Abstract
The way that gaseous metabolite production changes along with biofilm architecture development is poorly understood. To address this question, we developed a novel flow reactor biofilm culture method that allows for simultaneous assessment of gaseous metabolite production and architecture visualization. In this report, we establish the utility of this method using denitrification by Pseudomonas aeruginosa biofilms as a model system. Using this method, we were able to collect and analyze gaseous metabolites produced by denitrification and also visualize biofilm architecture in a nondestructive manner. Thus, we propose that this novel method is a powerful tool to investigate potential relationships between biofilm architecture and the gas-producing metabolic activity of biofilms, providing new insights into biofilm ecology.
It is known that environmental biofilms often produce gaseous metabolites. For example, natural lake reed biofilms are reported to produce N2 or N2O (28). Furthermore, microbial biofilms are utilized in wastewater treatment and other industrial processes to convert soluble material into gaseous material, such as the conversion of nitrate into N2 or N2O by denitrification (2, 21, 30). Previous studies of biofilm architecture have reported correlations between metabolic substrates and biofilm architecture (11, 24, 27), and another study showed the importance of biofilm structures in the distribution of dissolved gases (4). However, such correlations between biofilm architecture and gaseous metabolite production remain poorly understood.
If one were able to show simultaneously occurring changes in architecture and gaseous metabolite production, the existence of a correlation between biofilm architecture and gaseous metabolite production would be indicated. To date, the analysis of gaseous metabolite production by biofilms has mainly been performed using test tubes or flasks sealed with butyl rubber stoppers (28). These airtight containers allow for collection of gaseous metabolites and are suitable for batch culture experiments. However, there are currently no nondestructive methods to simultaneously allow for both biofilm visualization and gaseous metabolite analysis. Such a method would provide information on how gaseous metabolite production activity changes along with biofilm development.
The flow reactor method (15, 18), combined with confocal laser scanning microscopy (CLSM), is preferred in studies of biofilm architecture and development. Both electron microscopy and CLSM are often used in visualizing biofilms (12); however, electron microscopy requires dehydration (and therefore termination of biofilm growth), which in some cases distorts or damages bacterial cells (7). In contrast, CLSM can be used to visualize the three-dimensional architecture of living biofilms and can be applied to the flow reactor method while maintaining continuous culture (15, 18). If collection of gaseous metabolites is achieved in a flow reactor system, both biofilm visualization and gaseous metabolite analysis can be performed without destruction of the biofilm. To accommodate this need, we developed a novel airtight flow reactor that allows for collection of gaseous metabolites. This novel reactor was termed AFGAS (for airtight flow reactor for nondestructive gaseous metabolite analysis and structure visualization), and a novel biofilm analysis method using the AFGAS was termed the AFGAS method. In order to assess the utility of the AFGAS method, we utilized Pseudomonas aeruginosa as a model organism. P. aeruginosa is a gram-negative ubiquitous bacterium that is well-established as a model for biofilm formation (5, 27) and also possesses a functional denitrification pathway (3, 9, 29). Denitrification is an anaerobic respiration process that utilizes N oxides instead of oxygen as terminal electron acceptors in the respiratory chain (31). In denitrification, concomitant with energy production, nitrate (NO3−) and nitrite (NO2−) are reduced to gaseous nitrous oxide (N2O) and gaseous nitrogen (N2) (31).
The main purpose of this study was to introduce a novel method and establish its utility. Here, we report that denitrifying conditions are achievable with the AFGAS method. We further establish that the AFGAS method can be used to collect and analyze gaseous denitrification metabolites, and finally, we demonstrate that P. aeruginosa biofilms grown with the AFGAS method can be nondestructively visualized. According to these results, we conclude that the AFGAS method is a biofilm culture method suitable for both nondestructive gaseous metabolite analysis and nondestructive biofilm visualization.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The strains and plasmids used in this study are listed in Table 1. For routine culture, bacterial strains were grown in LB medium or on LB agar plates. When necessary, gentamicin was added at a concentration of 15 μg/ml for Escherichia coli DH5α (TaKaRa Shuzo, Osaka, Japan) and S17-1 (23) and at 100 μg/ml for P. aeruginosa. For biofilm experiments, stationary-phase cultures were diluted to an optical density at 660 nm of 0.1 with LB or LB25N medium (LB medium supplemented with 25 mM KNO3), and 300 μl of the cell suspension was injected into a flow cell (Stovall Life Science, Greensboro, NC) with channel dimensions of 1 by 4 by 40 mm (height by width by length). After being held static for 1 h, the flow of LB or LB25N medium was initiated at the rate of 0.015 ml/min. Biofilms were grown at 37°C for 24 h. A multichannel peristaltic pump (model IPC; Ismatec, Glattbrug, Switzerland) was employed to continuously irrigate the flow cell with liquid medium.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant properties | Reference |
|---|---|---|
| P. aeruginosa strains | ||
| PAO1 | Wild-type strain | 13 |
| PS1700 | narG; 3,835 bp deleted from PA3874-PA3876 | This study |
| E. coli strains | ||
| DH5α | E. coli strain for transformation | TaKaRa Shuzo |
| S17-1 | Mobilizer strain | 23 |
| Plasmids | ||
| pG19II | pK19mobsac derivative carrying a Gmr gene as a marker | 14 |
| pG19-delnar | pG19II derivative carrying a narG deletion fragment | This study |
Construction of plasmids and a P. aeruginosa mutant.
DNA manipulations followed standard procedures (22). A narG deletion vector pG19-delnar was constructed by overlap extension PCR (22). DNA fragments of 874 bp and 1,033 bp of PAO1 chromosome were amplified with NarGF1(CGGGATCCCGTGCTTCTCGTACAGCTCGCG) and NarGR1 (CGGAACAGTTCGGCATGGTGCTGAACCTCG) PCR primers and with NarGF2 (CCATGCCGATCTGTGCCGGGCGTTACAACG) and NarGR2 (CCCAAGCTTGGGACTTCGCCTCGAGCATGTCC) PCR primers, respectively. The two DNA fragments of 874 bp and 1,033 bp were used in overlap extension PCR to amplify an 1,886-bp DNA fragment with the narG deletion. The amplified 1,886-bp DNA fragment was subcloned into a BamHI-HindIII site in a multicloning site of pG19II, yielding pG19-delnarG. To construct a narG mutant (PS1700), the pG19-delnar plasmid was transferred into P. aeruginosa PAO1 by conjugation, followed by homologous recombination (14). The deletions of narG in P. aeruginosa PAO1 were confirmed by PCR analyses and by the phenotypes of the mutants (data not shown).
Hungate tube planktonic bacterial culture method.
The Hungate tube planktonic culture experiment was performed as previously described (26). Briefly, P. aeruginosa cells were grown aerobically in 24-ml test tubes containing 4 ml of LB medium. For planktonic anaerobic culture, 10 μl of a cell suspension was inoculated into a 17-ml Hungate tube containing 5 ml of LB100N medium (LB medium supplemented with 100 mM KNO3) or LB medium, and argon gas was flashed for 5 min. The Hungate tubes were incubated at 37°C with shaking at 200 rpm.
Gas replacement and inoculation for biofilm culture under low-oxygen partial pressure conditions.
Prior to inoculation, an entire flow cell system including 100 ml of LB25N medium in a medium bottle was degassed by vacuum for 10 min and then purged with argon gas for 10 min. The degassing-purging process was repeated twice. Inoculation was performed as follows: 5 ml of the cell suspensions from LB-grown overnight cultures were transferred to 17-ml Hungate tubes. The Hungate tubes were then sealed with butyl rubber stoppers, and argon gas was flashed through a needle for 5 min. A 300-μl aliquot of the cell suspension was injected into a flow cell from an inoculation port, with argon gas flashed around the inoculation port. For aerobic cultures, argon gas flashing was not performed.
Gaseous metabolite sampling and analysis.
The atmosphere in the gas collector (Fig. 1D) or Hungate tube was sampled with a gas-tight sampling syringe (Shimadzu, Tokyo, Japan) and subjected to gas chromatography. N2O and N2 concentrations were measured with a gas chromatograph (model GC-8AIT; Shimadzu, Tokyo, Japan) equipped with a gas thermal conductivity detector. Helium was used as the carrier gas. Shincarbon ST or PropackQ columns (Shimadzu, Tokyo, Japan), operated at 200°C or 50°C, were used for the detection of N2O or N2, respectively.
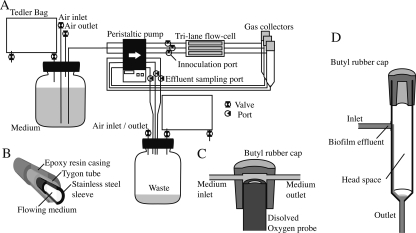
FIG. 1.
(A) AFGAS setup for biofilm experiments. AFGAS was designed to perform a triplicate experiment. Gas sampling bags enclose both ends and isolate the flow system from the external atmosphere. Three flow lanes are independently connected to the waste bottle in order to avoid mixture of waste between lanes and to allow for effluent sampling from individual lanes. (B) Cross-section of the multilayer tubing. Tygon R-3063 tubing is reinforced with stainless steel pipes and epoxy resin. In order to maintain flexibility, the pump connections and the joints are composed of only a monolayer of Tygon R-3063 tubing. (C) Cross-section of the oxygen probe flow cell. A dissolved oxygen probe is contained in a flow cell made of butyl rubber. The butyl rubber flow cells are equipped with an inlet and an outlet. For dissolved oxygen measurements, the standard flow cell was replaced with the oxygen probe flow cell. (D) Sectioned diagram of a gas collector. The duplicated pump connections, upstream of the inlet and downstream of the outlet, prevent free spillage of gas from the gas collector and help to maintain a constant minimum volume of effluent in the gas collector.
Biofilm effluent sampling and analysis.
A 300-μl aliquot of biofilm effluent was sampled from the effluent sampling port (Fig. 1A). NO3− concentrations were determined by the brucine-sulfanic acid method (16). NO2− concentrations were determined by the sulfanilamide-naphthylethylene diamine method (16). Since the volume of the medium that exists between the inoculation port and the gas collector is approximately 700 μl, the total CFU or total protein amount in 700 μl of biofilm effluent was assumed to be the total planktonic biomass. Cells in biofilm effluent were homogenized using an ultrasonic processor (model UP50H; Stahnsdorf, Germany), and protein concentrations of the homogenates were determined by the Bradford method with serum albumin as the standard.
Calculation of conversion efficiency.
We assessed the rate of conversion of substrate to the gaseous metabolites collected by calculating the conversion efficiency for these reactions. In this study, conversion efficiency was defined as follows: E = {[2(P1 + P2)]/S} × 100, where E is the conversion efficiency (a percentage), S is the total consumed NO3− (in micromoles), P1 is the total N2 (in micromoles) accumulated in the headspace, and P2 is the total N2O (in micromoles) accumulated in the headspace. For biofilm experiments, the integral of the NO3− concentration curve was assumed to be total consumed NO3−.
Dissolved oxygen concentration measurement.
For dissolved oxygen (DO) measurements, a standard flow cell was replaced with a DO metering flow cell (Fig. 1C). A DO metering flow cell contains an oxygen probe, and the oxygen concentration in the flowing medium was measured using a Metler Toredo (Greifensee, Switzerland) SG6 dissolved oxygen meter.
Biofilm quantification.
For total protein quantification, a flow cell was removed from the flow system, and each lane of the flow cell was washed with Tris-HCl buffer. The chambers were then filled with 300 μl of Tris-HCl buffer, and the inlet and outlet of the flow cell were tightly closed. The entire flow cell was placed in an ultrasonic processing tank (Insonator 201 M; Kubota, Tokyo, Japan) filled with water and processed by ultrasound at maximum power for 30 min. Buffers containing fragmented biofilm in each lane were separately collected, and residual fragments were eluted with 700 μl of Tris-HCl buffer. The two fractions of 300 μl and 700 μl were combined to make 1 ml of biofilm fragment suspension, which was then homogenized using an ultrasonic processor (model UP50H; Stahnsdorf, Germany). Protein concentrations of the homogenates were quantified by the Bradford method with serum albumin as the standard. For measurement of the total CFU in the biofilm, cells were recovered from the flow cell by weak sonication.
Biofilm visualization and image analysis.
A Carl Zeiss Pascal laser scanning microscope equipped with a 63×, 1.4 numerical aperture Plan-Apochromat objective (Carl Zeiss, Jena, Germany) was used to acquire confocal microscopic images. For destructive visualization of all cells in a biofilm, biofilms were stained with Syto9 fluorescent dye (Molecular Probes, Eugene, OR). Syto9 fluorescent dye was excited by an argon laser (514 nm) and detected with a 530-nm long-pass filter. For biovolume quantification, images of Syto9-stained biofilms were analyzed using the COMSTAT computer program (10), which functions within the MATLAB software (Mathworks, Natick, MA). The confocal reflection microscopy technique was employed as an independent nonfluorescence-based method to nondestructively acquire three-dimensional images of biofilms (17). Biofilms were illuminated with a 514-nm argon laser, and reflected light was collected through a 505- to 530-nm band-pass filter.
RESULTS
Features of the AFGAS method.
As shown in Fig. 1A, the AFGAS is a flow system consisting of a medium bottle, a flow cell, a gas collector, a waste bottle, and tubing that connects these components. Since gaseous metabolites generated in the flow cell are conveyed with effluent flow toward the lower streams, analysis of gaseous metabolites necessitates a mechanism to separate and accumulate gaseous metabolites away from the effluent flow. To accommodate this need, a gas collector, which has a 12-ml inner volume and a conical bottom, was placed downstream of the flow cell (Fig. 1A and D). The gas collector was capped with a butyl rubber cap for gas sampling and equipped with an inlet at the upper part of a sidewall and an outlet at the bottom of the lower conical part (Fig. 1D). Biofilm effluent flows in from the inlet and out to the outlet. Because the biofilm effluent, which is heavier than gas, gathers at the lower conical part, only effluent will flow out, while gaseous metabolites will accumulate in the headspace of the gas collector. The accumulation of gaseous metabolites raises pressure in the gas collector, which could potentially cause spillage of gas from its outlet. To prevent this spillage, the flow system of the AFGAS was connected to a multichannel peristaltic pump at two points in the same direction, upstream of the flow cell and downstream of the gas collector. This duplicated pump connection equalizes influx and efflux of biofilm effluent in the gas collector and prevents the spillage of gas due to the positive pressure in the gas collector.
An additional concern in gaseous metabolite analysis is contamination of the gas collector with ambient air. Because gaseous metabolites are often molecules that exist abundantly in the ambient atmosphere, it is necessary to construct an airtight gas reactor that can be purged of ambient air. As shown in Fig. 1A, the entire flow system of the AFGAS is closed to the outer ambient atmosphere. In order to avoid a change in pressure while maintaining an airtight structure, both the medium and the waste bottles are connected to Tedlar gas sampling bags (AS ONE, Osaka, Japan), the volumes of which are variable. To reduce gas permeation, the tubing consists of multiple layers (Fig. 1B). Both the medium and waste bottles are equipped with air inlet and air outlet bulbs, which allow gas replacement by degassing and gas purging (Fig. 1A). The gas replacement feature also accommodates any need for growth of biofilms under low-oxygen partial pressure conditions. Gas replacement with oxygen-free gas would allow for studies addressing anaerobic metabolism, such as denitrification (1, 18).
For inoculation and nondestructive effluent analysis, the AFGAS was equipped with inoculation ports in the upper stream of the flow cell and effluent sampling ports between the waste bottle and the peristaltic pump (Fig. 1A). A three-way bulb was used to provide these ports. The physical dimensions of the entire flow system of the AFGAS are 40 by 40 cm, which allows for storage of the AFGAS in a standard incubator.
Biofilm culture under denitrifying conditions.
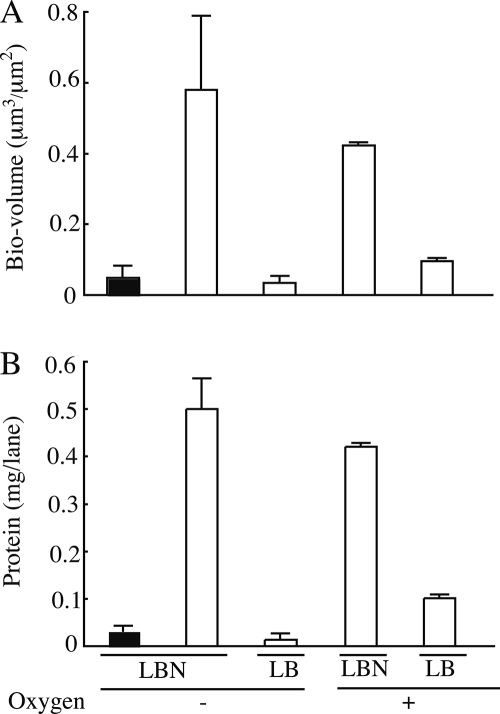
To examine whether denitrifying conditions were achievable with the AFGAS method, we first performed direct measurements of DO concentrations. When gas replacement was performed, the DO concentration in fresh flowing medium was maintained between 0.4 and 0.5 ppm during a 24-h period, indicating that low-oxygen, partial pressure conditions are achievable with the AFGAS method. Next, we examined whether our low-oxygen partial pressure condition was able to repress oxygen-dependent biofilm development of P. aeruginosa. As shown in Fig. 2A and B, when grown in LB medium, strain PAO1 formed significantly less biofilm under low-oxygen partial pressure than under aerobic conditions. We further examined whether nitrate could restore the biofilm development of P. aeruginosa strains. Strain PS1700 is a narG deletion mutant which is deficient in reducing nitrate to nitrite and exhibits significantly poor growth under planktonic denitrifying conditions (data not shown). As shown in Fig. 2A and B, a supplement of nitrate restored strain PAO1 biofilm development, whereas strain PS1700 showed poor biofilm development comparable to that of strain PAO1 grown under low-oxygen partial pressure conditions without nitrate. These results indicate that the low-oxygen partial pressure conditions achieved by the AFGAS method allowed a P. aeruginosa denitrifying biofilm to develop but did not allow a P. aeruginosa nondenitrifying biofilm to develop. Accordingly, we conclude that a denitrifying condition is achievable with the AFGAS method.
FIG. 2.
P. aeruginosa PAO1 (white bars) and PS1700 (black bars) biofilms were cultured for 24 h under low-oxygen partial pressure (-O2) or aerobic (+O2) conditions and supplemented with KNO3 (LBN) or without (LB). (A) Biofilm biovolumes measured by image analysis. The y axis of the graph shows the volume occupied by the biofilm on the lane of the flow cell. Nine images per condition were analyzed, and results are expressed as means and standard deviations. (B) Total protein amount per lane of the flow cell. Three independent biofilm cultures were measured, and results are expressed as means and standard deviations.
Nondestructive collection of gaseous denitrification metabolites.
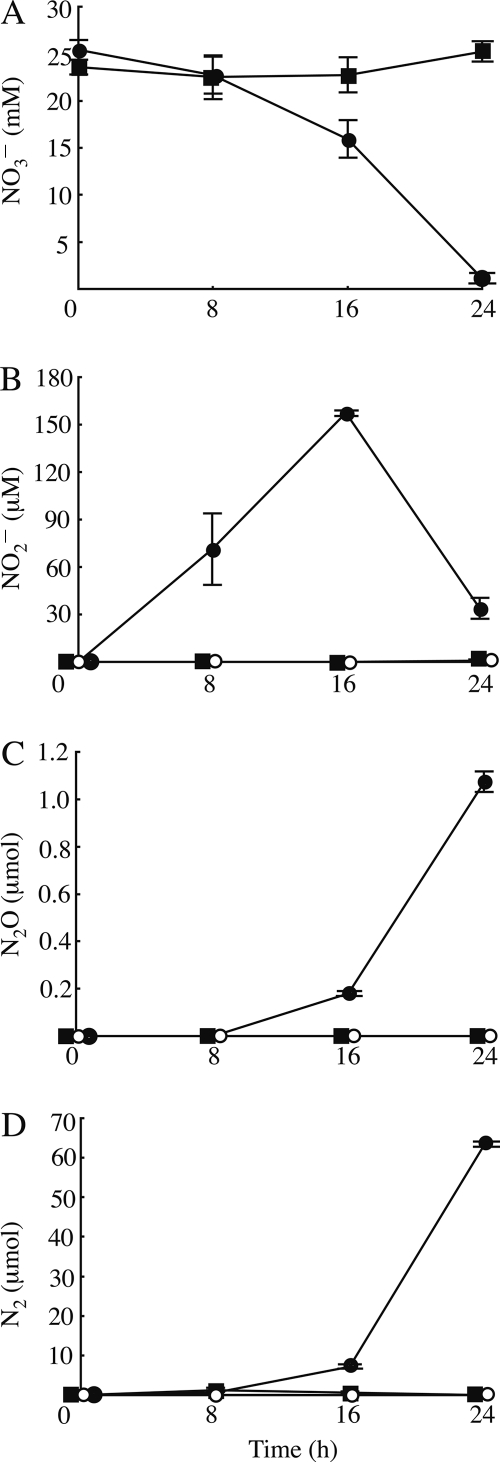
To establish the utility of the AFGAS method in gaseous metabolite analysis, we examined whether the AFGAS method could collect and analyze gaseous denitrification metabolites. First, we examined whether gaseous denitrification metabolites accumulated in the headspaces of the gas collectors when P. aeruginosa PAO1 biofilms were cultured under denitrifying conditions. As shown in Fig. 3, the total amount of both N2 and N2O in the headspaces increased with time, whereas the NO3− concentration decreased with time and the maximum NO2− concentration was observed at 16 h. Next, we examined whether the N2 and N2O that accumulated in the headspaces were generated by denitrification. To test this, we cultured the P. aeruginosa PS1700 biofilm with nitrate (LB25N medium) and PAO1 biofilm without nitrate (LB medium). The results showed that, in both cases, only background levels of N2 and N2O accumulated in the headspace, and NO3− or NO2− levels remained almost at initial concentrations. These results indicate that the accumulated N2 and N2O gases observed when PAO1 was cultured under low-oxygen partial pressure conditions in the presence of nitrate were denitrification metabolites (Fig. 3).
FIG. 3.
Denitrification activities of P. aeruginosa biofilms grown under low-oxygen partial pressure conditions. PAO1 (circles) and PS1700 (squares) were grown with (solid) or without (open) KNO3. (A and B) NO3− (A) and NO2− (B) concentrations in the effluent flow. (C and D) Amount of N2 (C) and N2O (D) that accumulated in the gas collector. The amount of N2 is calculated as the difference between total N2 and background accumulation of N2. Negative values of N2 are expressed as 0 μmol. Data are the averages of three biofilm cultures. Error bars indicate standard deviations.
When a strain PAO1 biofilm was grown under denitrifying conditions, the planktonic biomass leached its maximum at 24 h, and the total protein amount was estimated to be 0.13 mg, which is considerably smaller than the biomass of the biofilms (Fig. 3B). The total CFU in the effluent was approximately 3.9 × 107 at 24 h, whereas total CFU in biofilms was estimated to be 4.1 × 108, which is approximately 10 times higher than that of planktonic cells. These data show the biofilm biomass was the major portion of the total biomass in the system, indicating that the major portion of collected denitrification metabolites was generated by the biofilm. According to these results, we conclude that the AFGAS method allows for collection of gaseous metabolites.
To further assess the completeness of gaseous metabolite collection by the AFGAS method, we compared the rate of gaseous metabolites collected per consumed substrate (conversion efficiency) obtained using the AFGAS method with that obtained using the Hungate tube method, an established planktonic culture method for gaseous metabolite analysis (26). P. aeruginosa PAO1 was cultured under denitrifying conditions for 24 h using the AFGAS method and the Hungate tube method. With the AFGAS method, the estimated conversion efficiency was 50.9%, whereas the conversion efficiency using the Hungate tube method was 65.8%. These data indicate that the AFGAS method can achieve similar conversion efficiencies and suggest that the AFGAS method is capable of collecting a major portion of gaseous metabolites.
Nondestructive biofilm visualization under denitrifying conditions.
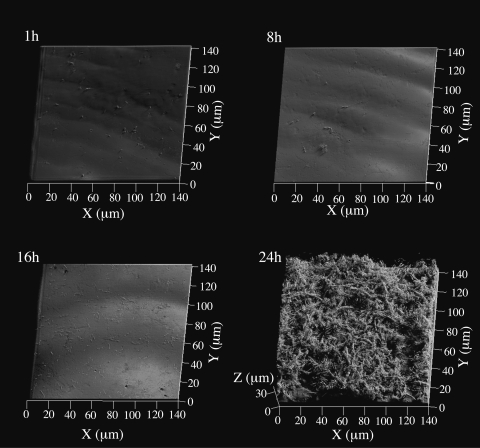
To demonstrate nondestructive biofilm visualization by the AFGAS method, we observed the developmental process of a P. aeruginosa PAO1 biofilm under denitrifying conditions using the confocal reflection microscopy technique. As shown in Fig. 4, two developmental stages were observed. In the first stage, observed at 1 h, 8 h, and 16 h, the majority of cells appeared to attach directly to the glass surface. Some small cell clusters were visible, and cells exhibited a normal rod-like morphology. In the second stage, observed at 24 h, cells formed a three-dimensional mesh-like structure with channels or spaces between large macrocolonies, and the majority of cells showed a filamentous morphology (Fig. 4).
FIG. 4.
Developmental process of P. aeruginosa PAO1 biofilm cultured with LB25N medium visualized by the confocal reflection microscopy technique. Images were acquired at 1, 8, 16, and 24 h after inoculation and are presented as simulated projections. Each projection shows fields of 140 by 140 μm (x-y), as indicated.
DISCUSSION
In this report, we establish that the AFGAS method permits the growth of biofilms under denitrifying conditions such that gaseous denitrification metabolite analysis and biofilm visualization can be performed simultaneously in a nondestructive manner.
We observed that the AFGAS method maintains DO concentrations in fresh flowing medium between 0.4 and 0.5 ppm for at least 24 h without continuous gas replacement, and the oxygen limitation is enough to repress oxygen-dependent biofilm development of P. aeruginosa (Fig. 2). Nevertheless, two limitations must be considered in applying the AFGAS method in anaerobic biofilm studies. First, the DO concentration in fresh medium tends to slowly increase with time. Therefore, for multiday experiments, we recommend that gas purges be performed continuously, as described in previous studies (1, 18). Use of more-gas-impermeable material, such as polyetheretherketone in the resin part of the flow system, such as the valve, is also recommended. Second, the DO concentration of 0.4 to 0.5 ppm is too high to be suitable for the study of strict anaerobes or metabolism, which requires extremely low redox potentials. To establish strict anaerobic conditions, the entire flow system should be placed in an anaerobic chamber, or medium should be supplemented with a reducing agent.
In our studies of denitrification, we have shown that the AFGAS method is able to collect both of the gaseous denitrification metabolites, N2 and N2O (Fig. 3C and D). The Hungate tube method exhibited slightly higher conversion efficiencies than the AFGAS method. At this time, we cannot determine the reason for this difference in conversion efficiencies between the two methods. One possible explanation for this difference is that gaseous metabolite collection by the AFGAS method was somehow incomplete relative to the Hungate tube method. A second (but not mutually exclusive) possibility is that there is a difference in denitrification activity between biofilm and planktonic cells. Therefore, at this time, it is not recommended that the AFGAS method be used to perform quantitative analyses. Nevertheless, with the AFGAS method, our data reveal a clear relationship between NO3− consumption and gaseous metabolite production (Fig. 3A, C, and D). Furthermore, our microscopic observations of biofilm development indicate that gaseous metabolites accumulate over the time course of biofilm development, and biofilm quantification indicates a clear relationship between biofilm growth and gaseous metabolite accumulation (Fig. 2, 3, and 4). Therefore, we conclude that the accumulation of gaseous metabolites in the gas collector reflects the output of gaseous products via metabolic activity of the biofilm. Accordingly, we propose that the AFGAS method can be applied to qualitative or semiquantitative gaseous metabolite analyses.
Most fluorescent proteins require oxygen in their functional maturation (8, 19). In contrast, as shown in Fig. 4, CLSM in combination with confocal reflection microscopy allows for visualization of P. aeruginosa biofilm development under denitrifying conditions. Therefore, we conclude that biofilms grown using the AFGAS method can be nondestructively visualized regardless of oxygen concentration. We are now preparing detailed information on the application of confocal reflection microscopy in biofilm visualization. Furthermore, the recent development of reporter proteins for in vivo fluorescence without oxygen would allow use of the AFGAS method with CLSM in combination with fluorescent protein-expressing cells under low-oxygen partial pressure conditions (6). The combination of CLSM and appropriate fluorescent reporter proteins would enable nondestructive analysis of protein localization or identification of distinct microorganism species in biofilms (5, 20, 25). Therefore, application of these fluorescent reporters for the AFGAS method would allow direct observation of changes in metabolite production and substrate consumption along with localization of such proteins or species in gaseous metabolite-producing biofilms.
In this study, the AFGAS method was used to perform time course observations of biofilm architecture, substrate consumption, soluble metabolite production, and gaseous metabolite production of P. aeruginosa biofilms. Under denitrifying conditions, our results show that N2 and N2O begin to accumulate between 8 h and 16 h of biofilm growth and reach a maximum at 24 h. The structure of the biofilm grown under these conditions is comprised of a characteristic three-dimensional mesh-like architecture consisting of filamentous cells (Fig. 4). Interestingly, both a characteristic three-dimensional mesh-like architecture and robust gaseous metabolite production appeared during the same period, between 16 h and 24 h (Fig. 3 and 4), indicating the possibility that biofilm architecture and metabolite production are correlated. In this manner, the AFGAS method is able to demonstrate how metabolite production or substrate consumption changes along with biofilm structural development. A detailed study on the effect of gas production on biofilm architecture will be included in our future work. We propose that the AFGAS method will allow for an assessment of potential correlations between the architecture and metabolism of gaseous metabolite-producing biofilms, providing new insights into biofilm ecology.
Acknowledgments
We thank Yuichi Suwa, Naoki Takaya, and Sherry L. Kuchma for advice. We thank Masanori Toyofuku for technical advice on determining denitrification activity. We also thank Arne Heydorn for kindly providing us with the COMSTAT program.
Footnotes
Published ahead of print on 7 July 2008.
REFERENCES
- 1.Alvarez-Ortega, C., and C. Harwood. 2007. Responses of Pseudomonas aeruginosa to low oxygen indicate that growth in the cystic fibrosis lung is by aerobic respiration. Mol. Microbiol. 65:153-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrade do Canto, C., J. Rodrigues, S. Ratusznei, M. Zaiat, and E. Foresti. 2008. Feasibility of nitrification/denitrification in a sequencing batch biofilm reactor with liquid circulation applied to post-treatment. Bioresource Technol. 99:644-654. [DOI] [PubMed] [Google Scholar]
- 3.Chen, F., Q. Xia, and L. Ju. 2006. Competition between oxygen and nitrate respirations in continuous culture of Pseudomonas aeruginosa performing aerobic denitrification. Biotechnol. Bioeng. 93:1069-1078. [DOI] [PubMed] [Google Scholar]
- 4.DeBeer, D., P. Stoodley, F. Roe, and Z. Lewandowski. 1994. Effects of biofilm structure on oxygen distribution and mass transport. Biotechnol. Bioeng. 43:1131-1138. [DOI] [PubMed] [Google Scholar]
- 5.De Kievit, T., M. Parkins, R. Gillis, R. Srikumar, H. Ceri, K. Poole, B. Iglewski, and D. Storey. 2001. Multidrug efflux pumps: expression patterns and contribution to antibiotic resistance in Pseudomonas aeruginosa biofilms. Antimicrob. Agents Chemother. 45:1761-1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drepper, T., T. Eggert, F. Circolone, A. Heck, U. Krauss, J. Guterl, M. Wendorff, A. Losi, W. Gärtner, and K. Jaeger. 2007. Reporter proteins for in vivo fluorescence without oxygen. Nat. Biotechnol. 25:443-445. [DOI] [PubMed] [Google Scholar]
- 7.Geesey, G., W. Richardson, H. Yeomans, R. Irvin, and J. Costerton. 1977. Microscopic examination of natural sessile bacterial populations from an alpine stream. Can. J. Microbiol. 23:1733-1736. [DOI] [PubMed] [Google Scholar]
- 8.Hansen, M., R. J. Palmer, C. Udsen, D. White, and S. Molin. 2001. Assessment of GFP fluorescence in cells of Streptococcus gordonii under conditions of low pH and low oxygen concentration. Microbiology 147:1383-1391. [DOI] [PubMed] [Google Scholar]
- 9.Hernandez, D., and J. Rowe. 1985. Gas chromatographic assay for in vitro complementation of Pseudomonas aeruginosa mutants deficient in nitrate reduction. Appl. Environ. Microbiol. 49:24-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heydorn, A., A. Nielsen, M. Hentzer, C. Sternberg, M. Givskov, B. Ersbøll, and S. Molin. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146:2395-2407. [DOI] [PubMed] [Google Scholar]
- 11.James, G., D. Korber, D. Caldwell, and J. Costerton. 1995. Digital image analysis of growth and starvation responses of a surface-colonizing Acinetobacter sp. J. Bacteriol. 177:907-915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lawrence, J., G. Swerhone, G. Leppard, T. Araki, X. Zhang, M. West, and A. Hitchcock. 2003. Scanning transmission X-ray, laser scanning, and transmission electron microscopy mapping of the exopolymeric matrix of microbial biofilms. Appl. Environ. Microbiol. 69:5543-5554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maseda, H., K. Saito, A. Nakajima, and T. Nakae. 2000. Variation of the mexT gene, a regulator of the MexEF-oprN efflux pump expression in wild-type strains of Pseudomonas aeruginosa. FEMS Microbiol. Lett. 192:107-112. [DOI] [PubMed] [Google Scholar]
- 14.Maseda, H., I. Sawada, K. Saito, H. Uchiyama, T. Nakae, and N. Nomura. 2004. Enhancement of the mexAB-oprM efflux pump expression by a quorum-sensing autoinducer and its cancellation by a regulator, MexT, of the mexEF-oprN efflux pump operon in Pseudomonas aeruginosa. Antimicrob. Agents. Chemother. 48:1320-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McLean, R., C. Bates, M. Barnes, C. McGowin, and G. Aron. 2004. Methods of studying biofilms, p. 379-413. In M. Ghannoum and G. A. O'Toole (ed.), Microbial biofilms. ASM Press, Washington, DC.
- 16.Nicholas, D. J. D., and A. Nason. 1957. Determination of nitrate and nitrite. Methods Enzymol. 3:981-984. [Google Scholar]
- 17.Paddock, S. 2002. Confocal reflection microscopy. BioTechniques 32:274-278. [PubMed] [Google Scholar]
- 18.Palmer, R. J. 1999. Microscopy flow cells: perfusion chambers for real-time study of biofilms. Methods Enzymol. 310:160-166. [DOI] [PubMed] [Google Scholar]
- 19.Remington, S. 2006. Fluorescent proteins: maturation, photochemistry and photophysics. Curr. Opin. Struct. Biol. 16:714-721. [DOI] [PubMed] [Google Scholar]
- 20.Riedel, K., M. Hentzer, O. Geisenberger, B. Huber, A. Steidle, H. Wu, N. Høiby, M. Givskov, S. Molin, and L. Eberl. 2001. N-Acylhomoserine-lactone-mediated communication between Pseudomonas aeruginosa and Burkholderia cepacia in mixed biofilms. Microbiology 147:3249-3262. [DOI] [PubMed] [Google Scholar]
- 21.Rittman, B. 2004. Biofilms in water industry, p. 359-378. In M. Ghannoum and G. A. O'Toole (ed.), Microbial biofilms. ASM Press, Washington, DC.
- 22.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 23.Simon, R., M. O'Connell, M. Labes, and A. Pühler. 1986. Plasmid vectors for the genetic analysis and manipulation of rhizobia and other gram-negative bacteria. Methods Enzymol. 118:640-659. [DOI] [PubMed] [Google Scholar]
- 24.Song, B., and L. Leff. 2006. Influence of magnesium ions on biofilm formation by Pseudomonas fluorescens. Microbiol. Res. 161:355-361. [DOI] [PubMed] [Google Scholar]
- 25.Sternberg, C., B. Christensen, T. Johansen, A. Toftgaard Nielsen, J. Andersen, M. Givskov, and S. Molin. 1999. Distribution of bacterial growth activity in flow-chamber biofilms. Appl. Environ. Microbiol. 65:4108-4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Toyofuku, M., N. Nomura, T. Fujii, N. Takaya, H. Maseda, I. Sawada, T. Nakajima, and H. Uchiyama. 2007. Quorum sensing regulates denitrification in Pseudomonas aeruginosa PAO1. J. Bacteriol. 189:4969-4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Werner, E., F. Roe, A. Bugnicourt, M. Franklin, A. Heydorn, S. Molin, B. Pitts, and P. Stewart. 2004. Stratified growth in Pseudomonas aeruginosa biofilms. Appl. Environ. Microbiol. 70:6188-6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamamoto, M., H. Murai, A. Takeda, S. Okunishi, and H. Morisaki. 2005. Bacterial flora of the biofilm formed on the submerged surface of the reed Phragmites australis. Microbes Environ. 20:14-24. [Google Scholar]
- 29.Yoon, S., R. Hennigan, G. Hilliard, U. Ochsner, K. Parvatiyar, M. Kamani, H. Allen, T. DeKievit, P. Gardner, U. Schwab, J. Rowe, B. Iglewski, T. McDermott, R. Mason, D. Wozniak, R. Hancock, M. Parsek, T. Noah, R. Boucher, and D. Hassett. 2002. Pseudomonas aeruginosa anaerobic respiration in biofilms: relationships to cystic fibrosis pathogenesis. Dev. Cell 3:593-603. [DOI] [PubMed] [Google Scholar]
- 30.Zhu, G., Y. Peng, B. Li, J. Guo, Q. Yang, and S. Wang. 2008. Biological removal of nitrogen from wastewater. Rev. Environ. Contam. Toxicol. 192:159-195. [DOI] [PubMed] [Google Scholar]
- 31.Zumft, W. 1997. Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 61:533-561. [DOI] [PMC free article] [PubMed] [Google Scholar]