Abstract
Clostridium perfringens is an important anaerobic pathogen causing food-borne gastrointestinal (GI) diseases in humans and animals. It is thought that C. perfringens food poisoning isolates typically carry the enterotoxin gene (cpe) on their chromosome, while isolates from other GI diseases, such as antibiotic-associated diarrhea, carry cpe on a transferable plasmid. However, food-borne GI disease outbreaks associated with C. perfringens isolates carrying plasmid-borne cpe (plasmid cpe isolates) were recently reported in Japan and Europe. To investigate whether retail food can be a reservoir for food poisoning generally, we evaluated Japanese retail meat products for the presence of two genotypes of enterotoxigenic C. perfringens. Our results demonstrated that approximately 70% of the Japanese retail raw meat samples tested were contaminated with low numbers of C. perfringens bacteria and 4% were contaminated with cpe-positive C. perfringens. Most of the cpe-positive C. perfringens isolates obtained from Japanese retail meat carried cpe on a plasmid. The plasmid cpe isolates exhibited lower spore heat resistance than did chromosomal cpe isolates. Collectively, these plasmid cpe isolates might be causative agents of food poisoning when foods are contaminated with these isolates from equipment and/or the environment after cooking, or they may survive in food that has not been cooked at a high enough temperature.
Clostridium perfringens type A food poisoning is among the most commonly identified food-borne illnesses in Japan, Europe, and the United States (15). The symptoms of diarrhea and abdominal cramping result from C. perfringens enterotoxin (CPE) (15). The vehicles of infection are typically meat and poultry. In Japan, approximately 20 to 40 outbreaks of C. perfringens food-borne diseases were identified from 2000 to 2005 and approximately 4,000 people became sick each year (http://www.mhlw.go.jp/topics/syokuchu/index.html [text in Japanese]). Only a small fraction (0 to 2%) of C. perfringens isolates, mainly type A, from Japanese retail food carry the CPE-encoding gene (cpe) (8, 10). It is thought that food poisoning isolates typically carry cpe on their chromosome while isolates from other gastrointestinal (GI) diseases, such as antibiotic-associated diarrhea, carry cpe on a transferable plasmid (12). A recent report from the United States showed that C. perfringens isolates in retail foods such as meat and poultry are chromosomal cpe strains (15). However, in Japan and Europe, food-borne GI disease outbreaks caused by plasmid cpe C. perfringens were reported recently, while several food-borne isolates carry chromosomal cpe (6, 14).
In previous Japanese surveys, the location of cpe in enterotoxigenic C. perfringens and the heat resistance of spores of these isolates were not investigated, even though these properties are important in establishing a link between plasmid cpe isolates and food poisoning (8, 10). Therefore, to investigate whether chromosomal cpe strains are the major population in Japanese retail meat, as they are in U.S. retail food, our present study included in-depth molecular genetic and phenotyping analyses of cpe-positive C. perfringens obtained from retail meat in Japan. We also measured the heat resistance of spores of plasmid cpe isolates and compared the results with those of spores of plasmid and chromosomal cpe isolates obtained in previous studies (11). Results of the present study indicate that plasmid cpe isolates are predominant in Japanese retail meat and that spores of these plasmid cpe isolates exhibit lower heat resistance compared to those of chromosomal cpe isolates.
MATERIALS AND METHODS
Bacterial strains.
A total of eight chromosomal cpe isolates were included in this study (9, 12, 15): two food poisoning isolates from Europe (NCTC8239 and NCTC8798), four food poisoning isolates from Japan (W4232, W5603, W6206, and OSAKA2), and two food isolates from the United States (P-1/09/03 and T-1/08/03). Three plasmid cpe food poisoning outbreak strains (T1, T16, and T102) belong to the cpe-IS1470-like genotype, while three cpe-positive outbreak strains (no. 2, no. 24, and no. 110) belonging to the cpe-IS1151 genotype were also included (9, 14). Isolates with cpe on a plasmid were also included (9, 12): three sporadic diarrhea isolates in Europe (F4969, F4013, and F5603) and a healthy human fecal isolate in Japan (MR2-4). A total of 10 plasmid cpe isolates were used in this study.
Collection of food samples.
Two hundred samples of retail raw meat were obtained from grocery stores and retail meat shops in Wakayama city between April and September 2006. A breakdown of these meat samples is shown in Table 1.
TABLE 1.
Isolation of C. perfringens strains from Japanese retail meats
| Food | No. of samples tested | MPN/g range | No. (%) of samples contaminated with C. perfringens | No. of samples with
|
||
|---|---|---|---|---|---|---|
| Type A | Type A-cpb2 | Type A-cpe | ||||
| Beef | 35 | <3 | 16 (45.7) | 19 | 9 | 2 |
| Ground beef | 22 | <3 | 18 (81.8) | 24 | 11 | 0 |
| Pork | 42 | <3 | 15 (35.7) | 22 | 9 | 0 |
| Ground pork | 21 | <3 | 17 (81.0) | 25 | 16 | 0 |
| Ground beef-pork mixture | 21 | <3 | 20 (95.2) | 26 | 12 | 0 |
| Chicken | 33 | 3∼3.0 | 32 (97.0) | 53 | 46 | 1 |
| Ground chicken | 22 | 3∼3.6 | 22 (100.0) | 40 | 38 | 0 |
| Duck | 1 | <3 | 1 (100.0) | 2 | 1 | 0 |
| Lamb | 3 | <3 | 1 (33.3) | 1 | 1 | 0 |
| Total | 200 | 142 (71.0) | 212 | 143 | 3 | |
Isolation and toxin genotype identification of C. perfringens strains from meat products.
A sample of 100 g or one block mass was aseptically put into a stomacher bag and blended with 100 ml of fluid thioglycolate (FTG) medium II (Eiken) and then incubated at 45°C overnight anaerobically in the bag. One loopful of each thioglycolate medium II culture was streaked onto SFP (Difco) agar containing 50% egg yolk-enriched saline (Kyokuto), 12 mg/ml kanamycin (Wako), and 30 U/ml polymyxin B (MP Biomedicals) and incubated at 37°C overnight anaerobically. From the SFP plates, colonies either showing black in the center or surrounding lecithinase activity were selected and put into 10 ml of TGY medium (3% Trypticase soy broth [Difco], 2% d-glucose [Wako], 1% yeast extract [Difco], 0.1% l-cysteine [Wako]) and incubated at 37°C overnight. Two hundred microliters of each TGY overnight culture was used for the preparation of DNA with InstaGene matrix (Bio-Rad). DNA preparations were used as templates for a C. perfringens genotype PCR assay (3). This PCR assay was used to identify C. perfringens isolates and also to detect the genes encoding beta-toxin (cpb), epsilon-toxin (etx), iota-toxin (ia), enterotoxin (cpe), and beta2-toxin (cpb2).
Determination of MPN of C. perfringens bacteria per gram in meat samples.
When 10 g of ground meat or meat samples chopped into small pieces was available (approximately 140 meat samples were surveyed in this study), a three-tube most-probable-number (MPN) method was used to investigate the C. perfringens amounts in the samples (15). Briefly, 10-g aliquots of meat samples were put into a stomacher bag and then 20 ml of 0.1% peptone (Difco) was added. An aliquot was diluted in 10-fold increments (from 10−1 to 10−3) in FTG (Difco). Next, 0.2 ml of each dilution from a single sample was inoculated into three tubes containing 5 ml of differential reinforced clostridial broth medium (Merck) and then incubated at 37°C for 16 to 24 h. Cultures testing positive for C. perfringens produced a unique black precipitation in this differential reinforced clostridial broth medium. Statistical analyses were performed according to the FDA web page (http://www.cfsan.fda.gov).
PCR detection of cpa and cpe in enriched cultures.
To confirm the presence of C. perfringens and enterotoxigenic C. perfringens, DNA was prepared from 400-μl enriched-culture samples with InstaGene matrix. For the cpa gene assay, the PCR primers used to detect cpa (3) and cpe, respectively, were cpe-F3 (5′ACATCTGCAGATAGCTTAGGAAT3′) and cpe-B3 (5′CCAGTAGCTGTAATTGTTAAGTGT3′). The 50-μl PCR volume included 10 μl of GoTaq DNA polymerase buffer (Promega), 4 μl of template DNA preparation, a 1 μM concentration of each primer, 0.2 mM concentrations of deoxynucleoside triphosphates, 2.0 mM MgCl2, and 2.5 U of Taq DNA polymerase (Promega). Amplification was carried out in a MiniCycler (MJ Research), with 1 cycle of 2 min at 94°C; 35 cycles of 30 s at 94°C, 30 s at 55°C, and 30 s at 68°C; and a final dwell time of 8 min at 68°C. The PCR-amplified products were analyzed by electrophoresis of 4-μl PCR samples in a 1.5% agarose gel and staining with ethidium bromide. cpe and cpa PCR products of 248 and 324 bp, respectively, were detected. The specificity of the cpe PCR assay was tested with Clostridium botulinum types A, B, C, E, and F; four food poisoning isolates of Bacillus cereus; and six isolates from meat samples (these six meat isolates were identified as Streptococcus species and four Clostridium species based on 16S rRNA sequences).
PCR assay for genotyping of cpe-positive C. perfringens isolates.
To investigate whether cpe is present on the chromosome or on a large plasmid in food isolates, we performed a cpe-genotyping PCR assay as described previously (9). For cpe-positive control strains NCTC8239, F4969, and F4013, DNAs prepared with the InstaGene matrix kit were used as the templates. For enriched-culture samples assayed by PCR, DNAs prepared with the InstaGene matrix kit were further purified by phenol-chloroform-isoamyl alcohol extraction before use.
SLST analysis of the superoxide dismutase gene (sod) of C. perfringens isolates.
Sixty-one C. perfringens food isolates (both cpe positive and cpe negative; this survey), eight chromosomal cpe strains, and six plasmid cpe strains (F4969, F4013, F5603, no. 2, T16, and MR2-4) (9, 14, 15) were used for sod gene single-locus sequence typing (SLST) analysis. PCR amplification was performed with primers sodF (5′AGCCTTTAGACTATCCTTATGATGCCC3′) and sodR2 (5′GAACCAAAGGTAGAGATTCCACATTGC3′). The PCR was performed with the following regimen: 1 cycle of 2 min at 94°C; 35 cycles of 30 s at 94°C, 60 s at 55°C, and 60 s at 68°C; and a final dwell time of 8 min at 68°C in a MiniCycler (MJ Research). PCR products were then used as sequence templates. Sequence data were analyzed by ClustalW with LaserGene software.
Detection of sod sequence in cpe-positive enriched-culture samples.
In our first attempt to isolate and detect the chromosomal cpe-positive strain, there was no evidence of chromosomal cpe-positive strains in retail meat. The low sensitivity of the cpe-genotyping assay might have affected the results, because the estimated size of the PCR products in this assay could be more than 1 kb. From the results of sod sequence analysis, all of the chromosomal cpe isolates belonged to a distinct cluster which could easily differ from other plasmid cpe-positive and cpe-negative isolates; also, the sod sequences of chromosomal cpe isolates showed the same sequence variations. To detect chromosomal cpe isolates with improved sensitivity, a new PCR assay, detecting ∼350 bp, was developed with a newly constructed inner primer pair (sodFPF, 5′-AGCCTTTAGACTATCCTTATGATGCCC-3′; sodFPR, 5′-CTTTTCTTTAAATTCTTCAAAAGAACC-3′) based on the common nucleotide differences between sod sequences from chromosomal cpe isolates and other isolates. With this new primer pair, we performed a PCR assay with DNAs from eight cpe-positive enriched cultures of food. To detect sod on chromosomal cpe isolates, a 50-μl PCR solution including 10 μl of GoTaq DNA polymerase buffer (Promega), 4 μl of a template DNA preparation, each primer at 1 mM, deoxynucleoside triphosphates at 0.2 mM each, 2.0 mM MgCl2, and 2.5 U of Taq DNA polymerase (Promega) was used. The PCR was performed under the following conditions: 1 cycle of 2 min at 94°C; 35 cycles of 30 s at 94°C, 30 s at 65°C, and 30 s at 68°C; and a final dwell time of 8 min at 68°C in a MiniCycler. When PCR products at the expected band sizes were detected, those PCR products were used as templates for sequencing analysis.
Determination of heat resistance of spores of plasmid cpe C. perfringens type A food poisoning outbreak isolates and plasmid cpe food isolates.
To evaluate the heat resistance of spores from cpe-positive C. perfringens isolates, the D100 value (the time taken to reduce spore survival by a factor of 10 at 100°C) for each isolate's spores was measured as described previously (11). Sporulating cultures of C. perfringens were prepared by inoculating a 0.2-ml aliquot of FTG culture into 10 ml of Duncan-Strong (DS) sporulating medium. After 24 h of incubation at 37°C, those DS cultures were heated at 75°C for 20 min to kill any remaining vegetative cells and also to facilitate spore germination (11). Each heat-shocked DS culture (0.2 ml) was serially diluted from 10−1 to 10−8 with FTG medium. Two 0.2-ml aliquots of each dilution were plated on brain heart infusion (Difco) agar plates to establish the number of viable spores.
The remainder of each heat-shocked DS culture was then heated at 100°C for various times (3 to 10 min for plasmid cpe isolates and 15 to 60 min for chromosomal cpe strains). At each time point, the boiled DS culture was diluted from 10−1 to 10−5 with FTG medium. Two 0.2-ml aliquots of each dilution were then plated on brain heart infusion agar plates and incubated anaerobically at 37°C overnight, and then the viable spores were counted. Strains NCTC8239 and F4969 were used as controls for chromosomal and plasmid cpe isolates, respectively.
CPE production by cpe-positive C. perfringens isolates.
Isolates confirmed as cpe-positive C. perfringens were tested for CPE production with a perfringens enterotoxin reverse passive latex agglutination (PET-RPLA) CPE toxin detection kit (Denka Seiken) (10). Briefly, a 0.2-ml aliquot of an FTG culture of each cpe-positive type A C. perfringens food poisoning isolate and food isolate was inoculated into two tubes of 10 ml of DS sporulating medium. After 24 h of incubation at 37°C, one tube of DS culture was used to determine the heat resistance of the spores. The remaining DS culture tube was incubated at 37°C for another 24 h and then stored at −40°C. Same-lot samples, confirmed by the formation of spores in heat resistance experiments, were used for the CPE production assay. Frozen culture samples were applied to a PET-RPLA kit in accordance with the instructions of the manufacturer.
Nucleotide sequence accession numbers.
The sequences determined in this study have been submitted to GenBank and assigned accession numbers AB377403 to AB377501.
RESULTS
Isolation of C. perfringens from retail meat products.
Our survey was focused on foods most commonly implicated as vehicles for C. perfringens type A food poisoning outbreaks, such as pork, beef, and poultry (15). Of the 200 meat samples tested, 142 (71%) were found to be contaminated with C. perfringens and a total of 212 strains were isolated (Table 1). All meat products showed C. perfringens contamination rates ranging from 33.3% (lamb) to 100% (ground chicken and duck) (Table 1). Most of the meat samples surveyed in this study had very low MPN-per-gram values of less than 3 (Table 1).
Toxin genotype of C. perfringens isolates in retail meat products.
Multiplex PCR is now routinely used to assign C. perfringens isolates to one of five toxinotypes (A to E) based upon whether an isolate carries the gene encoding alpha-, beta-, epsilon-, or iota-toxin (3, 15). Since it has been suggested that beta2-toxin may play a role in enteric disease caused by cpe-positive isolates (2), a revised version of the multiplex PCR toxin genotyping assay detecting cpe and cpb2 has been developed (3). All 212 isolates collected in this survey were subjected to a revised multiplex PCR assay to determine their toxin genotypes (A to E) and whether they carry cpe and cpb2. All of the C. perfringens isolates assayed by the multiplex PCR genotyping assay were classified as type A. From the results of this multiplex PCR genotyping assay, three isolates (1.5%) carrying cpe were identified and none of these three isolates carried cpb2. On the other hand, 143 isolates carried cpb2. Approximately half of the isolates from chicken meat products contained cpb2, but only one chicken meat product contained cpe. Despite the relationship between plasmid cpe and cpb2 for some plasmid cpe isolates in a previous report (2), no linkage of these two genes was detected in the present study among isolates from Japanese retail meat.
Detection of enterotoxigenic C. perfringens in enriched-culture samples from retail meat products by PCR assay.
Because molecular detection methods for many pathogens in food materials have been developed, we investigated the presence of cpe-positive C. perfringens in DNA preparations of enriched-culture samples from retail meat products with a standard PCR assay. We first tested whether our DNA preparation procedure was sufficient to detect the cpa gene encoding alpha-toxin that should be produced by all C. perfringens bacteria. In this assay, a primer pair against cpa that is routinely used and has known specificity for C. perfringens was used in the multiplex PCR genotyping assay (3). Our cpa PCR assay, with DNA prepared with a Bio-Rad kit, detected cpa in 147 (73.5%) of 200 Japanese retail meat product samples (Table 2). These results indicate that our DNA materials from enriched-culture samples are useful for detecting C. perfringens. To directly detect cpe-positive C. perfringens in enriched-culture samples, we used a cpe PCR assay that uses newly constructed primers in this study and confirmed its specificity with several species of Clostridium and Bacillus cereus isolates (data not shown). Eight (4%) of these enriched-culture meat product samples were positive in the cpe PCR assay, and cpe-positive C. perfringens strains (TM111C1, TM138, and TM178) were isolated from three of them (Table 2).
TABLE 2.
Detection of C. perfringens in Japanese retail meats by PCR assay
| Food | No. of samples tested | No. (%) of samples positive by
|
|
|---|---|---|---|
| cpa PCR | cpe PCR | ||
| Beef | 35 | 19 (54.3) | 2 (5.7) |
| Ground beef | 22 | 19 (86.4) | 0 (0.0) |
| Pork | 42 | 14 (33.3) | 1 (2.4) |
| Ground pork | 21 | 18 (85.7) | 0 (0.0) |
| Ground beef-pork mixture | 21 | 21 (100.0) | 1 (4.8) |
| Chicken | 33 | 32 (97.0) | 2 (6.1) |
| Ground chicken | 22 | 22 (100.0) | 2 (9.1) |
| Duck | 1 | 1 (100.0) | 0 (0.0) |
| Lamb | 3 | 1 (33.3) | 0 (0.0) |
| Total | 200 | 147 (73.5) | 8 (4.0) |
cpe-genotyping PCR assay of cpe-positive C. perfringens isolates to determine the chromosomal or plasmid location of cpe.
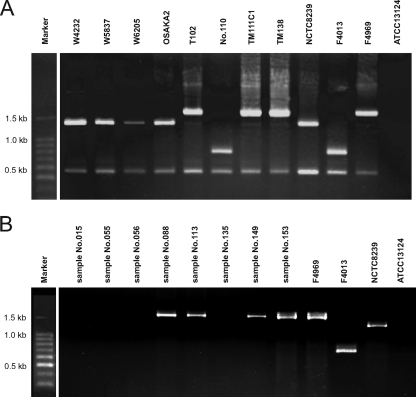
Our recent study indicated that a conventional PCR assay detecting the sequence downstream of cpe could identify whether the cpe gene was on the chromosome or on a plasmid (9). To identify the location of cpe, three cpe-positive strains obtained in this study and 10 cpe-positive food-borne disease isolates from three outbreaks in Japan (11) were assayed by a cpe-genotyping PCR assay (9). Three food isolates (TM111C1, TM138, and TM178) and three plasmid cpe outbreak strains (T1, T16, and T102) belong to the cpe-IS1470-like genotype, while the other plasmid cpe outbreak strains (no. 2, no. 24, and no. 110) belong to the cpe-IS1151 genotype and are cpb2 positive, as previously reported (Fig. 1A) (14). However, four strains (W4232, W5837, W6205, and OSAKA2) from a food poisoning outbreak in Japan belonged to the chromosomal cpe genotype, as described previously (Fig. 1A) (9).
FIG. 1.
PCR cpe-genotyping assay of food poisoning isolates and food isolates and cpe-positive enriched-culture samples. A PCR cpe-genotyping assay of isolates is shown in panel A, and the same assay with DNA preparations of enrichment culture samples is shown in panel B. In panel A, food poisoning chromosomal cpe isolates in Japan (W4232, W5837, W6205, and OSAKA2) and food poisoning plasmid cpe isolates in Japan (T102 and no. 110) are shown. Food isolates with cpe on a plasmid (TM111C1 and TM138) are also shown. Strains NCTC8239 (chromosomal cpe isolate), F4969 (plasmid cpe isolate with a downstream IS1470-like sequence), and F4013 (plasmid cpe isolate with a downstream IS1151 sequence) were used as controls. ATCC 13124 was a cpe-negative type strain. In panel B, DNA preparations from enriched-culture samples (no. 015, no. 055, no. 056, no. 088, no. 113, no. 135, no. 149, and no. 153) were positive by cpe PCR assay. Plasmid cpe strains TM111C1, TM138, and TM178 were isolated from sample no. 088, no. 113, and no. 153, respectively.
Interestingly, of eight DNA preparations from enriched-culture samples which showed cpe positivity by the cpe-specific PCR assay, four also showed a positive reaction in the cpe-genotyping PCR assay. Enriched-culture sample no. 149 was contaminated with cpe-IS1470-like genotype C. perfringens, although no plasmid cpe strain was isolated. cpe-IS1470-like genotype C. perfringens strains (TM111C1, TM138, and TM178) were isolated from three of those four positive samples (no. 088, no. 113, and no. 153, respectively) (Fig. 1B). However, four enriched-culture samples were not found to be contaminated with known cpe genotype C. perfringens strains.
Detection of sod from possible chromosomal cpe isolates in cpe-positive enriched-culture samples.
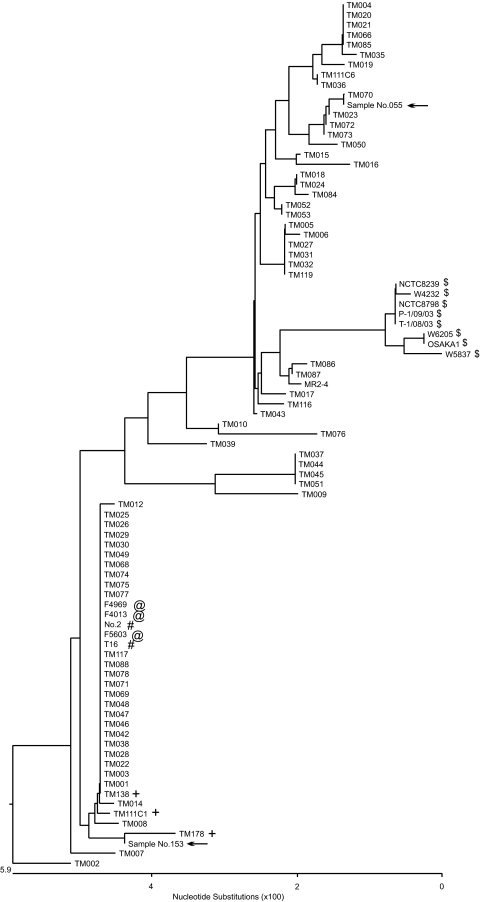
In the cpe-genotyping PCR assay, four of eight enriched-culture samples from retail meat products did not show any PCR products. These samples might have been contaminated with small populations of a chromosomal cpe strain(s) because the cpe-genotyping assay is less sensitive than the cpe-detecting PCR assay. In the cpe-genotyping assay, the size of PCR products from chromosomal cpe strains is more than 1 kb, compared to the ∼200-bp size of PCR products in the cpe-detecting PCR assay. To further investigate the presence or absence of chromosomal cpe strains in all eight cpe assay-positive enriched samples including four cpe-genotyping assay-negative samples, we performed a sod sequence analysis. In the sod sequence phylogenetic assay, chromosomal cpe isolates belonged to one definitive cluster, which was easily differentiated from other isolates such as plasmid cpe strains (Fig. 2).
FIG. 2.
Phylogenetic tree analysis of the superoxide gene (sod) of chromosomal cpe strains, plasmid cpe strains, and cpe-negative food isolates. The phylogenetic tree was constructed with 505 bp of sequence information from the sod open reading frame (full length, 760 bp) from food and food poisoning isolates and with 347 bp of sequence information from sod-based assay-positive samples. Symbols: $, chromosomal cpe strains recovered from food poisoning outbreaks in Japan and Europe and chromosomal cpe strains recovered from food isolates in the United States; @, plasmid cpe-positive European diarrhea isolates; #, plasmid cpe-positive strains recovered from food poisoning outbreaks in Japan; +, plasmid cpe-positive food isolates (strain MR2-4 is from a healthy human fecal isolate with cpe on a plasmid; the other strains are cpe-negative food isolates); arrows, sod PCR-positive enriched-culture samples. Chromosomal cpe strains (indicated by the symbol $) formed a distinct cluster which could be easily differentiated from other strains such as plasmid cpe strains. From the sequence information of PCR-positive products from a sod-based assay (indicated with arrows), the PCR products could not have come from a chromosomal cpe strain.
From sod sequence alignment analyses, we could identify nucleotide differences relatively specific for chromosomal cpe isolates. To sensitively detect sod in chromosomal cpe isolates, a primer pair was constructed based on the alignment analysis results that could produce ∼350-bp PCR products, smaller than those of the cpe-genotyping assay. In the PCR for detecting sod in chromosomal cpe strains, two of eight food samples were positive and yielded PCR products of the expected size (Fig. 3). These two sod PCR products (sample no. 055 and sample no. 153) might have come from sod of chromosomal cpe strains. To investigate whether these PCR products were from sod of a chromosomal cpe strain(s), we performed a sequence analysis. Based on the sequence information and subsequent phylogenetic analysis of the sequences of these sod PCR products, these two samples did not contain chromosomal cpe strains but might have been contaminated with cpe-positive strains of an unknown genotype, as has been reported elsewhere (4) (indicated by the arrow in Fig. 2).
FIG. 3.
Sequence-based PCR assay of sod from chromosomal and plasmid cpe isolates. PCR assay with a primer pair based on the sod sequence for detecting chromosomal cpe strains. Of eight cpe PCR assay-positive enriched-culture samples, two (no. 055 and no. 153) were positive by the sod-based assay.
Heat resistance properties of spores of plasmid cpe food isolates and plasmid cpe food poisoning strains.
Type A food poisoning isolates carrying cpe on the chromosome typically produce more heat-resistant spores than type A isolates carrying cpe on a plasmid (11). Possession of this heat resistance phenotype should be favorable for causing typical C. perfringens type A food poisoning, since this disease usually results from improper temperatures during the cooking or storage of foods (15).
To investigate the heat-resistant spore formation by the three cpe-positive type A food isolates and two plasmid cpe type A food poisoning outbreak isolates in Japan (T16 and no. 2), D values at 100°C were determined for sporulating cultures of these isolates. As expected from previously reported results that spores of strain NCTC8239 (a control chromosomal cpe isolate) exhibited higher heat resistance than spores of strain F4969 (a control plasmid cpe isolate) (Table 3) (11), spores of our surveyed isolates exhibited heat resistance lower than that of NCTC8239 spores (average D100 value of 2.0 versus 16.5) but similar to that of F4969 spores (average D100 value of 2.0 versus 2.2) (Table 3).
TABLE 3.
Heat resistance and CPE production of enterotoxigenic C. perfringens isolates from food poisoning outbreaks and Japanese retail meats
| Strain | Origin | cpe genotype | D value at 100°C (min) | CPE production |
|---|---|---|---|---|
| No. 2 | Food poisoning | Plasmid (IS1151) | 2.4 | + |
| T16 | Food poisoning | Plasmid (IS1470 like) | 2.5 | + |
| TM111C1 | Food | Plasmid (IS1470 like) | 1.9 | + |
| TM138 | Food | Plasmid (IS1470 like) | 1.3 | + |
| TM178 | Food | Plasmid (IS1470 like) | 1.9 | + |
| NCTC8239 | Food poisoning | Chromosome | 16.5 | + |
| F4969 | Sporadic diarrhea | Plasmid (IS1470 like) | 2.2 | + |
| ATCC 3624 | cpe negative | NTa | − |
NT, not tested.
CPE production.
All of our cpe-positive isolates, including plasmid cpe food poisoning isolates (no. 2 and T16), plasmid cpe food isolates (TM111C1, TM138, and TM178), and control strains (NCTC8239 and F4969), produced CPE, but a cpe-negative strain (ATCC 3624) did not show any positive reaction (Table 3).
DISCUSSION
C. perfringens is one of the most important food-borne GI pathogens and is also ubiquitous in nature. CPE is known to be the most important of the toxins of this bacterium for food poisoning. In a survey of retail food in the United States, ∼1.4% of the food samples tested were contaminated with enterotoxigenic C. perfringens and all of those isolates carried cpe on their chromosome and exhibited extremely high spore heat resistance (15). However, recent studies suggest that C. perfringens strains from several food poisoning outbreaks carry cpe on a plasmid (6, 14).
In this survey, approximately 70% of retail meat products were positive for C. perfringens by our bacterial isolation procedures. Previous surveys showed incidences of 8 to 37% in retail meat in Japan (8, 10) and 30 to 80% in American retail meat (7, 15). In the present study, cpe-positive C. perfringens strains were isolated from 3 out of 200 meat product samples. This rate is similar to the contamination rates from two previous surveys in Japan (8, 10). In those surveys, 0 and 2% of the tested meat samples were contaminated with CPE-positive C. perfringens by a PET-RPLA assay (8, 10). Since insufficient heating is thought to be a cause of C. perfringens food poisoning, the heat resistance of spores of CPE-positive isolates is likewise important. From this viewpoint, the significance of those previous Japanese surveys was limited because neither the location of cpe nor the heat resistance of spores was examined.
This study, for the first time, demonstrated the presence of plasmid cpe isolates in Japanese retail meat products, while all of the isolates in retail food in the United States carry cpe on their chromosome (15). Interestingly, these food isolates (TM111C1, TM138, and TM178) belong to the cpe-IS1470-like genotype, as observed with plasmid cpe outbreak isolates (T1, T16, and T102) (Fig. 1A). However, other plasmid cpe outbreak strains (no. 2, no. 24, and no. 110) belong to the cpe-IS1151 genotype, as previously reported (Fig. 1A) (14). Spores of chromosomal cpe isolates (previously reported control strain NCTC8239 [11] and Japanese food poisoning chromosomal cpe strains W4232 and W5837 [data not shown]) showed extremely high heat resistance, with a D value of more than 15 min at 100°C, as expected. In contrast, spores of two different food poisoning strains with cpe on a plasmid (no. 2 and T16) were relatively heat sensitive, with a D value of less than 2 min at 100°C. The food isolates obtained in this survey (TM111C1, TM138, and TM178) also produced relatively heat-sensitive spores, similar to plasmid cpe food poisoning strains and previously reported plasmid cpe strain F4969 (Table 3). Food isolates with cpe on a plasmid could produce CPE. Collectively, these three CPE-producing food isolates have the potential to cause food poisoning, as some previously isolated food poisoning strains carry cpe on a plasmid (14).
To further investigate the presence of chromosomal cpe strains in Japanese retail meat products, we applied sequence-based molecular epidemiology by sod sequence analysis. We used sod for sequence analysis because (i) sod is a representative housekeeping gene; (ii) sod is not a very large gene, and we can analyze ∼80% of the entire open reading frame; (iii) sod is a pathogenic gene and is thought to be important in retail food (5); and finally, (iv) sod is located far away from cpe and thus might not be affected by so-called cpe movement (1). Polymorphological analyses with the ClustalW program showed that chromosomal cpe food poisoning and retail food isolates in the United States comprised one distinct cluster, and this cluster is distant from plasmid cpe-positive and cpe-negative isolates (Fig. 2). By our newly constructed molecular method, our sod sequence analyses could not identify chromosomal cpe isolates in two cpe PCR-positive enriched samples (no. 055 and no. 153) that showed positive reactions in a chromosomal cpe strain-detecting sod-based PCR assay (Fig. 3). Collectively, our present study could not isolate any C. perfringens strains with cpe on their chromosome from Japanese retail meat by three different approaches (bacterial isolation, direct PCR cpe genotyping, and indirect sod SLST analysis). In our study, in-depth molecular analyses to detect chromosomal cpe isolates, in addition to bacterial isolation procedures, were performed. However, no Japanese retail meat products were contaminated with a chromosomal cpe strain. While chromosomal cpe strains were isolated in food poisoning outbreaks in Japan, the rate of presence of chromosomal cpe strains in Japanese retail meat products was lower, less than 0.5%, than in U.S. foods (15).
Enterotoxigenic C. perfringens strains with chromosomal cpe are still considered one of the important pathogens of food poisoning outbreaks, because those strains were almost always isolated from patients suffering from diarrhea secondary to C. perfringens in Europe, the United States, and Japan (9); were present in retail foods; and could produce highly heat-resistant spores (15). Recently, it was reported that plasmid cpe strains were isolated from several food poisoning outbreaks in Japan and Europe (6, 14). However, plasmid cpe food poisoning strains, at least in Japan, produced relatively heat-labile spores, similar to sporadic and antibiotic-associated diarrheal strains. These findings suggest that plasmid cpe isolates in food products could be a causative agent of C. perfringens food poisoning.
Because enterotoxigenic C. perfringens in retail foods in the United States usually carry cpe on their chromosome and produce heat-resistant spores, the contaminated food material itself could be a cause of food poisoning (15). However, plasmid cpe food poisoning strains and strains carrying cpe on a plasmid found in food could produce relatively heat-labile spores, as is the case with sporadic and antibiotic-associated diarrheal strains. Thus, these plasmid cpe strains in meat products could be easily killed during cooking. In food poisoning outbreaks, strains with cpe on a plasmid might be a causative agent when heating during cooking is inadequate (13) or when food might be contaminated during processing. From this point of view, the well-known food poisoning with heat-resistant-spore-forming strains carrying cpe on their chromosome could be called a classical type and food poisoning by plasmid cpe strains might be called an emerging type.
Acknowledgments
We thank Mahfuzur R. Sarker for helpful discussion. We thank Daisuke Tanaka, Teizo Tsukamoto, and Akemi Kai for providing Japanese food poisoning strains.
Footnotes
Published ahead of print on 7 July 2008.
REFERENCES
- 1.Brynestad, S., B. Synstad, and P. E. Granum. 1997. The Clostridium perfringens enterotoxin gene is on a transposable element in type A human food poisoning strains. Microbiology 143:2109-2115. [DOI] [PubMed] [Google Scholar]
- 2.Fisher, D. J., K. Miyamoto, B. Harrison, S. Akimoto, M. R. Sarker, and B. A. McClane. 2005. Association of β2 toxin production with Clostridium perfringens type A human gastrointestinal disease isolates carrying a plasmid enterotoxin gene. Mol. Microbiol. 56:747-762. [DOI] [PubMed] [Google Scholar]
- 3.Garmory, H. S., N. Chanter, N. P. French, D. Bueschel., J. G. Songer, and R. W. Titball. 2000. Occurrence of Clostridium perfringens β2-toxin amongst animals, determined using genotyping and subtyping PCR assays. Epidemiol. Infect. 124:61-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heikinheimo, A., M. Lindstrom, P. E. Granum, and H. Korkeala. 2006. Humans as reservoir for enterotoxin gene-carrying Clostridium perfringens type A. Emerg. Infect. Dis. 12:1724-1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jean, D., V. Briolat, and G. Reysset. 2004. Oxidative stress response in Clostridium perfringens. Microbiology 150:1649-1659. [DOI] [PubMed] [Google Scholar]
- 6.Lahti, P., A. Heikinheimo, T. Johansson, and H. Korkeala. 2008. Clostridium perfringens type A isolates carrying the plasmid-borne enterotoxin gene (genotype IS1151-cpe or IS1470-like-cpe) are a common cause of food poisonings. J. Clin. Microbiol. 46:371-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin, Y., and R. Labbe. 2003. Enterotoxigenicity and genetic relatedness of Clostridium perfringens isolates from retail foods in the United States. Appl. Environ. Microbiol. 69:1642-1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miwa, N., T. Nishina, S. Kubo, M. Atsumi, and H. Honda. 1998. Amount of enterotoxigenic Clostridium perfringens in meat detected by nested PCR. Int. J. Food Microbiol. 42:195-200. [DOI] [PubMed] [Google Scholar]
- 9.Miyamoto, K., Q. Wen, and B. A. McClane. 2004. Multiplex PCR genotyping assay that distinguishes between isolates of Clostridium perfringens type A carrying a chromosomal enterotoxin gene (cpe) locus, a plasmid cpe locus with an IS1470-like sequence, or a plasmid cpe locus with an IS1151 sequence. J. Clin. Microbiol. 42:1552-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saito, M. 1990. Production of enterotoxin by Clostridium perfringens derived from humans, animals, foods, and the natural environment in Japan. J. Food Prot. 53:115-118. [DOI] [PubMed] [Google Scholar]
- 11.Sarker, M. R., R. P. Shivers, S. G. Sparks, V. K. Juneja, and B. A. McClane. 2000. Comparative experiments to examine the effects of heating on vegetative cells and spores of Clostridium perfringens isolates carrying plasmid versus chromosomal enterotoxin genes. Appl. Environ. Microbiol. 66:3234-3240. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12.Sparks, S. G., R. J. Carman, M. R. Sarker, and B. A. McClane. 2001. Genotyping of enterotoxigenic Clostridium perfringens fecal isolates associated with antibiotic-associated diarrhea and food poisoning in North America. J. Clin. Microbiol. 39:883-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sutton, R. G. A., and B. C. Hobbs. 1968. Food poisoning caused by heat-sensitive Clostridium welchii. A report of five recent outbreaks. J. Hyg. 66:135-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tanaka, D., K. Kimata, M. Shimizu, J. Isobe, M. Watahiki, T. Karasawa, T. Yamagishi, S. Kuramoto, T. Serikawa, F. Ishiguro, M. Yamada, K. Yamaoka, M. Tokoro, T. Fukao, M. Matsumoot, R. Hiramatsu, C. Monma, and Y. Nagai. 2007. Genotyping of Clostridium perfringens isolates collected from food poisoning outbreaks and healthy individuals in Japan based on the cpe locus. Jpn. J. Infect. Dis. 60:68-69. [PubMed] [Google Scholar]
- 15.Wen, Q., and B. A. McClane. 2004. Detection of enterotoxigenic Clostridium perfringens type A isolates in American retail foods. Appl. Environ. Microbiol. 70:2685-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]