Abstract
About 22,000 1-methyl-3-nitro-1-nitrosoguanidine- and UV-induced mutants of the rubber-degrading bacterium Streptomyces sp. strain K30 were characterized for the ability to produce clear zones on natural rubber latex overlay agar plates. Thirty-five mutants were defective solely in cleavage of rubber and were phenotypically complemented with the wild-type lcp (latex clearing protein) gene. Sixty-nine mutants exhibited a pleiotropic phenotype and were impaired in utilization of rubber and xylan, indicating that the enzymes responsible for the initial cleavage of these polymers are exported by the same secretion pathway (Q. K. Beg, M. Kapoor, L. Mahajan, and G. S. Hoondal, Appl. Microbiol. Biotechnol. 56:326-3381, 2001; U. K. Laemmli, Nature 227:680-685, 1970). Analysis of the amino acid sequence encoded by lcp revealed a twin-arginine motif, indicating that Lcp is a substrate of the twin-arginine translocation (Tat) pathway (K. Dilks, W. Rose, E. Hartmann, and M. Pohlschröder, J. Bacteriol. 185:1478-1483, 2003). A tatC disruption mutant of Streptomyces lividans 10-164 harboring lcp from Streptomyces sp. strain K30 was not capable of forming clear zones on rubber overlay agar plates. Moreover, Lcp and enhanced green fluorescent protein fusion proteins were detected in the supernatant. Using Escherichia coli having the twin-arginine motif in the signal peptide upstream of Lcp, clear evidence that Lcp is secreted was obtained. Transcriptional analysis revealed basal expression of Lcp in glucose-grown cells and that transcription of lcp is obviously induced in the presence of poly(cis-1,4-isoprene). In contrast, oxiB and oxiA, which are located directly downstream of lcp and putatively encode a heteromultimeric aldehyde dehydrogenase oxidizing the primary cleavage products generated by Lcp from poly(cis-1,4-isoprene), were expressed only in the presence of poly(cis-1,4-isoprene). Expression of lcp at a low level is thus required for sensing the polymer in the medium. Rubber degradation products may then induce the transcription of genes coding for enzymes catalyzing the later steps of poly(cis-1,4-isoprene) degradation and the transcription of lcp itself. lcp, oxiB, and oxiA seem to constitute an operon, as a polycistronic mRNA comprising these three genes was detected. The transcriptional start site of lcp was mapped 400 bp upstream of the lcp start codon.
The microbial degradation of natural and synthetic poly(cis-1,4-isoprene) rubber is currently being intensively investigated (for a review, see reference 24), and two different strategies for degradation of isoprene rubber have been described (18). Members of one group of organisms form clear zones on natural rubber latex agar plates and generally belong to the mycelium-forming actinomycetes, such as Actinoplanes, Streptomyces, and Micromonospora. Members of the second group comprise nocardioform actinomycetes like Gordonia, Mycobacterium, and Nocardia and do not produce translucent halos; instead, they require direct contact with the rubber substrates (37). Xanthomonas sp. strain 35Y is the only known rubber-degrading bacterium that does not belong to the actinomycetes but is a gram-negative bacterium (34); however, in terms of its strategy for rubber degradation, it belongs to the first group and forms halos on rubber-containing agar plates.
Whereas no proteins involved in rubber degradation in the nocardioform actinomycetes are known so far, the rubber-cleaving dioxygenase RoxA occurring in culture supernatants of Xanthomonas sp. strain 35Y (16, 34) and the lcp gene encoding Lcp (latex clearing protein) in Streptomyces sp. strain K30 have recently been identified and characterized by Braaz et al. (6) and Rose and Steinbüchel (27), respectively. Both of these bacteria belong to the so-called clear-zone-forming group of rubber-degrading bacteria, and obviously, both RoxA and Lcp are secreted into the extracellular medium, leading to the formation of translucent halos on natural rubber latex. Sequence analysis of Lcp and characterization of pleiotropic mutants of Streptomyces sp. strain K30 defective in the formation of clear zones on latex and on xylan obtained in this study indicated that secretion of Lcp occurs via the twin-arginine translocation (Tat) pathway (19).
Tat substrates are secreted in a folded conformation (3, 4, 15, 24, 29, 32, 33, 35, 36), and Tat signal peptides contain a highly conserved twin-arginine motif with the consensus sequence (S/T)RRXFLK (3, 7, 20, 39). Originally, the bacterial Tat pathway was identified as translocation machinery for redox proteins that bind cofactors or catalytic metal ions in the cytoplasm prior to export.
In the last 15 years green fluorescent protein (Gfp) has become a frequently used biological marker for proteins (8, 36). The stability and solubility of Gfp and the fact that this chromophor does not need a cofactor allow quite convenient studies of the localization, structure, and dynamics of macromolecules (40). For example, protein expression is followed simply by fluorescence microscopy.
MATERIALS AND METHODS
Bacterial strains, plasmids, and cultivation conditions.
Bacteria and plasmids used in this study are listed in Table 1. Unless indicated otherwise, cells of Streptomyces sp. were grown at 30°C in tryptic soy broth (Merck, Darmstadt, Germany), whereas cells of Escherichia coli were cultivated at 37°C in Luria-Bertani (LB) broth. Antibiotics were used as described by Sambrook et al. (30) and as indicated below. For growth experiments with natural and synthetic polyisoprene, cells were cultivated in mineral salt medium (MSM) (30). Carbon sources were added to MSM as indicated below. Liquid cultures were grown in Erlenmeyer flasks and were incubated on a horizontal rotary shaker. Solid media were prepared by addition of agar-agar (18 g/liter). Purified natural rubber latex from Hevea braziliensis (Neotex Latz; 60% natural rubber latex and 40% water) was a gift from Weber & Schaer, Hamburg, Germany, and was used for overlay plates as described previously (17). These plates were used to screen for mutant strains with an altered clear-zone-forming phenotype, as indicated by the formation or size of translucent halos appearing around a colony.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics(s) | Reference |
|---|---|---|
| Strains | ||
| Streptomyces sp. strain K30 | Wild type; producing clear zones on natural rubber latex overlay agar plates | 25 |
| Streptomyces lividans TK23 | Wild type; clear zone negative | 16 |
| Streptomyces lividans 10-164 | Wild type; clear zone negative | 12 |
| Streptomyces lividans 10-164 ΔtatC | tatC disruption mutant of S. lividans 10-164; gentamicin resistant | 9 |
| Streptomyces lividans TK23/pIJ702::lcp-egfp+linker | pIJ702 harboring lcp-egfp fusion gene connected by a 5-amino-acid-linker (GSIAT) | This study |
| Streptomyces lividans TK23/pIJ702::lcp-egfp | pIJ702 harboring lcp-egfp fusion gene without linker | This study |
| Streptomyces lividans TK23/pIJ702::lcpsig-egfp | pIJ702 harboring a fusion of the first 150 nucleotides of lcp in the 5′ region to egfp | This study |
| Streptomyces lividans TK23/pIJ702::egfp | pIJ702 harboring egfp | This study |
| Plasmids | ||
| pIJ702 | 16 | |
| pIJ702::lcp | pIJ702 harboring wild-type lcp from Streptomyces sp. strain K30 | 25 |
| pIJ702::lcpR6A6 | pIJ702 harboring R6A mutation of lcp | This study |
| pIJ702::lcpR7A7 | pIJ702 harboring R7A mutation of lcp | This study |
| pET23a::hyaAlcpK30 | pET23a harboring the twin-arginine motif as a signal peptide (hyaA, hydrogenase 1 small subunit) and wild-type lcp from Streptomyces sp. strain K30 | This study |
Protoplast formation and regeneration.
Protoplasts of Streptomyces strains were prepared from cells grown in modified YEME medium (3% [wt/vol] yeast extract, 5% [wt/vol] Bacto peptone, 3% [wt/vol] malt extract, 34% [wt/vol] sucrose) (18). R5 agar plates were used for protoplast regeneration (18).
Isolation, analysis, and manipulation of DNA.
Plasmid DNA was prepared from crude cell lysates by the alkaline extraction method (5). Before lysis, cells of Streptomyces were incubated in the presence of lysozyme (2 mg/ml) for at least 2 h at 37°C. Recombinant DNA techniques for Streptomyces were performed as described by Kieser et al. (18). Total DNA from Streptomyces was isolated by the versatile quick-prep method for gram-positive bacteria as described by Pospiech and Neumann (25). DNA was restricted with restriction endonucleases (MBI Fermentas, St. Leon-Rot, Germany) as mentioned below using the conditions recommended by the manufacturer. All other genetic procedures and manipulations were conducted as described by Sambrook et al. (30).
Staining with Schiff's reagent.
Aldehyde groups resulting from poly(cis-1,4-isoprene) cleavage during clear zone formation on natural rubber latex overlay agar plates were stained with Schiff's reagent using the procedure described previously (21).
Isolation of mutants defective in clear zone formation on natural rubber latex overlay agar plates. (i) UV-induced mutagenesis.
A suspension of Streptomyces sp. strain K30 spores (109 to 1010 spores ml−1 in 15% [wt/vol] glycerol) was irradiated with UV light in a glass petri dish, which resulted in a survival rate of 1%. The mutagenized spores were directly plated onto MSM latex overlay agar plates without any enrichment, and the phenotypes were analyzed for the ability to produce translucent halos.
(ii) NMG-induced mutagenesis.
Five hundred microliters of a suspension of Streptomyces sp. strain K30 cells was sedimented by centrifugation, and the cells were resuspended in 1 ml of a 25-μg ml−1 1-methyl-3-nitro-1-nitrosoguanidine (NMG) solution. The suspension was incubated for 50 min at 30°C, which resulted in a survival rate of approximately 40%, and this was followed by two washes with a saline solution (0.9% [wt/vol] NaCl). After suspension in 1 ml of the saline solution, appropriate dilutions were plated onto MSM agar plates containing glucose to obtain auxotrophic mutants. After this, colonies were transferred with toothpicks onto natural rubber latex overlay agar plates and were characterized for the ability to form translucent halos.
Site-directed mutagenesis.
Site-directed mutagenesis was performed by using two separate PCRs to generate two DNA fragments having overlapping ends (22). One reaction was used to amplify the 5′-terminal region of lcp and to introduce the desired mutation with a mismatch oligonucleotide primer. The other reaction was used to amplify the 3′-terminal region of lcp, again introducing the desired mutation. The two PCR products were then fused by the overlap extension method described by Ho et al. (13).
DNA sequencing.
DNA sequences of mutated lcp genes were determined using IRD800-labeled primers, a SequiTherm EXCEL II Long-Read L-C kit (Epicentre, Madison, WI), and a Li-COR model 4200 sequencer (LI-COR Biosciences, Lincoln, NE). Sequences were compared with the previously described sequence of the wild-type lcp gene (GenBank accession number AY387589) of Streptomyces sp. strain K30. Nucleotide or amino acid sequences were aligned by using the ClustalX software (32; http://www-igbmc.u-strasbg.fr/BioInfo/). Tat signals were predicted with the TatP 1.0 software (2).
Primer extension experiments.
The lcp oligonucleotide (5′CCGCCCCTAACGCCCCGACG-3′ ) was 5′ end labeled with the fluorescent dye IRD800 (MWG Biotech, Ebersberg, Germany). Twenty micrograms of total RNA isolated from Streptomyces sp. strain K30 cells was mixed with 5 μl of a 1 μM lcp primer solution, heated for 10 min at 70°C, and immediately transferred to 50°C. The extension reaction was carried out at 50°C for 50 min by using a 50-μl (total volume) mixture containing 200 U of SuperScript II reverse transcriptase (RT) (GIBCO BRL Life Technologies, Karlsruhe, Germany). After digestion with RNase A for 30 min at 37°C, the sample was precipitated with ethanol, dissolved in Tris-EDTA, and analyzed on a 6% polyacrylamide sequencing gel containing 8 M urea with a Li-COR 4000L sequencer. A sequencing reaction mixture containing the same labeled primer was run alongside to determine the size of the primer extension product.
PCR and RT-PCR.
All PCR amplifications of plasmid DNA or genomic DNA were carried out as described by Sambrook et al. (30) using Platinum Pfx DNA polymerase (GIBCO BRL Life Technologies, Karlsruhe, Germany) and an Omnigene HBTR3CM DNA thermal cycler (Hybaid, Heidelberg, Germany). RT-PCR was performed using a Qiagen one-step RT-PCR kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions and the following primers: P0_lcp, P1_oxiB, P2_ oxiAB, P3_oxiA, and _lcp-oxiB (Table 2). Contaminating DNA was hydrolyzed by addition of 37.7 U DNase (GIBCO BRL Life Technologies, Karlsruhe, Germany) and 15 min of incubation at room temperature. For control of DNA contamination, an RNA template was always added to one reaction mixture after inactivation of the RT and activation of the HotStarTaq DNA polymerase. The absence of RT-PCR products in the control indicated that the RT-PCR products were not derived from contaminating DNA.
TABLE 2.
Oligonucleotides used in this study
| Oligonucleotide | Sequence (5′→3′) | Function |
|---|---|---|
| P0_lcpf (lanes 1-2) | 5′AGCCAGGCCGACATCATGGTC3′ | Degenerate PCR primers based on homologous sequences for |
| P0_lcpr (lanes 1-2) | 5′GCAGGACGAACTTACGGAGCGC3′ | amplification of a 500-bp region of lcp by PCR |
| P1_oxiBf (lanes 3-4) | 5′GCTCTCCGCGTGTCATGGAAG3′ | Degenerate PCR primers based on homologous sequences for |
| P1_oxiBr (lanes 3-4) | 5′TGCCGTTGCTTGTAGCTCAGG3′ | amplification of a 500-bp region of oxiB by PCR |
| P2_ oxiABf (lanes 9-10) | 5′CCCGATGCAGCAGGCATG3′ | Degenerate PCR primers based on homologous sequences for |
| P2_ oxiABr (lanes 9-10) | 5′CGCAGCGACAGATGTTGCG3′ | amplification of a 500-bp region of oxiB into oxiA by PCR |
| P3_oxiAf (lanes 5-6) | 5′CATCGTCATCATGCCGGACACC3′ | Degenerate PCR primers based on homologous sequences for |
| P3_oxiAr (lanes 5-6) | 5′GGACACCGTGTCCGCGAC3′ | amplification of a 160-bp region of oxiA by PCR |
| _lcp-oxiBf (lanes 7-8) | 5′GACTGGCCAAGCAGCCGG3′ | Degenerate PCR primers based on homologous sequences for |
| _lcp-oxiBr (lanes 7-8) | 5′CCAGTACGTATCCCAGGAAACGTC3′ | amplification of a 500-bp region of lcp into oxiB by PCR |
| A_P0_lcpf | 5′AGCCAGGCCGACATCATGGTC3′ | Degenerate PCR primers based on homologous sequences for |
| A_P1_oxiBr | 5′TGCCGTTGCTTGTAGCTCAGG3′ | amplification of a 778-bp region of lcp into oxiB by PCR |
| B_P1_oxiBf | 5′GCTCTCCGCGTGTCATGGAAG3′ | Degenerate PCR primers based on homologous sequences for |
| B_P3_oxiAr | 5′GGACACCGTGTCCGCGAC3′ | amplification of a 1.9-kb region of oxiB into oxiA by PCR |
| C_P2_ oxiABf | 5′CCCGATGCAGCAGGCATG3′ | Degenerate PCR primers based on homologous sequences for |
| C_P3_oxiAr | 5′GGACACCGTGTCCGCGAC3′ | amplification of a 1.8-kb region of oxiB into oxiA by PCR |
| D_lcp-oxiBf | 5′GACTGGCCAAGCAGCCGG3′ | Degenerate PCR primers based on homologous sequences for |
| D_P1_oxiBr | 5′TGCCGTTGCTTGTAGCTCAGG3′ | amplification of a 1.9-kb region of lcp into oxiB by PCR |
| E_P1_oxiBf | 5′GCTCTCCGCGTGTCATGGAAG3′ | Degenerate PCR primers based on homologous sequences for |
| E_P2_oxiABr | 5′CGCAGCGACAGATGTTGCG3′ | amplification of a 1.8-kb region of oxiB into oxiA by PCR |
| Lcp_primer extension | 5′CCGCCCCTAACGCCCCGACG-3′ | 5′ end labeled with the fluorescence dye IRD800 to determine the size of the primer extension product |
Preparation of crude extracts.
Cells from 20-ml cultures were washed in 10 mM Tris-HCl (pH 7.4) and resuspended in 1 ml of this buffer containing 10 μg DNase I. The cells were disrupted by sonication with a Sonopuls GM 200 apparatus (Bandelin, Berlin, Germany), using an amplitude of 16 μm (1 min ml−1). During ultrasonication the samples were cooled in an NaCl/ice bath. Soluble protein fractions of the crude extracts were obtained by 5 min of centrifugation at 13,000 rpm and 4°C.
Preparation of cell extracts.
Cells of strain Streptomyces lividans TK23 were grown in MSM containing the indicated carbon sources plus 500 μg thiostrepton ml−1 for 48 h at 30°C. The cells were harvested by centrifugation for 20 min at 4°C and 4,000 rpm with a Megafuge 1.0R (Heraeus Sepatech Gmbh, Osterode, Germany), washed with 100 mM Tris-HCl buffer (pH 7.6), and resuspended in the same buffer. Lysozyme and DNase I were added to the cell suspension, which was then incubated for 30 min on ice. The cells were disrupted by three passages through a French press cell (Amicon, Silver Spring, MD), and the cell debris was removed by centrifugation (1 h, 4°C, 100,000 × g). The resulting supernatant was referred to as the crude extract and was used for further purification.
Purification of Lcp or a fusion protein.
The Streptomyces sp. strain K30 crude extract was loaded onto a Procion Red A column (flow rate, 1 ml min−1; bed volume, 35 ml), and the column was washed with 3 bed volumes of 100 mM Tris-HCl buffer (pH 7.6). For elution an NaCl gradient (0 to 1 M NaCl; flow rate, 1 ml min−1) was used, and 8.0-ml fractions were collected. The fractions were monitored for the occurrence of the (fusion) protein by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE), and the fractions containing this protein were pooled and concentrated by ultrafiltration (Vivascience, Satorius Group, Göttingen, Germany) to a volume of 1 ml. The (fusion) protein was eluted during the wash step from the column. For further purification the pool was diluted in gel loading buffer (1% [wt/vol] SDS, 1.25% [wt/vol] β-mercaptoethanol, 0.25 mM EDTA, 10% [vol/vol] glycerol, 0.001% [wt/vol] bromophenol blue, 12.5 mM Tris-HCl [pH 6.8]), denatured for 10 min at 95°C, and separated by preparative SDS-PAGE (12% [wt/vol] polyacrylamide) using a PrepCell 491 apparatus (Bio-Rad, Richmond, CA). The fractions were monitored for the occurrence of the protein by SDS-PAGE, and fractions containing the protein were pooled and concentrated by ultrafiltration (Vivascience, Satorius Group, Göttingen, Germany) to a volume of 150 μl.
Expression of LcpK30 in E. coli strains C41(DE3) and C43(DE3) and isolation of inclusion bodies.
E. coli strains C41(DE3) and C43(DE3) are mutants of E. coli BL21(DE3). E. coli strains C41(DE3) and C43(DE3) harboring plasmid pET23a::hyaAlcpK30 were cultivated in LB medium at 37°C until the optical density at 600 nm was 0.5, and then expression was induced by addition of isopropyl-β-d-thiogalactopyranoside (IPTG) to a final concentration of 1 mM for 3 h, which yielded cells with inclusion bodies (IBs). For isolation of IBs, the cells in a 100-ml culture were harvested, resuspended in 4 ml of 20 mM Tris-HCl (pH 8.0) buffer, and disrupted by two passages in a French press at 1,000 MPa. The disrupted cells were centrifuged at 25,000 × g for 15 min at 4°C. The pellet obtained was resuspended in 3 ml of cold IB wash buffer (2 M urea, 20 mM Tris-HCl, 0.5 M NaCl, 2% Triton X-100; pH 8.0) by sonication (1 min/ml with an amplitude of 40 μm) with a Bandelin Sonopuls GM200 ultrasonic disintegrator. After 15 min of centrifugation at 4°C and 25,000 × g, treatment with IB wash buffer, resuspension by sonication, and centrifugation were repeated three times. The purified IBs were dissolved in SDS denaturation buffer (18). A sample consisting of the dissolved IBs containing the extracted Lcp protein was separated by SDS-PAGE.
Western blotting.
To determine the N-terminal amino acid sequence of the protein, the concentrated pool from preparative SDS-PAGE harboring the protein was subjected to Western blotting. The protein was blotted from an SDS-PAGE gel onto a polyvinylidene difluoride (PVDF) membrane (Millipore, Bedford, MA) using the method of Towbin et al. (35a) and a Semidry Fast Blot B33 apparatus (Biometra, Göttingen, Germany).
Antibodies.
Anti-enhanced Gfp (eGfp) antibodies were purchased from BD Bioscience (Heidelberg, Germany). Anti-Lcp antibodies generated by Eurogentec (Seraing, Belgium) were used as described by Bröker et al. (6a).
Immunoblotting.
Protein detection was performed with anti-eGfp and anti-Lcp antibodies. PVDF membranes with blotted proteins were placed for 1 h in skim milk (5% [wt/vol]) to block nonspecific binding domains. After a membrane was washed with Tris-buffered saline (TBS)-Tween buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.025% [vol/vol] Tween 20), it was incubated in an antibody solution (antibodies diluted 1:2,000 in TBS-Tween buffer; 200 μl cm−2 membrane) and shaken overnight at room temperature. The membrane was washed three times for 10 min with TBS-Tween and then incubated with secondary antibodies (alkaline phosphatase-conjugated goat anti-rabbit immunoglobulin [IgG] [Sigma-Aldrich GmbH, Munich, Germany] diluted 1:20,000 in TBS-Tween buffer; 200 μl cm−2 membrane) and shaken for 1 h at room temperature. The membrane was then washed three times for 10 min with TBS-Tween buffer and stained using 5-bromo-4-chloro-3-indolylphosphate (BCIP)/nitroblue tetrazolium tablets dissolved in 10 ml H2O (Sigma, Deisenhofen, Germany).
Electrophoretic methods.
Proteins were separated under denaturing conditions in polyacrylamide gels and stained with Coomassie brilliant blue R250 as described by Osborn and Weber (38) or with silver stain (12).
Northern blot hybridization and analysis.
DNA probes were prepared and purified as described above for DNA. The purified cDNA fragment was labeled using a digoxigenin probe synthesis kit (Roche Applied Science, Mannheim, Germany) and the random priming method. Samples of total RNA (15 μg/lane) were separated on a 1.1% formaldehyde-agarose gel and blotted onto a Hybond N nylon membrane (Roche Applied Science, Mannheim, Germany). After UV cross-linking, the membrane was prehybridized at 42°C for at least 3 h in 50% formamide, 5× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.4]), 5× Denhardt's solution, 0.5% (wt/vol) SDS, and 200 μg/ml salmon sperm DNA. After prehybridization, the purified labeled probe was added to the prehybridization buffer, and the membrane was hybridized at 42°C for 12 to 18 h. Posthybridization washing was done twice using 2× SSPE-0.1% SDS at room temperature for 20 min and then once using 0.1× SSPE-0.1% SDS at 65°C for 20 min. The membrane was stripped by boiling it in 0.1% SDS before rehybridization. The membrane was also hybridized with an 18S rRNA housekeeping gene probe for normalization of differences in the RNA quantities loaded in the lanes. The membrane was exposed to a phosphor storage screen for 2 to 24 h, scanned, and analyzed.
Fluorescence measurement.
The fluorescence in culture supernatants was determined with a fluorescence photometer (SFM25; Kontron Instruments, Zürich, Switzerland). For this, 3 ml of cell-free supernatant was collected and examined in a crystal cuvette using an emission wavelength of 488 nm and an excitation wavelength of 507 nm.
RESULTS
Isolation of clear-zone-negative mutants.
Approximately 12,000 UV-induced and 10,000 NMG-induced mutants of Streptomyces sp. strain K30 were characterized with regard to their abilities to generate clear zones on natural rubber latex overlay agar plates, which indicated their abilities to degrade natural poly(cis 1,4-isoprene) (natural rubber). A total of 131 mutants that exhibited a natural-rubber-negative phenotype and were defective in formation of translucent halos were found; 86 of these mutants occurred after UV mutagenesis, and 45 occurred after NMG mutagenesis. All 131 mutants were also examined for the ability to degrade starch, Tween, xylan, skim milk, casein, pectin, lichenan, and DNA (Table 3). Interestingly, only 35 of the 131 mutants were specifically defective in clear zone formation on latex overlay agar plates; the other 96 mutants exhibited additional defects in utilization of at least one of the other substrates tested. Strikingly, a majority of these mutants (69 of the 96 mutants) had a pleiotropic phenotype and were negative for clear zone formation on latex overlay plates, and they were also defective for utilization of xylan. This suggested that there was dysfunction of a component of the protein secretion machinery in these mutants since the initial steps of poly(cis-1,4-isoprene) and xylan degradation should be catalyzed by independent extracellular enzymes and since the further catabolism of the degradation products of the two polymers does not occur via a common pathway. However, the possibility that this phenotype may be a regulation defect cannot be excluded.
TABLE 3.
Characterization of mutants obtained after NMG and UV mutagenesis
| Utilization defect(s) | No. of mutants |
|---|---|
| Latex | 35 |
| Latex, xylan | 69 |
| Latex, xylan, skim milk | 11 |
| Latex, skim milk | 9 |
| Latex, skim milk, casein, pectin | 2 |
| Latex, xylan, skim milk, Tween | 2 |
| Latex, starch, casein, lichenan | 1 |
| Latex, xylan, Tween | 1 |
| Latex, xylan, skim milk, casein, pectin, lichenan, DNA | 1 |
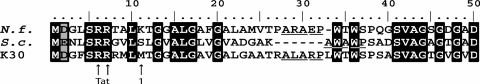
Since it was recently found that endo-1,4-β-xylanase C was secreted exclusively via the Tat protein translocation pathway in S. lividans (10), the amino acid sequence deduced from the lcp sequence was analyzed for the presence of a Tat secretion signal employing the TatP 1.0 server software (2; http://www.cbs.dtu.dk./services/TatP/). Surprisingly, no Tat motif was found. However, detailed sequence analysis of the native DNA fragment comprising lcp led to the conclusion that the previously published Lcp amino acid sequence (28, 29) was based on an incorrectly postulated start codon. Thirty base pairs upstream of the previously postulated start codon, an alternative ATG codon was identified in frame, leading to a hypothetical translational product consisting of 407 amino acids. The additional 10 amino acids at the N terminus contained a twin-arginine motif at positions 6 and 7. This region exhibited high levels of homology to the N termini of proteins homologous to Lcp in Nocardia farcinica IFM 10152 and Streptomyces coelicolor A3(2), and the TatP 1.0 software identified a hypothetical Tat signal peptide (Fig. 1).
FIG. 1.
Multiple alignment of the N-terminal amino acid sequences of Lcp from Streptomyces sp. strain K30 (K30) and homologous proteins from S. coelicolorA3(2) (S. c.) (accession no. CAB39713) and N. farcinica IFM 10152 (N. f.) (accession no. BAD57020). Arrow 1 indicates the amino acid previously described as the first amino acid of Lcp. The putative Tat motif predicted by TatP 1.0 is indicated by arrows (Tat). The most likely putative cleavage sites of the Tat signal peptides predicted by TatP 1.0 are underlined [K30, ALA-RP; N. farcinica, ARA-EP; S. coelicolorA3(2), AWA-WP].
Genetic complementation of clear-zone-negative mutants.
All 35 mutants specifically negative for clear zone formation on natural rubber latex overlay agar plates and nine randomly chosen pleiotropic mutants defective for formation of translucent halos both on latex and on xylan were characterized in more detail. Plasmid pIJ702::lcp, harboring the wild-type lcp gene and also the native promoter region of the gene isolated from Streptomyces sp. strain K30, was transformed by protoplast transformation into the corresponding mutants. This restored the wild-type phenotype in the 35 mutants, and the recombinants were able to produce clear zones on latex overlay plates and to produce aldehydes, as revealed by staining with Schiff's reagent. In contrast, none of the pleiotropic mutants were able to produce halos on latex agar plates or on xylan agar plates. This indicated that the pleiotropic mutants may have a defect in the protein secretion apparatus, causing deficiencies in both degradation capabilities.
Site-directed mutagenesis.
The revised N-terminal amino acid sequence deduced from lcp had a twin-arginine motif, indicating that there was Tat-dependent secretion of Lcp. Additionally, the TatP 1.0 software predicted that Lcp and homologous hypothetical proteins of N. farcinica IFM 10152 and S. coelicolor A3(2) are substrates of the Tat protein secretion machinery. Site-directed lcp mutants were constructed to prove this prediction. The start codon previously postulated by Rose et al. (28) and the start codon based on the reassessed sequence were replaced with codons for alanine. Additionally, each of the highly conserved arginine residues was changed to alanine to study the effect of these residues on secretion of Lcp. Mutants with site-directed mutations of lcp were cloned into pIJ702, and the resulting plasmids were transformed into S. lividans TK23 since clear zone formation by this recombinant strain generally was observed to occur more quickly and more distinctly than clear zone formation by recombinant Streptomyces sp. strain K30 mutants with defects in clear zone formation. If it harbored a pIJ702 derivative containing the wild-type lcp gene of Streptomyces sp. strain K30, an S. lividans TK23 recombinant formed translucent halos on natural rubber latex agar plates. Therefore, the abilities of recombinant strains of S. lividans harboring the different mutated lcp genes to produce visible clear zones and to form aldehydes detectable by staining with Schiff's reagent were compared.
The Met11Ala mutation of Lcp did not impair clear zone formation, indicating that translation does not start from this position. In contrast, the Met1Ala mutation impaired clear zone formation and also aldehyde formation. This provided strong evidence that translation starts at the corresponding ATG codon.
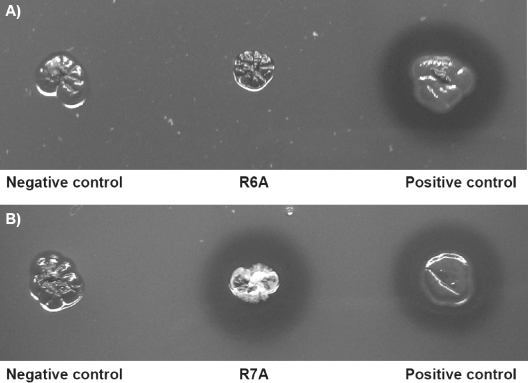
The recombinant strain of S. lividans TK23 harboring an Arg6Ala mutation was unable to form a clear zone or aldehydes, as revealed by staining with Schiff's reagent (Fig. 2) after 5 days of incubation. This strain produced very small translucent halos only after extended cultivation periods. Interestingly, the recombinant strain of S. lividans TK23 harboring an Arg7Ala mutation was not altered with regard to clear zone and aldehyde formation.
FIG. 2.
Effects of mutation of the “invariant” arginine residues. The mutants are shown in the center of the panels. S. lividans harboring pIJ702 was the negative control, and S. lividans/pIJ702::lcp was the positive control. (A) S. lividans/pIJ702::lcpR6A6 (Arg6Ala) did not produce clear zones stainable with Schiff's reagent. (B) S. lividans/pIJ702::lcpR7A7 (Arg7Ala) obviously secreted a functional Lcp, as indicated by the formation of a halo. After incubation for 5 days, agar plates were stained with Schiff's reagent to visualize aldehydes resulting from poly(cis-1,4-isoprene) cleavage (originally red).
Effect of tatC inactivation on clear zone formation.
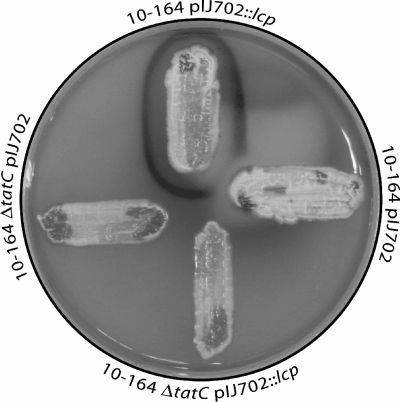
Since the data indicated that Lcp is a Tat substrate, the effect of tatC inactivation on clear zone formation mediated by the wild-type lcp gene was investigated using S. lividans 10-164, which is unable to degrade rubber. Plasmid pIJ702::lcp and plasmid pIJ702 (as a vector control) were transferred to a tatC disruption mutant, S. lividans 10-164 ΔtatC (10), and the parental strain S. lividans 10-164 (13) by protoplast transformation. Recombinant clones were then characterized to determine their abilities to produce translucent halos on natural rubber latex overlay plates and aldehydes (Fig. 3).
FIG. 3.
Effect of tatC inactivation on clear zone formation and production of aldehydes mediated by Lcp. S. lividans 10-164 harboring pIJ702::lcp (top) or pIJ702 (right) and S. lividans 10-164 harboring pIJ702::lcp (bottom) or pIJ702 (left) were cultivated for 5 days on a natural rubber latex overlay agar plate at 30°C. Aldehydes resulting from rubber degradation (clear zone formation) were visualized by staining with Schiff's reagent as described in Materials and Methods, which resulted in dark red staining.
The presence of plasmid pIJ702::lcp enabled S. lividans strain 10-164 to form clear zones and aldehydes, whereas formation of clear zones and aldehydes did not occur with the tatC disruption mutant or with the control strains harboring only pIJ702. These results clearly demonstrated that Lcp-mediated formation of clear zones on latex overlay agar plates, which is associated with the occurrence of aldehydes, is Tat dependent. Interestingly, the amount of stainable aldehydes surrounding a colony of S. lividans 10-164/pIJ702::lcp was clearly reduced if a colony of S. lividans 10-164 pIJ702 was nearby (Fig. 3). In contrast, this effect was not visible if S. lividans 10-164 ΔtatC harboring pIJ702 was grown in the neighborhood of S. lividans 10-164/pIJ702::lcp.
Transcription of lcp, oxiB, and oxiA.
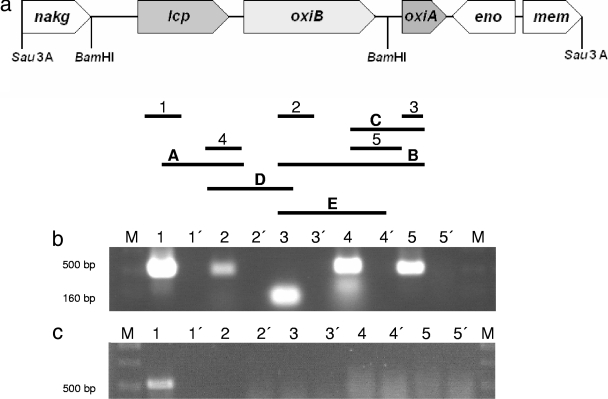
Previous studies led to identification of three genes (lcp, oxiB, and oxiA) coding for enzymes that are probably involved in the initial steps of rubber degradation in Streptomyces sp. strain K30. In order to analyze the physiological conditions under which these genes are transcribed, one-step RT-PCR assays were performed. Cells of Streptomyces sp. strain K30 were cultivated in MSM containing different carbon sources, including 0.5% natural rubber poly(cis-1,4-isoprene), 0.5% synthetic rubber poly(cis-1,4-isoprene), or 0.5% glucose. Total RNA from cells in the late logarithmic growth phase was subjected to RT-PCR using oligonucleotides specific for lcp, oxiB, or oxiA. Transcription of lcp, oxiB, and oxiA was strongly induced in the presence of natural or synthetic rubber poly(cis-1,4-isoprene), while in the presence of glucose only a faint RT-PCR product of lcp was detected; RT-PCR products of oxiB and oxiA could not be detected at all (Fig. 4a, b, and c), indicating that lcp was weakly transcribed during cultivation on glucose, whereas oxiB and oxiA were not transcribed under these conditions.
FIG. 4.
Transcriptional analysis of the lcp gene region. (a) Molecular organization of the 8-kbp Sau3A fragment from plasmid pR harboring lcp and adjacent genes. nakg encodes N-acetylglutamate kinase, lcp encodes latex-clearing protein (putative secreted protein), oxiB encodes oxidoreductase β-subunit, oxiA encodes oxidoreductase α-subunit, eno encodes enolase 2-phosphoglycerate dehydratase, and mem encodes a putative membrane transporter. The lines with numbers or letters represent the RT-PCR products obtained with mRNA from Streptomyces sp. strain K30; the numbers indicate the specific cDNA transcription products in the gel, and the letters indicate products of the other RT-PCRs. (b and c) Transcription analysis of the lcp region in Streptomyces sp. strain K30. Expression of lcp was analyzed by RT-PCR performed with samples containing total RNA isolated from cells in the logarithmic growth phase. The resulting PCR products were separated by agarose gel electrophoresis and stained with ethidium bromide, and negative images are shown. Cells were grown in MSM with either 0.5% (wt/vol) poly(cis-1,4-isoprene) (b) or 0.5% (wt/vol) glucose (c) as the carbon source. Lanes 1′, 2′, 3′, 4′, and 5′ contained the controls used to detect DNA contamination, whereas lanes 1, 2, 3, 4 and 5 contained the RT-PCR assay mixtures. A 100-bp DNA ladder (MBI Fermentas, Germany) (lanes M) was used as a molecular weight standard. Transcription of lcp (panel b, lane 1), oxiB (panel b, lane 2), oxiA (panel b, lane 3), Lcp into the OxiB PCR product (panel b, lane 4), and OxiB into the OxiA PCR product (panel b, lane5) was strongly induced in the presence of natural or synthetic rubber poly(cis-1,4-isoprene), while in the presence of glucose only a faint RT-PCR product of lcp (panel c, lane 1) was detected; RT-PCR products of oxiB and oxiA could not be detected at all.
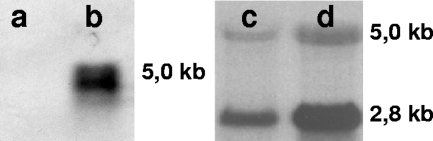
The Northern analysis produced similar results and confirmed the data obtained by RT-PCR. A specific signal was detected in RNA samples from cells grown in the presence of natural or synthetic rubber poly(cis-1,4-isoprene); such a signal was absent in the samples from glucose-grown cells (Fig. 5). The size of the signal was consistent with the presence of a polycistronic mRNA comprising lcp, oxiB, and oxiA.
FIG. 5.
Northern analysis of mRNA from Streptomyces sp. strain K30 cells cultivated in the presence of poly(cis-1,4-isoprene) and glucose as carbon sources. (Left panel) Northern blot. Lane a, mRNA isolated from glucose-grown cells; lane b, mRNA isolated from poly(cis-1,4-isoprene)-grown cells. (Right panel) Formaldehyde-agarose gel of total RNA. Lane c, mRNA isolated from glucose-grown cells; lane d, mRNA isolated from poly(cis-1,4-isoprene)-grown cells. For the panel on the right, total RNA samples (15 μg/lane) were separated in a 1.1% formaldehyde-agarose gel and blotted onto a Hybond N nylon membrane (Roche Applied Science, Mannheim, Germany). Agarose gel electrophoresis demonstrating the integrity of total RNA was used for the Northern analysis. The membrane was exposed to a phosphor storage screen for 2 to 24 h, scanned, and analyzed (left panel).
Identification of the lcp promoter.
After it was shown that expression of lcp is strongly induced during growth on poly(cis-1,4 isoprene), the transcriptional start site of lcp was mapped by primer extension. This site was located 400 bp upstream of the start codon of lcp.
Detection of Lcp-eGfp fusion proteins.
In order to localize Lcp in the cells and to demonstrate secretion of this protein, various fusions of Lcp with eGfp were generated and transferred to S. lividans TK23. S. lividans TK23 harboring pIJ702::lcp-egfp+linker, S. lividans TK23 harboring pIJ702::lcp-egf, S. lividans TK23 harboring pIJ702::lcpsig-egfp, and S. lividans TK23 harboring pIJ702::egfp were investigated. Cells were examined microscopically; in addition, antibodies against eGfp or Lcp were employed.
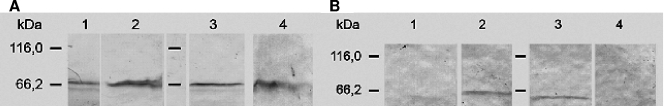
When supernatants and crude extracts from cells of S. lividans TK23 harboring pIJ702::lcp-egfp+linker or pIJ702::lcp-egfp were analyzed by immunoblotting with anti-eGfp antibodies, a signal for the fusion protein was found in the crude extract, as well as in the supernatant (Fig. 6). When anti-Lcp antibodies were used instead of anti-eGfp antibodies, the same signals were observed. The fluorescence in the cell-free supernatants increased strongly during cultivation of S. lividans TK23 harboring pIJ702::lcpsig-egfp; no increase occurred with the control.
FIG. 6.
(A) Detection of fusion proteins in crude extracts and in the medium by immunoblotting using anti-eGfp antibodies. Lane 1, supernatant from cultures of S. lividans TK23/pIJ702::lcp-egfp+linker; lane 2, crude extract from cells of S. lividans TK23/pIJ702::lcp-egfp+linker; lane 3, supernatant from cultures of S. lividans TK23/pIJ702::lcp-egfp; lane 4, crude extract from cells of S. lividans TK23/pIJ702::lcp-egfp. Cells were grown in MSM with 0.5% (wt/vol) glucose as the carbon source. (B) Detection of fusion proteins in crude extracts and in the medium by immunoblotting using anti-Lcp antibodies. Lane 1, supernatant from cultures of S. lividans TK23/pIJ702::lcp-egfp+linker; lane 2, crude extract from cells of S. lividans TK23/pIJ702::lcp-egfp+linker; lane 3, supernatant from cultures of S. lividans TK23/pIJ702::lcp-egfp; lane 4, crude extract from cells of S. lividans TK23/pIJ702::lcp-egfp. Cells were grown in MSM with 0.5% (wt/vol) glucose as the carbon source.
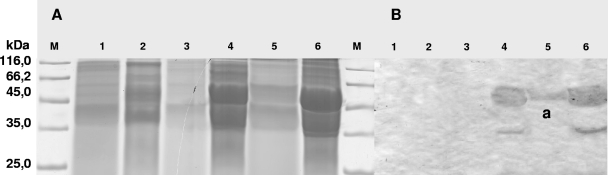
To obtain further direct evidence that Lcp is expressed and then secreted via the Tat pathway into the extracellular medium, plasmid pET23a::hyaAlcpK30 was constructed (Geneart's synthetic gene) and transferred to E. coli strains C41(DE3) and C43(DE3), which are mutants of BL21(DE3). HyaA (hydrogenase 1 small subunit) is a signal peptide with a twin-arginine motif, suggesting that the Tat pathway is involved in translocation of this protein. After cultivation of the cells in LB medium and addition of IPTG, IBs occurred. The IBs were isolated, dissolved in 4 ml of 20 mM Tris-HCl buffer (pH 8.0), separated in an SDS-polyacrylamide gel, and blotted onto a PVDF membrane. Incubation with anti-Lcp antibodies resulted in an intense signal for a fusion protein having a molecular mass of about 48 kDa in this Western blot (Fig. 7). Using the anti-Lcp IgGs with the extracellular protein fraction of E. coli C43(DE3) harboring pET23a::hyaAlcpK30 resulted in an intense immunoreaction signal corresponding to an apparent molecular mass of 43 kDa, indicative of cleavage of the fusion protein's HyaA moiety and the release of Lcp, as confirmed by matrix-assisted laser desorption ionization-time of flight analysis of the fusion protein (43-kDa Lcp plus 5-kDa HyaA). In contrast, using crude cell extracts of the negative controls (E. coli harboring pET23a) did not result in any signal, demonstrating that the anti-Lcp IgGs were specific for Lcp.
FIG. 7.
Immunological detection of Lcp from E. coli/pET23a::hyaAlcpK30 by Western blotting. (A) Electropherogram of an SDS-polyacrylamide gel after separation of proteins from crude cell extracts and extracellular protein fractions. Proteins in the gel were stained with Coomassie brilliant blue R250. (B) Western blot employing anti-LcpK30 IgGs prepared from an SDS-polyacrylamide gel. Lane M, molecular mass standard; lane 1, extracellular protein fraction of E. coli harboring pET23a; lane 2, crude cell extract of E. coli harboring pET23a; lane 3, extracellular protein fraction of E. coli C41(DE3) harboring pET23a::hyaAlcpK30; lane 4, crude cell extract of E. coli C41(DE3) harboring pET23a::hyaAlcpK30; lane 5, extracellular protein fraction of E. coli C43(DE3) harboring pET23a::hyaAlcpK30; lane 6, crude cell extract of E. coli C43(DE3) harboring pET23a::hyaAlcpK30. The E. coli cultures were grown in LB medium at 37°C to an optical density at 600 nm of 0.5, and then expression was induced by addition of IPTG to a final concentration of 1 mM for 3 h, which resulted in cells with IBs. In the Western blot the anti-LcpK30 IgGs recognized the approximately 43-kDa LcpK30 protein (band a).
DISCUSSION
Approximately 22,000 individual mutants of Streptomyces sp. strain K30 derived from chemical- or radiation-induced mutagenesis were examined, and 131 of these mutants exhibited a defect in the utilization of poly(cis-1,4-isoprene), as indicated by the lack of clear zone formation on natural rubber latex overlay agar plates. Thirty-five of these mutants were specifically defective in the formation of clear zones and were able to utilize all other substrates tested for growth like the wild type. Since all these mutants were phenotypically complemented by the wild-type lcp gene from Streptomyces sp. strain K30, it was concluded that the initial cleavage of poly(cis-1,4-isoprene) is dependent solely on Lcp in Streptomyces sp. strain K30. It is therefore unlikely that another protein is involved in rubber cleavage in this bacterium. Otherwise, not all of these mutants should have been complemented by lcp.
Most other mutants (69) had a pleiotropic phenotype and were defective not only in clear zone formation on latex overlay plates but also in the utilization of xylan and some other carbon sources. Xylans are degraded by endo-1,4-β-xylanases, which catalyze the hydrolytic endocleavage of β-1,4-d-xylopyranose linkages of the backbone, releasing xylose, xylobiose, xylotriose, and higher oligomers (1). This cleavage must occur outside the cells, and the endo-1,4-β-xylanases have to be secreted. Rubber and, to our knowledge, all other substrates require extracellular enzymes for initiation of degradation. A defect in the protein secretion machinery for extracellular enzymes therefore most likely explains the phenotypes of the pleiotropic mutants. Since it has been demonstrated that in S. lividans the export of endo-1,4-β-xylanase C is Tat dependent (10, 14) and since the TatP 1.0 software also predicted that Lcp is a Tat substrate, we concluded that both enzymes are secreted by a twin-arginine transporter. Consequently, the pleiotropic mutants should exhibit defects in the Tat-dependent protein secretion pathway.
Secretion of Lcp was proven by detection of Lcp-eGfp fusion proteins in the supernatant employing anti-eGfp and anti-Lcp antibodies, whereas the cells still formed clear zones on latex overlay agar plates. Further evidence was obtained when it was shown that the signal peptide nucleotide sequence of lcp is responsible for direction of eGfp out of the cells of S. lividans TK23/pIJ702lcpsig-egfp, whereas eGfp remained in the cytosol of cells of S. lividans TK23/pIJ702egfp. Direct proof for secretion of Lcp via the Tat pathway into the extracellular medium was obtained by immunological analyses using the Lcp-specific antibodies for Lcp (Fig. 7). Corresponding results were previously obtained by Bröker et al. (6a).
Two approaches verified that Lcp secretion occurs via the Tat pathway in Streptomyces sp. strain K30. (i) Like S. lividans 10-164 without a plasmid and in contrast to the parental strain and to S. lividans 10-164/pIJ702::lcp, the tatC disruption mutant S. lividans 10-164 ΔtatC harboring plasmid pIJ702::lcp did not produce clear zones on natural rubber latex overlay agar plates. (ii) Mutations in the highly conserved arginine residues confirmed that Lcp is a Tat substrate. S. lividans TK23/pIJ702::lcp harboring the wild-type lcp gene from Streptomyces sp. strain K30 cleaved poly(cis-1,4-isoprene), as visualized by production of clear zones on natural rubber latex overlay plates, whereas the Arg6Ala mutation resulted in an almost complete loss of this extracellular activity. In contrast, an Arg7Ala mutation did not result in a negative phenotype. However, this is consistent with previous studies which showed that a single mutation of an “invariant” arginine residue is not obligatorily associated with a lack of protein secretion and that there are even a few naturally occurring Tat substrates that have only a single arginine residue (23). Little is known about cofactors required for a functional active Lcp. However, histidine residues H198 and H203, which are highly conserved among Lcp homologues, are probably involved in the binding of iron (26). Iron may be incorporated into the folded protein, and the protein is then secreted in the folded state by the Tat transporter.
So far, heterologous protein secretion in bacterial expression systems has been investigated mostly by using signal peptides directing the proteins to secretion by the Sec pathway (31). However, for heterologous expression of extracellular proteins, knowledge of the secretion pathway is particularly important. When the signal peptide of xylanase C from S. lividans was replaced by the signal peptide of the hypothetical protein SCO6780 from S. coelicolor A3(2), the extracellular xylanase activity was increased more than twofold (20). In addition, simultaneous use of the Sec pathway and the Tat pathway also increased xylanase production (11). The knowledge concerning Lcp secretion obtained in this study will be used to optimize the heterologous expression of Lcp and its secretion.
Transcriptional analysis with RT-PCR revealed that basal expression of lcp occurred in glucose-grown cells and that transcription of lcp is obviously strongly induced in the presence of poly(cis-1,4-isoprene), whereas oxiB and oxiA, which are located directly downstream of lcp, were transcribed only in the presence of poly(cis-1,4-isoprene). These results indicated that a low level of expression of Lcp is required to generate cleavage products of the polymer that can then be sensed by the cells, thereby indicating that poly(cis-1,4-isoprene) is present in the habitat. Such product induction frequently regulates the expression of extracellular enzymes degrading polymers. In this example it includes the induction of lcp transcription from the lcp promoter identified 400 bp upstream of the lcp start codon and of the genes coding for enzymes catalyzing later steps of poly(cis-1,4-isoprene) degradation (i.e., transcription of oxiB and oxiA). In silico analysis identified 6 bp upstream of this transcriptional start site a “−10” box sequence and then further upstream a “−35” box sequence with reasonable similarity to other σ70 promoters. It seems that lcp, oxiB, and oxiA constitute an operon, because a polycistronic mRNA comprising the three genes (lcp, oxiB, and oxiA) was detected independently by two different methods (RT-PCR and Northern blot analysis). This is important evidence for the polycistronic regulation of lcp, oxiB, and oxiA. Further studies are necessary to determine rubber degradation in the gram-positive organism Streptomyces sp. strain K30 in more detail in order to isolate Lcp in an active form and to establish an activity assay.
Acknowledgments
Financial support for this study by the Deutsche Forschungsgemeinschaft is acknowledged (grant STE-386/10-1).
We are indebted to Rolf Morosoli (INRS-Institut Armand-Frappier, Universite du Quebec, Canada) for providing S. lividans 10-164 and the tatC disruption mutant S. lividans 10-164 ΔtatC. We thank D. Bröker for provision of antibodies raised against Lcp of Streptomyces sp. strain K30.
Footnotes
Published ahead of print on 7 July 2008.
REFERENCES
- 1.Beg, Q. K., M. Kapoor, L. Mahajan, and G. S. Hoondal. 2001. Microbial xylanases and their industrial application: a review. Appl. Microbiol. Biotechnol. 56:326-338. [DOI] [PubMed] [Google Scholar]
- 2.Bendtson, J. D., H. Nielsen, D. Widdick, T. Palmer, and S. Brunak. 2005. Prediction of twin-arginine signal peptides. BMC Bioinformatics 6:167-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berks, B. C. 1996. A common export pathway for proteins binding complex redox cofactors? Mol. Microbiol. 22:393-404. [DOI] [PubMed] [Google Scholar]
- 4.Berks, B. C., T. Palmer, and F. Sargent. 2005. Protein targeting by the bacterial twin-arginine translocation (Tat) pathway. Curr. Opin. Microbiol. 8:174-181. [DOI] [PubMed] [Google Scholar]
- 5.Birnboim, H. C., and J. Doly. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7:1513-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braaz, R., W. Armbruster, and D. Jendrossek. 2005. Heme-dependent rubber oxygenase RoxA of Xanthomonas sp. cleaves the carbon backbone of poly(cis-1,4-isoprene) by a dioxygenase mechanism. Appl. Environ. Microbiol. 71:2473-2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6a.Bröker, D., D. Dietz, M. Arenskötter, and A. Steinbüchel. 2008. The genomes of non-clearing-zone-forming and natural-rubber-degrading species Gordonia polyisoprenivorans and Gordonia westfalica harbor genes expressing Lcp activity in Streptomyces strains. Appl. Environ. Microbiol. 74:2288-2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chaddock, A. M., A. Mant, I. Karnauchov, S. Brink, R. G. Herrmann, R. B. Klosgen, and C. Robinson. 1995. A new type of signal peptide: central role of a twin-arginine motif in transfer signals for the delta pH-dependent thylakoid protein translocase. EMBO J. 14:2715-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chalfie, M., Y. Tu, G. Euskirchen, W. W. Ward, and D. C. Prasher. 1994. Green fluorescent protein as a marker for gene expression. Science 263:802-805. [DOI] [PubMed] [Google Scholar]
- 9.Dilks, K., W. Rose, E. Hartmann, and M. Pohlschröder. 2003. Prokaryotic utilization of the twin-arginine translocation pathway: a genomic survey. J. Bacteriol. 185:1478-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Faury, D., S. Saidane, H. Li, and R. Morosoli. 2004. Secretion of active xylanase C from Streptomyces lividans is exclusively mediated by the Tat protein export system. Biochim. Biophys. Acta 1699:155-162. [DOI] [PubMed] [Google Scholar]
- 11.Gauthier, C., H. Li, and R. Morosoli. 2005. Increase of xylanase production by Streptomyces lividans through simultaneous use of the Sec- and Tat-dependent protein export systems. Appl. Environ. Microbiol. 71:3085-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heukeshofen, J., and R. Dernick. 1985. A simplified method for silver staining of proteins in polyacrylamide gels and the mechanism of silver staining. Electrophoresis 6:103-112. [Google Scholar]
- 13.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 14.Hurtubise, Y., F. Shareck, D. Kluepfel, and R. Morosoli. 1995. A cellulase xylanase-negative mutant of Streptomyces lividans 1326 defective in cellobiose and xylobiose uptake is mutated in a gene encoding a protein homologous to ATP-binding proteins. Mol. Microbiol. 17:367-377. [DOI] [PubMed] [Google Scholar]
- 15.Hynds, P. J., D. Robinson, and C. Robinson. 1998. The sec-independent twin-arginine translocation system can transport both tightly folded and malfolded proteins across the thylakoid membrane. J. Biol. Chem. 273:34868-34874. [DOI] [PubMed] [Google Scholar]
- 16.Jendrossek, D., and S. Reinhardt. 2003. Sequence analysis of a gene product synthesized by Xanthomonas sp. during growth on natural rubber latex. FEMS Microbiol. Lett. 224:61-65. [DOI] [PubMed] [Google Scholar]
- 17.Jendrossek, D., G. Tomasi, and R. M. Kroppenstedt. 1997. Bacterial degradation of natural rubber: a privilege of actinomycetes? FEMS Microbiol. Lett. 150:179-188. [DOI] [PubMed] [Google Scholar]
- 18.Kieser, T., M. J. Bipp, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. John Innes Foundation, Norwich, United Kingdom.
- 19.Laemmli, U. K. 1970. Cleavage of structural proteins during assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 20.Li, H., P.-É. Jacques, M. G. Ghinet, R. Brzezinski, and R. Morosoli. 2005. Determining the functionality of putative Tat-dependent signal peptides in Streptomyces coelicolor A3(2) by using to different reporter proteins. Microbiology 151:2189-2198. [DOI] [PubMed] [Google Scholar]
- 21.Linos, A., M. M. Berekaa, R. Reichelt, U. Keller, J. Schmitt, H. C. Flemming, R. M. Kroppenstedt, and A. Steinbüchel. 2000. Biodegradation of cis-1,4-polyisoprene rubbers by distinct actinomycetes: microbial strategies and detailed surface analysis. Appl. Environ. Microbiol. 66:1639-1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Niviere, V., S. L. Wong, and G. Voordouw. 1992. Site-directed mutagenesis of the hydrogenase signal peptide consensus box prevents export of a β-lactamase fusion protein. J. Gen. Microbiol. 138:2173-2183. [DOI] [PubMed] [Google Scholar]
- 23.Palmer, T., F. Sargent, and B. C. Berks. 2005. Export of complex cofactor-containing proteins by the bacterial Tat pathway. Trends Microbiol. 13:175-180. [DOI] [PubMed] [Google Scholar]
- 24.Pohlschröder, M., K. Dilks, N. J. Hand, and W. Rose. 2004. Translocation of proteins across archaeal cytoplasmic membranes. FEMS Microbiol. Rev. 28:3-24. [DOI] [PubMed] [Google Scholar]
- 25.Pospiech, A., and B. Neumann. 1995. A versatile quick-prep of genomic DNA from gram-positive bacteria. Trends Genet. 11:217-218. [DOI] [PubMed] [Google Scholar]
- 26.Rodrigue, A., A. Chanal, K. Beck, M. Müller, and L.-F. Wu. 1999. Cotranslocation of a periplasmatic enzyme complex by a hitchhiker mechanism through the bacterial tat pathway. J. Biol. Chem. 274:13223-13228. [DOI] [PubMed] [Google Scholar]
- 27.Rose, K., and A. Steinbüchel. 2005. Biodegradation of natural rubber and related compounds: recent insights into a hardly understood catabolic capability of microorganisms. Appl. Environ. Microbiol. 71:2803-2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rose, K., K. B. Tenberge, and A. Steinbüchel. 2005. Identification and characterization of genes from Streptomyces sp. strain K30 responsible for clear zone formation on natural rubber latex and poly(cis-1,4-isoprene) rubber degradation. Biomacromolecules 6:180-188. [DOI] [PubMed] [Google Scholar]
- 29.Rose, R. W., T. Brüser, J. C. Kissinger, and M. Pohlschröder. 2002. Adaption of protein secretion to extremely high-salt conditions by extensive use of the twin-arginine translocation pathway. Mol. Microbiol. 45:943-950. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 31.Sargent, F., E. G. Bogsch, N. R. Stanley, M. Wexler, C. Robinson, B. C. Berks, and T. Palmer. 1998. Overlapping function of components of a bacterial Sec-independent protein export pathway. EMBO J. 17:3640-3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schaerlaekens, A., E. Lammertyn, N. Geukens, S. De Keersmaeker, J. Anné, and L. Van Mellaert. 2004. Comparison of the Sec and Tat secretion pathway for heterologous protein production by Streptomyces lividans. J. Biotechnol. 112:279-288. [DOI] [PubMed] [Google Scholar]
- 33.Schlegel, H. G., H. Kaltwasser, and G. Gottschalk. 1961. Ein Submersverfahren zur Kultur wasserstoffoxidierender Bakterien: Wachstumsphysiologische Untersuchungen. Arch. Mikrobiol. 38:209-222.13747777 [Google Scholar]
- 34.Thomas, J. D., R. A. Daniel, J. Errington, and C. Robinson. 2001. Export of active green fluorescent protein to the periplasm by the twin-arginine translocase (Tat) pathway in Escherichia coli. Mol. Microbiol. 39:47-53. [DOI] [PubMed] [Google Scholar]
- 35.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The ClustalX Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35a.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76:4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsien, R. Y. 1998. The green fluorescent protein. Annu. Rev. Biochem. 67:509-544. [DOI] [PubMed] [Google Scholar]
- 37.Tsuchii, A., and K. Takeda. 1990. Rubber-degrading enzyme from a bacterial culture. Appl. Environ. Microbiol. 56:269-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weber, K., and M. Osborn. 1969. The reliability of molecular weight determinations by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. J. Biol. Chem. 244:4406-4412. [PubMed] [Google Scholar]
- 39.Weiner, J. H., P. T. Bilous, G. M. Shaw, S. P. Lubitz, L. Frost, G. H. Thomas, J. A. Cole, and R. J. Turner. 1998. A novel and ubiquitous system for membrane targeting and secretion of cofactor-containing proteins. Cell 93:93-101. [DOI] [PubMed] [Google Scholar]
- 40.Zimmer, M. 2002. Green fluorescent protein (Gfp): applications, structure and related photophysical behavior. Chem. Rev. 102:759-781. [DOI] [PubMed] [Google Scholar]