Abstract
The availability of the complete genome sequence of Bdellovibrio bacteriovorus provides an opportunity for investigating genes that play a significant role in predation. Using two independently derived facultatively predatory Bdellovibrio strains, we have designed a method to cultivate and screen transposon insertion mutants in 96-well microtiter dishes. Transposon insertion mutants were produced by introducing the plasposon pRL27, which carries a mini-Tn5. Mutants have been screened for predatory activity using 96-well plates. Seventeen independent nonpredatory mutants have been isolated, and DNA flanking the insertion has been sequenced. BLAST analysis revealed that most of these interrupted DNA sequences do not code for known proteins or functions. Two of the inactivated genes were analyzed further: one was found to code for a putative serine protease and the other a probable protein involved in secretion through the outer membrane. The methods described here are the first for the generation and isolation of predation-deficient mutants using random-transposon-insertion mutagenesis. As more mutants are isolated and their gene products analyzed, more light will be shed on how this predator carries out its exclusive life processes and perhaps how these products, or the organism itself, can be used for therapeutic, agricultural, and/or other purposes.
The prokaryotic predatory lifestyle has been known and studied for more than 4 decades. The most prevalent species of intraperiplasmic predators studied has been Bdellovibrio bacteriovorus. As an obligate intraperiplasmic predator of other gram-negative bacteria, these organisms exemplify a very unique style of life, even for the prokaryotes. Beginning as individual free-living, highly motile, attack-phase cells, they attack and invade susceptible prey bacteria (29). Following successful penetration through the prey outer membrane, the Bdellovibrio cell begins an intraperiplasmic growth phase. The predator resides in the periplasmic space, where it grows at the expense of the prey and ultimately fragments into progeny attack-phase cells that are released to locate new prey (for a recent review, see reference 28). Much research has gone into the physiology, ecology, and structural interactions of this interesting bacterium with its prey. However, investigations into the genetics and genetic control of the Bdellovibrio developmental cycle have been hampered by the lack of a consistent, reliable mechanism for DNA transfer. Additionally, the fact that they are obligate predators makes it difficult to examine genetic regulation of predatory processes, since mutations in genes essential for predation would be lethal.
Prey-independent, commonly called host-independent (HI), mutants of a number of Bdellovibrio strains are easily isolated, and many have been described in the literature (20, 24). These HI mutants can grow axenically on complex media in the absence of prey cells. Even though their growth seems to follow a pattern somewhat similar to that of intraperiplasmic growth (i.e., elongation of the cells into filaments that fragment into individual progeny of unit length), most have lost the ability to efficiently grow as predators and generally have not proven to be very useful for examining predation and predatory genes.
Over a decade ago, Cotter and Thomashow (4) demonstrated that mobilizable plasmids of the IncQ family could be conjugated into B. bacteriovorus from Escherichia coli. Very little has been done since then to utilize this technology, with the exception of a few studies moving individual genes into the bdellovibrios (8, 14, 27), resulting in limited information concerning the genetics of the predatory lifestyle. The publication of the genome sequence of B. bacteriovorus HD100 (21) has spurred a renewal of interest in these interesting microbes and a search for new genetic methods to probe the genes responsible for their predatory behavior (7, 10, 23).
Mutagenesis by transposon insertion gene inactivation is a proven technique for generating mutants that can easily be isolated by the detection of antibiotic resistance encoded by a gene located within the transposon. Recently, a transposon delivery vector, pRL27 (12), that can be recovered from the mutants has been designed in a one-step cloning procedure similar to that of plasposons (6). This delivery vector has been shown to be mobilizable into a number of different phylogenetic groups of gram-negative bacteria, including HI mutants of Bdellovibrio (12, 14). A recent report (Medina et al. [15]) describes the use of transposon mutagenesis to generate mutants that fail to prey on prey biofilms. However, to date, no system has been developed to specifically target genes necessary for the general predatory lifestyle.
We report here the development of a system of transposon mutagenesis of two facultatively predatory strains of B. bacteriovorus utilizing the plasposon-like vector pRL27 (12). Mutagenized cultures can be selected on complex media and then screened for predatory activity in order to identify genes essential for predation.
(We first reported this method at the 2004 General Meeting of the American Society for Microbiology [5]).
MATERIALS AND METHODS
Bacteria and culture methods.
The bacterial strains used in this study are listed in Table 1. Escherichia coli K-12 was used as prey bacteria in this study, and E. coli DH5α/λpir (17) served as the recipient of the recovered plasposon. Conjugation experiments were carried out using E. coli strain BW20767 as the donor. Routine cultures of E. coli K-12 and DH5α/λpir were grown in Luria broth (Difco) or on Luria agar (L agar) plates overnight at 35°C. E. coli strain BW20767(pRL27) was maintained on L agar containing 50 μg/ml kanamycin sulfate (Km). Cultures to be used as prey were centrifuged at 5,000 × g, washed in dilute nutrient broth (DNB [25]), and suspended in the original culture volume in DNB. Suspensions of E. coli in DNB were stored at 4°C for up to 1 week prior to use. Bdellovibrio bacteriovorus 109JA (20) and B. bacteriovorus 109J-SJ are both facultative predatory mutants derived from B. bacteriovorus 109J (22). Strain 109J-SJ was isolated according to previously published methods for isolating HI mutants (24). Further characterization of this HI strain demonstrated that it retained the ability to grow in a predatory manner, in both liquid and solid medium cultures, as well as axenically on complex medium. Both strains were routinely grown in peptone-yeast extract (PYE [24]) broth or on PYE agar plates at 32°C. Transposon insertion mutants of B. bacteriovorus were maintained in PYE broth or on PYE agar supplemented with 25 μg/ml Km. Broth cultures were shaken at 240 rpm.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Strains | ||
| B. bacteriovorus 109JA | Facultatively HI mutant of 109J | 20 |
| B. bacteriovorus 109J-SJ | Facultatively HI mutant of 109J | This study |
| B. bacteriovorus 109JA-E7 | Predation-deficient mutant, Kmr | This study |
| B. bacteriovorus 109JA-C5 | Predation-deficient mutant, Kmr | This study |
| E. coli K-12 | Wild type | ATCC |
| E. coli DH5α λpir | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 deoRΔ (lacZYA-argF)U169 λpir+ | 17 |
| E. coli BW20767 | RP4-2-Tc::Mu-1 kan::Tn7 integrant, leu-63::IS10 recA1 zbf-5 creB510 hsdR17 endA1 thi uidA(ΔMluI)::pir+ | 16 |
| Plasmid pRL27 | Tn5-RL27 (Kmr-oriR6K) | 12 |
Plasmid and chromosomal DNA isolation.
The plasmid pRL27 was purified from E. coli BW20767 using the QIAprep spin miniprep kit (Qiagen, Valencia, CA) and stored at 4°C. Frozen cultures of E. coli BW20767 kept at −75°C were streaked onto fresh L agar-Km plates and incubated overnight. Plasmid isolations were routinely performed on these cultures taken directly from frozen stocks. Chromosomal DNA from mutagenized bdellovibrios was isolated using the Wizard genomic DNA purification kit (Promega Corp., Madison, WI) and stored at 4°C.
Transposon mutagenesis.
The plasmid pRL27, which carries the transposon Tn5 with a Km resistance gene, was chosen as the vector for transferring the transposon to the two facultative B. bacteriovorus strains. The origin of replication for this plasmid is not recognized by B. bacteriovorus, and therefore, selection of Kmr bdellovibrios is indicative of insertion of the transposon into the Bdellovibrio genome. B. bacteriovorus 109JA and 109J-SJ received the vector pRL27 either through conjugation or electroporation. Conjugation was carried out on sterile nitrocellulose squares (Schleicher and Schuell, Keene, NH) on plates of PYE agar as described by Cotter and Thomashow (4). Recipient bdellovibrios were grown axenically overnight in PYE broth, and 100 μl was spread onto the nitrocellulose filter, followed by incubation overnight at 32°C. Overnight cultures of donor E. coli BW20767 carrying pRL27 were concentrated 10-fold and spread over the recipient bdellovibrios on the nitrocellulose filter. Plates with the mating mixtures were incubated overnight at 32°C, after which the filters were removed, and then the mixtures were vortexed in 2 ml PYE, diluted, and plated onto PYE plus 25 μg/ml Km plus 500 μg/ml streptomycin. The streptomycin served as a counterselection against the donor E. coli strain. Plates were sealed with Parafilm, and Kmr colonies of B. bacteriovorus usually arose within 4 to 6 days of incubation at 32°C. B. bacteriovorus 109JA and 109-SJ were made electrocompetent by growth in PYE to an optical density at 600 nm of 0.4, washed twice in ice-cold double-distilled H2O, washed once with ice-cold 10% glycerol, suspended in a 0.01× volume of 10% glycerol, and stored at −75°C in 40-μl aliquots. Plasmid preps containing from 20 to 40 ng of pRL27 were electroporated into electrocompetent bdellovibrios at 1.3 kV/cm in 1-mm-gap electroporation cuvettes (BTX, Holliston, MA). Cells were recovered in PYE broth without antibiotic with shaking at 32°C for 1 to 2 h to allow expression of the antibiotic resistance gene. Transposon insertion mutants were then selected by plating cells onto PYE agar plates containing 25 μg/ml Km. Plates were sealed with Parafilm and incubated at 32°C for 4 days or until visible Bdellovibrio colonies developed.
Southern blots.
Ten mutant colonies were selected at random, inoculated into 5 ml PYE-Km broth, and grown overnight with shaking. Chromosomal DNA was isolated as described above, an aliquot was digested with BamHI, and fragments were separated on 0.8% agarose gels. DNA was blotted onto charged nylon membranes (Roche Diagnostics GmbH, Mannheim, Germany) and probed with pRL27 labeled with a chemiluminescent probe (AlkPhaos direct labeling reagents; Amersham Biosciences, Buckinghamshire, England).
Screening for predation-deficient mutants.
Transposon insertion mutants of B. bacteriovorus that had been selected on PYE-Km plates were screened for their ability to grow predaceously using E. coli K-12 as prey. Entire colonies of Kmr bdellovibrios were removed using sterile disposable pipettes (Perfector Scientific, Atascadero, CA) and placed in sterile 96-well microtiter dishes containing 150 μl of PYE plus 25 μg/ml Km per well (one colony per well). Microtiter dishes were sealed using Breathe-Easy strips (Diversified Biotech, Boston, MA) to prevent evaporation and incubated overnight at 32°C with shaking at 250 rpm. Following overnight incubation, microtiter cultures were replicated onto PYE agar plus 25 μg/ml Km in 150-mm plates using a 96-prong replicator (Boekel Scientific, Feasterville, PA). The microtiter dish cultures were then centrifuged at 5,000 × g for 30 min to pellet the Bdellovibrio cells. The medium was carefully removed from each well, and each pellet was suspended in 150 μl of DNB containing E. coli K-12. Microtiter dishes were again sealed as before and incubated overnight with shaking at 32°C. Dishes were examined visually for lysis of the E. coli, and wells exhibiting turbidity (no visible lysis) were sampled and examined using phase-contrast microscopy to confirm the absence of predatory growth. Wells containing turbidity were scored, and suspected predation mutants were picked from the replica plate and grown overnight in 1-ml cultures in PYE plus Km. These suspected mutants were subcultured in fresh PYE plus Km, grown overnight, centrifuged at 5,000 × g, suspended in 1 ml of DNB plus E. coli K-12, and incubated overnight to confirm that the mutants were not capable of growing predaceously.
Plasmid rescue and recovery of interrupted genes.
Larsen et al. (12) have shown that one-step cloning of the transposon with its associated flanking DNA can be accomplished using pRL27. Predation-deficient mutants were grown overnight in PYE plus 25 μg/ml Km, and 1.5 ml of the cultures was transferred to microcentrifuge tubes. Cells were harvested by centrifugation at 10,000 × g for 45 s. Chromosomal DNA was isolated from the pelleted cells as described above and digested with the restriction enzyme BamHI, which does not cut within the transposon sequence of pRL27. The restriction enzyme was inactivated by heating the reaction mixtures to 65°C for 5 min. Digested DNA was self-ligated using the Fast-Link DNA ligation kit (Epicentre, Madison, WI) at room temperature for 15 min. Material from ligated mixtures was electroporated into electrocompetent E. coli DH5α λpir (1.25 kV/cm using a 1-mm-gap cuvette). Cells were recovered for 1 hour in SOC (Invitrogen Corp., Carlsbad, CA) broth and plated onto L agar-Km plates to select for cells transformed with the ligated pRL27 plasmid containing flanking Bdellovibrio DNA. The plasposon with its flanking DNA was isolated in minipreps using the QIAprep spin miniprep kit (Qiagen Inc., Valencia, CA). Primers tpnRL17-1, 5′-AACAAGCCAGGGATGTAACG-3′, and tpnRL13-2, 5′-CAGCAACACCTTCTTCACGA-3′ (12), which anneal to the transposon sequence in pRL27 and read outwards into flanking DNA regions, were prepared by Invitrogen (Carlsbad, CA). Sequencing of the DNA interrupted by the transposon was carried out by Northwoods DNA, Inc. (Solway, MN).
Sequencing of predation-specific genes.
Two mutants were selected for further analysis: 109JA-C5 and 109JA-E7. Homologous sequences in the B. bacteriovorus HD100 genome were determined by BLASTN analysis (1). Primer pairs were designed in order to obtain entire gene sequences for the two selected genes. At-least-threefold coverage of each gene sequence was obtained.
Nucleotide sequence accession numbers.
DNA sequences of B. bacteriovorus 109JA representative of putative genes coding for a protein involved in secretion through the outer membrane (strain 109JA-E7) and a protease (strain 109JA-C5) have been deposited in GenBank under the accession numbers EU344891 and EU344892, respectively.
RESULTS
Mutagenesis.
Both conjugation and electroporation were effective techniques of introducing the transposon-containing plasmid pRL27 into axenically grown B. bacteriovorus 109JA and 109J-SJ cells. The efficiency of transformation and transposition were determined to be between 2 × 103 and 5 × 103 CFU per μg of transforming DNA for electroporation. The transfer and transposition frequencies for conjugation experiments were calculated as the number of Km-resistant colonies per total number of recipients and were about 1 × 10−3. This frequency is similar to that observed for other conjugative plasmids into Bdellovibrio (4). Given the similar efficiencies observed for these two delivery methods, transposon mutagenesis was routinely carried out using electroporation, since it is less time-consuming. Kmr colonies appeared on PYE-Km plates within 4 to 5 days and were routinely incubated for 6 days to obtain colonies of 2 to 3 mm in diameter.
Confirmation that these predatory mutants had been constructed by random transposon mutagenesis was shown through Southern blot analysis (26). Blots of DNA digests from several mutants demonstrated that a single copy of the transposon had been integrated at different positions of the chromosome (data not shown).
Effectiveness of screen for predatory mutants.
One of the difficulties in inducing and detecting mutants for Bdellovibrio predation has been the development of an effective method to generate relatively large numbers of potential mutants and screening them for predation. Using two independently derived facultatively predatory Bdellovibrio strains, we have designed a method to cultivate and screen transposon insertion mutants in 96-well microtiter dishes. Bdellovibrios do not routinely grow very well axenically without using a large inoculum. Picking entire colonies and transferring them to small volumes in microtiter dishes yielded good turbidity with overnight incubation. Even though HI mutants are also not very efficient in converting to the predatory lifestyle, they are quite effective in preying on and lysing prey cells in overnight cultures (7, 11). We determined that a quick overnight screen for predation was possible if the entire pellet from a microtiter well culture was suspended in the presence of susceptible prey. Wild-type B. bacteriovorus 109JA and 109J-JS were shown to be capable of producing complete lysis of the E. coli in such overnight cultures. Microtiter plates were examined visually, and lysis was indicated by a decrease in turbidity relative to that of control wells containing only E. coli. Microscopic examination of these cultures confirmed that the HI bdellovibrios were indeed growing as periplasmic predators, with the formation of invaded bdelloplasts, followed by lysis and release of actively motile attack-phase cells. Potential mutants defective in predation were scored as cultures that showed no decrease in turbidity compared to that of control wells. Microscopic examination of a number of cultures demonstrated that the visual screen of the microtiter dishes was effective in detecting potential predatory mutants.
The initial screen with 10 microtiter dishes (960 cultures) yielded 17 putative predation mutants. The majority of mutants were found in cultures of strain 109J, since only one mutagenesis in this initial screen was prepared using strain 109J-SJ. Following rescue of the plasposon with its flanking DNA from these mutants, the location of the insertion was determined by identifying the Tn5 inverted repeat sequence and target 9-bp duplication (12) in the sequences obtained from each primer. The site of integration of the plasposon was unique for each mutant isolated (Table 2). These 17 predation-deficient mutants have plasposon insertions located in 12 open reading frames (ORFs) and four intergenic regions of the B. bacteriovorus HD100 genome (21). Even though mutants JA-G6 and SJ-G8 had insertions in the same ORF, the site of integration for each was unique.
TABLE 2.
Transposon insertion mutant stains obtained
| Mutant strain(s) | ORF designation/location in HD100 genome | Predicted gene product | Top additional BLASTX hit (expect value) |
|---|---|---|---|
| JA-A1 | Between Bd3518 and Bd3519 | Unknown | None |
| JA-A8 | Bd2428 | Putative serine protease | Protease of Bacillus sp. strain PD498 (9e−30) |
| JA-A10 | Bd1580 | Hypothetical protein | None |
| JA-B5 | Bd3170 | Hypothetical protein | None |
| JA-B8 | Bd2093 | Hypothetical protein | Hypothetical SRU-0795 protein of Salinibacter spp. (2e−10) |
| JA-B9 | Between Bd0542 and Bd0543 | Putative CsrA regulatory protein | None |
| JA-C1 | Bd1131 | Putative amino acid aminotransferase | Branched-chain amino acid transferase of Clostridium spp. (3e−95) |
| JA-C4 | Between Bd3061 and Bd3062 | Unknown | None |
| JA-C5 | Bd3534 | Putative carboxy-terminal protease | Carboxy-terminal protease of Oceanospirillum spp. (8e−39) |
| JA-E1 | Bd2325 | Conserved hypothetical protein with a putative HD-GYP hydrolase domain | Conserved hypothetical signal transduction HD-GYP protein of “Candidatus” spp. (2e−36) |
| JA-E7 | Bd1915 | Putative protein involved in secretion through the outer membrane | Outer membrane efflux protein of Optutus spp. (5e−25) |
| JA-F1 | Between Bd2739 and Bd2740 | Unknown | None |
| JA-G1 | Bd2615 | Conserved hypothetical protein | Conserved hypothetical protein of “Candidatus” spp. (4e−72) |
| JA-G6 and SJ-G8 | Bd3489 | Hypothetical protein | None |
| JA-G9 | Bd2033 | Conserved hypothetical protein | GA21854 of Drosophila persimilis (6e−04) |
| SJ-VE4 | Bd3375 | Putative prolipoprotein diacylglycerol transferase | None |
Recovery, sequencing, and analysis of interrupted DNA.
Two of the suspected mutants (JA-E7 and JA-C5) were grown overnight in PYE plus Km, and genomic DNA was isolated. In order to recover the transposon with its flanking DNA, the DNA preparations were cut with BamHI, self-ligated, and electroporated into a permissive E. coli host. Resulting Km-resistant E. coli organisms were grown overnight in L broth plus 50 μg/ml Km, transposon-containing plasmids were recovered, and DNA adjacent to the transposon was sequenced.
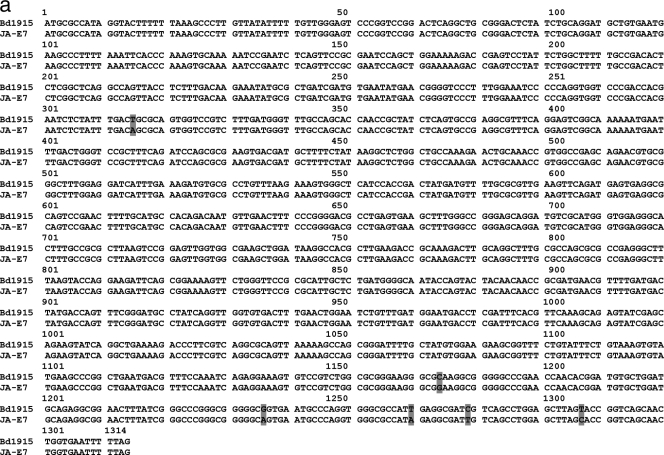
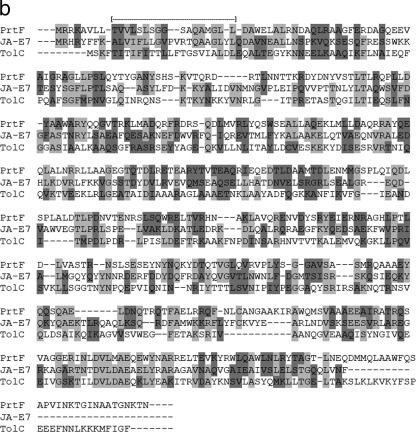
Sequences derived from these presumptively predation-deficient mutants of B. bacteriovorus 109JA were compared to sequences in the DNA sequence database at the National Center for Biotechnology Information (NCBI) using BLASTN. Both mutant sequences were very similar to segments of the deposited genome sequence of B. bacteriovorus HD100. Sequence analysis of the flanking regions of the transposon insertions located the insertions at 1,175 bp (for JA-E7) and 811 bp (for JA-C5) downstream from the translation start sites for these genes. Figure 1 shows the relative locations of the transposon insertions in each of the putative genes. The genes' positions relative to flanking genes in strain HD100 are also shown. The DNA sequence from mutant JA-E7 matched a gene coding for a putative protein involved in secretion through the outer membrane (Bd1915). The mutant JA-E7 nucleotide sequence was 99% identical to the putative Bd1915 gene for B. bacteriovorus HD100, with 1,308 identical nucleotides out of 1,314 nucleotides (Fig. 2a). All six substitutions were at the third position of the codon, resulting in a predicted protein product of 437 amino acids, identical to that of Bd1915. The B. bacteriovorus HD100 Bd1915 gene encodes a putative outer membrane protein representing a conserved orthologous gene, COG1538 (expect value, 5e−36), which represents a type I secretion protein of the TolC family, a large family of proteins that are found in many different gram-negative bacteria. These proteins can interact with a number of different membrane transporters and are involved in the export of a variety of molecules (19), including large proteins such as the α-hemolysin of E. coli (30). A recent analysis of B. bacteriovorus transport proteins using the Transporter Classification Database (2) showed a similarity of Bd1915 to the PrtF protein of E. coli. Analysis using PSORTb (9) predicts that the JA-E7 protein is an integral outer membrane protein with a localization score of 9.93 out of 10 and matches an outer membrane protein of Rickettsia prowazekii with an expect value of 2e−13. Alignment of the amino acid sequence of JA-E7 with these two proteins (PrtF and TolC) is shown in Fig. 2b.
FIG. 1.
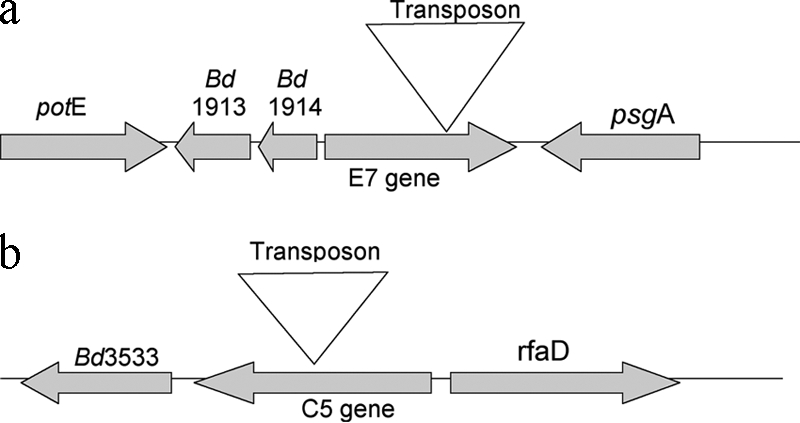
Relative transposon insertion locations in putative predatory genes of the JA-E7 mutant (a) and JA-C5 mutant (b). Genes flanking these loci and their orientations are also shown.
FIG. 2.
(a) Alignment of the B. bacteriovorus 109JA-E7 gene with its homolog in strain HD100 (Bd1915). Shaded areas indicate nucleotide differences between the genes. (b) Alignment of the amino acid sequence of the B. bacteriovorus 109JA-E7 protein with its homologs in Erwinia chrysanthemi (PrtF; expect value, 1e−11; 22% identity, 44% similarity) and Rickettsia prowazekii (TolC; expect value, 1e−11; 21% identity, 43% similarity). Lightly shaded areas signify identical amino acids, and darkly shaded areas represent similar amino acids. The bracketed dashed line at the top indicates the position of a probable transmembrane region (amino acids 8 to 28).
The transposon-interrupted gene in 109JA-C5 is homologous to the B. bacteriovorus HD100 gene Bd3534, as determined by BLASTN analysis of the nucleotide database at the NCBI. The DNA sequence of the JA-C5 mutant gene is 97% identical to that of the putative carboxy-terminal peptidase gene (Bd3534) of B. bacteriovorus HD100, with 1,548 of 1,581 nucleotides being the same. The predicted amino acid sequence of the protein from 109JA-C5 is also 97% identical to the Bd3534 protein (Fig. 3). Most of the amino acid substitutions are in the midportion of the sequence, and BLASTP analysis shows that the amino-terminal and carboxy-terminal thirds of the predicted proteins are probably highly conserved, containing either identical or similar amino acids. Both Pfam (18) and SMART (13) analyses of this putative protein indicate that it contains both PDZ and serine protease domains and a signal sequence likely making it a membrane-anchored periplasmic protein. Analysis of the predicted topology of these regions using TopPred analysis (3) shows that there are three potential transmembrane domains, two within 170 residues of the amino terminus and one within the 175 residues of the carboxy terminus.
FIG. 3.
Sequence alignment of the predicted proteins from B. bacteriovorus 109JA-C5 and HD100 Bd3534. Shaded areas indicate differences in amino acids between the proteins. Underlines indicate the three predicted domains: the signal peptide, amino acids 1 to 28; the PDZ domain, amino acids 189 to 264; and the serine protease domain, amino acids 264 to 458.
DISCUSSION
This report and that of Medina et al. (15) are the first to describe a general methodology for the large-scale production and isolation of transposon insertion mutants of facultative predatory bdellovibrios that are deficient in predation. The only other report of transposon mutagenesis in bdellovibrios (14) utilized an HI strain and demonstrated only the expression of genes delivered with the transposon. In order to detect insertional inactivation of predatory genes, it is necessary to use HI Bdellovibrio strains that retain the ability to attack and grow on susceptible bacterial prey. The use of 96-well microtiter plates allows for the screening of a large number of transformants for the ability to prey on E. coli. Wild-type B. bacteriovorus 109JA and 109J-SJ effectively prey on and lyse E. coli cultures overnight in similar assays. Visual examination of dual cultures in microtiter plates has proved to be sufficient to detect suspected predatory mutants. Subsequent tests for predation demonstrated that this visual check resulted in fewer than 20% false positives. The selection procedure used eliminates all mutants in essential genes for growth; therefore, the probability of finding predation mutants among the transformants is quite high, about 1 to 2 per 100 insertion mutants. Additional screens of mutagenized cultures should result in the recovery of many more predatory mutants. Based on our limited number of screens, we predict that from 75 to 100 genes that play essential roles in predation may be found.
There have been only a few reports in the literature of genes identified as essential for predation (10, 11). Each of these mutants has been isolated by starting with a cloned gene used to knock out targeted genes. To date, the only reported method for delivery of plasmid DNA into Bdellovibrio recipients has been conjugation with donor E. coli strains (4, 10, 11, 14, 15, 27). We report here the first use of electroporation as an effective means of transforming bdellovibrios and of introducing an active transposon capable of inserting at random into the Bdellovibrio genome. The efficiency of transformation utilizing electroporation was similar to that obtained with the conjugation procedure and requires less time and material, making it a more useful approach.
Sequence analysis of the 17 isolated mutant strains indicates that all of the transposon insertions are unique, even though two are in the same ORF. Additionally, Southern blots demonstrated that transposon insertion occurred randomly and only once in each of the 10 strains tested, suggesting that multiple transposon insertions are probably rare events. Only five of the predation genes identified have homologs coding for putative known proteins in the HD100 genome. The other predation mutants have insertions in ORFs for putative hypothetical proteins of unknown function or in intergenic regions. Phenotypes due to interruptions in sequences between genes may be due to polar effects or perhaps could code for regulatory RNAs. Two of the mutants with insertional inactivation of putative known proteins were selected for further analysis. Figure 1 shows the portion of the genome into which the transposon inserted and the approximate location of the transposon within these two predatory genes. Nucleotide sequences for complete genes showed very close similarity between sequences from 109JA and HD100 strains, specifically, 97% and 99% at the nucleotide level for mutant strains 109JA-C5 and 109JA-E7, respectively.
The amino acid sequence predicted for the putative protein encoded by the predation gene of 109JA-E7 is identical to that of Bd1915. This protein shows homology to type I secretory proteins, such as TolC and PrtF. Both of these proteins have been shown to be involved in the export of proteases (30). It is impossible at this point to predict what role this protein may play in predation by the bdellovibrios. However, given the massive arsenal of hydrolytic enzymes, especially proteases (21), that the bdellovibrios potentially may deliver to the prey protoplast during predatory growth, it is intriguing to speculate that Bd1915 might encode one of the secretory proteins needed during this process. BLASTP analysis of the JA-C5 protein shows that the amino-terminal and carboxy-terminal thirds of the predicted protein are probably highly conserved, containing amino acids either identical or similar to those of its HD100 homolog. In addition to the protease domain, JA-C5 contains a putative PDZ domain. PDZ domains are commonly responsible for protein-protein interactions and potentially play a role in selectivity and targeting proteins for proteolysis by the serine protease domain. Analysis of the amino acid sequence of JA-C5 using pSORTb (9) predicts that there is likely a signal sequence at the amino terminus, with a possible cleavage site at amino acid 23. It is therefore likely that this putative protease resides in the periplasmic space of the predator. However, topology prediction using the TopPred program (3) shows potential transmembrane segments between residues 120 and 170 and between residues 375 and 395. The presence of additional transmembrane segments suggests potential insertion into the predator's outer membrane. As an outer membrane protein, this protease could be involved in proteolysis of prey outer membrane or periplasmic proteins. If this protease is located in the Bdellovibrio strain's periplasm, it possibly could be secreted into the prey's periplasm using a Sec-dependent secretory pathway and be involved in the proteolysis of a variety of prey proteins.
The more interesting proteins essential for predation by the bdellovibrios may well be the hypothetical unknown proteins identified by this assay. It is not surprising that more than half of the identified genes necessary for the predatory lifestyle code for genes of unknown function. One expects to find unique gene products in an organism exhibiting such a unique lifestyle. As more mutants are isolated and their gene products analyzed, more light will be shed on how this predator carries out its exclusive life processes and perhaps how these products, or the organism itself, can be used for therapeutic, agricultural, and/or other purposes.
Acknowledgments
This work was supported by NIH grant R15 GM52640-01 and by Saint Joseph's University.
E. coli strain BW20767(pRL27) was supplied by W. Metcalf. We acknowledge helpful discussions by Michael McCann throughout this study.
Footnotes
Published ahead of print on 11 July 2008.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Barabote, R. D., S. Rendulic, S. C. Schuster, and M. H. Saier. 2007. Comprehensive analysis of transport proteins encoded within the genome of Bdellovibrio bacteriovorus. Genomics 90:424-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Claros, M. G., and F. von Heijne. 1994. TopPred II: an improved software for membrane protein structure predictions. Comput. Appl. Biol. Sci. 10:685-686. [DOI] [PubMed] [Google Scholar]
- 4.Cotter, T. W., and M. F. Thomashow. 1992. A conjugation procedure for Bdellovibrio bacteriovorus and its use to identify DNA sequences that enhance the plaque-forming ability of a spontaneous host-independent mutant. J. Bacteriol. 174:6011-6017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davis, J. J., M. A. Panichella, A. Zwolak, and J. J. Tudor. 2004. Isolation and analysis of transposon-insertion mutants of Bdellovibrio bacteriovorus, abstr. H-206. In Abstr. 104th Gen. Meet. Am. Soc. Microbiol. American Society for Microbiology, Washington, DC.
- 6.Dennis, J. J., and G. J. Zylstra. 1998. Plasposons: modular self-cloning minitransposon derivatives for rapid genetic analysis of gram-negative bacterial genomes. Appl. Environ. Microbiol. 64:2710-2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Evans, K. J., C. Lambert, and R. E. Sockett. 2007. Predation by Bdellovibrio bacteriovorus HD100 requires type IV pili. J. Bacteriol. 189:4850-4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flannagan, R. S., M. A. Valvano, and S. F. Koval. 2004. Downregulation of the motA gene delays the escape of the obligate predator Bdellovibrio bacteriovorus 109J from bdelloplasts of bacterial prey cells. Microbiology 150:649-656. [DOI] [PubMed] [Google Scholar]
- 9.Gardy, J. L., M. R. Laird, F. Chen, S. Rey, C. J. Walsh, M. Ester, and F. S. L. Brinkman. 2005. PSORTb v. 2.0: expanded prediction of bacterial protein subcellular localization and insights gained from comparative proteome analysis. Bioinformatics 21:617-623. [DOI] [PubMed] [Google Scholar]
- 10.Lambert, C., M. C. M. Smith, and R. E. Sockett. 2003. A novel assay to monitor predator-prey interactions for Bdellovibrio bacteriovorus 109J reveals a role for methyl-accepting chemotaxis proteins in predation. Environ. Microbiol. 5:127-132. [DOI] [PubMed] [Google Scholar]
- 11.Lambert, C., K. J. Evans, R. Till, L. Hobley, M. Capeness, S. Rendulic, S. C. Schuster, S.-I. Aizawa, and R. E. Sockett. 2006. Characterizing the flagellar filament and the role of motility in bacterial prey-penetration by Bdellovibrio bacteriovorus. Mol. Microbiol. 60:274-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Larsen, R. A., M. M. Wilson, A. M. Guss, and W. W. Metcalf. 2002. Genetic analysis of pigment biosynthesis in Xanthobacter autotrophicus Py2 using a new, highly efficient transposon mutagenesis system that is functional in a wide variety of bacteria. Arch. Microbiol. 178:193-201. [DOI] [PubMed] [Google Scholar]
- 13.Letunic, I., R. R. Copley, B. Pils, S. Pinkert, J. Schultz, and P. Bork. 2006. SMART 5: domains in the context of genomes and networks. Nucleic Acids Res. 34:D257-D260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martin, M. 2002. Predatory prokaryotes: an emerging research opportunity. J. Mol. Microbiol. Biotechnol. 4:467-477. [PubMed] [Google Scholar]
- 15.Medina, A. A., R. M. Shanks, and D. E. Kadouri. 2008. Development of a novel system for isolating genes involved in predator-prey interactions using host independent derivatives of Bdellovibrio bacteriovorus 109J. BMC Microbiol. 8:33-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Metcalf, W. W., W. Jiang, L. L. Daniels, S. K. Kim, A. Haldimann, and B. L. Wanner. 1996. Conditionally replicative and conjugative plasmids carrying lacZ alpha for cloning, mutagenesis, and allele replacement in bacteria. Plasmid 35:1-13. [DOI] [PubMed] [Google Scholar]
- 17.Miller, V. L., and J. J. Mekaloanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacterial. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakai, K., and M. Kanehisa. 1991. Expert system for predicting protein localization sites in Gram-negative bacteria. Proteins Struct. Funct. Genet. 11:95-110. [DOI] [PubMed] [Google Scholar]
- 19.Paulsen, I. T., J. H. Park, P. S. Choi, and M. H. Saier, Jr. 1997. A family of Gram-negative bacterial outer membrane factors that function in the export of proteins, carbohydrates, drugs and heavy metals from Gram-negative bacteria. FEMS Microbiol. Lett. 156:1-8. [DOI] [PubMed] [Google Scholar]
- 20.Pritchard, M. A., D. Langley, and S. C. Rittenberg. 1975. Effects of methotrexate on intraperiplasmic and axenic growth of Bdellovibrio bacteriovorus. J. Bacteriol. 121:1131-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rendulic, S., P. Jagtap, A. Rosinus, M. Eppinger, C. Baar, C. Lanz, H. Keller, C. Lambert, K. J. Evans, A. Goesmann, F. Meyer, R. E. Sockett, and S. C. Schuster. 2004. A predator unmasked: life cycle of Bdellovibrio bacteriovorus from a genomic perspective. Science 303:689-692. [DOI] [PubMed] [Google Scholar]
- 22.Rittenberg, S. C. 1972. Nonidentity of Bdellovibrio bacteriovorus strains 109D and 109J. J. Bacteriol. 109:432-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schwudke, D., A. Bernhardt, S. Beck, K. Madela, M. W. Linshceid, B. Appel, and E. Strauch. 2005. Transcriptional activity of the host-interaction locus and a putative pilin gene of Bdellovibrio bacteriovorus in the predatory life cycle. Curr. Microbiol. 51:310-316. [DOI] [PubMed] [Google Scholar]
- 24.Seidler, R. J., and M. P. Starr. 1969. Isolation and characterization of host-independent bdellovibrios. J. Bacteriol. 100:769-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seidler, R. J., and M. P. Starr. 1969. Factors affecting the intracellular parasitic growth of Bdellovibrio bacteriovorus developing within Escherichia coli. J. Bacteriol. 97:912-923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Southern, E. M. 1975. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98:503-517. [DOI] [PubMed] [Google Scholar]
- 27.Steyert, S. R., and S. A. Pieiro. 2007. Development of a novel genetic system to create markerless deletion mutants of Bdellovibrio bacteriovorus. Appl. Environ. Microbiol. 73:4717-4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tudor, J., and M. McCann. 2007. Genomic analysis and molecular biology of predatory prokaryotes, p. 153-189. In E. Jurkevitch (ed.), Predatory prokaryotes—biology, ecology and evolution. Springer-Verlag, Heidelberg, Germany.
- 29.Tudor, J. J., M. P. McCann, and I. A. Acrich. 1990. A new model for the penetration of prey cells by bdellovibrios. J. Bacteriol. 172:2421-2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wandersman, C. 1993. The general secretory pathway in bacteria. Trends Microbiol. 1:249-250. [DOI] [PubMed] [Google Scholar]