Abstract
This study assessed the potential effects of transgenic aspen overexpressing a polyphenol oxidase gene on diversity in rhizosphere communities. Cultivation-independent methods were used to better delineate bacterial and fungal populations associated with transgenic and nontransgenic trees. Gene libraries for the bacterial component of the rhizosphere were established using 16S rRNA and chaperonin-60 (CPN-60) gene sequences, while the fungal community was characterized using 18S rRNA gene sequences. The 16S rRNA gene libraries were dominated by alphaproteobacterial sequences, while the CPN-60 gene libraries were dominated by members of the Bacteroidetes/Chlorobi group. In both the CPN-60 and 16S rRNA libraries, there were differences in only minor components of the bacterial community between transgenic and unmodified trees, and no significant differences in species diversity were observed. Compared to the bacterial gene libraries, greater coverage of the underlying population was achieved with the fungal 18S rRNA libraries. Members of the Zygomycota, Chytridiomycota, Ascomycota, and Basidiomycota were recovered from both libraries. The dominant groups of fungi associated with each tree type were very similar, although there were some qualitative differences in the recovery of less-abundant fungi, likely as a result of the underlying heterogeneity of the fungal population. The methods employed revealed only minor differences between the bacterial and fungal communities associated with transgenic and unmodified trees.
The engineering of trees to produce antimicrobial or insecticidal peptides represents one approach to reduce the significant losses caused by disease and insect predation in nurseries and plantations (41). Despite the potential advantages of genetic modification, this technology also demands well-defined risk assessment for potentially undesired ecological side effects on associated communities (15, 63). Since root-associated fungi and bacteria are primary consumers of plant exudates and materials (21), rhizosphere communities are likely to respond to changes in plant activity and metabolism that could well be impacted by genetic modification of the crop tree species.
Potential impact can most accurately be measured by a thorough survey of the resident populations in the immediate ecosystem. Recent advances in cultivation-independent analyses of microbial communities have improved the detection of organisms that cannot be cultured in vitro and have allowed better assessment of highly diverse communities. The most commonly used gene targets for studying broad diversity have been the 16S rRNA gene for bacteria and the 18S rRNA gene for fungi (32, 51).
In order to reduce potential biases associated with using a single genetic target to depict community diversity, we generated profiles based on partial chaperonin-60 (CPN-60) sequences in conjunction with the rRNA markers. The gene encoding the 60-kDa protein subunit of type I chaperonins (23) has been used as an effective marker for microbial identification and phylogenetic studies. The CPN-60 gene is found in most prokaryotes and eukaryotes (23), and degenerate primers have been designed that allow amplification of a 0.5-kb portion of the gene from microorganisms (19), including microorganisms present in environmental samples.
In order to screen for changes in both rare and abundant taxonomic groups, diversity indices were calculated from sequence abundance data. While it is commonly accepted that such estimates do not address the true total richness of soil communities, they do allow comparison between treatments (25, 26).
The transgenic hybrid aspen analyzed in our study overexpressed a hybrid poplar leaf polyphenol oxidase (PPO) (EC 1.10.1.3) and was previously shown to be more resistant to herbivory by forest tent caterpillars (60). Despite numerous examples supporting the hypothesis that PPO has a role in plant defense against fungal and microbial pathogens (35, 40, 50), the mechanisms linking PPO to disease resistance are unclear, which makes it difficult to predict potential nontarget effects of overexpression on the rhizosphere community. PPOs catalyze the oxidation of o-diphenols to o-quinones (37), and most biological effects appear to be due to the highly reactive o-quinone products (48), suggesting that a broad range of organisms may be impacted. The persistence of PPOs in the soil is unknown, although these enzymes are known to be very stable (60, 62). Given the evidence linking PPO to plant defense, we hypothesized that release of transgenic products from sloughed or damaged root cells or an otherwise altered root exudate may influence the structure of rhizosphere communities. Our primary objective was to screen for broad changes in community composition and diversity.
MATERIALS AND METHODS
Plant material and soil sampling.
Transformations of hybrid aspen (Populus tremula × Populus alba clone INRA 717I-B4) were carried out using genetic constructs, as described by Wang and Constabel (60). Briefly, the plasmids used contained a double cauliflower mosaic virus 35S rRNA promoter, an avian myeloblastosis virus RNA4 trans/enhancer sequence, the hybrid poplar (Populus trichocarpa × Populus deltoides) PPO1 coding sequence, and a nopaline synthase (nos) terminator region. The neomycin phosphotransferase II (nptII) gene was used as a selectable marker.
Rooted plantlets overexpressing the hybrid poplar PPO1 gene were planted in 4-in. pots containing Sunshine Mix #4 (Sungro, Seba Beach, AB, Canada). Following acclimation in a mist chamber, transgenic and nontransgenic plants (which had the parental genotype) were transplanted into 11-liter pots containing a thoroughly homogenized soil mixture composed of 2 parts premium sterilized potting soil (Professional Gardener Series, Island's Finest; Cinnibar Valley Farms Ltd., Nanaimo, BC, Canada), 1 part peat moss (Sunshine; Sun Gro Horticulture, Canada Ltd.), 0.5 part premium grade vermiculite (Ultra Tech, Richmond, CA), and 0.5 part perlite (Dutch Treat Products, Surrey BC, Canada). Plants were maintained at 18°C in the greenhouse at the Centre for Forest Biology at the University of Victoria under ambient light conditions. Plants were watered uniformly each day, and necrotic leaves were pruned so that they did not contact the soil. After 5 months of growth, root tissue and rhizosphere soil were collected from two individual plants of transgenic line 19 (60) and two individual control plants of line 717. Each tree was given an identifying number and defined as a replicate. The two transgenic trees were designated 19-4 (replicate A) and 19-9 (replicate B), and the two control trees were designated 717-10 (replicate A) and 717-11 (replicate B). Soil collected from each tree was used to create a replicate gene library.
The rhizosphere soil was defined as the material collected by soaking roots after the bulk soil had been rigorously shaken from them. For each tree, 50 g of full-length roots with adhering soil was soaked in 0.1% sodium pyrophosphate diluent (200 ml) for 20 min with occasional agitation. The soil was then separated from the diluent by centrifugation (2,900 rpm, 10 min) and was lyophilized prior to storage at −80°C and DNA extraction.
For each tree, a 250- to 300-mg root tissue sample was collected for the PPO assays. After the adhering bulk soil was removed, the root tissue, including attached lateral roots, was soaked in sterile water to remove any residual soil, blotted dry, and aseptically collected. All tissue and soil samples were stored at −80°C.
Assays for PPO activity.
Frozen root tissue (300 mg) was ground with sand and polyvinylpolypyrrolidone in extraction buffer (100 mM NaPO4 [pH 7.0], 0.1% [vol/vol] Triton X-100) and clarified by centrifugation. The supernatant was immediately assayed for PPO activity by measuring the conversion of dl-dihydroxyphenylalanine (dl-DOPA) to dopaquinone as previously described by Constabel et al. (13). The protein concentration was determined with the Bradford reagent using bovine serum albumin as a standard (6). As a control, the reaction was inhibited by tropolone, confirming that DOPA oxidation was due to a PPO rather than to other oxidative enzymes.
Statistical analysis of PPO activity.
For each tree, the PPO activity per milligram of protein was reported as the mean value determined for five root samples. A one-way analysis of variance was performed with log-transformed data, followed by a Student-Newman-Keuls post hoc test for differences between the mean PPO activities for each tree (57). Differences between the activities for each tree were considered to be statistically significant if the P value was ≤0.05. Activity values were then back-transformed and were reported as geometric means with 95% confidence intervals.
Extraction of bacterial and fungal DNA from soil and PCR amplification.
Total genomic DNA was extracted from 150 mg of a lyophilized soil subsample. Soil was mixed with 750 μl of 0.1 M sodium phosphate buffer (pH 8) and 2 g of 0.1-mm zirconia/silica beads (Fisher Scientific). To aid in cell lysis, 370 μl of 100 mM NaCl-500 mM Tris-HCl (pH 8)-10% sodium dodecyl sulfate was added, and the tubes were shaken at high speed for 5 min with a mini bead beater. After centrifugation for 3 min (11,200 rpm), the supernatant was chilled for 10 min on ice with 0.4 volume of 7.5 M ammonium acetate. Cellular debris was pelleted by centrifugation for 3 min (11,200 rpm), and the crude DNA extract was filtered through microspin columns packed with polyvinylpolypyrrolidone to reduce humic acid contamination (5). The concentration was determined spectrophotometrically, and the purified DNA was stored in water at −20°C prior to PCR amplification.
PCR amplification of community DNA. (i) Bacterial 16S rRNA gene amplification.
A 433-bp portion of the bacterial 16S rRNA gene was PCR amplified with primers 968f (AACGCGAAGAACCTTAC) and 1401r (CGGTGTGTACAAGACCC), which encompassed variable regions V6 to V8 of the eubacterial 16S rRNA gene and corresponded to positions 968 to 1401 in Escherichia coli (43). The reaction mixtures included a negative control mixture containing no template DNA. Amplification was carried out using 50 pmol of each primer, 1 U of Taq polymerase (Invitrogen), and 1 ng of template DNA in a Perkin Elmer thermal cycler with the following parameters: (i) an initial denaturation step consisting of 3 min at 94°C, (ii) 28 cycles consisting of denaturation for 1 min at 94°C, annealing for 1 min at 61°C, and extension for 1 min at 72°C, and then (iii) a final extension step consisting of 4 min at 72°C.
(ii) CPN-60 gene amplification.
For amplification of the “universal target” within the CPN-60 gene, primers H279 (GAIIIIGCIGGIGA[T/C]GGIACIACIAC) and H280 ([T/C][T/G]I[T/C][T/G]ITCICC[AG]AAICCIGGIGC[T/C]TT) were used (modified from the primers used by Goh et al. [19]). Inosine was used to reduce the degeneracy of the sequences (46). These primers were designed to amplify the region between codons 92 and 277 based on the E. coli CPN-60 sequence (accession number X07850). The components of the reaction mixtures were the same as those described above. The PCR program consisted of an initial denaturation step of 2 min at 94°C, followed by 35 cycles of 30 s at 94°C, 30 s at 42°C, and 30 s at 72°C and then a final 2-min extension at 72°C.
(iii) Fungal 18S rRNA gene amplification.
Primers EF4 (GGAAGGG[G/A]TGTATTTATTAG) and Fung5 (GTAAAAGTCCTGGTTCCCC) were used to amplify a 550-bp fragment within the fungal 18S rRNA coding region (56). The components of the reaction mixtures were the same as those described above for 16S rRNA amplification, except that 10 ng template DNA was used. The PCR program consisted of an initial denaturation step of 2 min at 94°C, followed by 35 cycles of 1 min at 94°C, 1 min at 56°C, and 2 min at 72°C and then a final 10-min extension at 72°C.
Construction of gene libraries and sequencing.
A gene library was created for each of two individual transgenic plants and two individual control plants. The resulting libraries were designated using the line, plant identifying number, and primers used for PCR amplification. Following electrophoreses, PCR products were extracted from agarose gel slices using a QIAquick gel extraction kit (Qiagen, Mississauga, Ontario, Canada), ligated into pGEM-T cloning vectors (Promega, Nepean, Ontario, Canada), and transformed into electrocompetent E. coli DH5-α (Invitrogen). The DNA sequence was determined for 192 clones for each of the three primer sets (16S rRNA, 18S rRNA, and CPN-60) by the Centre for Biomedical Research at the University of Victoria. Sequencing reactions were conducted by using the 96-well microtiter plate format with 200 to 300 ng of template in a 5-μl BigDye Terminator sequencing reaction mixture (ABI). Completed-reaction mixtures were sequenced in a single direction by using an ABI 3730 DNA analyzer. Raw sequence data were processed using Phred software (16), which assigns quality values to the bases and trims poor-quality regions.
Sequence analysis and assignment of clones to phylogenetic groups.
Sequences were oriented using Orientation Checker, a program for checking and altering the orientation of 16S rRNA sequences (http://www.cf.ac.uk/biosi/research/biosoft/Squirrel/index.html), and they were aligned using Mega 3.1 software (http://megasoftware.net/) (34). The sequences were then screened for chimeric properties using Mallard software (http://www.cf.ac.uk/biosi/research/biosoft/Mallard/index.html) (3). Sequences for the 16S rRNA libraries were grouped into “operational taxonomic units” (OTUs) sharing 97% sequence similarity using the furthest-neighbor clustering algorithm in the DOTUR computer program (53). Sequences for the 18S rRNA libraries were grouped into OTUs sharing 99% sequence similarity. Input files for DOTUR were constructed by aligning sequences in ClustalW using default parameters and converting the data to a distance matrix using a Jukes-Cantor model in the DNADist program of the Phylip analysis package (17).
Once the 16S rRNA sequences were grouped into OTUs, the Library Compare tool at the Ribosomal Database Project website (12) (http://rdp.cme.msu.edu/) was used to assign library sequences with sequences of bacterial taxa and to compare the taxonomic compositions of replicate libraries. Replicate libraries from the same tree genotype were then pooled, and the comparison was repeated. The Library Compare tool used the Ribosomal Database Project naïve Bayesian classifier to rapidly classify library sequences in bacterial taxa. Each library sequence was assigned to a set of hierarchical taxa (as proposed in Bergey's Manual of Systematic Bacteriology [7]), and a bootstrap confidence estimate for each rank assignment was obtained.
Unique CPN-60 nucleotide sequences were compared to approximately 8,000 sequences in the CPN-60 database using the FASTA search algorithm (47) and were assigned to major phylogenetic groups based on the most homologous reference sequences (24).
Once fungal clones were grouped into OTUs, a BLAST-n search (2) was carried out for one representative sequence from each group.
Statistical analysis.
The taxonomic compositions of libraries were compared by performing a χ2 test of homogeneity for major phylogenetic groups. Comparisons between control and transgenic libraries were made using pooled data sets. The variability between replicate libraries derived from individual trees was also assessed for 16S rRNA and 18S rRNA libraries. For χ2 analysis, 2 × 2 contingency tables were constructed for the phylogenetic group being compared using frequency data, and Yates' correction for continuity was applied. In cases where the frequency of recovery was too low to allow meaningful χ2 analysis (if any observed or expected frequency in the contingency table was <5), Fisher's exact test (two sided) was used to test for statistical significance (57). P values of ≤0.05 were considered statistically significant and were reported. Calculations for χ2 and Fisher's exact test were performed online using S.I.S.A. (Simple Interactive Statistical Analysis) at http://home.clara.net/sisa/.
Estimation of species richness and evenness.
Once bacterial (16S rRNA and CPN-60) and fungal (18S rRNA) sequences were assigned to OTUs, the frequency data were used to calculate a number of diversity indices to describe and compare the richness (defined as the number of OTUs present) and level of evenness (the relative abundance of OTUs) within each pooled data set. The indices and 95% confidence intervals were calculated using DOTUR, which performed a random sampling without replacement procedure (53). The richness indicators used were (i) the total number of OTUs recovered from each library, (ii) the Shannon diversity index (36), (iii) the Chao richness estimator (9), and (iv) the abundance-based coverage estimator (10).
The indices used to describe the degree of evenness or dominance were (i) the Shannon evenness index, (ii) Simpson's index of diversity, and (iii) the Berger-Parker index (36).
Nucleotide sequence accession numbers.
The nucleotide sequences determined in this study have been deposited in the National Center for Biotechnology Information database under accession numbers EU695226 to EU696735.
RESULTS
PPO activity in root tissue.
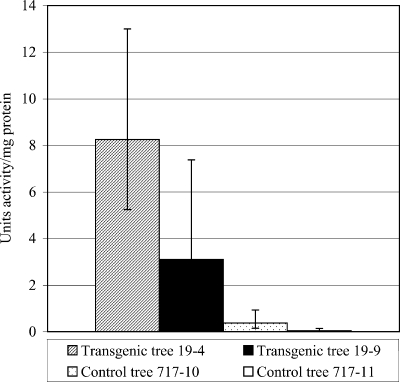
To confirm that the transgenic hybrid aspen did indeed express PPO in its root system, the conversion of DOPA to PPO by transgenic plants was compared to the conversion of DOPA to PPO by control plants. Elevated levels of PPO activity were observed in the root tissue of transgenic PPO-overexpressing trees relative to the root tissue of control trees (Fig. 1). Although the activity levels of root samples from the transgenic plants were highly variable, the average PPO activity in each of the PPO-overexpressing transgenic trees was at least 10-fold greater than that in control trees. Analysis of variance suggested that there were significantly different levels of PPO activity in all of the individual trees tested (P < 0.05).
FIG. 1.
PPO activity in transgenic PPO-overexpressing and nontransgenic hybrid aspen root tissue. Geometric means and 95% confidence intervals based on five root samples per tree are shown. Analysis of variance of log-transformed data indicated that the PPO activity in each tree was significantly different (P < 0.05).
Taxonomic composition of bacterial communities associated with transgenic and wild-type plants.
In both the 16S rRNA and CPN-60 libraries, duplicate OTUs were recovered infrequently, reflecting the highly diverse community. For the 16S rRNA libraries, 98 OTUs were recovered only from control rhizosphere libraries, 58 OTUs were recovered from both the transgenic and control libraries, and 108 OTUs were recovered only from the transgenic libraries. In many cases, each OTU contained only one sequence and comprised less than 1% of the clone library.
Analysis of 430 CPN-60 sequences revealed the presence of 337 unique nucleotide sequences, which were 62 to 90% identical to reference nucleotide sequences in the CPN-60 database. Interestingly, almost all library sequences were most closely associated with bacterial reference sequences; the only exception was one sequence whose nearest database neighbor had a fungal origin. This sequence was recovered from the control library and shared a low level of identity (67%) with Candida albicans.
In contrast to the 16S rRNA libraries, which were dominated by proteobacterial sequences (Table 1), most CPN-60 clone sequences were most closely associated with the Bacteroidetes/Chlorobi group, and such sequences accounted for 79 and 83% of the transgenic and control rhizosphere libraries, respectively (Table 2).
TABLE 1.
Relative abundance of 16S rRNA clones for different bacterial taxa from PPO-overexpressing and nontransgenic hybrid aspen rhizospheresa
| Phylogenetic group | Relative bacterial clone abundance (%)
|
|||||
|---|---|---|---|---|---|---|
| rRNA gene library from transgenic rhizosphere
|
rRNA gene library from control rhizosphere
|
|||||
| Replicate A | Replicate B | Pooled data | Replicate A | Replicate B | Pooled data | |
| Genera incertae sedis BRC1 | 0 | 1 | <1 | 0 | 0 | 0 |
| Thermomicrobia | 1 | 0 | <1 | 0 | 0 | 0 |
| Genera incertae sedis TM7 | 2 | 0 | 1 | 1 | 0 | <1 |
| Spirochaetes | 0 | 0 | 0 | 1 | 0 | <1 |
| Chlamydiae | 0 | 0 | 0 | 0 | 2 | 1 |
| Cyanobacteria | 0 | 2 | 1 | 1 | 2 | 1 |
| Acidobacteria | 2 | 2 | 2 | 2 | 1 | 2 |
| Verrucomicrobia | 5 | 2 | 3 | 2 | 2 | 2 |
| Firmicutes | 3 | 2 | 3 | 2 | 2 | 2 |
| Chloroflexi | 2 | 1 | 1 | 1 | 5 | 3 |
| Gemmatimonadetes | 4 | 8 | 6 | 3 | 3 | 3 |
| Actinobacteria | 9 | 8 | 9 | 12b | 2 | 7 |
| Bacteroidetes | 0 | 3 | 2 | 8 | 6 | 7c |
| Flavobacteria | 0 | 2 | 1 | 6 | 5 | 6c |
| Sphingobacteria | 0 | 1 | <1 | 2 | 1 | 2 |
| Proteobacteria | 73 | 71 | 72 | 68 | 76 | 72 |
| Alphaproteobacteria | 35 | 38 | 36 | 34 | 40 | 37 |
| Betaproteobacteria | 11 | 22 | 16 | 13 | 17 | 15 |
| Deltaproteobacteria | 12 | 4 | 8 | 6 | 5 | 6 |
| Gammaproteobacteria | 15 | 8 | 12 | 15 | 14 | 14 |
Clone sequences (130 sequences/replicate library) were classified using the Ribosomal Database naïve Bayesian classifier.
Replicates within the library type are statistically significantly different.
There is a statistically significant difference (P ≤ 0.05) in clone abundance, as determined using pooled data sets. P values were based on a χ2 test with Yates' correction or Fisher's exact test.
TABLE 2.
Relative abundance of clones recovered from CPN-60 libraries for different taxa from soil samples obtained from the rhizospheres of transgenic and nontransgenic hybrid aspena
| Phylogenetic group | Relative CPN-60 clone abundance (%)
|
|
|---|---|---|
| Transgenic plants | Control plants | |
| Fungi | 0 | <1 |
| Deinococcus-Thermus | <1 | 0 |
| Acidobacteria | <1 | 0 |
| Chloroflexi | 1 | 0 |
| Planctomycetes | 1 | 0 |
| Fusobacteria | 1 | 3 |
| Chlamydiae | 2 | 4 |
| Firmicutes | 5 | 3 |
| Proteobacteria | 11 | 6 |
| Alphaproteobacteria | 2 | 2 |
| Betaproteobacteria | <1 | <1 |
| Deltaproteobacteria | 3 | 3 |
| Gammaproteobacteria | 5 | 1b |
| Bacteriodetes/Chlorobi | 79 | 83 |
| Bacteroides | 1 | <1 |
| Sphingobacteria | 14 | 20 |
| Flavobacteria | 62 | 57 |
| Chlorobi | 2 | 5 |
Taxonomic assignment of clone sequences (215 sequences/library) was based on the nearest neighbors obtained using a FASTA search of DNA sequences catalogued in the CPN-60 database.
There was a statistically significant difference (P ≤ 0.05) in clone abundance, as determined using pooled data sets. P values were based on a χ2 test with Yates' correction or Fisher's exact test.
Taxonomic composition of fungal 18S rRNA libraries.
Phylogenetic analysis indicated that DNA from a diverse array of fungi was amplified from the rhizospheres. Basidiomycete, ascomycete, zygomycete, and chytridiomycete sequences were recovered from both transgenic and control libraries (Table 3). The majority of clones derived from soil obtained from both transgenic (68%) and control (61%) libraries grouped most closely with the Ascomycota.
TABLE 3.
Relative abundance of 18S rRNA clones for different fungal taxa from duplicate soil samples obtained from the rhizospheres of PPO-overexpressing and control hybrid aspena
| Phylogenetic group | Relative fungal clone abundance (%)
|
|||||
|---|---|---|---|---|---|---|
| rRNA gene library from transgenic seedlings
|
rRNA gene library from control seedlings
|
|||||
| Replicate A | Replicate B | Pooled data | Replicate A | Replicate B | Pooled data | |
| Unidentified fungi | 13b | 0 | 6c | 2 | 1 | 2 |
| Zygomycota | 4 | 1 | 3 | 6 | 4 | 5 |
| Chytridiomycota | 8 | 6 | 7 | 23b | 8 | 15c |
| Spizellomycetes | 4 | 0 | 2 | 23b | 6 | 14c |
| Chytridiomycetes | 4 | 6 | 5c | 0 | 2 | 1 |
| Basidiomycota | 17 | 16 | 16 | 12 | 23 | 18 |
| Hymenomycetes | 17 | 16 | 16 | 12 | 23b | 18 |
| Ascomycota | 58 | 77b | 68 | 57 | 64 | 61 |
| Chaetothyriomycetes | 0 | 5b | 3 | 0 | 0 | 0 |
| Incertae sedis | 0 | 1 | <1 | 0 | 1 | <1 |
| Dothiomycetes | 0 | 0 | 0 | 0 | 1 | <1 |
| Pezizomycetes | 1 | 1 | 1 | 0 | 1 | <1 |
| Orbiliomycetes | 10 | 4 | 7c | 1 | 0 | <1 |
| Leotiomycetes | 2 | 3 | 3 | 4 | 1 | 3 |
| Mitosporic Ascomycota | 8 | 10 | 9 | 7 | 4 | 5 |
| Eurotiomycetes | 2 | 13b | 8 | 4 | 13 | 9 |
| Saccharomycetes | 5 | 3 | 4 | 14 | 12 | 13c |
| Sordariomycetes | 30 | 38 | 34 | 28 | 32 | 30 |
Taxonomic assignment was based on the most homologous sequences in the GenBank database obtained with a BLAST-n search (280 18S rRNA clones/pooled library).
Replicates within the library type are statistically significantly different.
There is a statistically significant difference (P ≤ 0.05) in clone abundance, as determined using pooled data sets. P values were based on a χ2 test with Yates' correction or Fisher's exact test.
Within the Ascomycota, clones were assigned to 11 groups, the most abundant of which was the Sordariomycetes, for both the transgenic and control soils. Within the Ascomycetes, significantly more saccharomycete sequences (13%) were recovered from the pooled control library than from the pooled transgenic library (4%). In contrast, significantly more clones assigned to the Orbiliomycetes were recovered from the pooled transgenic library. For these two groups, no significant differences were observed in comparisons of replicate libraries from the same tree type.
Bacterial diversity in transgenic and control soils.
Calculated diversity parameters revealed that the 16S rRNA and CPN-60 libraries were characterized by high levels of species richness, defined as the number of different OTUs in a sample (Table 4). In general, slightly greater richness was found in the transgenic libraries than in the control libraries, as reflected by the richness indices; however, it was not possible to state with confidence that the two soils differed significantly, since the values for each richness index calculated to describe the transgenic library fell within the 95% confidence interval for the same estimate for the control library. The level of dominance in the bacterial libraries was low, and the levels of evenness were high (Table 4), as reflected by the Shannon evenness and Simpson's indices. The control libraries were characterized by a slightly higher level of dominance than the libraries derived from the rhizospheres of the transgenic trees. This was also reflected by the Berger-Parker index, which showed that the most dominant OTU was recovered infrequently from each library but was slightly more abundant in the control library.
TABLE 4.
Diversity indices for fungal and bacterial OTUs associated with PPO-overexpressing and nontransgenic hybrid aspen rhizospheresa
| Diversity parameter | Diversity index or richness estimator | Marker | Rhizosphere soil type
|
|
|---|---|---|---|---|
| Transgenic | Control | |||
| No. of OTUs | 16S rRNA | 163 | 151 | |
| CPN-60 | 84 | 88 | ||
| 18S rRNA | 44 | 35 | ||
| Species richness | Shannon diversity index | 16S rRNA | 4.87 (4.75-4.99) | 4.73 (4.61-4.85) |
| CPN-60 | 3.93 (3.78-4.09) | 3.89 (3.72-4.07) | ||
| 18S rRNA | 3.06 (2.92-3.20) | 2.65 (2.51-2.79) | ||
| Chao richness estimator | 16S rRNA | 451 (331-654) | 369 (277-529) | |
| CPN-60 | 162 (121-250) | 183 (134-286) | ||
| 18S rRNA | 101 (61.0-235) | 75.0 (46.3-176) | ||
| Abundance-based coverage | 16S rRNA | 502 (377-700) | 447 (336-626) | |
| richness estimator | CPN-60 | 182 (137-260) | 180 (137-266) | |
| 18S rRNA | 67.2 (52.8-105) | 56.0 (42.2-96.2) | ||
| Evenness or dominance | Shannon evenness index | 16S rRNA | 0.956 | 0.943 |
| CPN-60 | 0.887 | 0.869 | ||
| 18S rRNA | 0.808 | 0.745 | ||
| Simpson's index of diversity | 16S rRNA | 0.007 | 0.009 | |
| CPN-60 | 0.029 | 0.037 | ||
| 18S rRNA | 0.078 | 0.107 | ||
| Berger-Parker index | 16S rRNA | 0.031 | 0.046 | |
| CPN-60 | 0.112 | 0.130 | ||
| 18S rRNA | 0.225 | 0.225 | ||
Replicate libraries were pooled for a total of 260 16S rRNA clones, 215 CPN-60 clones, and 280 18S rRNA clones. The following OTU definitions were used: for 16S rRNA sequences, 97% sequence similarity; for CPN-60 sequences, 80% similarity; and for 18S rRNA sequences, 99% similarity. Where appropriate, 95% confidence intervals are indicated in parentheses.
Diversity indices for fungal 18S rRNA libraries.
Slightly greater richness of fungal OTUs was observed for the transgenic rhizosphere samples than for the control samples. The Shannon index also suggested that greater richness was associated with the transgenic library, and the 95% confidence intervals for each library did not overlap. The Chao and abundance-based coverage richness estimators also suggested that the transgenic library harbored greater richness, but the confidence intervals for the estimates overlapped with those of the control library.
The Shannon evenness index and Simpson's index of dominance suggested that a higher level of dominance of some fungal species was associated with the rhizospheres of control trees than with the rhizospheres of transgenic trees.
The Berger-Parker index, which indicated the proportional abundance of the most dominant species, was the same for both the transgenic and control libraries (0.225). The most abundant OTU recovered from both libraries was most closely related to Chaetomium globosum, a sordariomycete.
Collector's curves.
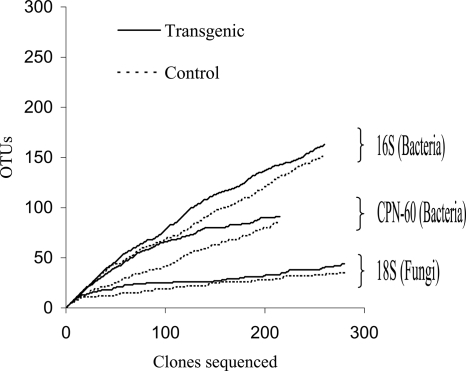
Collector's curves were constructed to evaluate sampling progress and to compare the relative diversities in the control and transgenic libraries. Once the diversity within a community has been sampled to its full extent, a collector's curve would be expected to plateau. The collector's curves for the bacterial libraries in this study were nearly linear, indicating that only part of the underlying sequence diversity had been sampled and that unique sequences would continue to be discovered if more clones were sequenced (Fig. 2).
FIG. 2.
Collector's curves for bacterial and fungal gene libraries derived from soil samples pooled from the rhizospheres of transgenic PPO-overexpressing and control aspen trees. The following definitions were used to group sequences into OTUs: for 16S rRNA, 97% similarity; for CPN-60, 80% similarity; and for 18S rRNA, 99% similarity.
In contrast to the collector's curves obtained for the bacterial libraries, the collector's curves for the fungal libraries approached a plateau, indicating that the sequence diversity of the actual fungal population was more thoroughly covered by the clone libraries. Since a high level of redundancy was present in the library, sequencing more 18S rRNA clones would not provide substantial information.
DISCUSSION
Numerous attempts have been made to monitor the impact of transgenic crop species, but the present study is one of the few studies aimed at assessing the effects of transgenic trees on the soil microbial community. The data set generated provided insight into the natural variability in the composition of the bacterial and fungal communities associated with the rhizospheres of transgenic and nontransgenic hybrid aspen.
It was anticipated that transgenic trees could significantly affect the rhizosphere community if the product of the transgene, in this case PPO, could come into contact with the soil community. Although the cauliflower mosaic virus promoter used in the construction of the transgenic plants generally drives constitutive transgene expression in most tissue types (4), confirmation of elevated PPO activity in the root tissue was of particular interest, since this is the portion of the plant most likely to influence the rhizosphere community. The trees used in our experiments had previously been well characterized, and Wang and Constabel (60) reported a 50-fold increase in the PPO activity in the leaves. Although our measurements were variable, we found that there was a significant increase in PPO activity in the root tissue compared to the activity in the nontransgenic plants. This confirmed the potential for differential release of PPO or reactive o-quinones into the soil matrix as PPO is localized within plastids (37) and thus may be released from the roots into the rhizosphere following the discharge of damaged or dead root cells. To overcome the effects of variable PPO activity in the roots or in different parts of the roots, soil was pooled and was homogenously mixed prior to analysis of the associated communities.
The composition of rhizosphere communities has been shown to vary with several factors, including tree species (49), plant health (18), and environmental conditions (20). In some cases, transgenic plants have also been shown to alter community composition (45, 58). In a previous study aimed at determining the impact of flooding on community composition, approximately 26% of the clones recovered from the rhizospheres of greenhouse-grown hybrid aspen trees were representatives of the Alphaproteobacteria, and clones affiliated with the Bacillales were the most prominent (20). Bacillales have also been found to dominate the rhizospheres of chrysanthemum (14) and barley (42), whereas the greatest abundance of alphaproteobacterial clones was recovered from the rhizospheres of grass (38) and lodgepole pine (11). In the present study, the 16S rRNA sequence libraries derived from both transgenic and unmodified trees were also largely affiliated with the Alphaproteobacteria, which accounted for 33 to 38% of each library; however, the Bacillales accounted for less than 2% of the libraries.
The phylogenetic representation in the CPN-60 libraries was significantly different than that in the 16S rRNA libraries, and this demonstrated the bias potentially resulting from use of a single gene locus to profile microbial diversity. In contrast to the 16S rRNA libraries, which were dominated by proteobacterial sequences, the majority of clones in the CPN-60 libraries were most closely associated with the Bacteroidetes/Chlorobi group. It should be noted that a higher level of uncertainty surrounded the taxonomic assignment of CPN-60 sequences. Each taxonomic assignment was an estimate based on the similarity of the unknown sequences to reference sequences with known identities, and the number of known 16S rRNA sequences (489,840) currently is far greater than the number of catalogued CPN-60 sequences (approximately 8,000). As the CPN-60 database expands in the future, it should be possible to estimate the identity of environmental sequences with increased precision, and this should permit identification at a higher level of resolution.
Together, the CPN-60 and 16S rRNA methods allowed a very good comparison of the rhizosphere communities associated with the transgenic and unmodified trees. Although some major bacterial groups were differentially recovered from transgenic and control soils, these taxa represented minor components of the libraries. Based on 16S rRNA sequence analysis, a decrease in Flavobacteria was observed in the transgenic library. Within the rhizosphere, Flavobacteria are likely involved in the breakdown of proteins and carbohydrates (54), and they also appear to actively decompose pesticides and insecticides (55). Flavobacteria have been recovered from diverse habitats, including freshwater and sediments (54), and their abundance in 16S rRNA libraries derived from soil is generally low (28).
In the CPN-60 libraries, more sequences assigned to the Gammaproteobacteria were recovered from transgenic rhizosphere soils than from control rhizosphere soils. The Gammaproteobacteria is the largest proteobacterial group and encompasses a broad range of physiological diversity (28), and the functional consequences of this difference are therefore unclear. The overall lack of variation in the frequency of recovery of major bacterial groups derived from transgenic and control soils, coupled with the lack of a difference in diversity, likely indicates that the impact of the transgenic plants on community composition was so low that it was undetectable by the approaches used.
In some cases, minor changes in soil bacterial diversity have been detected in association with transgenic plants producing antibacterial compounds (22, 52), but there is no clear evidence linking these changes with the antibiotic properties of the transgenic products. In common with other investigations aimed at comparing the richness of microbial species in soil and sediments (18, 38), we recovered numerous species at a low frequency.
It has not yet been possible to generate a well-defined catalogue of the dominant fungal taxa commonly associated with the rhizospheres of plants, because relatively few culture-independent studies have been performed. In the present study, sequences assigned to the four major fungal groups, Zygomycota, Chytridiomycota, Basidiomycota, and Ascomycota, were represented in the 18S rRNA libraries. Members of the Ascomycota were clearly dominant and accounted for over 60% of the sequences recovered from both the transgenic PPO-overexpressing and control rhizosphere soils. Ascomycetes include a wide range of root-associated saprobic taxa (30), which can often assimilate cellulose (31), so the recovery of this group from rhizosphere soils was not surprising. Analysis of the rhizosphere of wheat using multiple primer sets also revealed the presence of all four major fungal groups (56), while basidiomycete sequences were found to dominate grassland soils (27, 30). It should be noted that the 18S rRNA coding region is a commonly used phylogenetic target for community studies, but it shares high homology with regions of other eukaryotes, including plants and soil invertebrates, and coamplification of DNA from these organisms often occurs. All of the 18S rRNA sequences recovered in the present study were fungal in origin.
At the phylum level, some differences between replicate libraries derived from trees of the same type were observed. Most of the observed variation between replicate libraries likely reflected the discontinuous distribution of fungal populations rather than the influence of genetic modification of the trees. In a similar vein, fungal taxa were shown to occur sporadically on the roots of nontransgenic and transgenic potato plants expressing the antimicrobial peptide magainin and were not associated with any particular plant line (44). Moreover, qualitative differences in fungal community composition were also observed between transgenic potato plants with an altered starch composition and unmodified plants, but these differences were minor compared to differences observed between nontransgenic potato cultivars (39). Within the Ascomycota, data from our replicate libraries revealed a difference in the recovery of groups from transgenic soils and from control soils and supported the observation that transgenic modification may have resulted in minor qualitative changes in community composition. Significantly more saccharomycete sequences were recovered from the control soils in the present study, along with more nematode-trapping fungi belonging to the Orbiliomycetes. There was no expectation that the transgenic plants would influence the distribution of the Orbiliomycetes, but it is possible that unexpected changes in the abundance of these fungi may indirectly affect plant health since this fungal group may influence the density of plant-parasitic nematodes in the surrounding soil (29).
No important differences in fungal species richness between transgenic and control libraries were detected. The results presented here agree with the results of a study using denaturing gradient gel electrophoresis analysis that did not detect an influence of transgenic potatoes with an altered starch composition on fungal diversity (39). Most abundant OTUs were present in both the transgenic and control libraries, suggesting that the dominant populations within the fungal communities associated with each tree type were highly similar. Interestingly, the abundance of the most dominant OTU was the same in both the transgenic and control libraries. Sequences in this group were most closely related to C. globosum, a cellulolytic sordariomycete that has been used as a biocontrol fungus to reduce a range of leaf and soilborne diseases caused by fungal pathogens (1, 59).
The collector's curves for both the transgenic and control data sets suggested that the libraries thoroughly represented the underlying sequence diversity of the fungal population. Since our goal was to perform a preliminary assessment of the entire community, we targeted the relatively conserved 18S rRNA gene sequence rather than hypervariable gene sequences, such as rRNA internal transcribed spacer regions (61). The main strength of this approach was that it allowed comparison of dominant populations belonging to a broad range of taxa.
Conclusions.
Since the potential effects of overproduction of PPO were unpredictable, we screened for broad changes in the rhizosphere community. Change in diversity is a common measure of impact (8) and a first step that may be used to identify indicator species or narrower groups of organisms for continued monitoring (8, 33). In summary, our methods detected relatively minor differences in the abundance of fungal and bacterial taxa associated with PPO-overexpressing and unmodified trees. While further investigation is required to determine the biological consequences of minor changes in diversity, our data provide a good starting point for further assessment of the impact of PPO overexpression on the rhizosphere.
Acknowledgments
This research was supported by an NSERC Discovery grant to W. Hintz and by funds from the Canadian Forest Service Canadian Biotech Strategic Fund to R. Hamelin, A. Seguin, R. Brousseau, W. Hintz, and P. Hebert.
We thank P. Constabel for providing the trees used in this study and B. Koop, Center for Biomedical Research at the University of Victoria, for providing sequencing services.
Footnotes
Published ahead of print on 13 June 2008.
REFERENCES
- 1.Aggarwall, R., A. K. Tewari, K. D. Srivastava, and D. V. Singh. 2004. Role of antibiosis in the biological control of spot blotch (Cochliobolus sativus) of wheat by Chaetomium globosum. Mycopathologia 157:369-377. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Ashelford, K. E., N. Chuzhanova, J. C. Fry, A. J. Jones, and A. J. Weightman. 2006. New screening software shows that most recent large 16S rRNA gene clone libraries contain chimeras. Appl. Environ. Microbiol. 72:5734-5741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benfey, P. N., and N. H. Chua. 1990. The cauliflower mosaic virus 35S promoter: combinatorial regulation of transcription in plants. Science 250:959-966. [DOI] [PubMed] [Google Scholar]
- 5.Berthelet, M., L. G. Whyte, and C. W. Greer. 1996. Rapid, direct extraction of DNA from soils for PCR analysis using polyvinylpolypyrrolidone spin columns. FEMS Microbiol. Lett. 138:17-22. [DOI] [PubMed] [Google Scholar]
- 6.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Chem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 7.Brenner, D. J., N. R. Krieg, J. T. Staley, and G. M. Garrity (ed.). 2004. Bergey's manual of systematic bacteriology, vol. 2. Springer, New York, NY.
- 8.Cartwright, C. D., J. Kirton, and A. K. Lilley. 2004. Mechanisms for investigating changes in soil ecology due to GMO releases. Defra report EPG 1/5/214. Department for Environment, Food, and Rural Affairs, London, United Kingdom.
- 9.Chao, A. 1984. Non-parametric estimation of the number of classes in a population. Scan. J. Stat. 11:265-270. [Google Scholar]
- 10.Chao, A., and S. M. Lee. 1992. Estimating the number of classes via sample coverage. J. Am. Stat. Assoc. 87:210-217. [Google Scholar]
- 11.Chow, M. L., C. C. Radomski, J. M. McDermott, J. Davies, and P. E. Axelrood. 2002. Molecular characterization of bacterial diversity in lodgepole pine (Pinus contorta) rhizosphere soils from British Columbia forest soils differing in disturbance and geographic source. FEMS Microbiol. Ecol. 42:347-357. [DOI] [PubMed] [Google Scholar]
- 12.Cole, J. R., B. Chai, R. J. Farris, Q. Wang, S. A. Kulam, D. M. McGarrell, G. M. Garrity, and J. M. Tiedje. 2005. The Ribosomal Database Project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res. 33(Database Issue):D294-D296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Constabel, C. P., L. Yip, J. J. Patton, and M. E. Christopher. 2000. Polyphenol oxidase from hybrid poplar. Cloning and expression in response to wounding and herbivory. Plant Physiol. 124:285-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duineveld, B. M., G. A. Kowalchuk, A. Keijzer, J. D. van Elsas, and J. A. van Veen. 2001. Analysis of bacterial communities in the rhizosphere of chrysanthemum via denaturing gradient gel electrophoresis of PCR-amplified 16S rRNA as well as DNA fragments coding for 16S rRNA. Appl. Environ. Microbiol. 67:172-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dunfield, K. E., and J. J. Germida. 2004. Impact of genetically modified crops on soil and plant-associated microbial communities. J. Environ. Qual. 33:806-815. [DOI] [PubMed] [Google Scholar]
- 16.Ewing, B., L. Hillier, M. C. Wendl, and P. Green. 1998. Base-calling of automated sequencer traces using Phred. I. Accuracy assessment. Genome Res. 8:175-185. [DOI] [PubMed] [Google Scholar]
- 17.Felsenstein, J. 1989. PHYLIP—phylogeny inference package (version 3.2). Cladistics 5:164-166. [Google Scholar]
- 18.Filion, M., R. C. Hamelin, L. Bernier, and M. St-Arnaud. 2004. Molecular profiling of rhizosphere microbial communities associated with healthy and diseased black spruce (Picea mariana) seedlings grown in a nursery. Appl. Environ. Microbiol. 70:3541-3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goh, S. H., S. Potter, J. O. Wood, S. M. Hemmingsen, R. P. Reynolds, and A. W. Chow. 1996. HSP60 gene sequences as universal targets for microbial species identification: studies with coagulase-negative staphylococci. J. Clin. Microbiol. 34:818-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Graff, A., and R. Conrad. 2005. Impact of flooding on soil bacterial communities associated with poplar (Populus sp.) trees. FEMS Microbiol. Ecol. 53:401-415. [DOI] [PubMed] [Google Scholar]
- 21.Grayston, S. J., D. Vaughan, and D. Jones. 1996. Rhizosphere carbon flow in trees, in comparison with annual plants: the importance of root exudation and its impact on microbial activity and nutrient availability. Appl. Soil Ecol. 5:29-56. [Google Scholar]
- 22.Heuer, H., R. M. Kroppenstedt, J. Lottmann, G. Berg, and K. Smalla. 2002. Effects of T4 lysozyme release from transgenic potato roots on bacterial rhizosphere communities are negligible relative to natural factors. Appl. Environ. Microbiol. 68:1325-1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hill, J. E., R. P. Seipp, M. Betts, L. Hawkins, A. G. Van Kessel, W. L. Crosby, and S. M. Hemmingsen. 2002. Extensive profiling of a complex microbial community by high-throughput sequencing. Appl. Environ. Microbiol. 68:3055-3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hill, J. E., S. L. Penny, K. G. Crowell, S. Goh, and S. M. Hemmingsen. 2004. cpnDB: a chaperonin sequence database. Genome Res. 14:1669-1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hill, T. C., K. A. Walsh, J. A. Harris, and B. F. Moffett. 2003. Using ecological diversity measures with bacterial communities. FEMS Microbiol. Ecol. 43:1-11. [DOI] [PubMed] [Google Scholar]
- 26.Hughes, J. B., J. J. Hellmann, T. H. Ricketts, and B. J. Bohannan. 2001. Counting the uncountable: statistical approaches to estimating microbial diversity. Appl. Environ. Microbiol. 67:4399-4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hunt, J., L. Boddy, P. F. Randerson, and H. J. Rogers. 2004. An evaluation of 18S rDNA approaches for the study of fungal diversity in grassland soils. Microb. Ecol. 47:385-395. [DOI] [PubMed] [Google Scholar]
- 28.Janssen, P. H. 2006. Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl. Environ. Microbiol. 72:1719-1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jansson, H. 1982. Predacity by nematophagous fungi and its relation to the attraction of nematodes. Microb. Ecol. 8:233-240. [DOI] [PubMed] [Google Scholar]
- 30.Jumpponen, A., and L. C. Johnson. 2005. Can rDNA analyses of diverse fungal communities in soil and roots detect effects of environmental manipulations—a case study from tallgrass prairie. Mycologia 97:1177-1194. [DOI] [PubMed] [Google Scholar]
- 31.Kendrick, B. 1992. The fifth kingdom. Focus Information Group, Inc., Newburyport, MA.
- 32.Kent, A. D., and E. W. Triplett. 2002. Microbial communities and their interactions in soil and rhizosphere ecosystems. Annu. Rev. Microbiol. 56:211-236. [DOI] [PubMed] [Google Scholar]
- 33.Kowalchuk, G. A., M. Bruinsma, and J. A. van Veen. 2003. Assessing responses of soil microorganisms to GM plants. Trends Ecol. Evol. 18:403-410. [Google Scholar]
- 34.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 35.Li, L., and J. Steffens. 2002. Overexpression of polyphenol oxidase in transgenic tomato plants results in enhanced bacterial disease resistance. Planta 215:239-247. [DOI] [PubMed] [Google Scholar]
- 36.Magurran, A. 1988. Ecological diversity and its measurement. Princeton University Press, Princeton, NJ.
- 37.Mayer, A. M., and E. Harel. 1979. Polyphenol oxidases in plants. Phytochemistry 18:193-215. [DOI] [PubMed] [Google Scholar]
- 38.McCaig, A. E., L. A. Glover, and J. I. Prosser. 1999. Molecular analysis of bacterial community structure and diversity in unimproved and improved upland grass pastures. Appl. Environ. Microbiol. 65:1721-1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Milling, A., K. Smalla, F. X. Maidl, M. Schloter, and J. C. Munch. 2004. Effects of transgenic potatoes with an altered starch composition on the diversity of soil and rhizosphere bacteria and fungi. Plant Soil 266:23-39. [Google Scholar]
- 40.Mohammadi, M., and H. Kazemi. 2002. Changes in peroxidase and polyphenol oxidase activities in susceptible and resistant wheat heads inoculated with Fusarium graminearum and induced resistance. Plant Sci. 162:491-498. [Google Scholar]
- 41.Nevill, R. J., P. M. Hall, and J. Beale. 1995. Forest health research needs in British Columbia. For. Chron. 71:489-496. [Google Scholar]
- 42.Normander, B., and J. I. Prosser. 2000. Bacterial origin and community composition in the barley phytosphere as a function of habitat and presowing conditions. Appl. Environ. Microbiol. 66:4372-4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nübel, U., B. Engelen, A. Felske, J. Snaidr, A. Wieshuber, R. I. Amann, W. Ludwig, and H. Backhaus. 1996. Sequence heterogeneities of genes encoding 16S rRNAs in Paenibacillus polymyxa detected by temperature gradient gel electrophoresis. J. Bacteriol. 178:5636-5643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.O'Callaghan, M., E. M. Gerard, N. W. Waipara, S. D. Young, T. R. Glare, P. J. Barrell, and A. J. Conner. 2004. Microbial communities of Solanum tuberosum and magainin-producing transgenic lines. Plant Soil 266:47-56. [Google Scholar]
- 45.Oger, P., A. Petit, and Y. Dessaux. 1997. Genetically engineered plants producing opines alter their biological environment. Nat. Biotechnol. 15:369-372. [DOI] [PubMed] [Google Scholar]
- 46.Ohtsuka, E., S. Matsuki, M. Ikehara, Y. Takahashi, and K. Matsubara. 1985. An alternative approach to deoxyoligonucleotides as hybridization probes by insertion of deoxyinosine at ambiguous codon positions. J. Biol. Chem. 260:2605-2608. [PubMed] [Google Scholar]
- 47.Pearson, W., and D. Lipman. 1988. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA 85:2444-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peter, M. G. 1989. Chemical modifications of biopolymers by quinines and quinone methides. Angew. Chem. Int. Ed. Engl. 28:555-570. [Google Scholar]
- 49.Priha, O., S. J. Grayston, R. Hiukka, T. Pennanen, and A. Smolander. 2001. Microbial community structure and characteristics of the organic matter in soils under Pinus sylvestris, Picea abies and Betula pendula at two forest sites. Biol. Fertil. Soils 22:17-24. [Google Scholar]
- 50.Raj, S. N., B. R. Sarosh, and H. S. Shetty. 2006. Induction and accumulation of polyphenol oxidase activities as implicated in development of resistance against pearl millet downy mildew disease. Funct. Plant Biol. 33:563-571. [DOI] [PubMed] [Google Scholar]
- 51.Ranjard, L., F. Poly, and S. Nazaret. 2000. Monitoring complex bacterial communities by culture-independent molecular techniques: application to soil environment. Res. Microbiol. 151:167-177. [DOI] [PubMed] [Google Scholar]
- 52.Rasche, F., V. Hödl, C. Poll, E. Kandeler, M. H. Gerzabek, J. D. van Elsas, and A. Sessitsch. 2006. Rhizosphere bacteria affected by transgenic potatoes with antibacterial activities compared with the effects of soil, wild-type potatoes, vegetation stage and pathogen exposure. FEMS Microbiol. Ecol. 56:219-235. [DOI] [PubMed] [Google Scholar]
- 53.Schloss, P. D., and J. Handelsman. 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71:1501-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shewan, J. M., and T. A. McMeekin. 1983. Taxonomy (and ecology) of Flavobacterium and related genera. Annu. Rev. Microbiol. 37:233-252. [DOI] [PubMed] [Google Scholar]
- 55.Singh, B. K., and A. Walker. 2006. Microbial degradation of organophosphorus compounds. FEMS Microbiol. Rev. 30:428-471. [DOI] [PubMed] [Google Scholar]
- 56.Smit, E., P. Leeflang, B. Glandorf, J. D. van Elsas, and K. Wernars. 1999. Analysis of fungal diversity in the wheat rhizosphere by sequencing of cloned PCR-amplified genes encoding 18S rRNA and temperature gradient gel electrophoresis. Appl. Environ. Microbiol. 65:2614-2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sokal, R. R., and F. J. Rolf. 1995. Biometry: the principles and practice of statistics in biological research, 3rd ed. W. H. Freeman, New York, NY.
- 58.Tesfaye, M., N. S. Dufault, M. R. Dornbusch, D. L. Allan, C. P. Vance, and D. A. Samac. 2003. Influence of enhanced malate dehydrogenase expression by alfalfa on diversity of rhizobacteria and soil nutrient availability. Soil Biol. Biochem. 35:1103-1113. [Google Scholar]
- 59.Tomilova, O. G., and M. V. Shternshis. 2006. The effect of a preparation from Chaetomium fungi on the growth of phytopathogenic fungi. Appl. Biochem. Microbiol. 42:67-71. [PubMed] [Google Scholar]
- 60.Wang, J., and C. P. Constabel. 2004. Polyphenol oxidase overexpression in transgenic Populus enhances resistance to herbivory by forest tent caterpillar (Malacosoma disstria). Planta 220:87-96. [DOI] [PubMed] [Google Scholar]
- 61.White, T. J., T. Bruns, S. Lee, and J. W. Taylor. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p. 315-322. In M. A. Innis, D. H. Gelfand, J. J. Sninsky, T. J. White (ed.), PCR protocols: a guide to methods and applications. Academic Press Inc., New York, NY.
- 62.Wititsuwannakul, D., N. Chareonthiphakorn, M. Pace, and R. Wititsuwannakul. 2002. Polyphenol oxidases from latex of Hevea brasiliensis: purification and characterization. Phytochemistry 61:115-121. [DOI] [PubMed] [Google Scholar]
- 63.Wolfenbarger, L. L., and P. R. Phifer. 2000. The ecological risks and benefits of genetically engineered plants. Science 290:2088-2093. [DOI] [PubMed] [Google Scholar]