Abstract
Viruses induce pathogenic symptoms on plants but the molecular basis is poorly understood. Here, we show that transgenic Arabidopsis expressing the pathogenesis protein βC1 of Tomato yellow leaf curl China virus (TYLCCNV), a geminivirus, can phenocopy to a large extent disease symptoms of virus-infected tobacco plants in having upward curled leaves, radialized leaves with outgrowth tissues from abaxial surfaces, and sterile flowers. These morphological changes are paralleled by a reduction in miR165/166 levels and an increase in PHB and PHV transcript levels. Two factors, ASYMMETRIC LEAVES 1 (AS1) and ASYMMETRIC LEAVES 2 (AS2), are known to regulate leaf development as AS1/AS2 complex. Strikingly, βC1 plants phenocopy plants overexpressing AS2 at the morphological and molecular level and βC1 is able to partially complement as2 mutation. βC1 binds directly to AS1, elicits morphological and gene expression changes dependent on AS1 but not AS2, and attenuates expression of selective jasmonic acid (JA)-responsive gene. Our results show that βC1 forms a complex with AS1 to execute its pathogenic functions and to suppress a subset of JA responses.
Keywords: Geminivirus, asymmetric leaves 1, asymmetric leaves 2, jasmonic acid, silencing suppressor, whitefly
Host–virus interactions often involve defense mechanisms implemented by the host and counterdefense strategies developed by the successful, offending pathogen. To combat against virus infection, hosts have evolved signaling pathways to sense invading viruses and trigger appropriate defense responses. However, as virulent pathogens, viruses have developed counterdefense strategies that can suppress host defense system and usurp host cellular resources leading to disease symptoms. In mammals, for example, viruses can avoid host immune surveillance by triggering the down-regulation MHC class I molecules through the ubiquitin–proteasome system (Gao and Luo 2006). In other cases, viruses use molecular mimicry to manipulate host signaling pathway—e.g., the Notch and Wnt pathways—to up-regulate their own gene expression and weaken host cell defense responses (Hayward et al. 2006). The identification and elucidation of these host–virus interactions provide clues about the molecular basis of viral susceptibility and disease symptoms and also shed light on the regulation of host signaling pathways.
Following viral infections plants usually display disease symptoms such as developmental defects, chlorosis, and even necrosis (Hull 2001). The development of these disease symptoms require specific interactions between viral and plant components to disrupt physiological and developmental processes (Culver and Padmanabhan 2007). Although a number of virus–plant interactions have been characterized, few plant receptors for viral components have yet been identified and the molecular basis of disease symptom development remains largely unknown.
Recent studies indicate that viral RNA silencing suppressors, which were previously known to play important roles in viral infection and pathogenesis, appear to be responsible for a significant proportion of morphological and developmental alterations of host plants seen after virus infection (Li and Ding 2006). In plants, post-transcriptional gene silencing (PTGS) is an important antiviral response that suppresses viral gene expression through siRNA-mediated viral RNA degradation. To counteract the host PTGS response, viruses encode various suppressors of RNA silencing targeting different key steps of the host defense pathway. Because of the biochemical similarities between PTGS and the endogenous miRNA pathway, which regulates plant development, viral suppressors also interfere with the latter, thereby causing alterations in plant development (Kasschau et al. 2003).
Most characterized plant viral suppressors are derived from RNA viruses, although plant DNA viruses also encode suppressors and some of them have been characterized (Bisaro 2006). An example is βC1, an RNA silencing suppressor, encoded by the οnly ORF located on DNAβ of the Tomato yellow leaf curl China virus (TYLCCNV) (Cui et al. 2005). In addition to blocking PTGS, this viral suppressor can enhance DNA-A accumulation and induce disease symptoms on host plants including leaf curling, enations, shoot bending, vein thickening, and stunting (Cui et al. 2004). As the plant receptor for βC1 has not yet been identified, the mechanism by which this key pathogenesis protein elicits developmental abnormalities on host plants remains obscure.
The majority of plant viruses are vectored by insects and the tripartite relationship between virus, insect vector, and host plant is a subject of intense investigations (Stout et al. 2006). Several groups have reported increased aggregation of insect vectors on virus-infected plants (Maris et al. 2004; Belliure et al. 2005). In the case of insect vector for begomoviruses, Jiu et al. (2007) showed that type B whiteflies have developed mutualistic relationships with satellite DNAβ-associated TYLCCNV and TbCSV to improve its performance on virus-infected plants. This mutualism has apparently accelerated an increase in type B whitefly population, which promoted virus spreading and the occurrence of DNAβ-associated disease complex. Other studies have implicated jasmonic acid (JA) pathways in mediating plant defenses against insects (de Vos 2007; Howe and Jander 2008). However, it is not known whether attraction of insect vectors to virus-infected plants entails any attenuation in host JA responses, and if it does, which viral protein(s) are involved in causing such changes.
Here, we show that the TYLCCNV pathogenesis factor βC1 interacts with ASYMMETRIC LEAVES 1 (AS1) to cause alterations in leaf development resulting in the manifestation of disease symptoms. AS1 is needed for βC1 function as changes in leaf morphology elicited by this viral factor is largely attenuated in as1 mutant. Amazingly, βC1 is able to partially complement as2 mutation suggesting that βC1 is a molecular mimic of ASYMMETRIC LEAVES 2 (AS2). Finally, we showed that βC1 can suppress expression of several JA-responsive genes that are implicated in plant defenses against insects. These results advance our understanding of the molecular basis of disease symptoms elicited by viruses and provide fresh insights into the tripartite relationships between virus, insect, and host plant.
Results
Developmental defects induced by βC1
Nicotiana benthamiana plants coinfected with TYLCCNV and DNAβ displayed severe symptoms including curled leaves, bending shoots, and enations from abaxial leaf surfaces (Fig. 1A–D). Previous work has shown that these phenotypes are elicited by the viral pathogenesis factor, βC1 (Cui et al. 2004). Because Arabidopsis cannot be infected with TYLCCNV, we transferred the entire DNAβ that encodes only the βC1 protein into Arabidopsis. Transgenic plants expressing DNAβ displayed curled leaves and outgrowth tissues from abaxial leaves similar to the disease symptoms of tobacco plants infected with TYLCCNV and DNAβ (Fig. 1E; Supplemental Fig. S1A). These morphological alterations were also recapitulated in transgenic Arabidopsis plants expressing βC1 either from its native promoter (Supplemental Fig. S1B,C) or a 35S promoter (Fig. 1H). Together, these results indicate that the morphological alterations seen in Arabidopsis phenocopy to a large extent disease symptoms of tobacco coinfected with TYLCCNV and DNAβ, and can be attributed to βC1 expression. Moreover, compared with the flattened cotyledons in wild-type Arabidopsis (Supplemental Fig. S1G), C1p:βC1 as well as 35S:βC1 plants produced narrow and upward-curled cotyledons with outgrowth tissues from hypocotyls (Supplemental Fig. S1E,F). These results indicate that Arabidopsis can be used as a model system to investigate the mechanism of action of βC1.
Figure 1.
Symptoms of N. benthamiana plants infected with TYLCCNV and phenotypes of βC1 and AS2 transgenic plants. (A–D) Symptoms of N. benthamiana plants infected with TYLCCNV plus DNAβ. (A) Uninfected control N. benthamiana plant. (B) N. benthamiana plants infected with TYLCCNV plus DNAβ. Arrowhead shows the bending shoot. (C) Adaxial side of upward-curled leaves. (D) Abaxial side of upward-curled leaves. Arrowhead indicates enation tissues on the abaxial leaf surface. (E–L) Phenotypes of βC1 transgenic plants expressed from various promoters. (E) Severe phenotypes of 21-d-old pBINplus-2β transgenic plants with radial leaves and outgrowth tissues on the abaxial surface of leaves. Arrowhead indicates outgrowth tissues from abaxial leaf surfaces. (F) Twenty-eight-day-old class I 35S:HA-βC1 transgenic plants. Arrowhead indicates outgrowth tissues. (G) Phenotypic variability in leaves of different 35S:HA-βC1 lines. (H) Phenotypes of 28-d-old 35S:βC1 transgenic plant with radial leaves and outgrowth tissues on the abaxial surface of leaves. Arrowhead indicates outgrowth tissues from abaxial leaf surface. (I–L) Class II 35S:HA-βC1 transgenic plants. Note the altered inflorescence architecture with downward-pointed flowers (J), downward-pointed siliques (K). and bending nodes (L). (M–O) Phenotypes of AS2 transgenic plants expressed from various promoters. (M) Twenty-one-day-old XVE:AS2-myc transgenic plants with severe phenotypes. Plants were treated with inducer. Arrowhead indicates outgrowth tissues. (N) Twenty-eight-day-old XVE:AS2-myc transgenic plants with mild phenotype. Plants were treated with inducer. (O) 35S:AS2-myc transgenic plants with mild phenotypes having downward-pointed flowers and siliques. (P) Twenty-one-day-old wild-type Col-0 plants. Bars: A,B, 15 mm; C,D,I,N,O, 10 mm; E–H,M,P, 5 mm; J–L 2.5 mm.
We generated 35S:HA-βC1 plants in which βC1 was expressed as a HA fusion protein. Based on the severity of the leaf phenotype on seedlings (Fig. 1G), we divided the transgenic lines into two classes. Class I seedlings showed severe phenotypes with radial leaves and outgrowth tissues from the abaxial leaf surfaces (Fig. 1F; Supplemental Fig. S1D). These seedlings were stunted and leaves were usually narrow and failed to expand. At a later growth stage, some plants did not produce inflorescence, whereas others only produced short inflorescences bearing sterile flowers. Class II seedlings showed mild phenotypes with narrow, upward-curled leaves and long petioles (Fig. 1I; Supplemental Fig. S1H). These plants produced inflorescences with bent nodes (Fig. 1L) and carrying downward-oriented flowers with shorter pedicles (Fig. 1J) and downward-oriented siliques (Fig. 1K).
We analyzed the phenotypes by scanning electron microscopy (SEM). The small leaf-like enations were observed in the abaxial side of both virus-infected tobacco leaves (Supplemental Fig. S1J) and 35S:HA-βC1 transgenic Arabidopsis leaves (Supplemental Fig. S1K). In severe 35S:HA-βC1 plants, small abaxial protrusions appeared to be more common (Supplemental Fig. S1K). To further examine the morphological abnormalities induced by βC1 in Arabidopsis, transverse sections of rosette leaves were performed. Disorganized vascular bundles associated with abnormal cell division were observed in XVE:HA-βC1 transgenic plants (Supplemental Fig. S1M). By contrast, normal vascular bundles were found in wild type (Supplemental Fig. S1L). In wild-type leaves, xylem elements were located on the adaxial side, whereas phloem on the abaxial (Supplemental Fig. S1N). In XVE:HA-βC1 transgenic plants, the vascular polarity was disrupted, with xylem elements forming on both sides of the leaf (Supplemental Fig. S1O,P). Abnormal cell division associated with vascular bundles was also observed in virus-infected tobacco (Saunders et al. 2004).
βC1 transgenic plants phenocopy AS2-overexpressing plants
We noted phenotypic similarities between βC1-overexpressing plants described here and those of AS2-overexpressing plants reported earlier (Iwakawa et al. 2002; Lin et al. 2003; Xu et al. 2003). We confirmed that plants overexpressing AS2 displayed upward-curled leaves and downward-oriented flowers and siliques (Fig. 1O). For further investigations, we generated transgenic plants carrying an inducible XVE:AS2-Myc transgene. Upon treatment with inducer, plants displayed mild phenotypes with upward-curled leaves (Fig. 1N), and severe phenotypes with radial leaves, outgrowth tissues from abaxial leaf surfaces and sterile (Fig. 1M) as compared with wild-type plants (Fig. 1P). The perturbed vascular polarity with xylem on adaxial and abaxial side of leaves in XVE:HA-βC1 transgenic plants (Supplemental Fig. S1O,P) was also observed in 35S:AS2 transgenic plants (Lin et al. 2003). We also generated transgenic plants carrying 35S:HA-βC1 and 35S:AS2 transgenes in N. benthamiana. Both 35S:HA-βC1 and 35S:AS2 transgenic plants displayed upward-curled leaves (Supplemental Fig. S1I). The phenotypic similarities between βC1- and AS2-overexpressing plants suggest that βC1 may have some functions similar to those of AS2.
βC1 represses the accumulation of miR165/166
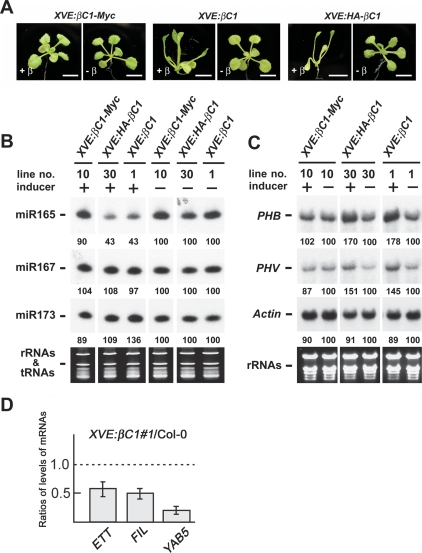
The presence of upward-curled leaves or even radial leaves in βC1 plants suggested that this viral pathogenesis protein may disrupt the formation of adaxial–abaxial polarity in leaves. Leaf polarity is known to be regulated by Class III HD-ZIP genes (McConnell et al. 2001; Emery et al. 2003). HD-ZIP III transcripts, in turn, are regulated at the post-transcriptional level by miRNA 165/166, which mediate cleavage of HD-ZIP III transcripts (Mallory et al. 2004). To investigate the effects of βC1 on the accumulation of miR165/166, we examined miRNA levels in XVE:HA-βC1 and XVE:βC1 plants in which βC1 expression was dependent on the β-estradiol inducer (Zuo et al. 2000). XVE:βC1-myc plants were used as negative control, as no phenotypic changes were observed in these plants upon inducer treatment (Fig. 2A). This was due to the fact that appending a tag at the C terminus of βC1 inactivates its function. By contrast, treatment of XVE:HA-βC1 or XVE:βC1 plants with inducer resulted in the production of upward-curled leaves (Fig. 2A). Figure 2B shows that miR165/166 levels in XVE:HA-βC1 or XVE:βC1 plants were reduced after inducer treatment. The depressed accumulation of miR165/166 was specific, because miR167 was unaltered in the same treated plants and miR173 levels were slightly increased. We also examined PHB and PHV transcript levels. Compared with Actin transcripts, PHV and PHB transcript levels were increased after inducer treatment (Fig. 2C). The accumulation of PHV and PHB transcripts was correlated with the decrease of miRNA 165/166. The same phenomenon has been reported in AS2-overexpressing plants in which the decrease in miR165/166 levels (Ueno et al. 2007) is accompanied by a increase in PHB transcript levels (Lin et al. 2003).
Figure 2.
βC1 reduces accumulation of miR165/166 but increases accumulation of PHB and PHV transcripts. (A) Phenotypes of 18-d-old βC1 transgenic plants treated with 25 μM β-estradiol. Treated XVE:βC1-Myc transgenic plants were like wild-type plants, whereas treated XVE:HA-βC1 and XVE:βC1 transgenic plants showed upward-curled leaves. Bars, 5 mm. (B) Expression of miR165/166, miR167 and miR173 in β-estradiol-treated or untreated seedlings of transgenic plants. Stained rRNAs and tRNAs bands were used as loading controls. (C) PHB, PHV, and Actin transcript levels in β-estradiol-treated or untreated seedlings of XVE:βC1-myc, XVE:HA-βC1, and XVE:βC1 transgenic plants. Stained rRNAs were used as a loading control. Expression levels of miRNAs and mRNAs were calculated using the program of Image Gauge version 3.12 (Fuji) and the values of β-estradiol-treated samples were normalized to untreated samples. (D) Levels of expression of indicated genes in shoot apices of XVE:βC1 plants. Data show expression levels in XVE:βC1 relative to wild-type levels. Levels of the ETT/ARF3, FIL, and YAB5 transcripts in shoot apices of 15-d-old plants were measured by real-time RT–PCR. XVE:βC1 and wild-type plants were grown on MS medium with 25μM β-estradiol. Each value was normalized by reference to the level of ACTIN2 transcripts. Error bars are indicated.
We also examined the transcript levels of ETTIN (ETT)/ARF3, FILAMENTOUS FLOWER (FIL), and YABBY5 (YAB5), which were known to be negatively regulated by AS1 and AS2 (Garcia et al. 2006; Iwakawa et al. 2007). We found that βC1-overexpressing plants reduced levels of ETT/ARF3, FIL, and YAB5 transcripts as was found in AS2-overexpressing plants (Fig. 2D).
βC1 directly interacts with AS1 but not AS2
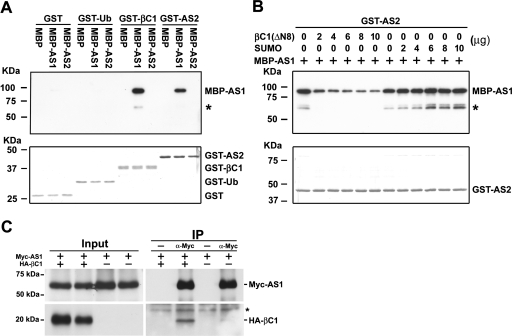
Previous genetic analysis and protein interaction results indicate that AS2 functions together with AS1 to regulate the development of leaf polarity (Lin et al. 2003; Xu et al. 2003). The similar morphological and molecular phenotypes between βC1- and AS2-overexpressing plants prompted us to examine whether βC1 can also directly interact with AS1. To this end, we performed in vitro pull-down assays with purified recombinant proteins. Figure 3A shows that MBP-AS1 was pulled down by GST-βC1 as well as GST-AS2, indicating that AS1 can directly interact with βC1 as well as AS2. By contrast, no signal was observed when the negative control protein GST or GST–Ub was used to pull down MBP–AS1. Moreover, we also found that GST-βC1 specifically interacted with MBP-AS1 but not MBP-AS2, indicating that, like AS2, βC1 can directly associate with AS1.
Figure 3.
βC1 Interacts with AS1 but Not AS2. (A) In vitro pull-down assays. Two micrograms of GST or GST fusion proteins were used to pull down 2 μg of MBP or MBP fusion proteins. (B) Competitive pull-down assays. Indicated protein amounts of βC1(ΔN8) or SUMO were mixed with 2 μg of MBP-AS1 and pulled down by 2 μg of GST-AS2. After being pulled down, Western blottings were performed using anti-MBP antibody to detect the associated proteins. Membranes staining with Coomassie Brilliant Blue were used to monitor input protein amounts. Asterisks indicate degradation products of MBP-AS1. (C) In vivo interaction of βC1 and AS1 in N. benthamiana. Agrobacterium cultures carrying 35S:Myc-AS1 and XVE:HA-βC1 were suspended in 50 μM β-estradiol and coinfiltrated into tobacco leaves. Transiently expressed Myc-AS1 and HA-βC1 were analyzed by coimmunoprecipitation. Crude extracts (Input) were used for immunoprecipitation (IP) with or without polyclonal anti-myc antibody and analyzed by Western blottings. Asterisk indicates the cross-reacting band.
To test the possibility that βC1 may compete with AS2 for binding with AS1 we performed competitive pull-down assays. Because full-length βC1 was insoluble in Escherichia coli when overexpressed, we purified a soluble βC1(ΔN8), in which the second ATG (codon number 9) in the ORF was used as a start codon. Although the βC1(ΔN8) has a deletion of eight amino acids from the N terminus of the βC1 protein, it can still elicit severe symptoms in N. bethamiana when coinfected with TYLCCNV (Cui et al. 2004). This result indicates that the N-terminal deletion has very little effect on βC1 biological activities. Figure 3B shows that the amounts of MBP-AS1 pull downed by GST-AS2 were reduced with increasing amounts of βC1(ΔN8) in the mix. By contrast, increasing amounts of SUMO, a negative control protein, did not affect the recovery of MBP-AS1. These results provide evidence that βC1 competes with AS2 for direct binding to AS1.
We used a tobacco transient expression system to test whether βC1 and AS1 can interact in vivo. Tobacco leaves were infiltrated with Agrobacterial cells carrying XVE:HA-βC1 and 35S:Myc-AS1, and leaves extracts were analyzed by coimmunoprecipitation. Figure 3C shows that the immunoprecipitation of Myc-AS1 pulled down HA-βC1. Although the recovery of HA-βC1 was low, the interaction was clearly specific and dependent on Myc-AS1, as HA-βC1 was not detected in the absence of polycolonal anti-myc antibody.
βC1 functions in Arabidopsis depend on endogenous AS1 activity
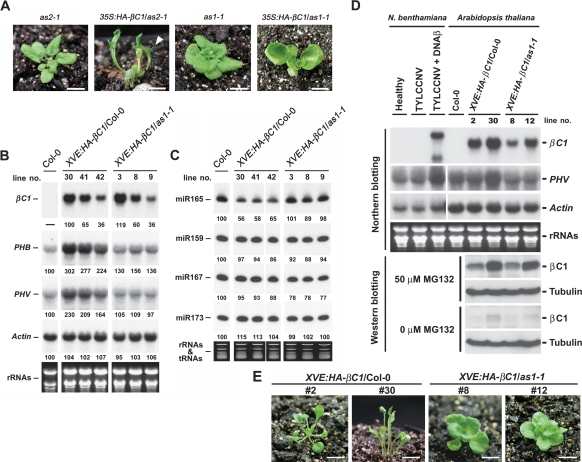
Based on the protein association results, we can propose two possibilities to account for βC1 actions in Arabidopsis. The first possibility is that βC1 may compete with AS2 for interaction with AS1 in vivo. The release of AS2 from the AS1/AS2 complex, and not the βC1/AS1 complex, is, in fact, responsible for the βC1 overexpression phenotypes. To test this hypothesis, we introduced 35S:HA-βC1 into as2-1 mutant (Fig. 4A) background. We found 35S:HA-βC1/as2-1 plants phenocopied 35:HA-βC1/Columbia (Col-0) plants. These phenotypes included upward-curled leaves or even radial leaves (Fig. 4A), outgrowths from abaxial leaf surfaces (Fig. 4A), and downward-oriented flowers (data not shown). These results rule out the possibility that AS2 activity is required for βC1 function in Arabidopsis, and therefore the phenotypes of 35S:HA-βC1 plants were not caused by AS2 released from the AS1/AS2 complex.
Figure 4.
Functions of βC1 in Arabidopsis depend on endogenous AS1 activity. (A) Morphology of mutants and transgenic plants. 35S:HA-βC1/as2-1 transgenic plant with radial leaves and outgrowth tissues on abaxial leaf surfaces. The arrowhead indicates outgrowth tissues. 35S:HA-βC1/as1-1 transgenic plant with mild upward-curled leaves. Bars, for as2-1 and as1-1 mutants, 9 mm; for 35S:HA-βC1/as2-1 and 35S:HA-βC1/as1-1 transgenic plants, 7.5 mm. (B, C) Expression levels of mRNAs and miRNAs in XVE:HA-βC1/Col-0 and XVE:HA-βC1/as1-1 transgenic plants treated with 25 μM β-estradiol. (B) Stained bands of rRNAs were used as a loading control for RNA gel blots. (C) Stained bands of rRNAs and tRNAs were used as a loading control for small RNA gel blots. Expression levels of miRNAs and mRNAs were calculated using the program of Image Gauge version 3.12 (Fuji) and the values (except βC1 transcripts) were normalized to those of the wild-type Col-0 sample. The values of βC1 transcripts were normalized to that of the XVE:HA-βC1/Col-0, line #30 sample. (D) Expression of βC1 transcripts and proteins in tobacco and Arabidopsis. Virus-infected tobacco samples were harvested from systemic leaves. βC1 transgenic plants were grown on MS medium containing 25 μM β-estradiol before being treated with MG132 or harvested for RNA. Specific probes against N. benthamiana and Arabidopsis PHV transcripts were prepared individually. Stained rRNAs bands were used as a loading control. Arabidopsis seedlings were treated with or without MG132 (50 μM) and βC1 proteins in extracts were detected with or without MG132 treatment by anti-HA antibody. Tubulin levels were used as loading control. (E) Phenotypes of 24-d-old βC1 transgenic plants treated with 25 μM β-estradiol. Bars, 7.5 mm.
Another possibility is that βC1 may mimic the functions of AS2 in Arabidopsis. In this case, the active component is the βC1/AS1 complex instead of the native AS1/AS2 complex. To test this second hypothesis, we introduced 35S:HA-βC1 into as1-1 mutant (Fig. 4A) background. We found that the phenotypes of 35S:HA-βC1 were reduced in as1-1 mutant as compared with wild type. Only mild upward-curled leaves with short petioles were seen in 35S:HA-βC1/as1-1 plants (Fig. 4A). Moreover, outgrowth tissues from abaxial leaf surfaces, which were common in 35:HA-βC1/Col-0 plants, were hardly observed in 35S:HA-βC1/as1-1 plants. These results show that the functions of βC1 in Arabidopsis depend to a large extent, but not entirely on the endogenous AS1 activity.
To correlate the phenotypic alterations with molecular changes, we analyzed PHB, PHV, and miR165/166 levels. Because severe lines of 35:HA-βC1/Col-0 transgenic plants were infertile, we used β-estradiol-inducible transgenic plants for these analyses. Figure 4B shows that in wild type, increasing βC1 levels elicited a corresponding increase in PHB and PHV transcript levels, whereas no such increase was seen in the as1-1 mutant background. For comparable βC1 expression levels, PHB and PHV transcript levels were lower in XVE:HA-βC1/as1-1 plants compared with XVE:HA-βC1/Col-0 plants. The expression levels of miR165/166 in XVE:HA-βC1/as1-1 plants were indistinguishable from wild-type plants, but much higher than those in XVE:HA-βC1/Col-0 plants (Fig. 4C).
To further correlate phenotypic alterations with βC1 protein levels, XVE:HA-βC1/Col-0 and XVE:HA-βC1/as1-1 plants were treated with or without MG132, a 26S proteosome inhibitor. Without MG132, βC1 protein was hardly detected (Fig. 4D), but in its presence, βC1 protein accumulated to levels correlating with βC1 transcript levels in both wild-type and as1-1 mutant plants (Fig. 4D). These results show that βC1 protein is highly unstable and degraded by 26S proteosomes in plant cells. Figure 4D shows that although βC1 transcript levels in XVE:HA-βC1/Col-0 #2 and XVE:HA-βC1/as1-1 #12 plants were comparable, βC1 protein accumulated to higher levels in as1-1 mutant (#12) than in wild type (#2). These results suggest that the absence of AS1 may further promote βC1 destabilization. Examination of βC1 protein expression in XVE:HA-βC1/Col-0 plants showed that βC1 protein levels correlated with phenotypic severity (Fig. 4D). Radial leaves with abaxial outgrowth tissues were observed with high βC1 expression levels in line #30 and mild upward-curled leaves were seen with low βC1 expression levels in line #2 (Fig. 4E). By contrast, with comparable βC1 expression levels, XVE:HA-βC1/as1-1 plants showed reduced phenotype with mild upward-curled leaves (Fig. 4E, #30 vs. #12). These results are consistent with the mild phenotype observed in 35S:HA-βC1/as1-1 plants.
βC1 expression levels in inducer-treated XVE:HA-βC1/Col-0 transgenic Arabidopsis plants were comparable with or two times higher than βC1 levels in virus-infected tobacco (Fig. 4D). Note that the length of βC1 transcript in virus-infected tobacco (samples collected from systemic leaves) was longer than those in Arabidopsis βC1 transgenic plants. This could be due to the use of a different 3′ poly A addition signal in the transgenic plants. Consistent with the function of βC1 in elevating PHV transcript levels in XVE:HA-βC1/Col-0 transgenic plants, an increase in PHV transcripts were also observed in tobacco plants infected with TYLCCNV plus DNAβ satellite, but not in plants infected with TYLCCNV alone (Fig. 4D). These results suggest that βC1 has similar functions in tobacco and Arabidopsis, and βC1 is solely responsible for the observed developmental symptoms during normal virus infection.
βC1 partially complements as2 mutation in leaf development
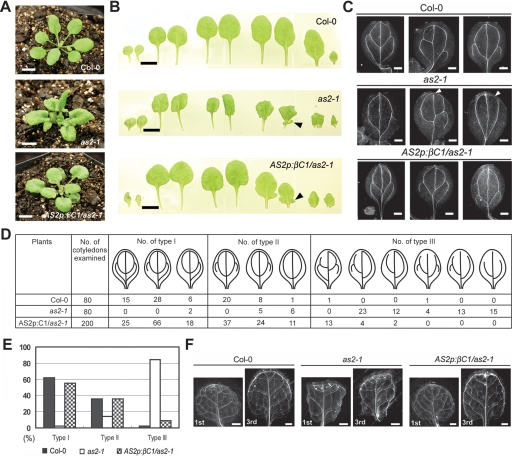
Both βC1 and AS2 overexpression can decrease miR165/166 levels and increase transcript levels of PHB and PHV, which disrupts the formation of adaxial–abaxial polarity in leaves. The morphological and molecular similarities between βC1- and AS2-overexpressing plants prompted us to examine whether βC1 can functionally replace AS2. To test this possibility, βC1 was placed under the control of a native AS2 promoter and introduced into the as2-1 mutant background. More than 30 independent lines of AS2p:βC1/as2-1 transgenic plants were generated. Majority of the lines (80%) showed a partial complementation phenotype with flatten and rounder leaves compared with downward-curled leaves found in as2-1 (Fig. 5A). In lines showing partial complementation, plump and humped lamina at the leaf base was not observed; instead, leaf lobes were still visible (Fig. 5B). The leaf lobes might be related to the ectopic expression of KNAT1 (Chuck et al. 1996; Ori et al. 2000). More than 50% of the partially complemented lines showed mild upward-curled cotyledons, but no curling was seen in rosette leaves (data not shown). Similarly, ∼50% of the partially complemented lines displayed abnormal flowers phenotypes with partially radialized floral organs (data not shown). The flatten leaves in AS2p:βC1/as2-1 indicated that βC1 can supplant AS2 to some extent in regulating the formation of adaxial–abaxial polarity in leaves.
Figure 5.
Partial complementation of as2 by AS2p:βC1. (A, B) Phenotypes of 21-d-old wild-type Col-0, as2-1 and AS2p:βC1/as2-1 transgenic plants. (A) Leaves in AS2p:βC1/as2-1 were flatten and rounder compared with the downward-curled leaves with plump and humped lamina at the leaf base of as2-1. (B) Morphology of cotyledons and rosette leaves. (From left to right) Two cotyledons, first two rosette leaves, second two rosette leaves, third two rosette leaves, and fourth two rosette leaves. Arrowheads indicate leaf lobes. (C–F) Venation patterns of 12-d-old cotyledons and 21-d-old rosette leaves in wild-type Col-0, as2-1 and AS2p:βC1/as2-1 transgenic plants. (C) Dark-field images of venation patterns in cotyledons. Wild-type Col-0 showed 2-4 loops with connection between primary and secondary veins. as2-1 showed 0-2 loops with connection defects in primary and secondary veins. AS2p:βC1/as2-1 transgenic plants showed wild-type venation patterns. Arrowheads indicate connection defect. (D) Patterns of veins were classified into three types according to the number of loops and connection defects in primary and secondary veins. In wild-type Col-0 and AS2p:βC1/as2-1, the largest population contained three loops with five NBPs. In as2-1, the largest population contained one loop with four NBPs and had connection defects. (E) Summary of the distribution of venation patterns in D. The percentage of each venation type in wild-type Col-0, as2-1 and AS2p:βC1/as2-1 was shown in bars with different shades. (F) Dark-field images of venation patterns in first and third rosette leaves. Bars: A,B, 5 mm; C, 0.25 mm; F, 1 mm.
A distinguishing phenotype of as2-1 is the retardation of vein development compared with wild type (Fig. 5C,F). Cotyledons of as2-1 displayed connection defects in primary and secondary veins; the mid-veins are less prominent, and there are fewer lateral veins in rosette leaves. We found the veins defects in as2-1 mutant were restored to wild type in AS2p:βC1/as2-1 transgenic plants (Fig. 5C,F). To facilitate a semiquantitative comparison, we classified the venation patterns in the cotyledons into three types (Fig. 5D). Type I has three to four loops with five to seven NBPs (number of branching points); type II has two loops with two to five NBPs; and type III has connection defects in the primary and secondary veins. We examined 80 wild-type cotyledons. Among them, 62% were type I, 36% type II, and 2% type III. The same numbers of cotyledons (80) were examined in as2-1 mutant. Among them, 2% were type I, 14% type II, and 84% type III. In AS2p:βC1/as2-1, four lines were chosen and 50 cotyledons from each line were examined. The distributions of venation pattern were 55% type I, 36% type II, and 9% type III (Fig. 5D,E). The rescue of vein defects in AS2p:βC1/as2-1 transgenic lines provide evidence that βC1 has similar functions as AS2 in regulating the formation of veins in leaves.
AS2 has two distinct and unrelated functions: the repression of KNOX gene and the regulation of adaxial–abaxial polarity (Lin et al. 2003). The appearance of leaf lobes in AS2p:βC1/as2-1 transgenic plants suggested that βC1 cannot complement the functions of AS2 with respect to KNOX gene repression. To examine the effects of βC1 on the expression pattern of KNAT1, XVE:HA-βC1 was introduced into KNAT1:GUS transgenic plants. Upon inducer treatment, XVE:HA-βC1/KNAT1:GUS transgenic plants still maintained strong GUS staining intensity in shoot meristems, and weak GUS staining intensity was detected in cotyledonary veins (Supplemental Fig. S2). These observations support the partial complementation phenotypes in AS2p:βC1/as2-1 transgenic plants, and suggest that βC1 and AS2 functions may only partially overlap in the establishment of adaxial–abaxial polarity of leaves and in vein development.
JA responses of βC1 plants
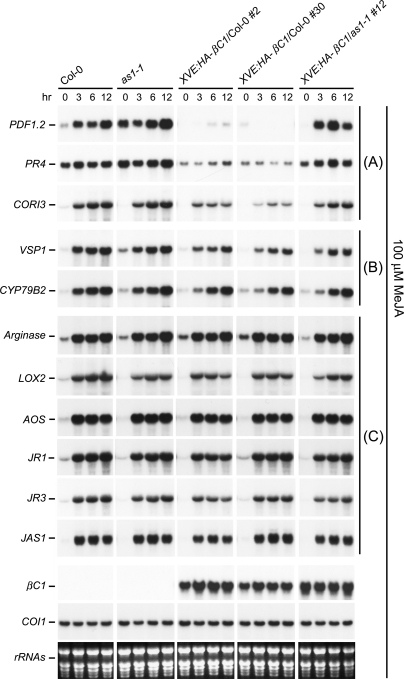
Recent molecular and genetic analysis of Nurmberg et al. (2007) provided evidence that AS1 is a negative regulator of plant defense response as as1 displays elevated expression of a group of JA-responsive genes, PDF1.2, PR3, and PR4, and is more resistant to fungal pathogens. Our finding that AS1 is a target protein for βC1 prompted us to investigate the consequence of βC1/AS1 interaction on JA-responsive gene expression. Neither βC1 expression (XVE:HA-βC1/Col-0 transgenic plants) nor AS1 deficiency (as1-1 mutant) had noticeable effect on expression of JA biosynthetic genes (LOX2 and AOS) and genes (Arginase, JR1, JR3, and JAS1) whose expression is sensitive to endogenous JA levels (Fig. 6). These results suggest that βC1 and AS1 are unlikely to modulate endogenous JA levels.
Figure 6.
βC1 suppresses JA responses. RNA samples from Col-0, as1-1, XVE:HA-βC1/Col-0, and XVE:HA-βC1/as1-1 treated with 100 μM MeJA were analyzed by Northern blots. XVE:HA-βC1/Col-0 and XVE:HA-βC1/as1-1 lines were pretreated with 50 μM β-estradiol to induce high-level expression of βC1. Each lane contained 15 μg of total RNA. Stained rRNAs were used as loading controls. COI1 transcripts showed no change with 100 μM MeJA. JA-responsive genes were grouped into A, B, and C depending on the effects of βC1. In A, JA-responsive transcript levels were reduced in XVE:HA-βC1/Col-0 lines but largely restored in the XVE:HA-βC1/as1-1 line. In B, JA-responsive transcript levels were only weakly repressed in XVE:HA-βC1/Col-0 lines and slightly recovered or no change were detected in the XVE:HA-βC1/as1-1 line. In C, no or little change was seen with JA-responsive transcripts in XVE:HA-βC1/Col-0 and XVE:HA-βC1/as1-1 lines.
Compared with wild-type plants, expression of several JA-responsive genes were suppressed in XVE:HA-βC1/Col-0 transgenic plants (Fig. 6). Transcript levels of PDF1.2, PR4, and CORI3 were reduced by βC1 in wild-type plants but largely restored in as1-1 mutant plants expressing βC1 (Fig. 6). In the case of VSP1 and CYP79B2, the transcript levels were only weakly repressed by βC1 in wild-type plants but slightly recovered or no change was seen in XVE:HA-βC1/as1-1 mutant plants (Fig. 6). These results show that βC1 can attenuate the plant defense system by inhibiting expression of several JA-responsive genes, and this repression is dependent to a large extent on the endogenous AS1.
Discussion
Arabidopsis as a model system to investigate βC1 function
The βC1 protein of geminivirus is a pathogenicity factor in host plants like tobacco, tomato, and petunia (Cui et al. 2005). Because of the importance of this viral protein in eliciting disease symptoms, identification of its host-interacting proteins/receptors will advance our knowledge of viral pathogenesis and help formulate strategies to combat the rapid spread of this devastating disease in the Old World. Using a transgenic approach, we found that βC1 can also induce similar disease symptoms in the model plant Arabidopsis, which is a nonhost. Several lines of evidence support our claim that the phenotypes found in Arabidopsis transgenic plants phenocopy to a large extent those observed in virus-infected tobacco. (1) Phenotypes including upward-curling leaves, bending shoot, and enations from abaxial side of leaves elicited by βC1 in Arabidopsis transgenic plants have been observed as disease symptoms of virus-infected tobacco (Fig. 1; Supplemental Fig. S1). (2) The abnormal cell division associated with vascular bundles was observed in both Arabidopsis βC1 transgenic plants (Supplemental Fig. S1M) and virus-infected tobacco (Saunders et al. 2004). (3) The capacity of βC1 to elevate PHV transcript levels in Arabidopsis transgenic plants were also observed in tobacco plants infected with TYLCCNV plus DNAβ satellite (Fig. 4D). Altered expression of PHV transcripts is known to cause development changes in leaf polarity (McConnell et al. 2001). These observation justifies the use of Arabidopsis to dissect the molecular mechanisms of disease symptom induction by βC1 and also suggests that homologous components likely exist in both host and nonhost plants, which can interact with βC1. Since nuclear targeting of βC1 is required for disease symptom induction (Cui et al. 2005), some of the βC1 interacting proteins/receptor(s) are likely to be nuclear localized.
AS1 is a nuclear receptor for βC1
We provide several lines of evidence to support the notion that AS1 is a receptor for βC1: (1) AS1 contains nuclear localization signals and is localized in discrete subnuclear bodies (Theodoris et al. 2003; Ueno et al. 2007). (2) βC1 can specifically interact with AS1 in vitro and in vivo (Fig. 3). (3) The capacity of βC1 to induce phenotypic changes and suppress selective JA responses in Arabidopsis depends to a large extent on endogenous AS1 levels. Phenotypic alterations associated with βC1 overexpression in wild-type plants—e.g., upward-curled leaves and outgrowth tissues from abaxial leaf surfaces—were suppressed by as1 but not as2 mutation. Moreover, reduction of miR165/166 levels and accumulation of PHB and PHV transcripts in βC1 overexpression plants were also reversed by as1 mutation. However, as1 plants overexpressing βC1 still displayed mildly curled leaves, indicating the existence of other plant factors that can interact with βC1 in the absence of AS1.
Many unrelated viral proteins have been identified as RNA silencing suppressors, and several of them have been introduced into Arabidopsis for molecular analyses. Transgenic plants overexpressing individual suppressor showed moderate to severe defects in leaf development depending on the suppressor. Interesting, many of these plants (e.g., P19, P1/HC-Pro, 2b, P15, P21, and CP transgenic plants) produced serrated and curled leaves (Chapman et al. 2004; Dunoyer et al. 2004). By contrast, leaves of βC1-overexpressing plants were not serrated. This interesting phenomenon may indicate that the function of βC1 is different from those of other silencing suppressors. Viral suppressors such as P19, P1/HC-Pro, 2b, P15, P21, and CP may cause defects in miRNA-mediated silencing pathway or miRNA biogenesis pathway by sequestering miRNA duplexes (Lakatos et al. 2006), blocking slicing activity of AGO1 (Zhang et al. 2006) or interfering with the dicing activity of DCL1 (Mlotshwa et al. 2005). βC1, on the other hand, only selectively regulates the accumulation of specific miRNAs. The precise mechanism by which βC1 regulates miRNA levels remains to be investigated.
βC1 phenocopies AS2
We showed that βC1 manipulates leaf polarity by mimicking certain functions of Asymmetric Leaves2 (AS2). Overexpression of βC1 in wild-type plants phenocopies AS2-overexpresing plants (Iwakawa et al. 2002; Lin et al. 2003; Xu et al. 2003), and strikingly, in both cases, the phenotypes were suppressed by as1 mutation (Lin et al. 2003; Xu et al. 2003). The perturbed vascular polarity with xylem on adaxial and abaxial sides of leaves in XVE:HA-βC1 transgenic plants (Supplemental Fig. S1O,P) was also observed in 35S:AS2 transgenic plants (Lin et al. 2003). Moreover, like AS2 overexpression, overexpression of βC1 also resulted in a reduction of miR165/166 and an accumulation of PHB transcripts (Lin et al. 2003; Ueno et al. 2007). Because no amino acid sequence similarity was detected between βC1 and AS2, the mimicry probably occurs through a similarity in the three-dimensional structures of the two proteins. Indeed, our in vitro results support this view. We showed that βC1, like AS2, can form a complex with AS1 in vitro, indicating a direct interaction. More important, the two proteins, βC1 and AS2, can compete for AS1 binding in complex formation, consistent with the hypothesis that they may bind to similar surfaces of AS1.
That βC1 can, at least in part, mimic AS2 functions in vivo is further supported by analysis of gene expression in βC1 plants and by complementation experiments. AS2 is known to negatively regulate, in the adaxial domain, transcript levels of ETTIN/ARF3 and FILAMENTOUS FLOWER, which are required for abaxial fate (Garcia et al. 2006; Iwakawa et al. 2007). We found that expression of βC1 in Arabidopsis reduced levels of these transcripts as well (Fig. 2D). Using a native AS2 promoter to express βC1 in the as2 mutant background, we found that this viral protein can at least partially complement AS2 functions. The downward curled leaves in as2 became flatten in AS2p:βC1/as2-1 transgenic lines. Furthermore, vein defects seen in as2 were rescued in these complemented lines. On the other hand, these transgenic lines still produced leaf lobes, which may be related to KNAT1 mis-expression. These observations suggest that the molecular mimicry of AS2 by βC1 is not complete, and βC1 and AS2 may share only partially overlapping functions in the establishment of adaxial–abaxial polarity of leaves and in vein development. Notwithstanding the partial overlapping functions of these two proteins, there is no amino acid sequence similarity between them. Nevertheless, they share some similar characteristics: Both βC1 (Mr 14,800) and AS2 (Mr 21,800) are small nuclear proteins (Cui et al. 2005; Ueno et al. 2007), and both are able to bind to DNA (Cui et al. 2005; Husbands et al. 2007).
Molecular mimicry has emerged as a common theme in virus–host interaction for several animal viruses (Grossman et al. 1994; Hsieh and Hayward 1995). Our results here with βC1 show that molecular mimicry is used by plant virus as well. Previous studies have shown that βC1 is not only responsible for symptom induction but also enhances the accumulation of begomovirus DNA-A (Cui et al. 2004). As a MYB-domain protein, AS1 may also function as a transcription regulator, and it is possible that the βC1/AS1 complex may regulate expression of begomovirus genes and modulate virus replication. Future work will be directed toward exploring this possibility.
Selective repression of JA-responsive genes by βC1
Infections by TYLCCNV and TbCSV carrying satellite DNAβ are not only associated with disease symptoms but also known to promote population increase of its vector, the type B invasive whitefly (Jiu et al. 2007). Although the mutualism between the two gemini viruses and whitefly has been shown to be indirect via the host plants, the precise mechanism remains to be elucidated. One attractive hypothesis is that TYLCCNV/TbCSV may somehow attenuate plant defenses against their vectors, leading to increased type B whitefly population, which in turn aids the spread of these viruses.
In plants, the jasmonate signaling pathway plays a central role in regulating defense responses to herbivores, and specific host defense responses are implemented for herbivores using different feeding strategies (Stout et al. 2006; de Vos 2007; Howe and Jander 2008). Usually provoking extensive tissue damage, chewing insects trigger production of plant defensive proteins, such as proteinase inhibitors (PIs), polyphenol oxidase (PPO), lipoxygenase (LOX), threonine deaminase (TD), and arginase, which bring about nutrient deprivation and disrupt vector digestive physiology. Unlike chewing insects, phloem-feeding insects, such as whiteflies and aphids, use specialized stylets to establish a feeding site in the phloem and cause minimal damage to plants. Kempema et al. (2007) showed that silverleaf whitefly (SLWF; Bemisia tabaci type B), a phloem-feeding insect, can suppress JA responses but activate SA responses. After SLWF feeding, JA-regulated genes PDF1.2, VSP1, and FAD3 were repressed more than twofold, with no detectable changes in THI2.1 and COI1 expression. In contrast to aphids, which are also phloem feeders but mobile, SLWF nymphs feed continuously from the same location on Arabidopsis leaves for more than 28 d during development. Hence, it is reasonable to assume that the down-regulation of these JA-responsive defense genes are necessary for and conducive to whitefly nymphal development on infected leaves. The same group further demonstrated that SLWF development on Arabidopsis correlates with the level of JA defenses but not with SA defenses (Zarate et al. 2007), and that SLWF development is delayed on Arabidopsis mutants with activated JA defenses and even on npr mutant plants with compromised SA defenses but treated with MeJA (Zarate et al. 2007).
The results of Jiu et al. (2007), Kempema et al. (2007), and Zarate et al. (2007) suggest that the increased aggregation of whitefly on TYLCCNV-infected plants may be due to possible suppression by the virus of specific JA-responsive plant genes that have defense function against the vector. Our results on the ability of βC1 to suppress expression of selective JA-responsive genes are consistent with this hypothesis. Among the five JA-responsive genes (PDF1.2, PR4, CORI3, VSP1, and CYP79B2) suppressed by βC1, PDF1.2, and VSP1 were down-regulated by SLWF feeding (Kempema et al. 2007). We suggest that βC1, along with other as yet unidentified viral proteins, are responsible for suppression of a subset of JA-responsive genes whose encoded proteins are needed for plant defense against whitefly, and these viral genes may accelerate the population increase of type B whitefly on TYLCCNV-infected plants. Although the origin of DNAβ of gemini viruses remains obscure, following its first report in 2000, many different DNAβs have been cloned and proven to be widespread throughout the Old World (Mansoor et al. 2006). This biological invasion may result from the indirect mutualism between viruses and the type B whiteflies. Our identification of AS1 as the main target for βC1 to attenuate plant defense response may help formulate strategies to slow down the biological invasion of B biotype whiteflies and curtail the spreading of begomoviruses among agricultural important crops.
Concluding remarks
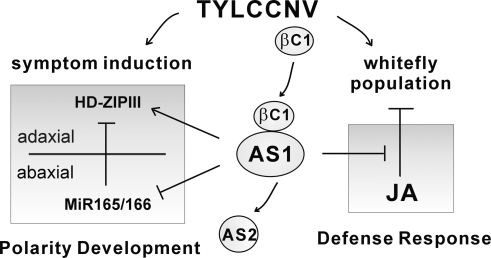
Figure 7 summarizes the proposed function of βC1 in altering leaf development and our working hypothesis on the tripartite relationship between virus–vector–host. AS1 binds to AS2 and regulates leaf development by suppressing the accumulation of miR165/166 and increasing HD-ZIPIII transcript levels. AS1 is also a conserved regulator of plant defense response, and it acts as negative regulator to suppress a subset of JA-induced genes such as PDF1.2, PR3, PR4, VSP1, and THI (Nurmberg et al. 2007). As a pathogenicity determinant, βC1 mimics AS2 function and competes with AS2 to form a complex with AS1. The βC1/AS1 complex, in turn, regulates leaf polarity by repressing miR165/166 and increasing HD-ZIPIII transcript levels. On the other hand, βC1 enhances the functions of AS1 to block selective JA responses by down-regulating the expression of PDF1.2, PR4, CORI3, and VSP1. The suppression of this subset of JA responses by the βC1/AS1 complex may attenuate plant defense response against whitefly. Consequently, satellite DNAβ-associated TYLCCNV elicits disease symptoms on infected plants and also benefits its vector by aiding an increase in vector population density via the host plants.
Figure 7.
A working model to explain the roles of AS1 and βC1 in virus–insect vector–plant interaction. AS1 is a conserved regulator for leaf polarity development and plant immune response. AS1 interacts with AS2 to down-regulate miR165/166 and up-regulate HD-ZIPIII genes, but AS1 alone acts to suppress JA responses. βC1 is encoded by the satellite DNAβ, which is associated with the helper virus, TYLCCNV. βC1 targets AS1 to manipulate leaf polarity by modulating miR165/166 and HD-ZIPIII transcripts resulting in disease symptoms. This viral protein also attenuates plant immune response by suppressing expression of several JA-responsive genes. This AS1-dependent suppression may account, in part, for the aggregation of whitefly population on TYLCCNV-infected plants.
Materials and methods
Plant materials, growth conditions, and transformation
Arabidopsis thaliana as1-1 and as2-1 mutants were used (Semiarti et al. 2001). The as1-1 was isolated from the Col-0 ecotype. The as2-1 was isolated from the Landsberg (Lan, with the wild-type allele for ERECTA gene) ecotype and outcrossed with Col-0 three times. After 2 d at 4°C in darkness, seeds were germinated on Murashing and Skoog (MS) medium at 22°C with 16 h light. Plasmids were introduced into Agrobacterium tumefaciens strain ABI or EHA105 by electrotransformation. Arabidopsis transformations were performed using the floral-dip method (Clough and Bent 1998).
Plant agroinfiltration
Agroinfiltration was performed with an overnight culture of A. tumefaciens strain EHA105 carrying a tandem repeat construct of TYLCCNV isolate Y10 and DNAβ in pBINplus (Cui et al. 2004). After agroinfiltration, N. benthamiana plants were grown in a greenhouse with 16 h light/8 h dark. For immunoprecipitation, Agrobacterium tumefaciens strain ABI carrying 35S:Myc-AS1 and XVE:HA-βC1 were suspended in 50 μM β-estradiol and coinfiltrated into tobacco leaves.
Constructs
Full-length cDNAs were amplified by PCR using AccuPrime Pfx DNA polymerase (Invitrogen) and subcloned into binary vectors pBA002-3HA and pBA002-6Myc to generate HA-tagged and Myc-tagged constructs under the control of a 35S promoter. For β-estradiol-inducible expression driven by the XVE promoter (Zuo et al. 2000), 3HA- or 6Myc-tagged cDNAs from pBA002 constructs were subcloned into pER8 vectors. A 955-bp fragment upstream of the βC1 ORF was used as the βC1 native promoter (Guan and Zhou 2006). This fragment was amplified from TYLCCNV DNAβ and subcloned into binary vector pBA002a to generate the C1p:βC1 construct. A 3262-bp fragment upstream of the AS2 open reading frame was amplified from Arabidopsis genomic DNA and used as the AS2 promoter.
β-Estradiol treatment and RNA gel blot analysis
Transgenic seeds were germinated on MS medium with or without 25 μM β-estradiol (Sigma). Total RNA was extracted from 12-d-old seedlings using Trizol reagent (Invitrogen) according to the manufacturer’s instructions. For RNA expression analysis, 15 μg of total RNA were fractionated on a 1.2% (w/v) agarose gel and then transferred to a Hybond-XL membrane (GE Biosciences). DNA probes were labeled with [α-32P]dCTP using the random prime labeling system (GE Biosciences). Hybridization was performed overnight at 65°C in hybridization buffer (0.3 M sodium phosphate at pH 7.0, 10 mM EDTA, 5% SDS, 10% dextran sulfate, 0.15 mg/mL salmon sperm DNA), and signals were detected by autoradiography. For small RNA analysis, 15 μg of total RNA were fractionated on a 15% polyacrylamide gel containing 8 M urea and then transferred to a Hybond-N+ membrane (GE Biosciences). DNA oligonucleotides were end-labeled with [γ-32P]ATP using T4 polynucleotide kinase (New England Biolabs). Hybridization was performed overnight at 42°C using the ULTRAHyb-Oligo hybridization buffer (Ambion) and signals were detected by autoradiography.
Real-time RT–PCR
XVE:βC1 line #1 and wild-type plants were grown on MS medium with 25 μM β-estradiol. Shoot apices of 15-d-old plants were harvested and Poly(A)+ RNA was purified from 10 μg of total RNA. Reverse transcription was performed using First-Strand cDNA synthesis kit (GE Healthcare). Real-time RT–PCR was performed as described by Iwakawa et al. (2007).
In vitro pull-down and competition assay
cDNAs encoding full-length βC1, AS1, and AS2 were amplified by PCR using AccuPrime Pfx DNA polymerase (Invitrogen) and subcloned into pGEX4T-1 or pMAL-c2 to generate GST fusion and MBP fusion constructs. All constructs were transformed into E. coli BL21(DE3) cells and cultured at 37°C. After the OD600 had reached ∼0.6, isopropyl β-D-thiogalactopyranoside was added to a final concentration of 0.2 mM and the culture incubated overnight at 24°C. Bacterial cells were collected by centrifugation and suspended in a lysis buffer containing proteinase inhibitor cocktail (Roche). After French press treatment, lysates were incubated with amylose resin (New England Biolads) or glutathione Sepharose 4B (GE Biosciences), and recombinant proteins were purified according to the manufacturer’s instructions. The eluted proteins were dialyzed against buffer (20 mM Tris-HCl at pH 7.4, 200 mM NaCl, 10 mM β-mercaptoethanol, 10% glycerol) and concentrated by Ultracel YM-30 (Millipore). In vitro pull-down assays were performed with 2 μg of GST fusion proteins and 2 μg of MBP fusion proteins. Proteins were incubated in a binding buffer (50 mM Tris-HCl at pH 7.5, 100 mM NaCl, 0.25% Triton X-100, 35 mM β-mercaptoethanol) for 2 h at 25°C, and 25 μL of glutathione sepharose 4B (GE Biosciences) were added and the mix incubated for an additional 1 h. After washing with binding buffer for six times, pulled-down proteins were separated on 10% SDS–polyacrylamide gel and detected by Western blotting using anti-MBP antibody. For competitive pull-down assay, sequence encoding a βC1(ΔN8) without the first eight amino acids was amplified by PCR and subcloned into pET29a to generate His fusion construct. βC1(ΔN8) was purified using Ni-NTA resin (Qiagen) according to the manufacturer’s instruction. Indicated amounts of βC1(ΔN8) were mixed with 2 μg of MBP-AS1 for 1 h before being incubated with 2 μg of GST-AS2 for pull-down assays.
Immunoprecipitation
Two days after infiltration, tobacco leaves were harvested and ground in liquid nitrogen. Proteins were extracted in extraction buffer (50 mM Tris-HCl at pH 7.5, 150 mM NaCl, 2 mM MgCl2, 1 mM DTT, 20% glycerol, 0.5% nonident P-40) containing protease inhibitor cocktail (Roche) and protease inhibitor mixture (Sigma). Cell debris was pelleted by centrifugation at 14,000g for 30 min. The supernatant was incubated with 10 μL of anti-Myc polycolonal antibody (Santa Cruz Biotechnologies) for 2 h at 4°C, then 20 μL of protein A agarose beads (GE Healthcare) were added. After 2 h of incubation at 4°C, the beads were centrifuged and washed six times with washing buffer (50 mM Tris-HCl at pH 7.5, 150 mM NaCl, 2 mM MgCl2, 1 mM DTT, 10% glycerol, 0.5% nonident P-40). Proteins were eluted by 30 μL of 2.5× sample buffer and analyzed by Western blotting using monoclonal anti-HA antibody (Santa Cruz Biotechnologies) and monoclonal anti-Myc antibody.
MG132 treatment
Transgenic seeds were germinated on MS medium with 25 μM β-estradiol (Sigma). Eighteen-day-old seedlings were transferred to liquid MS medium containing 25 μM β-estradiol and 50 μM MG132 (Calbiochem) for 12 h before harvested. Proteins were extracted and analyzed by Western blotting using monoclonal anti-HA antibody (Santa Cruz Biotechnologies).
Vasculature analysis
Twelve-day-old cotyledons and 21-d-old rosette leaves were fixed with 15% glacial acid and 85% ethanol for overnight. Samples were washed twice with 70% ethanol and cleared by chloral hydrate solution (trichloroacetoaldehyde monohydrate:glycerol:H2O = 8:1:2). Venation patterns were observed using a microscope under dark-field condition. The classification of venation patterns in cotyledons was according to Semiarti at al. (2001) with modification.
MeJA treatment
Plants were grown on soil under long-day conditions at 22°C and watered with 50 μM β-estradiol. Four-week-old plants were treated with 100 μM MeJA (Sigma) using foliar sprays. After spraying, plants were covered with a plastic cover and samples were harvested at indicated times.
Acknowledgments
We thank Dr. Sarah Hake for providing KNAT1:GUS seeds, Ms Li-Fang Huang for excellent technical assistance, Eleana Sphicas and Qi-Wen Niu for technical support on SEM and transverse sections, and Dr. Takashi Soyano for helpful suggestions. J.-Y.Y. was supported by a Taiwan Merit Scholarship from the Ministry of Education, the Council for Economic Planning and Development, and the National Science Council of Taiwan. This work was supported in part by NIH grant GM44640.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1682208.
References
- Belliure B., Janssen A., Maris P.C., Peters D., Sabelis M.W. Herbivore arthropods benefit from vectoring plant viruses. Ecol. Lett. 2005;8:70–79. [Google Scholar]
- Bisaro D.M. Silencing suppression by geminivirus proteins. Virology. 2006;344:158–168. doi: 10.1016/j.virol.2005.09.041. [DOI] [PubMed] [Google Scholar]
- Chapman E.J., Prokhnevsky A.I., Gopinath K., Dolja V.V., Carrington J.C. Viral RNA silencing suppressors inhibit the microRNA pathway at an intermediate step. Genes & Dev. 2004;18:1179–1186. doi: 10.1101/gad.1201204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck G., Lincoln C., Hake S. KNAT1 induces lobed leaves with ectopic meristems when overexpressed in Arabidopsis. Plant Cell. 1996;8:1277–1289. doi: 10.1105/tpc.8.8.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Cui X., Tao X., Xie Y., Fauquet C.M., Zhou X. A DNAβ associated with tomato yellow leaf curl China virus is required for symptom induction. J. Virol. 2004;78:13966–13974. doi: 10.1128/JVI.78.24.13966-13974.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X., Li G., Wang D., Hu D., Zhou X. A begomovirus DNAβ-encoded protein binds DNA, functions as a suppressor of RNA silencing, and targets the cell nucleus. J. Virol. 2005;79:10764–10775. doi: 10.1128/JVI.79.16.10764-10775.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culver J.N., Padmanabhan M.S. Virus-induced disease: Altering host physiology one interaction at a time. Annu. Rev. Phytopathol. 2007;45:221–243. doi: 10.1146/annurev.phyto.45.062806.094422. [DOI] [PubMed] [Google Scholar]
- de Vos M., Kim J.H., Jander G. Biochemistry and molecular biology of Arabidopsis–aphid interactions. Bioessays. 2007;29:871–883. doi: 10.1002/bies.20624. [DOI] [PubMed] [Google Scholar]
- Dunoyer P., Lecellier C.-H., Parizotto E.A., Himber C., Voinnet O. Probing the MicroRNA and small interfering RNA pathways with virus-encoded suppressors of RNA silencing. Plant Cell. 2004;16:1235–1250. doi: 10.1105/tpc.020719. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Emery J.F., Floyd S.K., Alvarez J., Eshed Y., Hawker N.P., Izhaki A., Baum S.F., Bowman J.L. Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr. Biol. 2003;13:1768–1774. doi: 10.1016/j.cub.2003.09.035. [DOI] [PubMed] [Google Scholar]
- Gao G., Luo H. The ubiquitin–proteasome pathway in viral infections. Can. J. Physiol. Pharmacol. 2006;84:5–14. doi: 10.1139/y05-144. [DOI] [PubMed] [Google Scholar]
- Garcia D., Collier S.A., Byrne M.E., Martienssen R.A. Specification of leaf polarity in Arabidopsis via the trans-acting siRNA pathway. Curr. Biol. 2006;16:933–938. doi: 10.1016/j.cub.2006.03.064. [DOI] [PubMed] [Google Scholar]
- Grossman S., Johannsen E., Tong X., Yalamanchili R., Kieff E. The Epstein-Barr virus nuclear antigen 2 transactivator is directed to response elements by the Jκ recombination signal binding protein. Proc. Natl. Acad. Sci. 1994;91:7568–7572. doi: 10.1073/pnas.91.16.7568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan C., Zhou X. Phloem specific promoter from a satellite associated with a DNA virus. Virus Res. 2006;115:150–157. doi: 10.1016/j.virusres.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Hayward S.D., Liu J., Fujimuro M. Notch and Wnt signaling: Mimicry and manipulation by γ herpesviruses. Sci. STKE. 2006;2006:re4. doi: 10.1126/stke.3352006re4. [DOI] [PubMed] [Google Scholar]
- Howe G.A., Jander G. Plant immunity to insect herbivores. Annu. Rev. Plant Biol. 2008;59:41–66. doi: 10.1146/annurev.arplant.59.032607.092825. [DOI] [PubMed] [Google Scholar]
- Hsieh J., Hayward S. Masking of the CBF1/RBPJ κ transcriptional repression domain by Epstein-Barr virus EBNA2. Science. 1995;268:560–563. doi: 10.1126/science.7725102. [DOI] [PubMed] [Google Scholar]
- Hull R. Matthews’ plant virology. Academic Press; San Diego, CA: 2001. [Google Scholar]
- Husbands A., Bell E.M., Shuai B., Smith H.M.S., Springer P.S. LATERAL ORGAN BOUNDARIES defines a new family of DNA-binding transcription factors and can interact with specific bHLH proteins. Nucleic Acids Res. 2007;35:6663–6671. doi: 10.1093/nar/gkm775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwakawa H., Ueno Y., Semiarti E., Onouchi H., Kojima S., Tsukaya H., Hasebe M., Soma T., Ikezaki M., Machida C., et al. The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana, required for formation of a symmetric flat leaf lamina, encodes a member of a novel family of proteins characterized by cysteine repeats and a leucine zipper. Plant Cell Physiol. 2002;43:467–478. doi: 10.1093/pcp/pcf077. [DOI] [PubMed] [Google Scholar]
- Iwakawa H., Iwasaki M., Kojima S., Ueno Y., Soma T., Tanaka H., Semiarti E., Machida Y., Machida C. Expression of the ASYMMETRIC LEAVES2 gene in the adaxial domain of Arabidopsis leaves represses cell proliferation in this domain and is critical for the development of properly expanded leaves. Plant J. 2007;51:173–184. doi: 10.1111/j.1365-313X.2007.03132.x. [DOI] [PubMed] [Google Scholar]
- Jiu M., Zhou X.-P., Tong L., Xu J., Yang X., Wan F.-H., Liu S.-S. Vector–virus mutualism accelerates population increase of an invasive whitefly. PLoS ONE. 2007;2:e182. doi: 10.1371/journal.pone.0000182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasschau K.D., Xie Z., Allen E., Llave C., Chapman E.J., Krizan K.A., Carrington J.C. P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA function. Dev. Cell. 2003;4:205–217. doi: 10.1016/s1534-5807(03)00025-x. [DOI] [PubMed] [Google Scholar]
- Kempema L.A., Cui X., Holzer F.M., Walling L.L. Arabidopsistranscriptome changes in response to phloem-feeding silverleaf whitefly nymphs. Similarities and distinctions in responses to aphids. Plant Physiol. 2007;143:849–865. doi: 10.1104/pp.106.090662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakatos L., Csorba T., Pantaleo V., Chapman E.J., Carrington J.C., Liu Y.P., Dolja V.V., Calvino L.F., Lopez-Moya J.J., Burgyan J. Small RNA binding is a common strategy to suppress RNA silencing by several viral suppressors. EMBO J. 2006;25:2768–2780. doi: 10.1038/sj.emboj.7601164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Ding S.-W. Virus counterdefense: Diverse strategies for evading the RNA-silencing immunity. Annu. Rev. Microbiol. 2006;60:503–531. doi: 10.1146/annurev.micro.60.080805.142205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W.-c., Shuai B., Springer P.S. The Arabidopsis LATERAL ORGAN BOUNDARIES-domain gene ASYMMETRIC LEAVES2 functions in the repression of KNOX gene expression and in adaxial–abaxial patterning. Plant Cell. 2003;15:2241–2252. doi: 10.1105/tpc.014969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory A.C., Reinhart B.J., Jones-Rhoades M.W., Tang G., Zamore P.D., Barton M.K., Bartel D.P. MicroRNA control of PHABULOSA in leaf development: Importance of pairing to the microRNA 5′ region. EMBO J. 2004;23:3356–3364. doi: 10.1038/sj.emboj.7600340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansoor S., Zafar Y., Briddon R.W. Geminivirus disease complexes: The threat is spreading. Trends Plant Sci. 2006;11:209–212. doi: 10.1016/j.tplants.2006.03.003. [DOI] [PubMed] [Google Scholar]
- Maris P.C., Joosten N.N., Goldbach R.W., Peters D. Tomato spotted wilt virus infection improves host suitability for its vector Frankliniella occidentalis. Phytopathology. 2004;94:706–711. doi: 10.1094/PHYTO.2004.94.7.706. [DOI] [PubMed] [Google Scholar]
- McConnell J.R., Emery J., Eshed Y., Bao N., Bowman J., Barton M.K. Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature. 2001;411:709–713. doi: 10.1038/35079635. [DOI] [PubMed] [Google Scholar]
- Mlotshwa S., Schauer S.E., Smith T.H., Mallory A.C., Herr J.M., Roth B., Merchant D.S., Ray A., Bowman L.H., Vance V.B. Ectopic DICER-LIKE1 expression in P1/HC-Pro Arabidopsis rescues phenotypic anomalies but not defects in microRNA and silencing pathways. Plant Cell. 2005;17:2873–2885. doi: 10.1105/tpc.105.036608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurmberg P.L., Knox K.A., Yun B.-W., Morris P.C., Shafiei R., Hudson A., Loake G.J. The developmental selector AS1 is an evolutionarily conserved regulator of the plant immune response. Proc. Natl. Acad. Sci. 2007;104:18795–18800. doi: 10.1073/pnas.0705586104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ori N., Eshed Y., Chuck G., Bowman J., Hake S. Mechanisms that control knox gene expression in the Arabidopsis shoot. Development. 2000;127:5523–5532. doi: 10.1242/dev.127.24.5523. [DOI] [PubMed] [Google Scholar]
- Saunders K., Norman A., Gucciardo S., Stanley J. The DNA β staellite component associated with ageratum yellow vein disease encodes an essentail pathogenicity protein (βC1) Virology. 2004;324:37–47. doi: 10.1016/j.virol.2004.03.018. [DOI] [PubMed] [Google Scholar]
- Semiarti E., Ueno Y., Tsukaya H., Iwakawa H., Machida C., Machida Y. The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana regulates formation of a symmetric lamina, establishment of venation and repression of meristem-related homeobox genes in leaves. Development. 2001;128:1771–1783. doi: 10.1242/dev.128.10.1771. [DOI] [PubMed] [Google Scholar]
- Stout M.J., Thaler J.S., Thomma B.P.H.J. Plant-mediated interactions between pathogenic microorganisms and herbivorous arthropods. Annu. Rev. Entomol. 2006;51:663–689. doi: 10.1146/annurev.ento.51.110104.151117. [DOI] [PubMed] [Google Scholar]
- Theodoris G., Inada N., Freeling M. Conservation and molecular dissection of ROUGH SHEATH2 and ASYMMETRIC LEAVES1 function in leaf development. Proc. Natl. Acad. Sci. 2003;100:6837–6842. doi: 10.1073/pnas.1132113100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno Y., Ishikawa T., Watanabe K., Terakura S., Iwakawa H., Okada K., Machida C., Machida Y. Histone deacetylases and ASYMMETRIC LEAVES2 are involved in the establishment of polarity in leaves of Arabidopsis. Plant Cell. 2007;19:445–457. doi: 10.1105/tpc.106.042325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Xu Y., Dong A., Sun Y., Pi L., Xu Y., Huang H. Novel as1 and as2 defects in leaf adaxial–abaxial polarity reveal the requirement for ASYMMETRIC LEAVES1 and 2 and ERECTA functions in specifying leaf adaxial identity. Development. 2003;130:4097–4107. doi: 10.1242/dev.00622. [DOI] [PubMed] [Google Scholar]
- Zarate S.I., Kempema L.A., Walling L.L. Silverleaf whitefly induces salicylic acid defenses and suppresses effectual jasmonic acid defenses. Plant Physiol. 2007;143:866–875. doi: 10.1104/pp.106.090035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Yuan Y.-R., Pei Y., Lin S.-S., Tuschl T., Patel D.J., Chua N.-H. Cucumber mosaic virus-encoded 2b suppressor inhibits Arabidopsis Argonaute1 cleavage activity to counter plant defense. Genes & Dev. 2006;20:3255–3268. doi: 10.1101/gad.1495506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo J., Niu Q.-W., Chua N.-H. An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J. 2000;24:265–273. doi: 10.1046/j.1365-313x.2000.00868.x. [DOI] [PubMed] [Google Scholar]