Abstract
The origin, structure, and composition of biofilms in various compartments of an industrial full-scale reverse-osmosis (RO) membrane water purification plant were analyzed by molecular biological methods. Samples were taken when the RO installation suffered from a substantial pressure drop and decreased production. The bacterial community of the RO membrane biofilm was clearly different from the bacterial community present at other locations in the RO plant, indicating the development of a specialized bacterial community on the RO membranes. The typical freshwater phylotypes in the RO membrane biofilm (i.e., Proteobacteria, Cytophaga-Flexibacter-Bacteroides group, and Firmicutes) were also present in the water sample fed to the plant, suggesting a feed water origin. However, the relative abundances of the different species in the mature biofilm were different from those in the feed water, indicating that the biofilm was actively formed on the RO membrane sheets and was not the result of a concentration of bacteria present in the feed water. The majority of the microorganisms (59% of the total number of clones) in the biofilm were related to the class Proteobacteria, with a dominance of Sphingomonas spp. (27% of all clones). Members of the genus Sphingomonas seem to be responsible for the biofouling of the membranes in the RO installation.
Membrane biofouling is an important problem for reverse-osmosis (RO) systems, in particular for RO membranes (13, 14, 17). The attachment of bacteria to membrane surfaces and subsequent biofilm growth in the spiral-wound RO membrane elements strongly influence RO system performance and RO plant productivity. Problems are due primarily to an increase in the differential pressures of the RO modules, the long-term membrane flux reduction of the RO plant, and the deterioration of product water quality as a result of high levels of biomass accumulation on RO membrane surfaces (37, 43, 45). Once in progress, biofouling regularly and persistently hampers the RO water treatment process (13, 15).
Presently, adequate measures to prevent or reduce biofouling are lacking. The microbiological and physical processes associated with biofilm formation and biofouling in these dynamic and high-pressure environments are poorly understood. The conditions change from an oligotrophic environment in the beginning to a heterotrophic environment when the biofilm is mature. The first indications that a variety of different microorganisms participate in biofilm development on RO membranes were obtained by traditional dissections of fouled RO membrane elements (autopsies) and the subsequent analysis of the membrane surface-fouling layers. The conventional plating and colony isolation methods showed the presence of a wide variety of species on the feed and permeative surfaces of biofouled cellulose acetate, polyetherurea thin-film composite, or polyamide thin-film-composite membranes (4, 9, 17, 19, 28, 38, 39). However, by cultivation-dependent methods, information about only 0.01 to 3% of the population in natural environments is obtained (2, 20, 23). In recent years, the microbial-community structure in RO membrane samples obtained from full-scale membrane-based water purification processes was examined using 16S rRNA gene clone libraries and fluorescence in situ hybridization methods (7) and using PCR-denaturing gradient gel electrophoresis (DGGE) and sequence analysis of constructed clone libraries containing larger PCR fragments of the 16S rRNA gene (6). Pang and Liu (33) investigated the microbial-community composition of a biofilm retrieved from a lab-scale RO membrane module by applying a 16S rRNA gene-based clone library and terminal restriction fragment length polymorphism (RFLP) analysis. Nevertheless, a complete picture of the bacterial population responsible for the biofouling of RO systems is still lacking. A molecular study of microbial populations in all compartments of a full-scale RO water purification plant had not yet been performed.
This study aims to gain insight into the origins and compositions of the biofilms in full-scale RO systems by investigating the bacterial communities in terms of species composition and species diversity, as part of the free-living communities in the feed and product water, and as part of the film-forming communities attached to surfaces. The bacterial-community structure in various compartments of a full-scale RO water purification plant, including the RO feed water (F) (fresh surface water), the wall of the ultrafiltration storage tank (UF), a cartridge filter (CF), a biofouled RO membrane (M), and RO product water (P) (process water) was determined by molecular techniques. A PCR-DGGE approach (31) combined with the analysis of constructed clone libraries containing larger PCR fragments of the 16S rRNA gene (44) and DGGE screening of the isolated clones were used to reveal the differences between the bacterial community of the RO membrane biofilm and the other different locations of an RO plant.
MATERIALS AND METHODS
Sampling locations and procedures.
Samples were collected in May 2006 from a full-scale RO water purification plant located in Veendam, The Netherlands. The plant used energy-saving polyamide (ESPA) RO membrane elements (ESPA 2; Hydranautics, CA) to produce process water. The F fed to the RO system of the plant was extensively treated by the sequential application of coagulation, flocculation, sand filtration, ultrafiltration, and cartridge filtration processes. An additional chemical treatment of the RO membrane elements with an acid-alkaline solution was applied to this system once a week to maintain a reasonable flux. The samples were taken from the F, UF, CF, M, and P when the RO installation suffered from a substantial pressure drop and decreased production. The F, P, and UF samples were obtained prior to plant shutdown and RO membrane element removal. The UF sample was scraped from the walls of the ultrafiltration storage tank. For the collection of the RO membrane samples, the first membrane element from the first stage of the investigated RO system was selected. The element, used for about 1 year in the water purification process, was retrieved from the RO unit after plant shutdown, wrapped in plastic sheeting, and transferred to the laboratory for an autopsy on the same day. The samples were taken directly after physical dissection and during the autopsy of the RO membrane by excising small sections from different locations in the membranes (the tightly associated membrane samples) or by scraping material from a known area on the surfaces of the membranes (the loose biofilm samples). All samples were collected in sterile tubes and kept on ice until further processing within 1 day.
Total DNA extraction.
The microbial biomass from the water samples (10 ml) was collected by centrifugation at 10,000 × g for 10 min and suspended in 0.5 ml of phosphate-buffered saline (pH 7.0). Approximately 0.5 mg material was transferred from the biofilm samples to a clean tube, mixed with 0.5 ml of phosphate-buffered saline (pH 7.0), and homogenized using a vortex. All samples were subjected to 20 min of sonication, and the total community DNA was extracted from 0.5 ml of homogenate using a minibead beater with the Fast DNA spin kit for soil (MP Biomedicals) in accordance with the manufacturer's instructions. The quality of the DNA was checked by agarose gel electrophoresis. Aliquots of each DNA extract were further purified and concentrated with a DNA Clean & Concentrator-5 kit (Zymo Research) according to the manufacturer's instructions.
PCR amplification and DGGE analysis of amplified 16S rRNA genes.
PCR amplification of bacterial 16S rRNA genes from total genomic DNA was performed using Go Taq DNA polymerase (Promega) with primers 954-f and 1369-r (MWG-Biotech AG), targeting the hypervariable V6-V8 region, as previously described by Zhongtang and Morrison (46). A 40-base GC clamp (5′-CGCCGGGGGCGCGCCCCGGGCGGGGCGGGGGCACGGG-GGG-3′) was attached to the forward primer at the 5′ end. A typical PCR mixture (50 μl) contained 10 ng template DNA, each deoxynucleoside triphosphate at a concentration of 200 μM (Invitrogen), each primer at 0.5 μM, 1.25 U of Go Taq DNA polymerase, and 1× PCR buffer containing 3 mM MgCl2 (Promega). The reactions were performed in an iCycler (Bio-Rad) with predenaturation at 94°C for 2 min, followed by 35 cycles consisting of 30 s at 94°C, 30 s at 56°C, and 60 s at 72°C. The cycles were completed with a final extension step of 7 min at 72°C. DGGE analysis of the generated amplicons was performed using a DCode System (Bio-Rad) as previously described by Heilig et al. (18). A mixture of the DGGE-PCR products from nine bacterial species was applied as a marker. The number of operational taxonomic units (OTUs) in each sample was defined as the number of DGGE bands with distinct electrophoretic mobilities.
Cloning of PCR-amplified products and sequence analyses.
Amplification of the almost-full-length bacterial 16S rRNA gene fragments with the 7-f and 1510-r universal bacterial primers (27) was performed with the iCycler as described previously (6). Amplified fragments were purified with the DNA Clean & Concentrator-5 kit, ligated into the pGEM-T easy vector (Promega kit), and cloned into Escherichia coli XL1-Blue according to the manufacturer's instructions. Vector-harboring clones were transferred with a sterile toothpick into 50 μl of Tris-EDTA buffer and were incubated at 95°C for 15 min to lyse the cells. The PCR amplification of cell lysates with T7 and Sp6 pGEM-T-specific primers and the selection of clones containing insertions of the appropriate sizes by the RFLP analysis were performed as described previously (6). Individual clones with a unique RFLP pattern were selected and screened by DGGE analysis with the V6-V8 primers (GC-954-f and 1369-r). Their DGGE bands were detected in the original DGGE fingerprint profile of the biofilm community by using BioNumerics software (BioSystematica). Unique inserts were bidirectionally sequenced with T7 and Sp6 primers (BaseClear, Leiden, The Netherlands). Checks for chimeric sequences were conducted by using the Chimera Check program at http://www.cme.msu. edu/RDP/html/index.html (29), and sequence similarity was analyzed by using the NCBI BLAST search tool at http://www.ncbi.nlm.nih.gov/BLAST (1) and the GenBank database.
Nucleotide sequence accession numbers.
The nucleotide sequence data reported in this study were deposited in GenBank under accession numbers EU428849 to EU428950.
RESULTS
Observations during autopsy.
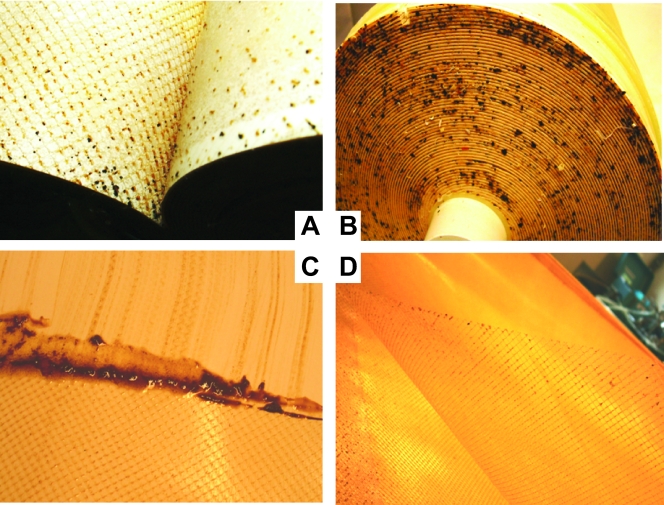
At the laboratory, the removed RO membrane element was unpacked and cut open, and the membrane packs were unfolded and visually examined. The visual inspection of the element showed the presence of a slimy, opaque, light-brown deposit on the surfaces of all membrane sheets and feed spacers (Fig. 1), indicating that the fouling layers were not eliminated by the routine (once-a-week) cleaning procedures of the RO units in this system. After 1 year of operation, the fouling layer was spread over the complete membrane and the feed spacer surfaces in the module. This fouling layer was quite loosely attached to the RO membrane and could be relatively easily scraped from the surface (Fig. 1C). It was also noted that the membrane surfaces were more intensely fouled than the feed spacer surfaces, and no visible fouling was observed on the surfaces of the product spacers.
FIG. 1.
Photographs of an autopsy of fouled RO ESPA-2 spiral-wound M elements (Hydranautics ESPA). The feed side of the membrane element (A and B), the feed side of the fouled membrane (C), and the fouled plastic feed channel spacer (D) are shown. The surfaces of the membrane and spacer were completely covered with fouling layers. The fouling layer could be relatively easily scraped from the membrane surfaces (C).
Clone library construction and analysis.
In total, five 16S rRNA gene clone libraries, containing a total of 635 clones, were constructed with the Bacteria primer set (7-f and 1510-r) by using total genomic DNA isolated from the F, UF, CF, M, and P. All the clones in the libraries were subjected to RFLP analysis, and clones with identical RFLP patterns were grouped together into clone families. One representative clone from each clone family was partially sequenced. Subsequently, the full sequence of the 16S rRNA gene was determined from those clones that contained a unique sequence and that corresponded with a dominant band in the DGGE community fingerprints.
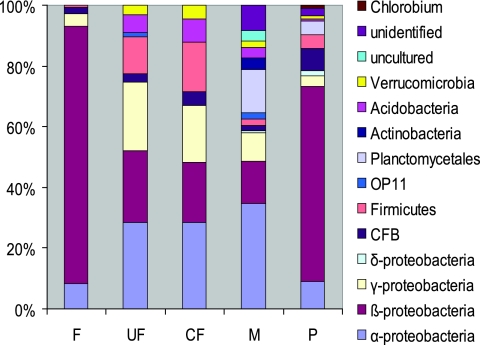
The nucleotide sequences of a total of 635 clones were determined. A total of 35 clones were detected as possible chimeras and were excluded from further community analysis. The nucleotide sequences of the remaining clones, which included 179, 67, 90, 152, and 112 clones from the F, UF, CF, M, and P libraries, respectively, were further analyzed for their phylogenetic affiliations and closest relatives by searching the GenBank database with the NCBI BLAST search tool. Different sequence types (or OTUs) affiliated with various phylogenetic lineages of the domain Bacteria (with a sequence similarity of >0.90) were obtained from the clone libraries (Table 1). Phylogenetic analysis indicated that the Proteobacteria division dominated all clone libraries in this study, in which the Betaproteobacteria subdivision was the largest bacterial group found in water samples of the F and P (85% and 65% of total clones, respectively), and members of the Alphaproteobacteria subdivision were numerically the most frequently encountered in the biofilms of the samples of the UF, CF, and M (28%, 29%, and 35% of total clones, respectively). Furthermore, members of the Betaproteobacteria subdivision made up the second-largest fraction in the UF, CF, and M (24%, 20%, and 14% of total clones, respectively), whereas members of the Alphaproteobacteria subdivision made up the second-largest fraction in the F and P (8 and 9% of total clones).
TABLE 1.
Phylogenetic affiliations and frequencies of cloned bacterial 16S rRNA gene ampliconsa retrieved from RO samples from the full-scale RO water treatment plant
| Closest relative in GenBank (accession no.) | Similarity (%)b | % of clones in indicated clone libraryc
|
||||
|---|---|---|---|---|---|---|
| F | UF | CF | M | P | ||
| Sphingomonas sp. strain MTR-71 (DQ898300.1) | 95 | 16.4 | 17.7 | 7.9 | 2.7 | |
| Sphingomonas subterranea (AB025014.1) | 98 | 1.7 | 1.5 | 2.2 | 3.3 | 0.9 |
| Sphingomonas sp. strain ORS 1497 (AJ968701.1) | 96 | 1.5 | 1.1 | 0.7 | ||
| Sphingomonas sp. strain DB-1 (AY947554.1) | 98 | 1.5 | 2.2 | 0.7 | ||
| Sphingomonas sp. strain BAC151 (EU131005.1) | 97 | 5.6 | 1.5 | 1.1 | 9.2 | 2.7 |
| Sphingomonas sp. strain HI-K4 (DQ205308.1) | 98 | 3.0 | 1.3 | 0.9 | ||
| Sphingomonas sp. strain BAC13P (EU131003.1) | 97 | 1.1 | 0.7 | 0.9 | ||
| Sphingomonas oligophenolica (AB365794.1) | 98 | 0.7 | ||||
| Sphingomonas sp. strain HTCC500 (AY584571.1) | 98 | 0.7 | ||||
| Sphingomonas suberifaciens (D13737.1) | 97 | 0.7 | ||||
| Sphingomonas sp. strain P2 (AB091683.1) | 97 | 0.7 | ||||
| Other Alphaproteobacteria (AY029562.1, AB271055.1, AM411913.1, DQ414680.1, AM286550.1, X97691.1, DQ177493.1, AY921677.1, and EU050759.1) | 94-99 | 1.1 | 3.0 | 3.3 | 8.6 | 0.9 |
| Acidovorax spp. (AM262110.1, Y18617.1, EF540489.1, and AF235013.1) | 99 | 1.1 | 1.5 | 1.1 | 2.1 | 0.9 |
| Burkholderia spp. (AB232330.1, AY752954.1, AB212237.1, and DQ156083.1) | 99 | 78.3 | 1.5 | 1.1 | 1.4 | 41.2 |
| Janthinobacterium spp. (AF174648.1, EF422171.1, and AJ551147.1) | 98-99 | 5.1 | 14.9 | 14.4 | 1.4 | 6.3 |
| Nitrosomonas spp. (AB000700.1, AY123811.1, and AY123797.1) | 95-99 | 0.6 | 1.5 | 1.1 | 4.6 | |
| Other Betaproteobacteria (AB120966.1, AB195750.1, AF236004.1, AJ556799.1, DQ413154.1, EU130968.1, AM236310.1, AF351219.1, AJ575695.1, AF204252.1, AB265946.2, and AF078758.1) | 93-99 | 4.5 | 2.2 | 4.7 | 16.2 | |
| Pseudomonas spp. (AM157452.1 and DQ178233.1) | 97-99 | 1.1 | 20.9 | 15.5 | 2.0 | |
| Lysobacter spp. (AB161360.1 and AB249682.1) | 95-96 | 0.6 | 6.0 | 0.9 | ||
| Legionella spp. (X73406.1 and AM747393.1) | 93-97 | 0.6 | 1.1 | 1.3 | 2.7 | |
| Other Gammaproteobacteria (AM396494.1, EF191354.1, AJ583181.1, and AM229325.1) | 97-99 | 1.7 | 1.5 | 2.2 | ||
| Deltaproteobacteria (AY921696.1, AF418174.1, and U41561.1) | 98 | 0.7 | 1.8 | |||
| Flavobacterium spp. (AM230485.1, DQ628949.1, EF520552.1, and EF540472.1) | 97-100 | 2.2 | 1.5 | 3.3 | 2.7 | |
| Other CFBd (AY780553.1, AB074940.1, DQ640688.1, and AY910857.1) | 92-98 | 1.5 | 1.1 | 2.0 | 4.5 | |
| Clostridium spp. (AY360624.1, X75909.1, AJ506120.1, AB288643.1, and AY935674.1) | 92-99 | 10.5 | 13.3 | 2.0 | 4.5 | |
| Other Firmicutes spp. (EF033503.1, AM745263.1, and AY766466.1) | 91-97 | 0.6 | 1.5 | 3.3 | ||
| Actinobacteria spp. (AB271048.1, AY368456.1, and AM410685.1) | 98 | 3.9 | ||||
| Chlorobium phaeobacteroides (AM050128.1) | 98 | 0.9 | ||||
| Uncultured candidate division OP11 (AF047573.1) | 92 | 1.5 | 2.0 | |||
| Acidobacteria spp. (DQ513986.1, AY921727.1, and EF032752.1) | 96-97 | 6.0 | 7.8 | 3.3 | 0.9 | |
| Planctomycetales sp. (AY942960.1, AY500064.1, AB116499.1, DQ676396.1, AJ290177.1, EF221226.1, and X81950.1) | 91-98 | 14.5 | 4.5 | |||
| Verrucomicrobiae spp. (AB305640.1, AB288576.1 and AB288579.1) | 92-97 | 3.0 | 4.4 | 2.1 | 0.9 | |
| Uncultured bacteria (AY917428.1, EF220517.1, EF663458.1, EU273223.1, AB288666.1, EF506959.1, EF688335.1, AB290357.1, and DQ241389.1) | 84-98 | 10.8 | 2.7 | |||
Amplicons were approximately 1.45 kb in size.
Percentage of similarity between the cloned 16S rRNA gene and its closest relative in the NCBI database.
F clone library, 179 clones; UF clone library, 67 clones; CF clone library, 90 clones; M clone library, 152 clones; P clone library, 112 clones.
CFB, Cytophaga-Flexibacter-Bacteroides spp.
The majority of the Alphaproteobacteria found in all samples were primarily affiliated with the genus Sphingomonas; 2 to 7% of the total number of clones in these libraries were related to known Sphingomonas species (>97% similarity) (Table 1). The remaining clones in this group were closely related to other known Alphaproteobacteria, like Afipia massiliensis and Hyphomicrobium sp. (present in three of the five samples), “Caulobacter ginsengisoli,” Mesorhizobium sp., uncultured “Nordella” sp., Pedomicrobium manganicum, and Sphingopyxis sp. Two OTUs (3% of the total number of clones) from the M sample were related to two uncultured species of Alphaproteobacteria.
Within the Betaproteobacteria lineage, Acidovorax, Burkholderia, and Janthinobacterium were the common bacterial genera in all samples. In this study, the genus Burkholderia represented the largest fraction in the Betaproteobacteria subgroup (Fig. 2) of the F and P libraries (78% and 41% of total clones, respectively). Janthinobacterium spp. were found mainly in the UF and CF samples (15% and 14% of total clones, respectively). The most dominant betaproteobacterium in the M sample was related to Nitrosomonas sp. strain Nm59 (4%). The remaining sequences identified as Betaproteobacteria were closely related to known species, such as “Aquamonas fontana,” the aquatic bacterium R1-B18, the betaproteobacterium A0637, Comamonadaceae, Hydrogenophaga spp., and Simplicispira spp. However, in P samples, 11% of the total clones were related to uncultured betaproteobacterium species.
FIG. 2.
Schematic diagram depicting the results of the clone library analyses performed on the samples obtained from the different functional parts of a full-scale RO plant. F was initially pretreated by the sequential application of coagulation, flocculation, sand filtration, UF, and CF processes. The two-stage RO system integrated ESPA-2 spirally wound M elements (Hydronautics ESPA) which were installed in a series inside pressure vessels.
All biofilm samples (UF, CF, and M) further comprised OTUs from the Gammaproteobacteria division (22%, 19%, and 9% of the total clones, respectively). Members of the phylum Firmicutes were found mostly in UF and CF biofilms (12% and 17% of the total clones, respectively) and consisted mainly of Clostridium species (UF, 11% of the total clones; CF, 13%). From the Gammaproteobacteria division, the most frequently encountered OTUs from the biofilm samples were closely related to Pseudomonas spp. (UF, 21% of the total clones; CF, 16%) or showed 95 to 96% similarity with Lysobacter spp. (M, 6% of the total clones). Bacteria related to members of the Cytophaga, Flexibacter, and Bacteroides groups were found in all samples in similar percentages. Furthermore, in biofilms of the UF, CF, and M, bacteria related to uncultured environmental clones from the Acidobacteria (6%, 8%, and 3% of total clones, respectively) and Verrucomicrobiae groups (3%, 4%, and 2% of total clones, respectively) were found. Species related to uncultured environmental clones from the Planctomycetacea group were found in the M and P samples only (15% and 5% of all clones, respectively). Differing from the rest of the biofilm samples, the M sample further comprised clones related to Actinobacteria (4% of the total clones). Nine other OTUs from M samples (11% of the total clones) were related to unknown uncultured bacteria, some with a homology of less than 95%, or showed no exact match (<90% similarity) with any of the known bacterial sequences found in the databases. Similarly, no exact match was found for 3% of the total clones in the P sample.
Fingerprinting of RO biofilm communities by DGGE.
DGGE analysis of PCR-amplified fragments of the hypervariable V6-V8 region of the bacterial 16S rRNA gene (approximately 415 bp) obtained from the F, UF, CF, M, and P samples revealed clearly discriminative “fingerprints” of bacterial communities from various compartments of the investigated RO plant (Fig. 3). The gel image shows distinct bands (or OTUs), indicating the presence of multiple species in all of the samples tested. At least two DGGE bands could clearly be discriminated as dominant (intense bands) within each single biofilm sample. The highest number of dominant bands was present in the M sample (seven bands) and the lowest in the F sample (two bands). The remainder of the samples each contained four dominant bands.
FIG. 3.
DGGE fingerprinting of RO biofilm samples collected from a full-scale water treatment plant. Arrows with numbers indicate the positions of the identified bands.
The complexity of the DGGE profiles (Fig. 3) of the microbial community in the F sample (lane F) is less than that of the free-living community in the P sample (lane P) and that of the biofilm-forming communities attached to the surfaces of the UF (lane UF), CF (lane CF), and M (lane M). In general, the bacterial communities from the F and P samples had similar community fingerprints (71% similarity) but were markedly different from those of the other three samples. The DGGE profiles obtained from the samples from the CF and UF showed similar community fingerprints (96% similarity), but these were also different from the fingerprints from the other three samples. The M sample had a unique fingerprint compared to those of the other samples. The feed spacer sample of the investigated RO membrane module had a very similar DGGE pattern (data not shown). Moreover, the samples of the tested product spacer showed the presence of bacteria that were also found in the RO membrane sample but in relatively low numbers according to their band intensities in the gel (vaguely visible bands [data not shown]). The bacterial species representing the visible bands of the DGGE profiles of the F, UF, CF, M, and P samples were identified by comparing the migration profiles of the PCR amplicons in the original biofilm community fingerprints with the migration profiles of the DGGE-PCR products from the individual clones. Except for the minor constituents, most members of biofilm communities detected by the cloning library approach could be associated with one of the visible bands in the DGGE profiles. In total, 35 distinct DGGE bands could be associated with at least one of the identified clones in the constructed libraries.
DISCUSSION
In this study, we describe the complex and diverse bacterial communities in various compartments of a full-scale RO water purification plant by using two culture-independent methods. The bacterial communities were investigated in terms of their species diversity and the relative abundances of the free-living communities in the F and P samples and of the film-forming communities attached to the UF, CF, and M. The biodiversity of these communities, as revealed by analysis of 16S rRNA genes from the total biofilm community, was larger than that found by the DGGE approach. The cloning method was more powerful than DGGE in evaluating the complexity and composition especially of biofilms on M, because 2.5-fold-more genetically different bacteria were identified than in the DGGE analysis. The DGGE fingerprints underestimated the diversity of the communities due to the comigration of several different 16S rRNA gene fragments observed in experiments where individual clones from the libraries were subjected to DGGE profiling. All OTUs from the species that were present in relative high numbers in the sample communities (Table 1; Fig. 2) were detected as visible DGGE bands (Fig. 3). Although DGGE analysis in this study did not visualize all the members of a complex microbial community as separate bands, both methods could detect the same dominant species in the communities. The inability to detect populations of low abundance and overlapping DGGE bands was also shown by Muyzer et al. (31) and Murray et al. (30).
A large difference exists between the bacterial-community composition of the M biofilm and the bacterial-community compositions at other locations in the RO plant. The M biofilm community was more complex than the bacterial populations in the other compartments of the RO plant (Table 1; Fig. 2 and 3), indicating the occurrence of different selection mechanisms at different compartments in the full-scale plant. These differences indicate that the biofilm was actively formed on the M surfaces and was not the result of a simple concentration of bacteria present in the F. Undoubtedly, a bacterial community adapted to this environment was present in the form of a biofilm on the M surface at the moment of sampling, when changes in plant performance were noted (an increased pressure drop over the RO module). This complex community was represented by bacterial species with different physiological traits, most likely selectively promoted under changing physical-chemical and microbiological conditions in the dynamic and high-pressure (12-bar) operating environment of the RO system. Apparently, the predominant bacterial species capable of handling these conditions were related to the genus Sphingomonas (27% of all clones), which is known to thrive in biofilms (7, 21, 24, 25). The Planctomycetacea, the second-largest group associated with the M biofilm (15% of all clones), are free-living aquatic oligotrophs that feed on algae or on their degradation products (16). Some of them contain a large number of open reading frames coding for enzymes necessary for polysaccharide degradation (www.regx.de), which are present in large amounts in biofilms (22, 26). A relatively low abundance of other different species in the M biofilm community found in this study (Table 1) cannot be interpreted as evidence that these minority populations are of little importance to the community as a whole. Even very low levels of bacterial species can maintain community activity (11).
The discovery of the typical freshwater phylotypes (47) in the M biofilm, as well as the detection of most of them (i.e., Proteobacteria, Cytophaga-Flexibacter-Bacteroides, and Firmicutes) in the F sample, suggests a feed water origin rather than a manufacturing contamination in the RO unit. Although a relatively small number of bacterial genera (approximately 12) appears to dominate the F community at the moment of the sampling, all of them, except Dyella, were also found in the film-forming communities on the UF, CF, or M surfaces in addition to the P community (Table 1). The observed dominance of the genus Sphingomonas in the UF, CF, and M samples (Table 1; Fig. 2 and 3) may be explained by the strong association of these organisms with surfaces (34), while the prevalence of the Betaproteobacteria in the water samples (the F and P samples) was consistent with their abundance in the freshwater as plankton (47). On the other hand, the absence of bacteria related to Acidobacteria, Actinobacteria, Deltaproteobacteria, Chlorobium, Planctomycetacea, Verrucomicrobiae, and some other bacterial groups in the F sample and their presence in the other samples suggest that these organisms entered the plant prior to the sampling. The logical explanation of the exclusive presence of some bacterial species at different locations is that the conditions inside the RO plant were optimal.
The detection of different bacterial sequences in the P sample was rather unexpected, since the passage of the bacteria through the RO membrane (8-in. Hydranautics ESPA 2) is theoretically impossible. Also, it is not clear why bacteria such as Aquamonas, Chlorobium, Desulfarculus, Geobacter, and Mesorhizobium were found in the P sample, since they were not detected in the other samples. The reason might be that these organisms are involved in the biofouling of the pipelines connecting the RO system with the permeate storage tank. The presence of Geobacter, an anaerobe involved in the reduction of Fe(III) (8), could indicate the corrosion processes of these pipelines on metallic surfaces. Also, the detection of the green sulfur bacteria from the genus Chlorobium indicates that the environment is anaerobic, because their photosynthesis can occur only in the complete absence of oxygen (32). However, it is not clear how these bacteria can survive and possibly even grow without light.
This is the first molecular study of microbial populations that has been performed on all units of a full-scale RO water purification plant. This approach allowed for the understanding of how bacterial communities are distributed throughout the RO plant and where they originate. The investigations suggest an important role of Sphingomonas in the biological-membrane fouling of spiral-wound membrane elements applied in the RO water purification processes. The members of this genus were the most prevalent organisms in the M biofilms in this study but also in our previously reported investigations, in which bacterial biofilms that developed on RO membranes of ∼5.5 year-old M elements were investigated (6). As the RO plant location, process configuration, cleaning type and frequency, membrane surface material, feed water, and the sampling time (May) of the M samples were the same as in our earlier study, the presence of Sphingomonas in all membrane biofilm communities confirms that these organisms are positively selected because of their competitive advantages for survival in this environment. As facultative oligotrophs, they are metabolically well adapted to a low-carbon environment (10, 41) and can proliferate under conditions of limited substrates for bacterial growth in the initially clean RO system. Sphingomonas organisms are able to utilize a broad range of naturally occurring organic compounds as well as many types of environmental contaminants (5). Apparently, they are also able to survive at high nutrient concentrations that occur close to the membrane surface in the RO units due to the concentration polarization effect in membrane separation processes and the accumulation of nutrients in the biofilm matrix. Furthermore, Sphingomonas species can change their planktonic state to sessile when the culture conditions, such as the level of aeration, are changed (35). Hence, a low-oxygen concentration, generally typical for the M modules, could stimulate their potential ability to form M biofilms. The transport of Sphingomonas to the membrane surfaces under continuous-flow conditions in spiral-wound M elements could be facilitated by their twitching and swarming motility (34). Their ability to produce different kinds of extracellular polysaccharides (12, 22, 35, 36) can help to initiate biofilm formation and to keep them attached to the membranes (3, 34). Moreover, the slimy extracellular polysaccharide matrix may protect the cells inside a biofilm matrix against the regular chemical cleaning procedures by acting as a chemically reactive barrier that inactivates the cleaning chemicals (40). Pang and coworkers (34) observed that one of the most dominant bacterial isolates previously retrieved by Chen et al. (7) from a biofouled M sample treating potable water, Sphingomonas sp. strain RO2, effectively colonized different RO membranes in continuous-flow cell systems regardless of their surface properties. Hence, Sphingomonas and other biofilm-associated slime producers, like Rhizobiales bacteria (33), are responsible for membrane surface colonization that facilitates the attachment of other bacteria and encourages the maturation of the biofilm. The formation and accumulation of exopolymeric substances, characteristic of growing biofilms (42), substantially decrease the water flux through membranes (21), one of the typical problems associated with biofouling in the actual practice of RO systems. Research is in progress to identify the nature of exopolymers formed by sphingomonas strains isolated from RO membranes.
Acknowledgments
This work was performed at Waterlaboratorium Noord (Kisuma, Veendam) and Wetsus, Centre of Excellence for Sustainable Water Technology. Wetsus is funded by the city of Leeuwarden, the Province of Fryslân, the European Union's European Regional Development Fund, the EZ/KOMPAS program of the Samenwerkingsverband Noord-Nederland, and the Ministry of Economic Affairs. We thank the participants of the theme “Biofouling” for their interest and financial contributions.
Footnotes
Published ahead of print on 11 July 2008.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann, R. I., W. Ludwig, and K.-H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Azeredo, J., and R. Oliveira. 2000. The role of exopolymers in the attachment of Sphingomonas paucimobilis. Biofouling 16:59-67. [Google Scholar]
- 4.Baker, J. S., and L. Y. Dudley. 1998. Biofouling in membrane systems—a review. Desalination 118:81-89. [Google Scholar]
- 5.Balkwill, D. L., J. K. Fredrickson, and M. F. Romine. 2003. Sphingomonas and related genera. In M. Dworkin (ed.), The prokaryotes: an evolving electronic resource for the microbiological community, 3rd ed., release 3.14. Springer-Verlag, New York, NY.
- 6.Bereschenko, L. A., A. J. M. Stams, G. H. J. Heilig, G. J. W. Euverink, M. M. Nederlof, and M. C. M. van Loosdrecht. 2007. Investigation of microbial communities on reverse osmosis membranes used for process water production. Water Sci. Technol. 55:181-190. [DOI] [PubMed] [Google Scholar]
- 7.Chen, C.-L., W.-T. Liu, M.-L. Chong, M.-T. Wong, S. L. Ong, H. Seah, and W. J. Ng. 2004. Community structure of microbial biofilms associated with membrane-based water purification processes as revealed using a polyphasic approach. Appl. Microbiol. Biotechnol. 63:466-473. [DOI] [PubMed] [Google Scholar]
- 8.Coates, J. D., E. J. P. Phillips, D. J. Lonergan, H. Jenter, and D. R. Lovley. 1996. Isolation of Geobacter species from diverse sedimentary environments. Appl. Environ. Microbiol. 62:1531-1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dudley, L. Y., and N. S. J. Christopher. 1999. Practical experiences of biofouling in reverse osmosis systems, p. 101-159. In C. W. Keevil, A. Godfree, D. Holt, and C. Dow (ed.), Biofilms in the aquatic environment. Royal Society of Chemistry, Cambridge, United Kingdom.
- 10.Eguchi, M., M. Ostrowski, F. Fegatella, J. Bowman, D. Nichols, T. Nishino, and R. Cavicchioli. 2001. Sphingomonas alaskensis strain AFO1, an abundant oligotrophic ultramicrobacterium from the North Pacific. Appl. Environ. Microbiol. 67:4945-4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Erb, R. W., C. A. Eichner, I. Wagner-Döbler, and K. N. Timmis. 1997. Bioprotection of microbial communities from toxic phenol mixtures by a genetically designed pseudomonad. Nat. Biotechnol. 15:378-382. [DOI] [PubMed] [Google Scholar]
- 12.Fialho, A. M., L. O. Martins, M.-L. Donval, J. H. Leitão, M. J. Ridout, A. J. Jay, V. J. Morris, and I. Sá-Correia. 1999. Structures and properties of gellan polymers produced by Sphingomonas paucimobilis ATCC 31461 from lactose compared with those produced from glucose and from cheese whey. Appl. Environ. Microbiol. 65:2485-2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flemming, H.-C. 2002. Biofouling in water systems—cases, causes and countermeasures. Appl. Environ. Biotechnol. 59:629-640. [DOI] [PubMed] [Google Scholar]
- 14.Flemming, H.-C., G. Schaule, T. Griebe, J. Schmitt, and A. Tamachkiarowa. 1997. Biofouling—the Achilles heel of membrane processes. Desalination 113:215-225. [Google Scholar]
- 15.Flemming, H.-C., T. Griebe, and G. Schaule. 1996. Antifouling strategies in technical systems—a short review. Water Sci. Technol. 34:517-524. [Google Scholar]
- 16.Fuerst, J. A. 1995. The planctomycetes: emerging models for microbial ecology, evolution, and cell biology. Microbiology 141:1493-1506. [DOI] [PubMed] [Google Scholar]
- 17.Goosen, M. F. A., S. S. Sablani, H. Al-Hinai, S. Al-Obeidani, R. Al-Belushi, and D. Jackson. 2004. Fouling of reverse osmosis and ultrafiltration membranes: a critical review. Sep. Sci. Technol. 39:2261-2297. [Google Scholar]
- 18.Heilig, H. G. H. J., E. G. Zoetendal, E. E. Vaughan, P. Marteau, A. D. L. Akkermans, and W. M. de Vos. 2002. Molecular diversity of Lactobacillus spp. and other lactic acid bacteria in the human intestine as determined by specific amplification of 16S ribosomal DNA. Appl. Environ. Microbiol. 68:114-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ho, L. C. W., D. D. Martin, and W. C. Lindemann. 1983. Inability of microorganisms to degrade cellulose acetate reverse-osmosis membranes. Appl. Environ. Microbiol. 45:418-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hugenholtz, P., B. M. Goebel, and N. R. Pace. 1998. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 180:4765-4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ivnitsky, H., I. Katz, D. Minz, G. Volvovic, E. Shimoni, E. Kesselman, P. Semiat, and C. G. Dosoretz. 2007. Bacterial community composition and structure of biofilms developing on nanofiltration membranes applied to wastewater treatment. Water Res. 41:3924-3935. [DOI] [PubMed] [Google Scholar]
- 22.Johnsen, A. R., M. Hausner, A. Schnell, and S. Wuertz. 2000. Evaluation of fluorescently labeled lectins for noninvasive localization of extracellular polymeric substances in Sphingomonas biofilms. Appl. Environ. Microbiol. 66:3487-3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kalmbach, S., W. Manz, B. Bendinger, and U. Szewzyk. 2000. In situ probing reveals Aquabacterium commune as a widespread and highly abundant bacterial species in drinking water biofilms. Water Res. 34:575-581. [Google Scholar]
- 24.Kelley, S. T., U. Theisen, L. T. Angenent, A. St. Amand, and N. R. Pace. 2004. Molecular analysis of shower curtain biofilm microbes. Appl. Environ. Microbiol. 70:4187-4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koskinen, R., T. Ali-Vehmas, P. Kampfer, M. Laurikkala, I. Tsitko, E. Kostyal, F. Atroshi, and M. Salkinoja-Salonen. 2000. Characterization of Sphingomonas isolates from Finnish and Swedish drinking water distribution systems. J. Appl. Microbiol. 89:687-696. [DOI] [PubMed] [Google Scholar]
- 26.Kuehn, M., M. Mehl, M. Hausner, H. J. Bungartz, and S. Wuertz. 2001. Time-resolved study of biofilm architecture and transport processes using experimental and simulation techniques: the role of EPS. Water Sci. Technol. 43:143-151. [PubMed] [Google Scholar]
- 27.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. J. Wiley & Sons Ltd., Chichester, United Kingdom.
- 28.LeChavellier, M. W., T. M. Babcock, and R. G. Lee. 1987. Examination and characterization of distribution system biofilms. Appl. Environ. Microbiol. 3:2714-2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maidak, B. L., J. R. Cole, T. G. Lilburn, C. T. Parker, Jr., P. R. Saxman, R. J. Farris, G. M. Garrity, G. J. Olsen, T. M. Schmidt, and J. M. Tiedje. 2001. The RDP-II (Ribosomal Database Project). Nucleic Acids Res. 29:173-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murray, A. E., J. T. Hollibaugh, and C. Orrego. 1996. Phylogenetic compositions of bacterioplankton from two California estuaries compared by denaturing gradient gel electrophoresis of 16S rDNA fragments. Appl. Environ. Microbiol. 62:2676-2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muyzer, G., E. C. De Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ollivier, B., P. Caumette, J.-L. Garcia, and R. A. Mah. 1994. Anaerobic bacteria from hypersaline environments. Microbiol. Rev. 58:27-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pang, C. M., and W.-T. Liu. 2007. Community structure analysis of reverse osmosis membrane biofilms and the significance of Rhizobiales bacteria in biofouling. Environ. Sci. Technol. 41:4728-4734. [DOI] [PubMed] [Google Scholar]
- 34.Pang, C. M., P. Hong, H. Guo, and W.-T. Liu. 2005. Biofilm formation characteristics of bacterial isolates retrieved from a reverse osmosis membrane. Environ. Sci. Technol. 39:7541-7550. [DOI] [PubMed] [Google Scholar]
- 35.Pollock, T. J., and R. W. Armentrout. 1999. Planktonic/sessile dimorphism of polysaccharide-encapsulated sphingomonads. J. Ind. Microbiol. Biotechnol. 23:436-441. [DOI] [PubMed] [Google Scholar]
- 36.Pollock, T. J. 1993. Gellan-related polysaccharides and the genus Sphingomonas. J. Gen. Microbiol. 139:1939-1945. [Google Scholar]
- 37.Ridgway, H. F., and H.-C. Flemming. 1996. Membrane biofouling, p. 6.1-6.62. In J. Mallevialle, P. E. Odendaal, and M. R. Wiesner (ed.), Water treatment membrane processes. McGraw-Hill, New York, NY.
- 38.Ridgway, H. F., C. A. Justice, C. Whittaker, D. G. Argo, and B. H. Olson. 1984. Biofilm fouling of RO membranes: its nature and effect on treatment of water for reuse. J. Am. Water Works Assoc. 76:94-102. [Google Scholar]
- 39.Ridgway, H. F., A. Kelly, C. Justice, and B. H. Olson. 1983. Microbial fouling of reverse-osmosis membranes used in advanced wastewater treatment technology: chemical, bacteriological, and ultrastructural analyses. Appl. Environ. Microbiol. 45:1066-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schulte, S., J. Wingender, and H.-C. Flemming. 2005. Efficacy of biocides against biofilms, p. 90-120. In W. Paulus (ed.), Directory of microbicides for the protection of materials and processes. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 41.Schut, F., J. C. Gottschal, and R. A. Prins. 1997. Isolation and characterization of the marine ultramicrobacterium Sphingomonas sp. strain RB2256. FEMS Microbiol. Rev. 20:363-369. [Google Scholar]
- 42.Venugopalan, V. P., M. Kuehn, M. Hausner, D. Springael, P. A. Wilderer, and S. Wuertz. 2005. Architecture of a nascent Sphingomonas sp. biofilm under varied hydrodynamic conditions. Appl. Environ. Microbiol. 71:2677-2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vrouwenvelder, J. S., and D. van der Kooij. 2001. Diagnosis, prediction and prevention of biofouling of NF and RO membranes. Desalination 139:65-71. [Google Scholar]
- 44.Ward, D. M., R. Weller, and M. M. Bateson. 1990. 16S rRNA sequences reveal numerous uncultured microorganisms in a natural community. Nature 345:63-65. [DOI] [PubMed] [Google Scholar]
- 45.Wiesner, M. R., and P. Aptel. 1996. Mass transport and permeate flux and fouling in pressure-driven membrane processes, p. 4.1-4.30. In J. Mallevialle, P. E. Odendaal, and M. R. Wiesner (ed.), Water treatment membrane processes. McGraw-Hill, New York, NY.
- 46.Zhongtang, Y., and M. Morrison. 2004. Comparisons of different hypervariable regions of rrs genes for use in fingerprinting of microbial communities by PCR-denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 70:4800-4806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zwart, G., B. C. Crump, M. P. Kamst-van Agterveld, F. Hagen, and S. Han. 2002. Typical freshwater bacteria: an analysis of available 16S rRNA gene sequences from plankton of lakes and rivers. Aquat. Microb. Ecol. 28:141-155. [Google Scholar]