for a novel protease governing regulated intramembrane proteolysis and resistance to antimicrobial peptides in Bacillus subtilis. Genes Dev. 20:1911-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fabret, C., S. D. Ehrlich, and P. Noirot. 2002. A new mutation delivery system for genome-scale approaches in Bacillus subtilis. Mol. Microbiol. 46:25-36. [DOI] [PubMed] [Google Scholar]
- 18.Guerout-Fleury, A. M., K. Shazand, N. Frandsen, and P. Stragier. 1995. Antibiotic-resistance cassettes for Bacillus subtilis. Gene 167:335-336. [DOI] [PubMed] [Google Scholar]
- 19.Härtl, B., W. Wehrl, T. Wiegert, G. Homuth, and W. Schumann. 2001. Development of a new integration site within the Bacillus subtilis chromosome and construction of compatible expression cassettes. J. Bacteriol. 183:2696-2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harwood, C. R., and S. M. Cutting (ed.). 1990. Molecular methods for Bacillus. John Wiley & Sons, Chichester, United Kingdom.
- 21.Horinouchi, S., and B. Weisblum. 1982. Nucleotide sequence and functional map of pE194, a plasmid that specifies inducible resistance to macrolide, lincosamide, and streptogramin type B antibiotics. J. Bacteriol. 150:804-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kasahara, Y., S. Nakai, N. Ogasawara, K. Yata, and Y. Sadaie. 1997. Sequence analysis of the groESL-cotA region of the Bacillus subtilis genome, containing the restriction/modification system genes. DNA Res. 4:335-339. [DOI] [PubMed] [Google Scholar]
- 23.Kobayashi, K., S. D. Ehrlich, A. Albertini, G. Amati, K. K. Andersen, M. Arnaud, K. Asai, S. Ashikaga, S. Aymerich, P. Bessieres, F. Boland, S. C. Brignell, S. Bron, K. Bunai, J. Chapuis, L. C. Christiansen, A. Danchin, M. Debarbouille, E. Dervyn, E. Deuerling, K. Devine, S. K. Devine, O. Dreesen, J. Errington, S. Fillinger, S. J. Foster, Y. Fujita, A. Galizzi, R. Gardan, C. Eschevins, T. Fukushima, K. Haga, C. R. Harwood, M. Hecker, D. Hosoya, M. F. Hullo, H. Kakeshita, D. Karamata, Y. Kasahara, F. Kawamura, K. Koga, P. Koski, R. Kuwana, D. Imamura, M. Ishimaru, S. Ishikawa, I. Ishio, D. Le Coq, A. Masson, C. Mauel, R. Meima, R. P. Mellado, A. Moir, S. Moriya, E. Nagakawa, H. Nanamiya, S. Nakai, P. Nygaard, M. Ogura, T. Ohanan, M. O'Reilly, M. O'Rourke, Z. Pragai, H. M. Pooley, G. Rapoport, J. P. Rawlins, L. A. Rivas, C. Rivolta, A. Sadaie, Y. Sadaie, M. Sarvas, T. Sato, H. H. Saxild, E. Scanlan, W. Schumann, J. F. Seegers, J. Sekiguchi, A. Sekowska, S. J. Seror, M. Simon, P. Stragier, R. Studer, H. Takamatsu, T. Tanaka, M. Takeuchi, H. B. Thomaides, V. Vagner, J. M. van Dijl, K. Watabe, A. Wipat, H. Yamamoto, M. Yamamoto, Y. Yamamoto, K. Yamane, K. Yata, K. Yoshida, H. Yoshikawa, U. Zuber, and N. Ogasawara. 2003. Essential Bacillus subtilis genes. Proc. Natl. Acad. Sci. USA 100:4678-4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuhn, R., and R. M. Torres. 2002. Cre/loxP recombination system and gene targeting. Methods Mol. Biol. 180:175-204. [DOI] [PubMed] [Google Scholar]
- 25.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, P. Bessieres, A. Bolotin, S. Borchert, R. Borriss, L. Boursier, A. Brans, M. Braun, S. C. Brignell, S. Bron, S. Brouillet, C. V. Bruschi, B. Caldwell, V. Capuano, N. M. Carter, S. K. Choi, J. J. Codani, I. F. Connerton, N. J. Cummings, R. A. Daniel, F. Denizot, K. M. Devine, A. Dusterhoft, S. D. Ehrlich, P. T. Emmerson, K. D. Entian, J. Errington, C. Fabret, E. Ferrari, D. Foulger, C. Fritz, M. Fujita, Y. Fujita, S. Fuma, A. Galizzi, N. Galleron, S. Y. Ghim, P. Glaser, A. Goffeau, E. J. Golightly, G. Grandi, G. Guiseppi, B. J. Guy, K. Haga, J. Haiech, C. R. Harwood, A. Henaut, H. Hilbert, S. Holsappel, S. Hosono, M. F. Hullo, M. Itaya, L. Jones, B. Joris, D. Karamata, Y. Kasahara, M. Klaerr Blanchard, C. Klein, Y. Kobayashi, P. Koetter, G. Koningstein, S. Krogh, M. Kumano, K. Kurita, A. Lapidus, S. Lardinois, J. Lauer, V. Lazarevic, S. M. Lee, A. Levine, H. Liu, S. Masuda, C. Mauel, C. Medigue, N. Medina, R. P. Mellado, M. Mizuno, D. Moestl, S. Nakai, M. Noback, D. Noone, M. O'Reilly, K. Ogawa, A. Ogiwara, B. Oudega, S. H. Park, V. Parro, T. M. Pohl, D. Portetelle, S. Porwollik, A. M. Prescott, E. Presecan, P. Pujic, B. Purnelle, G. Rapoport, M. Rey, S. Reynolds, M. Rieger, C. Rivolta, E. Rocha, B. Roche, M. Rose, Y. Sadaie, T. Sato, E. Scanlan, S. Schleich, R. Schroeter, F. Scoffone, J. Sekiguchi, A. Sekowska, S. J. Seror, P. Serror, B. S. Shin, B. Soldo, A. Sorokin, E. Tacconi, T. Takagi, H. Takahashi, K. Takemaru, M. Takeuchi, A. Tamakoshi, T. Tanaka, P. Terpstra, A. Tognoni, V. Tosato, S. Uchiyama, M. Vandenbol, F. Vannier, A. Vassarotti, A. Viari, R. Wambutt, E. Wedler, H. Wedler, T. Weitzenegger, P. Winters, A. Wipat, H. Yamamoto, K. Yamane, K. Yasumoto, K. Yata, K. Yoshida, H. F. Yoshikawa, E. Zumstein, H. Yoshikawa, and A. Danchin. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 26.Lambert, J. M., R. S. Bongers, and M. Kleerebezem. 2007. Cre-lox-based system for multiple gene deletions and selectable-marker removal in Lactobacillus plantarum. Appl. Environ. Microbiol. 73:1126-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mascher, T., N. G. Margulis, T. Wang, R. W. Ye, and J. D. Helmann. 2003. Cell wall stress responses in Bacillus subtilis: the regulatory network of the bacitracin stimulon. Mol. Microbiol. 50:1591-1604. [DOI] [PubMed] [Google Scholar]
- 28.Micka, B., N. Groch, U. Heinemann, and M. A. Marahiel. 1991. Molecular cloning, nucleotide sequence and characterization of the Bacillus subtilis gene encoding the DNA-binding protein HBsu. J. Bacteriol. 173:3191-3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oliver, S. G., M. K. Winson, D. B. Kell, and F. Baganz. 1998. Systematic functional analysis of the yeast genome. Trends Biotechnol. 16:373-378. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 31.Schallmey, M., A. Singh, and O. P. Ward. 2004. Developments in the use of Bacillus species for industrial production. Can. J. Microbiol. 50:1-17. [DOI] [PubMed] [Google Scholar]
- 32.Shevchuk, N. A., A. V. Bryksin, Y. A. Nusinovich, F. C. Cabello, M. Sutherland, and S. Ladisch. 2004. Construction of long DNA molecules using long PCR-based fusion of several fragments simultaneously. Nucleic Acids Res. 32:e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stragier, P., C. Bonamy, and C. Karmazyn-Campelli. 1988. Processing of a sporulation sigma factor in Bacillus subtilis: how morphological structure could control gene expression. Cell 52:697-704. [DOI] [PubMed] [Google Scholar]
- 34.Suzuki, N., H. Nonaka, Y. Tsuge, M. Inui, and H. Yukawa. 2005. New multiple-deletion method for the Corynebacterium glutamicum genome, using a mutant lox sequence. Appl. Environ. Microbiol. 71:8472-8480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wach, A. 1996. PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in S. cerevisiae. Yeast 12:259-265. [DOI] [PubMed] [Google Scholar]
- 36.Westers, H., R. Dorenbos, J. M. van Dijl, J. Kabel, T. Flanagan, K. M. Devine, F. Jude, S. J. Séror, A. C. Beekman, E. Darmon, C. Eschevins, A. de Jong, S. Bron, O. P. Kuipers, A. M. Albertini, H. Antelmann, M. Hecker, N. Zamboni, U. Sauer, C. Bruand, D. S. Ehrlich, J. C. Alonso, M. Salas, and W. J. Quax. 2003. Genome engineering reveals large dispensable regions in Bacillus subtilis. Mol. Biol. Evol. 20:2076-2090. [DOI] [PubMed] [Google Scholar]
- 37.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved m13 phage cloning vectors and host strains: nucleotide sequences of the m13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 38.Zhang, X. Z., X. Yan, Z. L. Cui, Q. Hong, and S. P. Li. 2006. mazF, a novel counter-selectable marker for unmarked chromosomal manipulation in Bacillus subtilis. Nucleic Acids Res. 34:e71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang, X. Z., Z. L. Cui, Q. Hong, and S. P. Li. 2005. High-level expression and secretion of methyl parathion hydrolase in Bacillus subtilis WB800. Appl. Environ. Microbiol. 71:4101-4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhangli, C., L. Shunpeng, and F. Guoping. 2001. Isolation of methyl parathion-degrading strain M6 and cloning of the methyl parathion hydrolase gene. Appl. Environ. Microbiol. 67:4922-4925. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.