Abstract
The common brushtail possum (Trichosurus vulpecula) is one of the most abundant native marsupials in urban Australia, having successfully adapted to utilize anthropogenic resources. The habituation of possums to food and shelter available in human settlements has facilitated interaction with people, pets, and zoo animals, increasing the potential for transmission of zoonotic Cryptosporidium pathogens. This study sought to examine the identity and prevalence of Cryptosporidium species occurring in possums adapted to urban settings compared to possums inhabiting remote woodlands far from urban areas and to characterize the health of the host in response to oocyst shedding. Findings indicated that both populations were shedding oocysts of the same genotype (brushtail possum 1 [BTP1]) that were genetically and morphologically distinct from zoonotic species and genotypes and most closely related to Cryptosporidium species from marsupials. The urban population was shedding an additional five Cryptosporidium isolates that were genetically distinct from BTP1 and formed a sister clade with Cryptosporidium parvum and Cryptosporidium hominis. Possums that were shedding oocysts showed no evidence of pathogenic changes, including elevated levels of white blood cells, diminished body condition (body mass divided by skeletal body length), or reduced nutritional state, suggesting a stable host-parasite relationship typical of Cryptosporidium species that are adapted to the host. Overall, Cryptosporidium occurred with a higher prevalence in possums from urban habitat (11.3%) than in possums from woodland habitat (5.6%); however, the host-specific nature of the genotypes may limit spillover infection in the urban setting. This study determined that the coexistence of possums with sympatric populations of humans, pets, and zoo animals in the urban Australian environment is unlikely to present a threat to public health safety.
Cryptosporidium is a genus of protozoan parasite that causes enteric disease in a range of vertebrates with potentially fatal consequences for young or immunocompromised individuals (17). While most species within the Cryptosporidium genus exhibit strong adaptation to the host (55), eight Cryptosporidium species and genotypes from animals (C. parvum, C. meleagridis, C. suis, C. baileyi, C. canis, C. felis, C. muris, and the cervine genotype) have zoonotic capacity and present a public health risk (8, 33, 41). To assess the threat of spillover infection, a growing number of studies have characterized Cryptosporidium species circulating in wildlife populations. To date, most studies have focused on rodents (5, 9, 13) and a range of animals that, by inhabiting natural watersheds, pose a risk of contaminating human drinking water supplies (2, 22, 31, 34, 36, 58). In contrast, few studies have investigated the epidemiology of Cryptosporidium in wildlife that live directly at the interface of urban settlements, despite their coexistence with high densities of humans and domestic pets, a situation that has proven conducive to spillover infection of a number of zoonotic pathogens (10, 14, 24, 27). With urbanization of natural landscapes intensifying worldwide, characterization of Cryptosporidium occurring in urban wildlife may be an important step to identify new reservoirs for the parasite that pose a threat to public health safety.
The common brushtail possum (Trichosurus vulpecula) is one of the most abundant and widespread native marsupial species in urban Australia. Possums have flourished in urban areas by adapting their diet to include food scraps and garden plants found in abundance in residential areas, public parks, botanic gardens, and metropolitan zoos. These aspects of possum ecology have promoted interaction with humans, domestic pets, and other introduced animals, facilitating the transmission of zoonotic pathogens, such as Toxoplasma gondii (15, 23) and Leptospira spp. (16); however, there has been no systematic investigation of Cryptosporidium in urban-environment-adapted possums. Studies of other marsupials have identified host-adapted Cryptosporidium species and genotypes, including Cryptosporidium fayeri from the red kangaroo (Macropus rufus) (40), Cryptosporidium macropodum from the eastern gray kangaroo (Macropus giganteus) (35), and opossum genotypes I and II from the Virginia opossum (Didelphis virginiana) (55). Recent species description of C. fayeri has classified opossum genotype I as a subtype belonging to the species (40). However, natural infection of captive bilbies (Macrotis lagotis) (49) with C. muris from the domestic mouse Mus musculus is evidence that conditions which place marsupials in close proximity to nonnative animal hosts can promote transmission of introduced Cryptosporidium species. Examination of possums that cohabitate with an array of nonnative animals and humans in an urban environment offers an ideal scenario to assess the potential of marsupials to act as a host for Cryptosporidium with zoonotic capacity.
The primary aim of this study was to determine the prevalence and identity of Cryptosporidium isolates from common brushtail possums that live at high density at the urban interface. This study also sought to characterize changes in host physiology associated with shedding of Cryptosporidium oocysts to gain insights into the pathogenicity of the host-parasite relationship and ultimately the level of host adaptation of the parasite. Finally, this study aimed to clarify whether the conditions of urbanization promote heightened transmission of Cryptosporidium among the possum population compared to a population that inhabited native, woodland habitat, remote from urban areas. These findings were used to assess the capacity of urban-environment-adapted possums to act as a reservoir host for Cryptosporidium spp. of public health significance.
MATERIALS AND METHODS
Study sites.
The principal study area was Taronga Zoo (33°50′S 151°14′E) and adjacent residences in the northern Sydney suburb of Mosman, Australia. The zoo is inhabited by an abundant, free-ranging common brushtail possum population that forage nightly on leftover foodstuffs in the animal exhibits, anthropogenic food from waste bins, and plants in the zoo gardens. Also included in the “urban” field site were the properties of Mosman residents who lived in proximity to the zoo (<1 km) and had experienced possum activity. Preliminary estimates suggested possums inhabiting this urban area achieved a population density of 5.03 ha−1, one of the highest recorded in Australia (N. J. Hill and E. M. Deane, unpublished data). For comparison, trapping took place at a nonurban site located within Jenolan Caves Reserve Trust land, Blue Mountains, Australia (33°48′S 125°29′E). The reserve was characterized by eucalyptus woodland and was inaccessible to the public. The site was bound by fencing to prevent incursion by feral cats and foxes and protect the endangered brush-tailed rock wallaby (Petrogale penicillata) that inhabited the reserve. Ethics approval for this project was obtained from the Macquarie University Animal Ethics Committee (2004/16), the Zoological Parks Board of New South Wales Animal Care and Ethics Committee (4c/08/04), and the New South Wales Department of Environment and Climate Change (S11107).
Trapping and fecal collection.
At the urban site, trapping was performed on a monthly basis between March 2005 and November 2006 (20 trapping surveys). At the woodland site, the remote location limited trapping to November 2005, May 2006, and October 2006 (3 trapping surveys). All possums were permanently implanted with a passive integrated transponder (Microchips Australia, Victoria, Australia) to enable identification of recaptured animals. Recaptured possums were included in the survey if the time since original capture exceeded 3 months (to avoid pseudoreplication). Fecal samples were collected directly from the floor of the traps. In total, 221 samples were collected from 133 individual possums in urban habitat and 39 samples were collected from 18 individual possums in woodland habitat.
Health assessment.
Possums were anesthetized with 5% isoflurane in oxygen via a face mask prior to the health assessment. Age was estimated from the degree of molar tooth wear (51) and then pooled into two groups, juveniles (tooth wear score of ≤2) and adults (tooth wear score of >3) corresponding to sexual maturity. Body condition (BC) was calculated by dividing body mass (BM) by the skeletal body length (BS) of possums (BC = BM/BS). Skeletal body length was measured from the tip of the nose to the cloaca, while body mass was measured on electronic scales to the nearest gram. This method provided an estimate of body condition previously validated for the closely related mountain brushtail possum, Trichosurus caninus (47). Approximately 5 ml of blood was drawn from the lateral tail vein and distributed between two vacuette tubes (Interpath Services, Sydney, Australia), one tube lined with EDTA for hematology analysis and a serum clot activator tube for biochemistry analysis. After the blood clotted, the serum clot activator tube was centrifuged at 200 × g for 10 min, and the serum was transferred to a microtube. All testing was conducted at Pacific Laboratory Medicine Services (New South Wales, Australia) as previously described (29) within 72 h of blood collection.
Sample purification and DNA extraction.
Fecal samples were concentrated using immunomagnetic separation (IMS) as previously described (37). In brief, a fecal slurry was mixed with paramagnetic beads conjugated with Cry104 (50), a monoclonal antibody that targets the Cryptosporidium oocyst wall (BTF Australia, Sydney, Australia). The resulting bead-oocyst complex was separated from the fecal debris by magnetic concentration, and the beads were resuspended in sterile water. DNA extraction was performed directly on the bead-oocyst complex using the FastDNA spin kit for soil (MP Biomedicals, Sydney, Australia) with minor modifications to the manufacturer's instructions to optimize DNA yield (57).
PCR screening at 18S rRNA locus.
DNA samples were screened using a previously published nested PCR (54) with minor modifications. The primers 5′-AACCTGGTTGATCCTGCCAGTAGTC-3′ (52) and 5′-CCCATTTCCTTCGAAACAGGA-3′ (54) were used to generate a 1,410-bp product, and primers 5′-AAGGAAGGCAGCAGGCG-3′ (this study) and 5′-AAGGAGTAAGGAACAACCTCCA-3′ (54) were used to obtain a 590-bp product. Each primary PCR mixture contained 1 U of RedTaq polymerase (Integrated Sciences, Sydney, Australia), 6 mM MgCl2, 1× PCR buffer, 200 μM (each) deoxyribonucleotide triphosphate, 100 nM (each) forward and reverse primer, bovine serum albumin at 50 μg/PCR mixture, and sterile water up to a final volume of 25 μl. The secondary PCR mixture was identical to the first except that 2 μl of the primary reaction mixture was used as DNA template and the MgCl2 concentration was 3 mM in a final volume of 50 μl.
PCR amplification of the actin locus.
All samples found to be positive for Cryptosporidium after 18S rRNA screening were also amplified at the actin locus using a nested PCR as previously described (42). In brief, each primary PCR mixture contained 1 U of RedTaq polymerase, 3 mM MgCl2, 1× PCR buffer, 200 μM (each) deoxyribonucleotide triphosphate, 100 nM (each) forward and reverse primer, bovine serum albumin at 50 μg/PCR mixture, and sterile water to a final volume of 25 μl. The secondary PCR mixture was identical to the first except that 2.5 μl of the primary reaction mixture was used as DNA template and the final volume was increased to 50 μl.
Verification of PCR-positive results and morphometrics.
To confirm the results of PCR screening, fecal samples identified as Cryptosporidium positive were processed by IMS and then by flow cytometry (IMS-FC) following a previously described method (37). In brief, IMS was carried out as described above; however, the bead-oocyst complex was suspended in antibody dissociation buffer for flow cytometric analysis. The bead-oocyst complex was labeled with Cry104-fluorescein isothiocyanate (BTF Australia, Sydney, Australia), sorted with a FACSCalibur flow cytometer (Becton Dickinson Biosciences, Sydney, Australia), and counted using an epifluorescence Nikon BH2 microscope. Images of oocysts (n = 50) were captured with a Nikon DXM digital camera (Coherent Scientific, Adelaide, Australia) at a magnification of ×100. Morphometric analysis was performed using ImageJ 1.38x software (National Institutes of Health, Bethesda, MD). To provide a comparison, C. parvum oocysts (n = 50) purified from cattle feces according to a modified sucrose gradient method (46) were examined using the same technique.
Minimum detection limits and oocyst recovery using IMS.
To establish the minimum number of oocysts that could be detected with the 18S rRNA PCR screening technique, C. parvum oocysts from cattle feces (purified as described above) were sorted using a FACSCalibur flow cytometer into aliquots of 10, 100, 1,000, 10,000, and 100,000 oocysts. Each aliquot was prepared in duplicate and added to 0.1 g of filtered feces obtained from captive common brushtail possums (previously confirmed to be negative using IMS-FC). Spiked fecal samples were processed with IMS, followed by DNA extraction and PCR (per the protocols described above). Quantities of oocysts that generated gel bands for duplicate PCR samples were considered inside the detection limits of the screening technique.
To determine the recovery of oocysts using IMS-FC, feces from a Cryptosporidium-negative possum were spiked with 10 and 100 oocysts (prepared as described above). After IMS, flow cytometry was performed, and the oocysts were sorted on a membrane and counted. Six replicates were processed for each quantity of oocysts, and the numbers of oocysts recovered were averaged. This value was used to calculate the oocyst shedding intensity.
Reamplification of PCR products for cloning.
Cryptosporidium-positive samples that produced a weak PCR signal were reamplified using a previously described nested PCR with primers 18SiCF2 (5′-GACATATCATTCAAGTTTCTGACC-3′) and 18SiCR2 (5′-CTGAAGGAGTAAGGAACAACC-3′) (39) to obtain a 763-bp product, and a secondary reaction was performed using primers 18SiF (5′-AGTGACAAGAAATAACAATACAG-3′) and 18SiR (5′-CCTGCTTTAAGCACTCTAATTTTC-3′) (32) to obtain a 310-bp product. To enhance amplification, 8 μl of Genereleaser (Integrated Sciences, Sydney, Australia) was heated with 2 μl of PCR template for 7 min at 500 W in a microwave. The resulting mixture was added to the reaction mix, prepared as described previously (39) and made up to a final volume of 25 μl. Secondary reaction mixtures were identical in composition to the first with the exception that 1 μl of primary PCR product was used as DNA template, Genereleaser was not used, and the final volume was increased to 50 μl. PCR products were purified using the QIAquick PCR purification kit (Qiagen, Victoria, Australia) per the manufacturer's instructions.
Cloning and sequencing.
Purified products were ligated into the pCR2.1 vector using the TA Cloning Kit (Invitrogen, Victoria, Australia), and Cryptosporidium-positive transformants were cultured overnight in liquid Luria-Bertani broth. The plasmid DNA was extracted using the QIAprep spin miniprep kit (Qiagen, Victoria, Australia) in preparation for sequencing. A minimum of two colonies per PCR product were sequenced. Automatic sequencing of DNA fragments was performed in both the forward and reverse directions using a 3130xl DNA capillary sequencer (Applied Biosystems, Foster City, CA).
Cryptosporidium species identification.
Sequences were analyzed with a BLASTN search (1) of the BioManager database at the Australian National Genomic Information Service (ANGIS) (www.angis.org.au). All phylogenetic analysis was thereafter performed using MEGA version 4.0 (43). Sequences were aligned with described Cryptosporidium species by using ClustalW (44) with default settings. Similarities between sequences were calculated using a pairwise distance matrix corrected with the Jukes-Cantor model (25). Two phylogenetic trees were generated using the neighbor-joining method. The 18S rRNA tree included seven isolates of C. parvum and six isolates of C. hominis to represent the interisolate sequence variation of these two species. The combined tree for the 18S rRNA and actin loci was generated for the small subset of samples for which both sequences were obtained. To statistically assess the reliability of the inferred trees, interior branch tests were used to generate confidence probabilities. This statistical test was chosen to provide a highly conservative estimate of the reliability of tree topology in view of the short sequence lengths obtained in the study. All trees were displayed using Eimeria tenella as the evolutionary outgroup. An additional two Cryptosporidium sequences (P111A and P111B) from a single possum host trapped in an urban locale of northern Sydney, Australia, were included in the phylogenetic analysis but were not treated as part of the study population for the purposes of statistical analysis.
Statistical analysis.
Prevalence was calculated by dividing the number of Cryptosporidium-positive individual possums by the total number of individual possums screened. The shedding intensity for each individual was calculated as the oocyst load multiplied by the inverse recovery rate of oocysts from IMS-FC. Possums were divided into two groups, those that were positive and those that were negative for Cryptosporidium, and a binary logistic regression was performed to determine whether age, gender, or season influenced prevalence. Possums were further divided into two groups: low (1 to 102) or high (101 to 106) shedders, and the number of oocysts shed by each possum was log transformed to minimize the scale of the data. Data were analyzed with a binary logistic regression to determine whether age, gender, or season influenced the shedding intensity of possums.
To determine any differences in the shedding pattern of the genotypes identified from possums, a range of nonparametric analyses was performed. Chi-square analysis was used to establish whether there was a significant difference in the prevalence of genotypes. To assess whether shedding of genotypes/isolates was associated with age, gender, or season, chi-square analysis was also performed. A one-way analysis of variance (ANOVA) was used to determine whether shedding intensity was associated with genotype.
To establish whether health parameters, such as body condition (body mass divided by skeletal body length), white blood cell counts (lymphocytes, neutrophils and eosinophils), and nutritional state (triglyceride, cholesterol, and lipase), were associated with shedding intensity, a linear regression was performed. To carry out the regression, shedding intensity values for the 16 Cryptosporidium-positive animals were log transformed and treated as the independent variable, while health parameters were treated as the dependent variable. To determine whether health parameters differed significantly between possums that were shedding oocysts and those that were not, a one-way ANOVA was performed.
A one-way ANOVA was performed to determine whether the length, width, and length-to-width ratio of oocysts shed by possums varied significantly from the those of the C. parvum control oocysts. For all analysis, an alpha value of ≤0.05 indicated a significant difference. Statistics were performed using SPSS version 15 software for Windows.
Nucleotide sequence accession numbers.
The partial Cryptosporidium sequences isolated from T. vulpecula for 18S rRNA and actin have been deposited in the GenBank database under accession numbers EU546846 to EU546869 and EU647729 to EU647733.
RESULTS
Sensitivity of IMS and PCR screening.
The 18S rRNA PCR screening technique produced a strong signal for duplicate samples containing 100 and 1,000 oocysts; however, 10 oocysts produced a weak signal for both duplicate samples. Consequently, the minimum detection limit was determined to be between 11 and 100 Cryptosporidium oocysts/g. The recovery of oocysts using IMS-FC was low, averaging 19% (±1.2%) for fecal samples spiked with 10 oocysts and 21% (±2.8%) for fecal samples spiked with 100 oocysts.
Sequence analysis.
Cloning and sequencing were attempted for all PCR-positive samples and were successful for 13 of the 16 samples. Partial nucleotide sequencing of the 18S rRNA locus revealed that possums shed a distinct Cryptosporidium genotype, brushtail possum 1 (BTP1), and another group of isolates henceforth referred to as brushtail possum 2 (BTP2). Distance matrices indicated that similarities among BTP1 clones ranged from 97.6 to 100% and similarities among BTP2 clones ranged from 95.4 to 100%. The two groups shared between 95.0% and 97.8% similarity and did not match identically with known Cryptosporidium species or genotypes from the GenBank database.
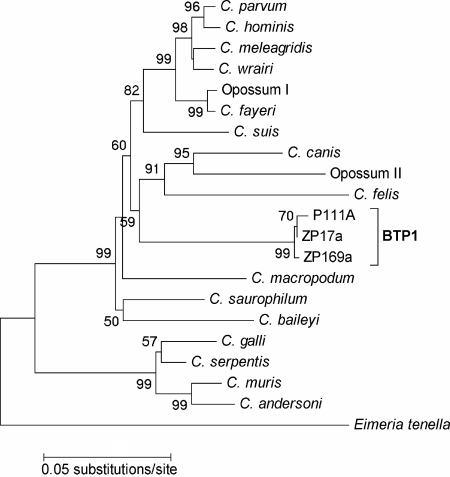
Distance matrices of the 18S rRNA locus indicated that BTP1 was most similar to C. fayeri subtype opossum genotype I (95.9 to 97.0% identity) and C. parvum (95.0 to 96.3%). At the actin locus, partial nucleotide sequences were obtained for three clones of BTP1. Similar to the 18S rRNA locus, distance analysis of the actin locus revealed that BTP1 was also most similar to C. fayeri opossum genotype I (86.5% identity) and C. parvum (85.9% identity). Inferred phylogenetic trees based on the combined 18S rRNA and actin loci grouped all BTP1 isolates in a monophyletic clade with a confidence probability of 99% (Fig. 1). The BTP1 cluster formed a sister clade with C. felis, opossum genotype II, and C. canis, which were encompassed in a monophyletic grouping by C. macropodum (49% confidence probability). Phylogenetic analysis of the 18S rRNA locus grouped the BTP1 isolates together as a sister clade of C. macropodum with a 32% confidence probability (Fig. 2).
FIG. 1.
Phylogenetic relationships of BTP1 genotypes and Cryptosporidium species inferred by the neighbor-joining method based on the combination of a 310-bp fragment of 18S rRNA and 510-bp fragment of actin. Evolutionary distances were computed using interior-branch analysis with confidence probabilities (≥50%) indicated at each node. Eimeria tenella is included as an evolutionary outgroup.
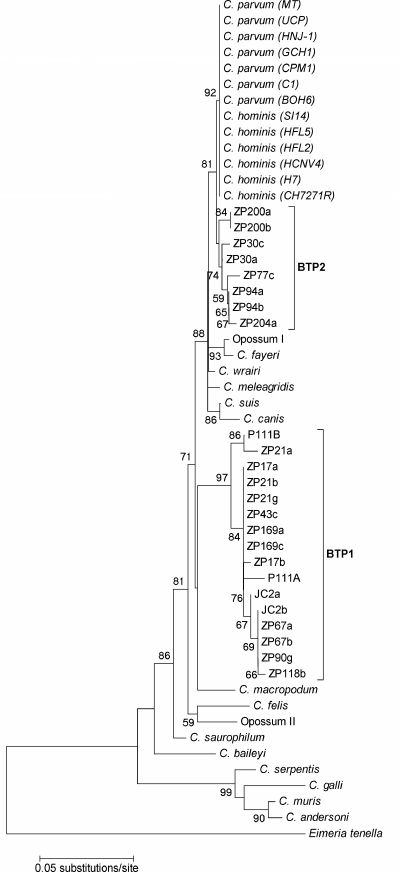
FIG. 2.
Phylogenetic relationships of BTP1 and BTP2 isolates and Cryptosporidium species inferred by the neighbor-joining method based on a 310-bp fragment of 18S rRNA. Evolutionary distances were computed using interior-branch analysis with confidence probabilities (≥50%) indicated at each node. The isolate designations are shown in italic type in parentheses. Eimeria tenella is included as an evolutionary outgroup.
For BTP2 isolates, distance matrices of the 18S rRNA locus revealed that the isolates were equally similar to C. parvum (98.5 to 99.3% identity) and C. hominis (98.5 to 99.3% identity). Sequences of BTP2 at the actin locus were not obtained due to the weak signal generated by PCR products, presumably caused by low oocyst burdens. Inferred phylogenetic trees of the 18S rRNA locus grouped the BTP2 isolates as a monophyletic cluster (34% confidence probability) that formed a sister clade with C. parvum and C. hominis with an 81% confidence probability (Fig. 2).
Prevalence and patterns of oocyst shedding.
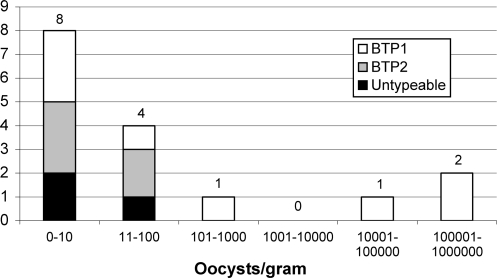
Approximately 11.3% (15/133) of possums from urban habitat were shedding Cryptosporidium compared to 5.6% (1/18) of the woodland population. Of the 15 Cryptosporidium-positive animals from urban habitat, a total of 7 (47%) were shedding BTP1 genotype isolates, 5 (33%) were shedding BTP2 genotype isolates, and sequence data for 3 animals (20%) were not obtained due to low DNA yield. The single Cryptosporidium-positive animal from a woodland habitat was shedding isolates with the BTP1 genotype. The BTP1 genotype was significantly more prevalent across both possum populations than the BTP2 genotype (χ2 = 6.250, df = 1, P = 0.012). The distribution of each genotype among the study population (urban and woodland pooled) is depicted in Fig. 3. Cryptosporidium prevalence was not significantly associated with age (β = 0.72, df = 1, P = 0.95), gender (β = 0.26, df = 1, P = 0.31), or season (β = 0.14, df = 3, P = 0.23). Nor was there any significant association between the genotype shed and age (χ2 = 0.68, df = 1, P = 062), gender (χ2 = 2.24 df = 1, P = 0.18), or season (χ2 = 0.87, df = 1, P = 0.36).
FIG. 3.
Distribution of Cryptosporidium oocysts (BTP1, BTP2, or untypeable) shed by the possum population (urban and woodland habitat pooled). Values above the bars indicate the number of possums belonging to each shedding category.
The shedding intensity of possums from urban habitats averaged 70,581 oocysts per gram (median value of 25 oocysts), while the single Cryptosporidium-positive animal from a woodland habitat shed 10 oocysts per gram. Shedding intensity was not significantly influenced by age (β = 0.76, df = 1, P = 0.97), gender (β = −0.13, df = 1, P = 0.66), or season (β = 22.30, df = 3, P = 0.95). Shedding intensity was significantly associated with genotype (F1,16 = 228.73, P = 0.04). Figure 3 illustrates that the majority of animals shed low numbers of oocysts (1 to 102), while only a few possums shed high numbers (101 to 106), reflecting an overdispersed distribution of parasites in the population.
Oocyst morphometrics.
The mean width of BTP1 oocysts was 3.92 μm (±0.25 μm), and the mean length was 4.12 μm (±0.34 μm). The dimensions of BTP1 oocysts were significantly different from the mean width (5.26 μm ± 0.28 μm) and mean length (5.35 μm ± 0.23) of C. parvum oocysts (for oocyst width, F1,100 = 228.73, P < 0.001; for oocyst length, F1,100 = 127.11, P < 0.001). The width-to-length ratio (or size index) of BTP1 oocysts was 1.06 (±0.10), which was not significantly different (F1,100 = 1.28, P = 0.26) from the width-to-length ratio of C. parvum, 1.02 (±0.08). Due to low oocyst shedding, an adequately large sample size of BTP2 oocysts could not be measured. Comparison of BTP1 oocyst morphometrics with C. parvum and close relatives C. macropodum and C. fayeri is presented in Table 1.
TABLE 1.
Comparison of oocyst morphometrics of Cryptosporidium genotype BTP1 isolates and C. parvum with C. fayeri and C. macropodum
Health of the host.
Shedding intensity did not significantly influence body condition (body mass divided by skeletal body length) (R2 = 1.77, P = 0.07), white blood cell counts, including neutrophils (R2 = 0.48, P = 0.37), lymphocytes (R2 = 0.15, P = 0.11), and eosinophils (R2 = 0.07, P = 0.30), or the nutritional state of the host as indicated by cholesterol (R2 = 0.03, P = 0.51) and triglyceride (R2 = 0.10, P = 0.20). Possums that shed oocysts did not differ significantly in their health parameters compared to possums that were negative for Cryptosporidium (Table 2).
TABLE 2.
Physiology of Cryptosporidium-positive possums compared to Cryptosporidium-negative possums
| Health parameter | Mean (±SE) for possums
|
P valuea | |
|---|---|---|---|
| Cryptosporidium positive | Cryptosporidium negative | ||
| Body condition (%)b | 66.64 (±3.06) | 67.43 (±0.70) | 0.77 |
| Neutrophils (109/liter) | 2.81 (±0.41) | 3.3 (±0.14) | 0.34 |
| Eosinophils (109/liter) | 0.07 (±0.03) | 0.41 (±0.07) | 0.19 |
| Lymphocytes (109/liter) | 3.68 (±0.52) | 3.3 (±0.17) | 0.57 |
| Triglyceride (mmol/liter) | 1.60 (±0.29) | 1.69 (±0.08) | 0.76 |
| Cholesterol (mmol/liter) | 3.09 (±0.18) | 3.08 (±0.05) | 0.94 |
| Lipase (mmol/liter) | 10.33 (±0.89) | 10.42 (±0.25) | 0.93 |
P values of >0.05 indicate that there was no significant difference between Cryptosporidium-positive and -negative hosts.
Body condition was calculated by dividing body mass by the skeletal body length of possums.
DISCUSSION
This investigation revealed that common brushtail possums inhabiting the urban areas of northern Sydney, Australia, were shedding Cryptosporidium isolates that were genetically and morphologically distinct from described species or genotypes. The dominant genotype, BTP1, exhibited the highest similarity with opossum genotype I hosted by the North America marsupial, Didelphis virginiana, the Virginia opossum (55). Opossum genotype I is considered a subtype of C. fayeri from Australian marsupials with the geographic distribution (North America versus Australia) being the only distinguishing factor (40). Phylogenetic analysis inferred that BTP1 isolates were closely related to C. macropodum hosted by the eastern gray kangaroo (Macropus giganteus) of Australia (35). Therefore, both distance and phylogenetic analyses were consistent in finding that BTP1 isolates grouped closely with Cryptosporidium hosted by marsupials. The four species/genotypes identified from marsupials thus far, opossum genotypes I and II, C. fayeri, and C. macropodum, are regarded as host specific: natural infections have been limited to marsupials, and there are no reported human cases (35, 40). The narrow host range of Cryptosporidium from marsupials suggests that the closely related BTP1 genotype may similarly pose limited infectivity to other groups of fauna or humans and is unlikely to be zoonotic.
A second group of Cryptosporidium isolates, BTP2, were most closely aligned with C. parvum and C. hominis, the two predominant species infectious to humans. Phylogenetic analysis of the 18S rRNA locus grouped BTP2 isolates as a sister clade of C. parvum and C. hominis with relatively high bootstrap support. However, the 98.5 to 99.3% genetic similarity between BTP2 isolates, C. parvum, and C. hominis is outside the range normally exhibited by isolates of these two species, which show high sequence uniformity at the hypervariable regions of the 18S rRNA locus (32). Preliminary findings imply that BTP2 isolates are genetically distinct from known zoonotic species. However, the identity of the BTP2 isolates could not be resolved due to low oocyst shedding by hosts, preventing sequencing of the actin locus and morphometric analyses. The inferred phylogeny of BTP2 isolates in relation to zoonotic species indicates that further investigation at other loci is necessary to assess the capacity of BTP2 isolates to infect humans.
The 18S rRNA gene is highly conserved across Cryptosporidium species; however, cloned PCR products from BTP1 and BTP2 clones showed marked variation at this locus. The genetic variation between BTP1 clones ranged from 97.6 to 100% with even greater variation exhibited by BTP2 clones (95.4 to 100%). Clonal variation can arise due to recombination of DNA during PCR amplification (59). However, polymorphisms in BTP1 and BTP2 clones were consistent between forward and reverse sequences and between samples from different hosts, eliminating PCR artifacts as a possible cause for the observed variation. Direct sequencing provides the genetic identity as a consensus of the entire clonal metapopulation and is favored over cloning for Cryptosporidium genotyping. In this study, an additional level of genetic information about the BTP1 and BTP2 genotypes was gained through cloning. Adopting this technique revealed heterogeneity at the 18S rRNA locus within the BTP1 genotype for two possum hosts (ZP21 and P111). The less predominant genotype B had an insertion of 25 bp at the hypervariable region from approximately nucleotide 681 of the 18S rRNA locus, evidence for heterogenous copies of the gene. The heterogeneity within the 18S rRNA was supported by minimal genetic variation within the actin locus, precluding the possibility that types A and B were two distinct genotypes infecting the host. Heterogeneity of the 18S rRNA has been reported for C. fayeri, C. felis, and C. parvum; however, the selective advantage of this feature to the parasite remains unresolved (28, 53).
Evidence for the host-specific nature of the genotypes isolated in this study is reflected by the health of possums in response to oocyst shedding. To date, the majority of Cryptosporidium surveys of wildlife carried out have been based on opportunistic sampling of feces, which unlike trapping rules out collection of information on the host biology. Although body condition (body mass divided by skeletal body length) has been previously investigated in feral pigs (4), this study is unique in examining multiple health parameters of the host to gain insights into the nature of the host-parasite association. Possums that shed oocysts were asymptomatic and did not display elevated white blood cell counts, reduced nutritional state, or diminished body condition, changes reported to occur in response to infection with helminths (48) and other coccidia (11). Such changes to the health of small mammals can have detrimental effects on host fitness, including diminished growth (21), survival, or reproductive success (18). The ability of the Cryptosporidium genotypes isolated in the study to parasitize possums without diminishing host fitness provides evidence of a nonpathogenic relationship with the host, as parasites pose little harm to hosts with which they have had time to form a stable relationship (45). This finding is consistent with a growing body of work that has suggested that coevolution and, consequently, host adaptation are common characteristics of the genus Cryptosporidium (55).
The stability of the host-parasite relationship was reflected in the overdispersed distribution of the parasite within the host population, whereby the majority of animals were shedding low numbers of oocysts and only a few shed high numbers. This distribution is typical in host populations which have achieved immunity as a result of coevolution with the parasite (45). Unable to mount resistance to infection, juveniles from populations of wild animals (4, 36) and livestock (19, 56) tend to be the demographic group most susceptible to Cryptosporidium infection. In addition, seasonal peaks of Cryptosporidium infection tend to coincide with weaning of young animals, when as a result of learning to forage independently, susceptibility to fecal-oral transmission is high in the population (3, 36). In possums, prevalence and shedding intensity were consistent across age groups and seasons, suggesting that age-related variation in immunity did not influence the distribution of Cryptosporidium in the host. This finding presumably reflects the low number of Cryptosporidium-positive possums in the study population, which may be too small to account for a significantly elevated prevalence of Cryptosporidium in juveniles.
Comparison of habitat types indicated a higher prevalence of Cryptosporidium in the urban possum population (11.3%) than in the woodland possum population (5.6%). Due to the small sample size of the woodland population, comparisons between the two habitats should be interpreted with caution. However, these findings parallel the results of a large number of studies which have found that suburban landscapes support densities of wildlife adapted to urban settings that are often higher than in their original habitat (20, 30, 38), inadvertently promoting the transmission of pathogens (7, 24, 26). Studies of Cryptosporidium have shown that host density and prevalence are positively correlated in wild animal populations, such as Soay sheep (Ovis aries) (12), feral pigs (4) and red squirrels (Sciurus vulgaris) (6). In urban areas, the aggregation of resources, such as refuse sites and water tanks, may encourage congregation of possums, potentially increasing the fecal-oral transmission of oocysts. Although urban-environment-induced changes in the biology of possums are conducive to elevated rates of Cryptosporidium transmission, host specificity may limit the potential for spillover of possum genotypes to sympatric populations of humans, domestic pets, and captive zoo animals. Consequently, possums adapted to urban settings pose minimal risk of disseminating Cryptosporidium of public heath concern.
Acknowledgments
This work was supported by an Australian Research Council Linkage Grant between Macquarie University and Taronga Zoo.
Ethics approval was granted by the Macquarie University Animal Ethics Committee (2004/16), the Zoological Parks Board of New South Wales Animal Care and Ethics Committee (4c/08/04), and the New South Wales Department of Environment and Climate Change (DECC) (S11107).
We thank all animal keepers at the Taronga Zoo, veterinarians and nurses at the Taronga Zoo Veterinary and Quarantine Centre, pathologists at the Pacific Laboratory Medicine Services, DECC field officers from Jenolan Caves, and residents of Mosman, Australia, who participated in the study. Thanks also to Jutta Eymann for contributing samples from an additional urban possum population.
Footnotes
Published ahead of print on 18 July 2008.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atwill, E. R., S. M. Camargo, R. Phillips, L. H. Alonso, K. W. Tate, W. A. Jensen, J. Bennet, S. Little, and T. P. Salmon. 2001. Quantitative shedding of two genotypes of Cryptosporidium parvum in California ground squirrels (Spermophilus beechyi). Appl. Environ. Microbiol. 67:2840-2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atwill, E. R., R. Phillips, M. D. G. C. Pereira, X. Li, and B. McCowan. 2004. Seasonal shedding of multiple Cryptosporidium genotypes in California ground squirrels (Spermophilus beecheyi). Appl. Environ. Microbiol. 70:6748-6752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Atwill, E. R., R. A. Sweitzer, M. D. G. C. Pereira, I. A. Gardner, D. Van Vuren, and W. M. Boyce. 1997. Prevalence of and associated risk factors for shedding Cryptosporidium parvum oocysts and Giardia cysts within feral pig populations in California. Appl. Environ. Microbiol. 63:3946-3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bajer, A., M. Bednarska, and E. Sinski. 1997. Wild rodents from different habitats as a reservoir for Cryptosporidium parvum. Acta Parasitol. 42:192-194. [Google Scholar]
- 6.Bertolino, S., L. A. Wauters, L. De Bruyn, and G. Canestri-Trotti. 2003. Prevalence of coccidia parasites (protozoa) in red squirrels (Sciurus vulgaris): effects of host phenotype and environmental factors. Oecologia 137:286-295. [DOI] [PubMed] [Google Scholar]
- 7.Bradley, C. A., and S. Altizer. 2007. Urbanization and the ecology of wildlife diseases. Trends Ecol. Evol. 22:95-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caccio, S. M., R. C. A. Thompson, J. McLauchlin, and H. V. Smith. 2005. Unravelling Cryptosporidium and Giardia epidemiology. Trends Parasitol. 21:430-437. [DOI] [PubMed] [Google Scholar]
- 9.Chalmers, R. M., A. P. Sturdee, S. A. Bull, A. Miller, and S. E. Wright. 1997. The prevalence of Cryptosporidium parvum and C. muris in Mus domesticus, Apodemus sylvaticus and Clethrionomys glareolus in an agricultural system. Parasitol. Res. 83:478-482. [DOI] [PubMed] [Google Scholar]
- 10.Comer, A. J., D. C. Paddock, and J. E. Childs. 2001. Urban zoonoses caused by Bartonella, Coxiella, Ehrlichia and Rickettsia species. Vector Borne Zoonotic Dis. 1:91-118. [DOI] [PubMed] [Google Scholar]
- 11.Cowan, P. E., D. D. Heath, and M. Stankiewicz. 2002. Effects of season, age, and sex on infection with endoparasites of brushtail possums, Trichosurus vulpecula, from a forest/farmland site, lower North Island, New Zealand. N. Z. J. Zool. 29:161-169. [Google Scholar]
- 12.Craig, B. H., J. G. Pilkington, L. E. B. Kruuk, and J. M. Pemberton. 2007. Epidemiology of parasitic protozoan infections in Soay sheep (Ovis aries L.) on St. Kilda. Parasitology 134:9-21. [DOI] [PubMed] [Google Scholar]
- 13.Daniels, M. J., M. R. Hutchings, and A. Greig. 2003. The risk of disease transmission to livestock posed by contamination of farm stored feed by wildlife excreta. Epidemiol. Infect. 130:561-568. [PMC free article] [PubMed] [Google Scholar]
- 14.Deplazes, P., D. Hegglin, S. Gloor, and T. Romig. 2004. Wilderness in the city: the urbanization of Echinococcus multilocularis. Trends Parasitol. 20:77-84. [DOI] [PubMed] [Google Scholar]
- 15.Eymann, J., C. A. Herbert, D. W. Cooper, and J. R. Dubey. 2006. Serologic survey for Toxoplasma gondii and Neospora caninum in the common brushtail possum (Trichosurus vulpecula) from urban Sydney, Australia. J. Parasitol. 92:267-272. [DOI] [PubMed] [Google Scholar]
- 16.Eymann, J., L. D. Smythe, M. L. Symonds, M. F. Dohnt, L. J. Barnett, D. W. Cooper, and C. A. Herbert. 2007. Leptospirosis serology in the common brushtail possum (Trichosurus vulpecula) from urban Sydney, Australia. J. Wildl. Dis. 43:492-497. [DOI] [PubMed] [Google Scholar]
- 17.Fayer, R. 2004. Cryptosporidium: a water-borne zoonotic parasite. Vet. Parasitol. 126:37-56. [DOI] [PubMed] [Google Scholar]
- 18.Fuller, C. A., and A. R. Blaustein. 1996. Effects of the parasite Eimeria arizonensis on survival of deer mice (Peromyscus maniculatus). Ecology 77:2196-2202. [Google Scholar]
- 19.Garber, L. P., M. D. Salman, H. S. Hurd, T. Keefe, and J. L. Schlater. 1994. Potential risk factors for Cryptosporidium infection in dairy calves. J. Am. Vet. Med. Assoc. 205:87-91. [PubMed] [Google Scholar]
- 20.Gloor, S., F. Bontadina, D. Hegglin, P. Deplazes, and U. Breitenmoser. 2001. The rise of urban fox populations in Switzerland. Mamm. Biol. 66:155-164. [Google Scholar]
- 21.Hakkarainen, H., E. Huhta, E. Koskela, T. Mappes, T. Soveri, and P. Suorsa. 2007. Eimeria parasites are associated with a lowered mother's and offspring's body condition in island and mainland populations of the bank vole. Parasitology 134:23-31. [DOI] [PubMed] [Google Scholar]
- 22.Heitman, T. L., L. M. Frederick, J. R. Viste, N. J. Guselle, U. M. Morgan, R. C. A. Thompson, and M. E. Olson. 2002. Prevalence of Giardia and Cryptosporidium and characterization of Cryptosporidium spp. isolated from wildlife, human, and agricultural sources in the North Saskatchewan River Basin in Alberta, Canada. Can. J. Microbiol. 48:530-541. [DOI] [PubMed] [Google Scholar]
- 23.Hill, N. J., J. P. Dubey, L. Vogelnest, M. L. Power, and E. M. Deane. 2008. Do free-ranging common brushtail possums (Trichosurus vulpecula) play a role in the transmission of Toxoplasma gondii within a zoo environment? Vet. Parasitol. 152:202-209. [DOI] [PubMed] [Google Scholar]
- 24.Jones, M. E., A. T. Curns, J. W. Krebs, and J. E. Childs. 2003. Environmental and human demographic features associated with epizootic raccoon rabies in Maryland, Pennsylvania, and Virginia. J. Wildl. Dis. 39:869-874. [DOI] [PubMed] [Google Scholar]
- 25.Jukes, T. H., and C. R. Cantor. 1969. Evolution of protein molecules, p. 21-123. In H. N. Munro (ed.), Mammalian protein metabolism. Academic Press, New York, NY.
- 26.Junge, R. E., K. Bauman, M. King, and M. E. Gompper. 2007. A serologic assessment of exposure to viral pathogens and Leptospira in an urban raccoon (Procyon lotor) population inhabiting a large zoological park. J. Zoo Wildl. Med. 38:18-26. [DOI] [PubMed] [Google Scholar]
- 27.Ko, A. I., M. Galvao Reis, C. M. Ribeiro Dourado, W. D. Johnson, Jr., L. W. Riley, and Salvador Leptospirosis Study Group. 1999. Urban epidemic of severe leptospirosis in Brazil. Lancet 354:820-825. [DOI] [PubMed] [Google Scholar]
- 28.Le Blancq, S. M., N. V. Khramtsov, F. Zamani, S. J. Upton, and T. W. Wu. 1997. Ribosomal RNA gene organization in Cryptosporidium parvum. Mol. Biochem. Parasitol. 90:463-478. [DOI] [PubMed] [Google Scholar]
- 29.McKenzie, S., E. M. Deane, and L. Burnett. 2002. Haematology and serum biochemistry of the Tammar wallaby, Macropus eugenii. Comp. Clin. Pathol. 11:229-237. [Google Scholar]
- 30.McKinney, M. L. 2002. Urbanization, biodiversity, and conservation. Bioscience 52:883-890. [Google Scholar]
- 31.Mendez-Hermida, F., H. Gomez-Couso, R. Romero-Suances, and E. Ares-Mazas. 2007. Cryptosporidium and Giardia in wild otters (Lutra lutra). Vet. Parasitol. 144:153-156. [DOI] [PubMed] [Google Scholar]
- 32.Morgan, U. M., C. C. Constantine, D. A. Forbes, and R. C. A. Thompson. 1997. Differentiation between human and animal isolates of Cryptosporidium parvum using rDNA sequencing and direct PCR analysis. J. Parasitol. 83:825-830. [PubMed] [Google Scholar]
- 33.Morgan, U. M., P. T. Monis, R. Fayer, P. Deplazes, and R. C. A. Thompson. 1999. Phylogenetic relationships among isolates of Cryptosporidium: evidence for several new species. J. Parasitol. 85:1126-1133. [PubMed] [Google Scholar]
- 34.Perz, J. F., and S. M. Le Blancq. 2001. Cryptosporidium parvum infection involving novel genotypes in wildlife from lower New York State. Appl. Environ. Microbiol. 67:1154-1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Power, M. L., and U. M. Ryan. A new species of Cryptosporidium (Apicomplexa: Cryptosporidiidae) from eastern grey kangaroos (Macropus giganteus). J. Parasitol., in press. [DOI] [PubMed]
- 36.Power, M. L., N. C. Sangster, M. B. Slade, and D. A. Veal. 2005. Patterns of Cryptosporidium shedding by eastern grey kangaroos inhabiting an Australian watershed. Appl. Environ. Microbiol. 71:6159-6164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Power, M. L., S. R. Shanker, N. C. Sangster, and D. A. Veal. 2003. Evaluation of a combined immunomagnetic separation/flow cytometry technique for epidemiological investigations of Cryptosporidium in domestic and Australian native animals. Vet. Parasitol. 112:21-31. [DOI] [PubMed] [Google Scholar]
- 38.Prange, S., S. D. Gehrt, and E. P. Wiggers. 2003. Demographic factors contributing to high raccoon densities in urban landscapes. J. Wildl. Manag. 67:324-333. [Google Scholar]
- 39.Ryan, U., L. Xiao, C. Read, L. Zhou, A. A. Lal, and I. Pavlasek. 2003. Identification of novel Cryptosporidium genotypes from the Czech Republic. Appl. Environ. Microbiol. 69:4302-4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ryan, U. M., M. L. Power, and L. H. Xiao. 2008. Cryptosporidium fayeri n. sp. (Apicomplexa: Cryptosporidiidae) from a red kangaroo (Macropus rufus). J. Eukaryot. Microbiol. 55:22-26. [DOI] [PubMed] [Google Scholar]
- 41.Sargent, K. D., U. M. Morgan, A. Elliot, and R. C. A. Thompson. 1998. Morphological and genetic characterisation of Cryptosporidium oocysts from domestic cats. Vet. Parasitol. 77:221-227. [DOI] [PubMed] [Google Scholar]
- 42.Sulaiman, I. M., A. A. Lal, and L. H. Xiao. 2002. Molecular phylogeny and evolutionary relationships of Cryptosporidium parasites at the actin locus. J. Parasitol. 88:388-394. [DOI] [PubMed] [Google Scholar]
- 43.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 44.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Toft, C. A., A. Aeschlimann, and L. Bolis. 1993. Parasite-host associations: coexistence or conflict? Oxford University Press, Oxford, United Kingdom.
- 46.Truong, Q., and B. C. Ferrari. 2006. Quantitative and qualitative comparisons of Cryptosporidium faecal purification procedures for the isolation of oocysts suitable for proteomic analysis. Int. J. Parasitol. 36:811-819. [DOI] [PubMed] [Google Scholar]
- 47.Viggers, K. L., D. B. Lindenmayer, R. B. Cunningham, and C. F. Donnelly. 1998. Estimating body condition in the mountain brushtail possum, Trichosurus caninus. Wildl. Res. 25:499-509. [Google Scholar]
- 48.Viggers, K. L., D. B. Lindenmayer, R. B. Cunningham, and C. F. Donnelly. 1998. The effects of parasites on a wild population of the mountain brushtail possum (Trichosurus caninus) in south-eastern Australia. Int. J. Parasitol. 28:747-755. [DOI] [PubMed] [Google Scholar]
- 49.Warren, K. S., R. A. Swan, U. M. Morgan-Ryan, J. A. Friend, and A. Elliot. 2003. Cryptosporidium muris infection in bilbies (Macrotis lagotis). Aust. Vet. J. 81:739-741. [DOI] [PubMed] [Google Scholar]
- 50.Weir, C., G. Vesey, M. Slade, B. C. Ferrari, D. A. Veal, and K. Williams. 2000. An immunoglobulin G1 monoclonal antibody highly specific to the wall of Cryptosporidium oocysts. Clin. Diagn. Lab. Immunol. 7:745-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Winter, J. W. 1980. Tooth wear as an age index in a population of the brush-tailed possums (Trichosurus vulpecula). Aust. Wildl. Res. 7:359-364. [Google Scholar]
- 52.Xiao, L., L. Escalante, C. Yang, I. Sulaiman, R. J. Montali, R. Fayer, and A. A. Lal. 1999. Phylogenetic analysis of Cryptosporidium parasites based on the small-subunit rRNA gene locus. Appl. Environ. Microbiol. 65:1578-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xiao, L., J. R. Limor, L. Li, U. Morgan, R. C. A. Thompson, and A. A. Lal. 1999. Presence of heterogeneous copies of the small subunit rRNA gene in Cryptosporidium parvum human and marsupial genotypes and Cryptosporidium felis. J. Eukaryot. Microbiol. 46:44S-45S. [PubMed] [Google Scholar]
- 54.Xiao, L., U. M. Morgan, J. Limor, A. Escalante, M. Arrowood, W. Shulaw, R. C. A. Thompson, R. Fayer, and A. A. Lal. 1999. Genetic diversity within Cryptosporidium parvum and related Cryptosporidium species. Appl. Environ. Microbiol. 65:3386-3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xiao, L., I. M. Sulaiman, U. M. Ryan, L. Zhou, E. R. Atwill, M. L. Tischler, X. Zhang, R. Fayer, and A. A. Lal. 2002. Host adaptation and host-parasite co-evolution in Cryptosporidium: implications for taxonomy and public health. Int. J. Parasitol. 32:1773-1785. [DOI] [PubMed] [Google Scholar]
- 56.Xiao, L. H., and R. P. Herd. 1994. Epidemiology of equine Cryptosporidium and Giardia infections. Equine Vet. J. 26:14-17. [DOI] [PubMed] [Google Scholar]
- 57.Yeates, C., and M. R. Gillings. 1998. Rapid purification of DNA from soil for molecular biodiversity analysis. Lett. Appl. Microbiol. 27:49-53. [Google Scholar]
- 58.Zhou, L., R. Fayer, J. M. Trout, U. M. Ryan, F. W. Schaefer III, and L. Xiao. 2004. Genotypes of Cryptosporidium species infecting fur-bearing mammals differ from those of species infecting humans. Appl. Environ. Microbiol. 70:7574-7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou, L., C. F. Yang, and L. H. Xiao. 2003. PCR-mediated recombination between Cryptosporidium spp. of lizards and snakes. J. Eukaryot. Microbiol. 50:563-565. [DOI] [PubMed] [Google Scholar]