Abstract
The response of bacteria in bulk soil and earthworm casts to carbon enrichment was studied by an RNA stable-isotope probing/terminal restriction fragment length polymorphism strategy with 13C-labeled glucose and acetate. Both the soil microsite status and the carbon enrichment selected rapidly for different active bacterial communities, which resulted in different degradation kinetics. Our study clearly illustrates the biases that are generated by adding C substrates to detect metabolically active bacteria in soil.
Soil comprises a variety of discontinuous microhabitats with different physicochemical characteristics (22). It thus sustains an immense diversity of microorganisms (18), but less than 5% of the overall available space in soil is occupied by active microorganisms (17). However, zones of increased biological activities defined as “hot spots” are present in soils (17). Earthworms, by subjecting the soil to bioturbation, are providers of such “hot spots” (4). Whereas most soil bacteria are in a state of dormancy (17, 20), soil bioturbation by earthworms has been shown to increase soil bacterial activity (1) by changing the pore space as well as the soil moisture and nutrient contents (9, 13, 14).
Although soil is a major reservoir of organic carbon (27), the available carbon concentrations are low and certainly among the major factors limiting bacterial growth (6) and structuring the bacterial community in soil (3). Thus, carbon amendments, generally with glucose and acetate, which are known to be metabolized through different metabolic pathways (26), have been used as a tool to (i) measure the size of the microbial biomass and its active and dormant fractions (20) or (ii) maximize soil bacterial diversity (25). However, to date, the active microbial fraction that directly consumes the added C substrate, i.e., the primary users, has rarely been investigated.
The aims of the present work were to examine the responses of soil bacteria to glucose and acetate enrichments by discriminating the primary users from the total active soil bacteria by using the RNA stable-isotope probing (SIP) strategy with 13C-labeled substrates. We hypothesized that (i) different C substrates initiate different short-term responses of the active soil bacterial communities but (ii) the short-term response to carbon enrichment depends also on the “hot spot” status of the soil microsite (i.e., bulk soil or earthworm casts).
Soil microcosms were (i) prepared as described previously (2, 15) with a clay-loam agricultural soil (19), (ii) inoculated with earthworms (Lumbricus terrestris), and (iii) incubated at 12°C for 26 days. Earthworm casts and bulk-soil samples (the soil not burrowed by earthworms) were sampled from soil microcosms (more details are given by Kersanté et al. [12] and Monard et al. [15]) and then stored at −20°C until used.
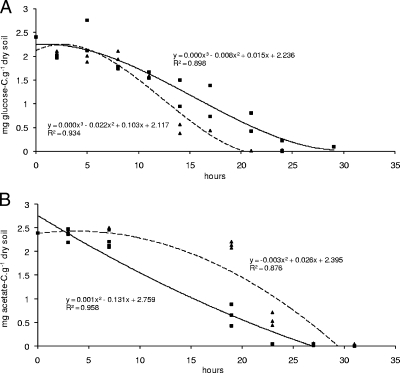
The kinetics of glucose and acetate degradation in bulk soil and casts were first measured (n = 3). Five grams of soil was suspended in 50 ml of a mineral salt medium (21) and placed at 30°C and 150 revolutions min−1 in the presence of glucose or sodium acetate (2.4 g of C per g of dry soil) as the sole C substrate. Glucose degradation was followed spectrophotometrically (EnzyPlus kit; Diffchamb AB), and acetate degradation was measured by ionic chromatography (Dionex DX 120). Both substrates were rapidly consumed (Fig. 1). Glucose degradation was faster in casts than in bulk soil (Fig. 1A). Despite a lag period of 19 h, the acetate degradation rate in casts was finally faster than in bulk soil, and acetate was consumed within an identical incubation period (Fig. 1B).
FIG. 1.
Glucose (A) and acetate (B) degradation curves in bulk soil (squares, unbroken line) and earthworm casts (triangles, dashed line) after the addition of 2.4 mg C substrate g−1 dry soil. Least-square regressions computed using Excel are presented.
For the 13C-RNA SIP experiments, [13C]glucose (>98%; Cambridge Isotope Laboratories) or [13C]acetate (>99%; Sigma-Aldrich) was added to bulk-soil and cast samples as described above. Three milliliters of soil solution was sampled regularly after 5 to 24 h of incubation (n = 3) and centrifuged for 10 min at 10,000 × g, and RNA was extracted from soil pellets according to Courty et al. (P. E. Courty, M. Poletto, F. Duchaussoy, M. Buée, J. Garbaye, and F. Martin, submitted for publication). Control experiments were performed with 12C-labeled substrates to check for the absence of 13C-RNA. 12C-RNA and 13C-RNA were separated by isopycnic ultracentrifugation and sampled as described previously (24). After RNA precipitation and purification, terminal restriction fragment polymorphism (T-RFLP) analyses were twice performed for each soil sample: reverse transcription-PCR was carried out using the Titan one-tube reverse transcription-PCR kit (Roche Applied Science) with the Eub_519f and Eub_1390r primers labeled with the 6-carboxyfluorescein and hexa-chloro derivatives, respectively (24). Labeled PCR products were independently digested with the restriction enzyme StyI or HinfI (Promega). The T-RFs were separated and accurately sized using an automated sequencer (model ABI3130xl; Applied Biosystems). The T-RF sizing and diversity signature analyses were performed using the Peak Scanner 1.0 software (Applied Biosystems). Only the lengths of the T-RFs were taken into account for data analysis, and each T-RF was coded as a discrete variable (0 for its absence or 1 for its presence) (11).
By using the RNA SIP/T-RFLP strategy, like others recently (16), we clearly showed a rapid differentiation within the active soil bacterial communities and distinguished two selective factors that interacted together.
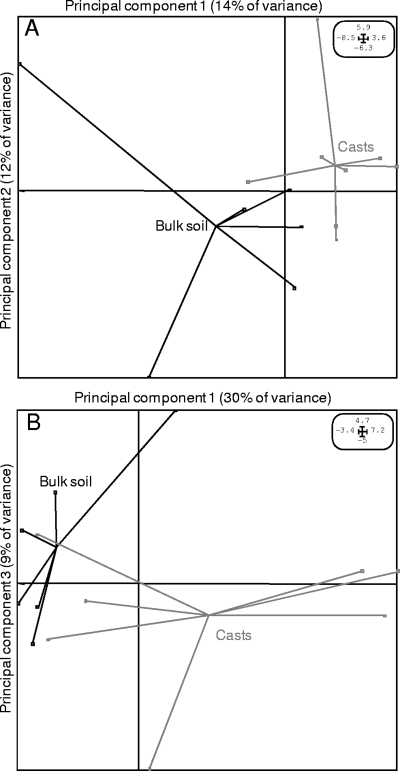
First, the active bacteria were selected differently in earthworm casts and bulk soil with both enrichments (Fig. 2), indicating that soil heterogeneity sustains bacterial diversity. The specific richness (S) and diversity (Shannon's information measure [H0]) of the total active bacteria (12C-RNA) were higher in bulk soil than in casts after the addition of glucose but not after the addition of acetate (Table 1). From the time of ingestion to the time of excretion by earthworms, ingested soil is subjected to different modifications which likely help to change the structures of active bacterial communities; (i) earthworms feed selectively on material rich in organic matter and bacteria (7); (ii) the earthworm gut contains large quantities of available acid organic compounds, such as acetate, which make it an ideal habitat for the anaerobic metabolism and fermentative processes of ingested soil microorganisms (7, 10); and (iii) passage through earthworm guts increases the culturability of certain microorganisms in ingested soil (7, 10). Despite the biases that might have been generated by our soil suspension enrichment methodology (destructured soil microsites), differences between active bacteria in bulk soil and those in casts were still observed, indicating that this selection factor might be even stronger than the one observed. Moreover, our results are the first to show a differentiation between active bacteria according to soil microsite status.
FIG. 2.
Principal-component analysis of the active bacterial communities in soil microsites after glucose (A) and acetate (B) enrichments, implemented with the ADE-4 software package. Each dot in the vectorial space represents the diversity signatures (T-RFLP profiles) of a given sample.
TABLE 1.
S estimates of the total number of T-RFs and H0 estimates of the bacterial diversity for 12C- and 13C-RNA from bulk soil and casts after acetate and glucose enrichmenta
| Substrate | Soil microsite |
12C-RNA
|
13C-RNA
|
||
|---|---|---|---|---|---|
| S | H0 | S | H0 | ||
| Glucose | Bulk soil | 303 | 8.13 | 184 | 7.4 |
| Casts | 238 | 7.76 | 181 | 7.34 | |
| Acetate | Bulk soil | 187 | 7.4 | 130 | 6.87 |
| Casts | 219 | 7.62 | 257 | 7.89 | |
Six samples were used for each enrichment.
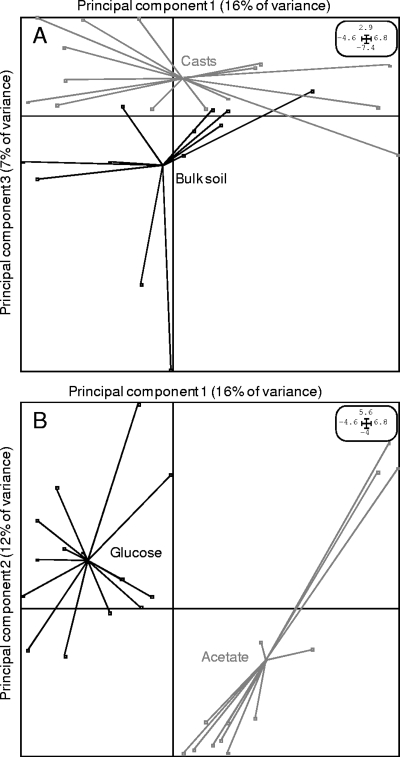
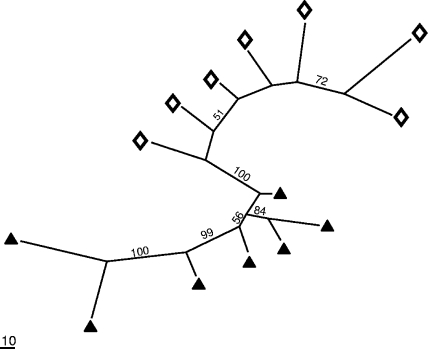
As a second selective factor, the nature of the C substrate added to the soil resulted in profound, specific, and rapid selection within the active bacteria despite the fact that the selection by the soil bioturbation was still observed (Fig. 3). This selection by the C substrate had a greater effect than the soil microsite, although an interaction between the two was detected. Parsimony analysis performed on 13C-RNA demonstrated solely that the composition of the primary users was specific to the added C substrate (Fig. 4). Both substrate enrichments, in addition to removing trophic limitation, created novel environments which selected divergent bacterial communities. Glucose and acetate carbon are not assimilated by the same metabolic pathways. Glucose can be used in the glycolysis pathway, the pentose phosphate pathway, and the Entner-Doudoroff pathway (26). Although glycolysis is the main pathway, some bacteria rely completely on the pentose phosphate pathway and others only on the Entner-Doudoroff pathway (26). Regarding acetate, only soil bacteria that harbor the key enzyme isocitrate lyase involved in the glyoxylate cycle can grow with acetate as the sole carbon source (5), although new alternative pathways of acetate assimilation have recently been described (8). While glucose and acetate are readily metabolized, the different catabolic pathways lead to different degradation kinetics and involve different primary users (Fig. 1 and 4). Furthermore, the highest S and H0 values found in the total active bacteria of glucose-enriched samples indicated that more-diverse soil bacteria consumed glucose rather than acetate (Table 1).
FIG. 3.
Principal-component analysis of the active bacterial communities according to the soil microsite status and irrespective of the substrate added (A) and according to the substrate added and irrespective of the status of the soil microsite (B). Principal-component analyses were implemented with the ADE-4 software package. Each dot in the vectorial space represents the diversity signatures (T-RFLP profiles) of a given sample.
FIG. 4.
Similarity among active microbial communities displayed as the shortest unrooted maximum parsimony tree. The tree was computed with PAUP 4.0β10 (Sinauer Associates) as previously reported (23). Each terminal corresponds to the microbial community of a given sample; the closer they are, the more similar are their diversity signatures. Diamonds and triangles represent soil bacterial communities in 13C-RNA from glucose and acetate enrichments, respectively. Bootstrap values indicated at the nodes were computed from 500 pseudo-replicates. The scale bar corresponds to a gain/loss of 10 informative T-RFs.
Comparison of S and H0 values in 12C-RNA and 13C-RNA revealed differences between the acetate and glucose enrichments in the two soil microsites (Table 1). The glucose-consuming bacteria were observed to be a fraction of the total active soil bacteria in both soil microsites. However, in the acetate-enriched casts, both the S and the H0 of the active consuming bacteria were higher than those of the total active soil bacteria (Table 1) and the kinetics of acetate degradation were also enhanced (Fig. 1B). Thus, acetate enrichment associated with the specific physicochemical properties of casts together with the enhanced culturability of bacteria generated by their passage through earthworm guts (7, 10) might have activated dormant bacteria (K state, i.e., nongrowing bacteria) and/or increased poorly represented bacteria. This new selected fraction of bacteria probably became more competitive and abundant when acetate was available.
Both soil status (i.e., “hot spot” or not) and carbon enrichment had a strong selective effect on the active bacterial community structure. RNA SIP gave direct access to the ecological opportunistic bacteria (r state, i.e., growing bacteria) which grew immediately after substrate addition, in response to the removal of trophic limitations. It is very clear from this study that the carbon addition strategy is not fully adequate to study microbial activity or diversity.
Acknowledgments
This work was supported by funding from the CNRS-INSU institute to F.B. and a by grant from the council of Région Bretagne to C.M. and F.B.
We thank D. Warwick for editing the manuscript and N. Bougon for help in T-RFLP data software acquisition and treatment. We also thank S. Coedel for her technical support at the genotyping/sequencing platform of Ouest-Génopole.
Footnotes
Published ahead of print on 11 July 2008.
REFERENCES
- 1.Binet, F., L. Fayolle, M. Pussard, J. J. Crawford, S. J. Traina, and O. H. Tuovinen. 1998. Significance of earthworms in stimulating soil microbial activity. Biol. Fertil. Soils 27:79-84. [Google Scholar]
- 2.Binet, F., and P. Trehen. 1992. Experimental microcosm study of the role of Lumbricus terrestris (oligochaeta:lumbricidae) on nitrogen dynamics in cultivated soils. Soil Biol. Biochem. 24:1501-1506. [Google Scholar]
- 3.Bossio, D. A., and K. M. Scow. 1995. Impact of carbon and flooding on the metabolic diversity of microbial communities in soils. Appl. Environ. Microbiol. 61:4043-4050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bundt, M., F. Widmer, M. Pesaro, J. Zeyer, and P. Blaser. 2001. Preferential flow paths: biological ‘hot spots’ in soils. Soil Biol. Biochem. 33:729-738. [Google Scholar]
- 5.Clark, D. P., and E. J. Cronan. 1996. Two-carbon compounds and fatty acids as carbon sources, p. 343-357. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.). 1996. Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, DC.
- 6.Demoling, F., D. Figueroa, and E. Baath. 2007. Comparison of factors limiting bacterial growth in different soils. Soil Biol. Biochem. 39:2485-2495. [Google Scholar]
- 7.Drake, H. L., and M. A. Horn. 2007. As the worm turns: the earthworm gut as a transient habitat for soil microbial biomes. Annu. Rev. Microbiol. 61:169-189. [DOI] [PubMed] [Google Scholar]
- 8.Ensign, S. A. 2006. Revisiting the glyoxylate cycle: alternate pathways for microbial acetate assimilation. Mol. Microbiol. 61:274-276. [DOI] [PubMed] [Google Scholar]
- 9.Haynes, R. J., P. M. Fraser, J. E. Piercy, and R. J. Tregurtha. 2003. Casts of Aporrectodea caliginosa (Savigny) and Lumbricus rubellus (Hoffmeister) differ in microbial activity, nutrient availability and aggregate stability. The 7th International Symposium on Earthworm Ecology, Cardiff, Wales, 2002. Pedobiologia 47:882-887. [Google Scholar]
- 10.Horn, M. A., A. Schramm, and H. L. Drake. 2003. The earthworm gut: an ideal habitat for ingested N2O-producing microorganisms. Appl. Environ. Microbiol. 69:1662-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson, D., P. J. Vandenkoornhuyse, J. R. Leake, L. Gilbert, R. E. Booth, J. P. Grime, J. P. W. Young, and D. J. Read. 2004. Plant communities affect arbuscular mycorrhizal fungal diversity and community composition in grassland microcosms. New Phytol. 161:503-515. [DOI] [PubMed] [Google Scholar]
- 12.Kersanté, A., F. Martin-Laurent, G. Soulas, and F. Binet. 2006. Interactions of earthworms with atrazine-degrading bacteria in an agricultural soil. FEMS Microbiol. Ecol. 57:192-205. [DOI] [PubMed] [Google Scholar]
- 13.Lavelle, P., D. Bignell, M. Lepage, V. Wolters, P. Roger, P. Ineson, O. W. Heal, and S. Dhillion. 1997. Soil function in a changing world: the role of invertebrate ecosystem engineers. Eur. J. Soil Biol. 33:159-193. [Google Scholar]
- 14.Marashi, A. R. A., and J. Scullion. 2003. Earthworm casts form stable aggregates in physically degraded soils. Biol. Fertil. Soils 37:375-380. [Google Scholar]
- 15.Monard, C., F. Martin-Laurent, C. Vecchiato, A.-J. Francez, P. Vandenkoornhuyse, and F. Binet. 2008. Combined effect of bioaugmentation and bioturbation on atrazine degradation in soil. Soil Biol. Biochem. doi: 10.1016/j.soilbio.2008.04.022. [DOI]
- 16.Murase, J., and P. Frenzel. 2007. A methane-driven microbial food web in a wetland rice soil. Environ. Microbiol. 9:3025-3034. [DOI] [PubMed] [Google Scholar]
- 17.Nannipieri, P., J. Ascher, M. T. Ceccherini, L. Landi, G. Pietramellara, and G. Renella. 2003. Microbial diversity and soil functions. Eur. J. Soil Sci. 54:655-670. [Google Scholar]
- 18.Rossello-Mora, R., and R. Amann. 2001. The species concept for prokaryotes. FEMS Microbiol. Rev. 25:39-67. [DOI] [PubMed] [Google Scholar]
- 19.Rousseaux, S., A. Hartmann, and G. Soulas. 2001. Isolation and characterisation of new Gram-negative and Gram-positive atrazine degrading bacteria from different French soils. FEMS Microbiol. Ecol. 36:211-222. [DOI] [PubMed] [Google Scholar]
- 20.Stenstrom, J., K. Svensson, and M. Johansson. 2001. Reversible transition between active and dormant microbial states in soil. FEMS Microbiol. Ecol. 36:93-104. [DOI] [PubMed] [Google Scholar]
- 21.Topp, E., H. Zhu, S. M. Nour, S. Houot, M. Lewis, and D. Cuppels. 2000. Characterization of an atrazine-degrading Pseudaminobacter sp. isolated from Canadian and French agricultural soils. Appl. Environ. Microbiol. 66:2773-2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Torsvik, V., and L. Ovreas. 2002. Microbial diversity and function in soil: from genes to ecosystems. Curr. Opin. Microbiol. 5:240-245. [DOI] [PubMed] [Google Scholar]
- 23.Vandenkoornhuyse, P., R. Husband, T. J. Daniell, I. J. Watson, J. M. Duck, A. H. Fitter, and J. P. W. Young. 2002. Arbuscular mycorrhizal community composition associated with two plant species in a grassland ecosystem. Mol. Ecol. 11:1555-1564. [DOI] [PubMed] [Google Scholar]
- 24.Vandenkoornhuyse, P., S. Mahe, P. Ineson, P. Staddon, N. Ostle, J.-B. Cliquet, A.-J. Francez, A. H. Fitter, and J. P. W. Young. 2007. Active root-inhabiting microbes identified by rapid incorporation of plant-derived carbon into RNA. Proc. Natl. Acad. Sci. USA 104:16970-16975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wawrik, B., L. Kerkhof, J. Kukor, and G. Zylstra. 2005. Effect of different carbon sources on community composition of bacterial enrichments from soil. Appl. Environ. Microbiol. 71:6776-6783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.White, D. 2007. The physiology and biochemistry of prokaryotes, 3rd ed. Oxford University Press, New York, NY.
- 27.Whitman, W. B., D. C. Coleman, and W. J. Wiebe. 1998. Prokaryotes: the unseen majority. Proc. Natl. Acad. Sci. USA 95:6578-6583. [DOI] [PMC free article] [PubMed] [Google Scholar]