Abstract
The diversity of type I modular polyketide synthase (PKS) was explored by PCR amplification of DNA encoding ketosynthase and acyltransferase domains in myxobacteria. The sequencing of the amplicons revealed that many PKS genes were distantly related to the published sequences. Thus, myxobacteria may be excellent resources for novel and diverse polyketides.
Myxobacteria are gram-negative, rod-shaped, gliding bacteria with a high G+C content whose unique characteristic is the process of multicellular development that leads to fruiting body formation. Furthermore, myxobacteria are considered to be a rich source of antibiotics that are rarely produced by other microorganisms. Interestingly, the majority of bioactive compounds isolated from myxobacteria contain polyketide structures (1, 2, 22). Therefore, myxobacterial strains seem to have many novel polyketide synthase (PKS) genes and to produce hitherto unknown polyketides. While the presence of novel PKS genes in an organism is a good indicator of the production of novel polyketide molecules in this organism, reports on analyses of PKS genes in taxonomically diverse myxobacterial strains have been limited (17). Here, we characterized PKS genes in various myxobacterial strains of nine different genera, which were derived not only from terrestrial but also from marine environments.
PCR amplification of PKS genes in myxobacteria.
Two primer sets, 5′-GCSATGGAYCCSCARCARCGSVT-3′/5′-GTSCCSGTSCCRTGSSCYTCSAC-3′, reported by Schirmer et al. (23), and 5′-TTCSTSTTYMCSGGVCAGG-3′/5′-GSGGGCYSABYTCSABGAA-3′, designed in this study, with conserved regions in mta (25), cta (4), mel (28), epo (27), sti (8), tub (GenBank accession no. EAU69663), mch (GenBank accession no. AJ698723), and chi (21) were used for the amplification of DNA encoding the ketosynthase (KS) and acyltransferase (AT) domains, respectively. The reaction mixture contained a 0.2 mM concentration of each deoxynucleoside triphosphate, 0.5% (vol/vol) dimethyl sulfoxide, a 1 μM concentration of degenerate primers, 12.5 ng/μl of genomic DNA, and 0.025 units/μl of EX-Taq Hot Start version polymerase (TaKaRa Bio, Shiga, Japan) in 1× EX-Taq PCR buffer. Amplification of KS sequences was performed with an initial denaturation step (94°C, 5 min), followed by 25 cycles of denaturation (94°C, 30 s), annealing (66°C, 30 s), and extension (72°C, 1 min). A final extension was performed at 72°C for 5 min. For the amplification of AT sequences, 35 cycles of denaturation (94°C, 30 s), annealing (64°C, 30 s), and extension (72°C, 1 min) were employed instead of the 25 cycles used in the PCR amplification of KS. KS and AT DNAs (about 680 bp and 810 bp, respectively) were amplified from all 20 strains listed in Table 1. The PCR products were cloned, and the sequences were compared to the published sequences by performing a BLAST search against sequences in the GenBank/EMBL/DDBJ databases. For convenience, they were classified into two categories: “known genes,” exhibiting significant similarity (>70% identity of amino acid sequences) to published sequences, and “novel genes,” exhibiting less significant similarity (<70%) to the published sequences. The cutoff value of 70% to separate “known” and “novel” PKS genes was chosen because the amino acid sequences of KS domains involved in the synthesis of structurally related polyketide molecules were, in almost all cases, more than 70% identical to each other in Streptomyces (our unpublished observations).
TABLE 1.
Tested myxobacterial strains and numbers of PKS sequences obtained in this study
| Taxonomic group | Strain/product(s) (reference[s])a | Isolation origin | No. of PKS sequencesb
|
|
|---|---|---|---|---|
| Novel | Known | |||
| Terrestrial Cystobacterineae | Myxococcus xanthus ATCC 25232T (3) | Soil (from ATCC) | 9 | |
| Myxococcus sp. strain M1017/myxothiazol | Soil, Kanagawa, Japan | 6 | 10 | |
| Myxococcus flavescens AJ 12298/myxovirescin (19, 26) | Soil, Kanagawa, Japan | 2 | 7 | |
| Myxococcus stipitatus AJ 12587/phenalamid (14) | Soil, Kanagawa, Japan | 3 | 7 | |
| Cystobacter fuscus AJ 13278/cystothiazole (20), myxalamid (16) | Soil, Kanagawa, Japan | 1 | 10 | |
| Melittangium lichenicola ATCC 25946 | Soil (from ATCC) | 3 | 7 | |
| Stigmatella aurantiaca ATCC 25190T | Soil (from ATCC) | 2 | 6 | |
| Terrestrial Sorangiineae | Chondromyces apiculatus HT-1 | Goat feces, Okinawa, Japan | 6 | 1 |
| Sorangium cellulosum KU-4/disorazol | Soil, Tokyo, Japan | 6 | 13 | |
| S. cellulosum IS-3/ambruticin | Soil, Yokohama, Japan | 1 | 6 | |
| S. cellulosum IS-4 | Soil, Yokohama, Japan | 3 | ||
| S. cellulosum YA-2/epothilone, spirangien | Soil, Yokohama, Japan | 6 | 9 | |
| S. cellulosum EW4/epothilone | Dead angleworm, Tokyo, Japan | 3 | 11 | |
| Sorangineae myxobacteria HB-1 | Bacterial mat, Hakuba Spring, Japan | 1 | 1 | |
| Marine Sorangiineae | Plesiocystis pacifica SIR-1T (10) | Seagrass, Iriomote Island, Japan | 3 | |
| Plesiocystis sp. strain SIS-2 | Sea sand, Ishigaki Island, Japan | 7 | ||
| Enhygromyxa salina SHK-1T (11) | Sludge, Saroma Lake, Japan | 5 | ||
| Enhygromyxa sp. strain SMH-02-3/nannochelin | Sea sand, Miura Peninsula, Japan | 7 | ||
| Enhygromyxa sp. strain SYM-1 | Sea sand, Izu Peninsula, Japan | 5 | ||
| Haliangium tepidum SMP-10T (7) | Seagrass, Miura Peninsula, Japan | 10 | ||
Strains: from the culture collection of Ajinomoto Co., Inc., Japan. Product: identified in the joint research by Ajinomoto Co., Inc. and Nagoya University.
Numbers of non-redundant sequences in each strain. Novel: identity to the best match of BLAST search was <70%. Known: >70% identity.
Among the strains used in this study, nine have been observed to produce 10 different polyketides (Table 1). Among the 10 polyketides, biosynthesis genes have been identified for 7 polyketides, namely, myxothiazol (mta), cystothiazole (cta), myxalamid (mxa/mmx) (5, 24), disorazol (dis/dsz) (2, 15), ambruticin (amb) (13), epothilone (epo), and spirangien (spi) (6). As expected, sequences similar to these genes were found in the corresponding producers (data not shown).
Novel PKS genes observed in myxobacteria.
In addition to the known PKS genes described so far, many PKS gene sequences whose sequence identities to known genes are less than 70% were also found in this analysis. All PKS genes detected in the Myxococcus xanthus type strain were almost identical (>98% identity) to those in M. xanthus DK 1622, whose genome project has been completed (9), and therefore no novel PKS gene was found. On the other hand, all 19 of the other strains possessed novel PKS genes (Table 1).
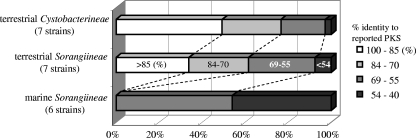
The percentages of novel genes in terrestrial Cystobacterineae and terrestrial Sorangiineae strains were 24% and 40%, respectively, while that in marine Sorangiineae was 100% (Fig. 1). Furthermore, the sequence similarities of the PKS genes found in the marine strains were quite low compared to those found in terrestrial strains: almost half of the marine PKS genes exhibited less than 55% identity to the reported sequences.
FIG. 1.
Percentages of novel PKS sequences in myxobacteria of different categories.
Phylogenetic diversity of PKS sequences.
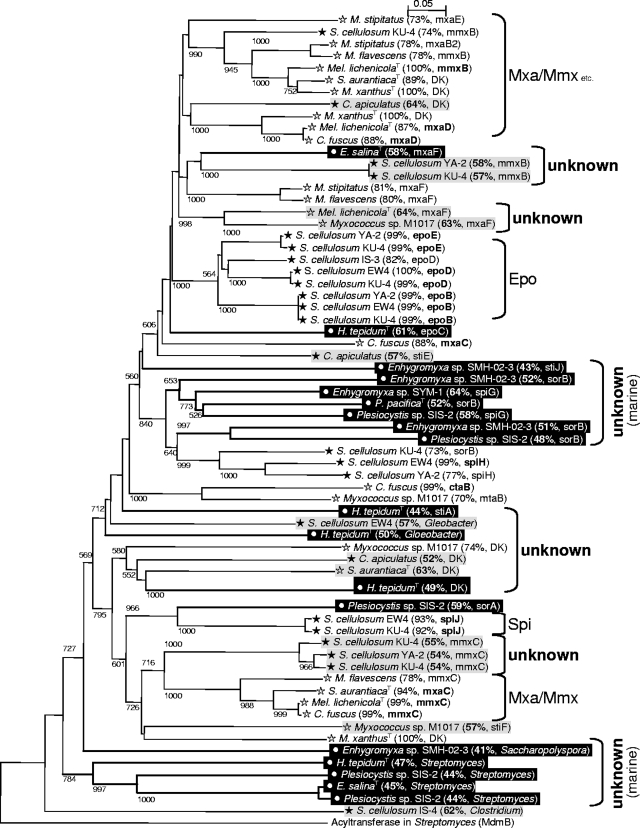
The phylogenetic relationship of myxobacterial PKS sequences was reconstructed by the NJ method (Fig. 2; see Fig. S1 in the supplemental material). Most of the sequences detected in Sorangium cellulosum EW-4 were similar to those in S. cellulosum YA-2. Often, but not always, PKS in one strain of Cystobacterineae was similar to that in another strain of Cystobacterineae; however, most other sequences were nonredundant, indicating that different strains have different PKS genes.
FIG. 2.
Phylogenetic trees of of AT domains in PKSs from myxobacteria. Bootstrap values of >500 calculated from 1,000 bootstrap trees (neighborhood-joining algorithm) are indicated at the nodes. Bars indicate 5% amino acid sequence divergence. Identity to the best-matched PKS genes (or their genomes) found in the BLAST search are indicated in brackets. DK, PKS genes in the genome of M. xanthus DK 1622 (9). Sequences with <70% identity are shaded in gray or black. Symbols: open star, terrestrial Cystobacterineae; filled star, terrestrial Sorangiineae; open circle (and black background), marine Sorangiineae.
PKS sequences were organized into many clades. In addition to the clades of reported PKS genes, many novel clades were constructed. These clades were often composed of only Cystobacterineae sequences or only Sorangiineae sequences, suggesting the relationship between the taxonomy and the distribution of PKS genes, although some PKS clades included sequences derived from different suborder strains. In general, the correlation between the phylogenetic positions based on 16S rRNA gene sequences and those based on PKS genes is low, and the distribution of PKS genes in different bacterial strains is often explained by horizontal gene transfer (12, 18). However, in myxobacteria, horizontal gene transfer between different suborders might not be so frequent, and different taxa of myxobacteria seem to possess different PKS genes.
Interestingly, some marine Sorangiineae sequences formed marine-specific clades which were well separated from the sequences of terrestrial strains (Fig. 2). Such phylogenetically novel clades found in this study would be especially interesting for exploring PKS genes involved in the synthesis of novel metabolites. The remarkable novelty and diversity of PKS genes in marine strains indicate the importance of marine myxobacteria as sources for exploring novel polyketide compounds.
Nucleotide sequence accession numbers.
DNA sequences obtained in this study were deposited in the DDBJ under accession numbers AB376371 to AB376541.
Supplementary Material
Acknowledgments
This work was supported by a grant from the New Energy and Industrial Technology Development Organization, Japan (P02038).
Footnotes
Published ahead of print on 7 July 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Bode, H. B., and R. Muller. 2006. Analysis of myxobacterial secondary metabolism goes molecular. J. Ind. Microbiol. Biotechnol. 33:577-588. [DOI] [PubMed] [Google Scholar]
- 2.Carvalho, R., R. Reid, N. Viswanathan, H. Gramajo, and B. Julien. 2005. The biosynthetic genes for disorazoles, potent cytotoxic compounds that disrupt microtubule formation. Gene 359:91-98. [DOI] [PubMed] [Google Scholar]
- 3.Dworkin, M. 1962. Nutritional requirements for vegetative growth of Myxococcus xanthus. J. Bacteriol. 84:250-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feng, Z., J. Qi, T. Tsuge, Y. Oba, T. Kobayashi, Y. Suzuki, Y. Sakagami, and M. Ojika. 2005. Construction of a bacterial artificial chromosome library for a myxobacterium of the genus Cystobacter and characterization of an antibiotic biosynthetic gene cluster. Biosci. Biotechnol. Biochem. 69:1372-1380. [DOI] [PubMed] [Google Scholar]
- 5.Feng, Z., J. Qi, T. Tsuge, Y. Oba, Y. Sakagami, and M. Ojika. 2006. Biosynthesis of 2′-O-methylmyxalamide D in the myxobacterium Cystobacter fuscus: a polyketide synthase-nonribosomal peptide synthetase system for the myxalamide D skeleton and a methyltransferase for the final O-methylation. Biosci. Biotechnol. Biochem. 70:699-705. [DOI] [PubMed] [Google Scholar]
- 6.Frank, B., J. Knauber, H. Steinmetz, M. Scharfe, H. Blocker, S. Beyer, and R. Muller. 2007. Spiroketal polyketide formation in Sorangium: identification and analysis of the biosynthetic gene cluster for the highly cytotoxic spirangienes. Chem. Biol. 14:221-233. [DOI] [PubMed] [Google Scholar]
- 7.Fudou, R., Y. Jojima, T. Iizuka, and S. Yamanaka. 2002. Haliangium ochraceum gen. nov., sp. nov. and Haliangium tepidum sp. nov.: novel moderately halophilic myxobacteria isolated from coastal saline environments. J. Gen. Appl. Microbiol. 48:109-116. [DOI] [PubMed] [Google Scholar]
- 8.Gaitatzis, N., B. Silakowski, B. Kunze, G. Nordsiek, H. Blocker, G. Hofle, and R. Muller. 2002. The biosynthesis of the aromatic myxobacterial electron transport inhibitor stigmatellin is directed by a novel type of modular polyketide synthase. J. Biol. Chem. 277:13082-13090. [DOI] [PubMed] [Google Scholar]
- 9.Goldman, B. S., W. C. Nierman, D. Kaiser, S. C. Slater, A. S. Durkin, J. A. Eisen, C. M. Ronning, W. B. Barbazuk, M. Blanchard, C. Field, C. Halling, G. Hinkle, O. Iartchuk, H. S. Kim, C. Mackenzie, R. Madupu, N. Miller, A. Shvartsbeyn, S. A. Sullivan, M. Vaudin, R. Wiegand, and H. B. Kaplan. 2006. Evolution of sensory complexity recorded in a myxobacterial genome. Proc. Natl. Acad. Sci. USA 103:15200-15205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iizuka, T., Y. Jojima, R. Fudou, A. Hiraishi, J. W. Ahn, and S. Yamanaka. 2003. Plesiocystis pacifica gen. nov., sp. nov., a marine myxobacterium that contains dihydrogenated menaquinone, isolated from the Pacific coasts of Japan. Int. J. Syst. Evol. Microbiol. 53:189-195. [DOI] [PubMed] [Google Scholar]
- 11.Iizuka, T., Y. Jojima, R. Fudou, M. Tokura, A. Hiraishi, and S. Yamanaka. 2003. Enhygromyxa salina gen. nov., sp. nov., a slightly halophilic myxobacterium isolated from the coastal areas of Japan. Syst. Appl. Microbiol. 26:189-196. [DOI] [PubMed] [Google Scholar]
- 12.Jenke-Kodama, H., A. Sandmann, R. Muller, and E. Dittmann. 2005. Evolutionary implications of bacterial polyketide synthases. Mol. Biol. Evol. 22:2027-2039. [DOI] [PubMed] [Google Scholar]
- 13.Julien, B., Z. Q. Tian, R. Reid, and C. D. Reeves. 2006. Analysis of the ambruticin and jerangolid gene clusters of Sorangium cellulosum reveals unusual mechanisms of polyketide biosynthesis. Chem. Biol. 13:1277-1286. [DOI] [PubMed] [Google Scholar]
- 14.Kim, Y. J., K. Furihata, S. Yamanaka, R. Fudo, and H. Seto. 1991. Isolation and structural elucidation of stipiamide, a new antibiotic effective to multidrug-resistant cancer cells. J. Antibiot. (Tokyo) 44:553-556. [DOI] [PubMed] [Google Scholar]
- 15.Kopp, M., H. Irschik, S. Pradella, and R. Muller. 2005. Production of the tubulin destabilizer disorazol in Sorangium cellulosum: biosynthetic machinery and regulatory genes. Chembiochem 6:1277-1286. [DOI] [PubMed] [Google Scholar]
- 16.Kundim, B. A., Y. Itou, Y. Sakagami, R. Fudou, S. Yamanaka, and M. Ojika. 2004. Novel antifungal polyene amides from the myxobacterium Cystobacter fuscus: isolation, antifungal activity and absolute structure determination. Tetrahedron 60:10217-10221. [Google Scholar]
- 17.Li, Z. F., J. Y. Zhao, Z. J. Xia, J. Shi, H. Liu, Z. H. Wu, W. Hu, W. F. Liu, and Y. Z. Li. 2007. Evolutionary diversity of ketoacyl synthases in cellulolytic myxobacterium Sorangium. Syst. Appl. Microbiol. 30:189-196. [DOI] [PubMed] [Google Scholar]
- 18.Lopez, J. V. 2003. Naturally mosaic operons for secondary metabolite biosynthesis: variability and putative horizontal transfer of discrete catalytic domains of the epothilone polyketide synthase locus. Mol. Genet. Genomics 270:420-431. [DOI] [PubMed] [Google Scholar]
- 19.Miyashiro, S., S. Yamanaka, S. Takayama, and H. Shibai. 1988. Novel macrocyclic antibiotics: megovalicins A, B, C, D, G and H. I. Screening of antibiotics-producing myxobacteria and production of megovalicins. J. Antibiot. (Tokyo) 41:433-438. [DOI] [PubMed] [Google Scholar]
- 20.Ojika, M., Y. Suzuki, A. Tsukamoto, Y. Sakagami, R. Fudou, T. Yoshimura, and S. Yamanaka. 1998. Cystothiazoles A and B, new bithiazole-type antibiotics from the myxobacterium Cystobacter fuscus. J. Antibiot. (Tokyo) 51:275-281. [DOI] [PubMed] [Google Scholar]
- 21.Perlova, O., K. Gerth, O. Kaiser, A. Hans, and R. Muller. 2006. Identification and analysis of the chivosazol biosynthetic gene cluster from the myxobacterial model strain Sorangium cellulosum So ce56. J. Biotechnol. 121:174-191. [DOI] [PubMed] [Google Scholar]
- 22.Reichenbach, H. 2001. Myxobacteria, producers of novel bioactive substances. J. Ind. Microbiol. Biotechnol. 27:149-156. [DOI] [PubMed] [Google Scholar]
- 23.Schirmer, A., R. Gadkari, C. D. Reeves, F. Ibrahim, E. F. DeLong, and C. R. Hutchinson. 2005. Metagenomic analysis reveals diverse polyketide synthase gene clusters in microorganisms associated with the marine sponge Discodermia dissoluta. Appl. Environ. Microbiol. 71:4840-4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Silakowski, B., G. Nordsiek, B. Kunze, H. Blocker, and R. Muller. 2001. Novel features in a combined polyketide synthase/non-ribosomal peptide synthetase: the myxalamid biosynthetic gene cluster of the myxobacterium Stigmatella aurantiaca Sga15. Chem. Biol. 8:59-69. [DOI] [PubMed] [Google Scholar]
- 25.Silakowski, B., H. U. Schairer, H. Ehret, B. Kunze, S. Weinig, G. Nordsiek, P. Brandt, H. Blocker, G. Hofle, S. Beyer, and R. Muller. 1999. New lessons for combinatorial biosynthesis from myxobacteria. The myxothiazol biosynthetic gene cluster of Stigmatella aurantiaca DW4/3-1. J. Biol. Chem. 274:37391-37399. [DOI] [PubMed] [Google Scholar]
- 26.Takayama, S., S. Yamanaka, S. Miyashiro, Y. Yokokawa, and H. Shibai. 1988. Novel macrocyclic antibiotics: megovalicins A, B, C, D, G and H. II. Isolation and chemical structures of megovalicins. J. Antibiot. (Tokyo) 41:439-445. [DOI] [PubMed] [Google Scholar]
- 27.Tang, L., S. Shah, L. Chung, J. Carney, L. Katz, C. Khosla, and B. Julien. 2000. Cloning and heterologous expression of the epothilone gene cluster. Science 287:640-642. [DOI] [PubMed] [Google Scholar]
- 28.Weinig, S., H. J. Hecht, T. Mahmud, and R. Muller. 2003. Melithiazol biosynthesis: further insights into myxobacterial PKS/NRPS systems and evidence for a new subclass of methyl transferases. Chem. Biol. 10:939-952. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.