Abstract
MicroRNAs (miRNAs) and transcription factors (TFs) are primary metazoan gene regulators. Whereas much attention has focused on finding the targets of both miRNAs and TFs, the transcriptional networks that regulate miRNA expression remain largely unexplored. Here, we present the first genome-scale Caenorhabditis elegans miRNA regulatory network that contains experimentally mapped transcriptional TF → miRNA interactions, as well as computationally predicted post-transcriptional miRNA → TF interactions. We find that this integrated miRNA network contains 23 miRNA ↔ TF composite feedback loops in which a TF that controls a miRNA is itself regulated by that same miRNA. By rigorous network randomizations, we show that such loops occur more frequently than expected by chance and, hence, constitute a genuine network motif. Interestingly, miRNAs and TFs in such loops are heavily regulated and regulate many targets. This “high flux capacity” suggests that loops provide a mechanism of high information flow for the coordinate and adaptable control of miRNA and TF target regulons.
Keywords: MicroRNA, transcription factor, regulatory network, network motif, feedback, C. elegans
Differential gene expression can be regulated at many levels and by various trans-acting factors. MicroRNAs (miRNAs) and transcription factors (TFs) are primary regulators of differential gene expression during organism development and function and in disease. While TFs physically interact with cis-regulatory DNA elements to activate or repress transcription of their target genes, miRNAs repress gene expression post-transcriptionally by interacting with complementary sequences located in the 3′ untranslated region (UTR) of their target mRNAs (Bartel 2004). Following the initial discovery of miRNAs in Caenorhabditis elegans (Lee et al. 1993; Wightman et al. 1993), much attention has focused on the identification of their target genes. miRNA targets are usually predicted computationally, and several algorithms have been developed for this purpose (Sethupathy et al. 2006). Target predictions revealed that miRNAs target TFs more frequently than other types of genes (Shalgi et al. 2007). This suggests that miRNAs and TFs could be intricately connected in the networks that control differential gene expression.
Transcription regulatory networks of protein-coding genes have been mapped and studied in yeast (Harbison et al. 2004), C. elegans (Deplancke et al. 2006a; Vermeirssen et al. 2007a), Drosophila melanogaster (Sandmann et al. 2007), and mammals (Boyer et al. 2005). While vastly incomplete, these networks have already provided insights into overall network architecture and have also revealed particular network subgraphs that are overrepresented in real networks compared with randomized networks. Such enriched subgraphs are referred to as network motifs (Milo et al. 2002; Shen-Orr et al. 2002). Since network motifs are recurrent regulatory circuits, they provide successful mechanisms of gene expression and, hence, play widespread roles in gene regulation. For instance, feed-forward loops provide a mechanism to ensure a robust transcriptional response to signals (e.g., environmental signals), and to protect against transcriptional noise (Milo et al. 2002; Shen-Orr et al. 2002). Feedback loops are important in homeostasis and cellular differentiation programs (Alon 2007). Surprisingly, whereas feed-forward loops are overrepresented in pure transcription regulatory networks, feedback loops were found to be less abundant. One explanation for the paucity of feedback loops is that they may be generated by a combination of transcriptional and post-transcriptional mechanisms, as opposed to being purely transcriptional (Shen-Orr et al. 2002; Yeger-Lotem et al. 2004). Interestingly, recent bioinformatic studies found that the expression of miRNAs and their targets is often highly correlated or anti-correlated (Farh et al. 2005; Stark et al. 2005; Sood et al. 2006; Tsang et al. 2007). Tsang et al. (2007) proposed that such (anti-)correlations can result from various types of feed-forward and feedback loops involving miRNAs, their predicted target genes, and upstream regulators (e.g., TFs, kinases). This study was exclusively based on a predicted miRNA → target network and did not use TF → miRNA transcriptional interactions because they were not available. Consequently, no actual loops were identified. However, several feedback loops involving miRNAs and TFs recently have been found experimentally in a variety of organisms (Fazi et al. 2005; Johnston et al. 2005; Kim et al. 2007; Varghese and Cohen 2007). Thus, we hypothesized that miRNAs may be a “missing post-transcriptional link” that, together with TFs, generate feedback loops in genome-scale gene regulatory networks.
miRNAs are transcribed as part of longer primary transcripts (pri-miRNAs), which are then processed in a stepwise manner by protein complexes that include the RNase III enzymes Drosha, to produce pre-miRNAs, and Dicer to produce mature 21–22-nt miRNAs (for review, see Kim 2005). miRNAs are transcribed by RNA polymerase II (Lee et al. 2004), which suggests that miRNA transcription is subject to similar control mechanisms as protein-coding genes. Although some TFs that regulate miRNA expression have been found (Fazi et al. 2005; Sylvestre et al. 2007), genome-scale transcriptional networks that control miRNA expression remain unexplored. Transcription regulatory networks have been mapped predominantly using protein–DNA interaction mapping methods that are TF-centered such as chromatin immunoprecipitation (ChIP) (Harbison et al. 2004). ChIP is impractical for the comprehensive mapping of TFs that regulate miRNA expression because it would require the testing of all predicted TFs of an organism in multiple tissue types and under different conditions. Moreover, many TFs are not sufficiently broadly or highly expressed to be detected by ChIP, and only a few suitable anti-TF antibodies are available (Walhout 2006).
We previously developed and applied a condition-independent yeast one-hybrid (Y1H) method that allows the identification of TFs that can bind a set of promoters of interest (Deplancke et al. 2004, 2006a; Vermeirssen et al. 2007a, b). Here, we use this method to experimentally map a genome-scale TF → miRNA transcription regulatory network in the nematode C. elegans. By integrating this network with a high-confidence miRNA → TF target network, we identify 23 miRNA ↔ TF composite feedback loops. Using rigorous network randomizations, we show that such miRNA ↔ TF feedback loops occur more frequently than expected by chance, and, hence, constitute a genuine network motif. We find that most miRNAs and TFs that participate in miRNA ↔ TF feedback loops are highly connected: The miRNAs regulate many TFs and are regulated by many TFs, and vice versa. We introduce a novel network parameter we named “flux capacity (Fc)” that captures the high flow of information that passes through many miRNAs and TFs involved in composite feedback loops. Finally, we propose a model in which feedback loops provide a mechanism for the highly coordinated and adaptable control of gene batteries, or regulons.
Results
A genome-scale C. elegans TF → miRNA transcription regulatory network
The C. elegans genome encodes 940 predicted TFs (Reece-Hoyes et al. 2005; Vermeirssen et al. 2007b) and 115 predicted miRNAs (Ambros et al. 2003; Lim et al. 2003; Griffiths-Jones et al. 2006). Of the 115 miRNA genes available in miRBase V4.0, 66 reside in intergenic regions and can be assigned to their own promoter. An additional 16 intergenic miRNAs are transcribed in a total of six intergenic operons, with a single promoter regulating each operon. The remaining 33 miRNAs are embedded within the intron of a protein-coding gene, either in the sense or antisense orientation. Thirteen of these intragenic miRNAs are transcribed in the antisense orientation in two operons. Twenty intragenic miRNAs are located in the sense orientation and are likely cotranscribed with their host gene, and may be controlled by the host gene promoter (Baskerville and Bartel 2005). These latter miRNAs were not included in our analyses.
Although the transcription start site of the majority of C. elegans miRNAs has not been mapped, it has been shown that fragments between 1 and 2 kb upstream of the pre-miRNA are sufficient to rescue lin-4, let-7, and lsy-6 mutant phenotypes (Lee et al. 1993; Johnson et al. 2003; Chang et al. 2004). In addition, reported expression of mir-84, mir-61, and mir-48 involved 1 to 2.2 kb of genomic sequence upstream of the annotated miRNA (Johnson et al. 2005; Li et al. 2005; Yoo and Greenwald 2005). For these reasons, we decided to use DNA fragments that correspond to the intergenic region upstream of the annotated miRNA with a minimum length of 300 bp and a maximum length of 2 kb as miRNA promoters. These fragments may not contain all the regulatory elements necessary for miRNA expression. For instance, Bracht et al. (2004) have shown that transcription of let-7 can start either ∼200 bp or ∼1 kb upstream of pre-let-7. However, we demonstrated recently that the vast majority of miRNA promoters as defined here are able to confer GFP expression in vivo, and >90% of these recapitulate known temporal expression as determined elsewhere by Northern blotting (N.J. Martinez, M.C. Ow, J. Reece-Hoyes, V. Ambros, and A.J. Walhout, in prep.). This indicates that the genomic fragments we used, indeed, encompass miRNA promoters. In total, we cloned 71 miRNA promoters (Supplemental Tables S1, S2).
To identify TFs that can interact with each miRNA promoter, we performed three Y1H assays: screens versus a C. elegans cDNA library (Walhout et al. 2000b) and a TF mini-library (Deplancke et al. 2004), and matrix assays of all promoters versus all TFs identified in the screens (Supplemental Table S3; Vermeirssen et al. 2007a). Thus, each promoter was directly tested against all TFs in our data set. We applied a stringent scoring and filtering system to minimize the inclusion of false positives (Vermeirssen et al. 2007a), and obtained 347 high-confidence interactions between 63 miRNA promoters and 116 proteins (Supplemental Table S4). These interactions are available in our EDGEdb database (http://edgedb.umassmed.edu; Barrasa et al. 2007). The most highly connected miRNAs belong to the let-7 and lin-4 families, implicated in developmental timing, as well as other miRNAs of unknown function such as mir-46, mir-355, and mir-243.
Interestingly, while the majority of the proteins retrieved are predicted TFs (Reece-Hoyes et al. 2005), some do not possess a known DNA-binding domain and may constitute novel TFs. We previously demonstrated (by ChIP in yeast) that nine of 11 tested novel putative TFs (∼80%) do interact with their target promoters, suggesting that they may possess an as-yet-unrecognized DNA-binding domain (Deplancke et al. 2006a; Vermeirssen et al. 2007a). The TFs that interact with miRNA promoters are diverse as they represent most of the known TF families in C. elegans. The most highly connected TFs include members of the ZF-C2H2 family (DIE-1 and ZTF-1), ZF-NHR (ODR-7), and MH-1 (DAF-3), as well as proteins with an unidentified DNA-binding domain (Y38C9A.1 and C32D5.1). Together, these observations indicate that there is no DNA-binding domain bias in the transcriptional miRNA network (Supplemental Table S4; data not shown).
It is possible that we identified multiple members of a TF family binding to the same promoter in Y1H assays simply because these TFs recognize similar DNA sequences. Alternatively, it may be that members of a TF family function redundantly in vivo, as has been shown for mammalian ETS proteins (Hollenhorst et al. 2007). Indeed, we demonstrated that FLH-1 and FLH-2, members of the FLYWCH family of TFs, redundantly regulate the expression of several miRNAs in the C. elegans embryo (Ow et al. 2008).
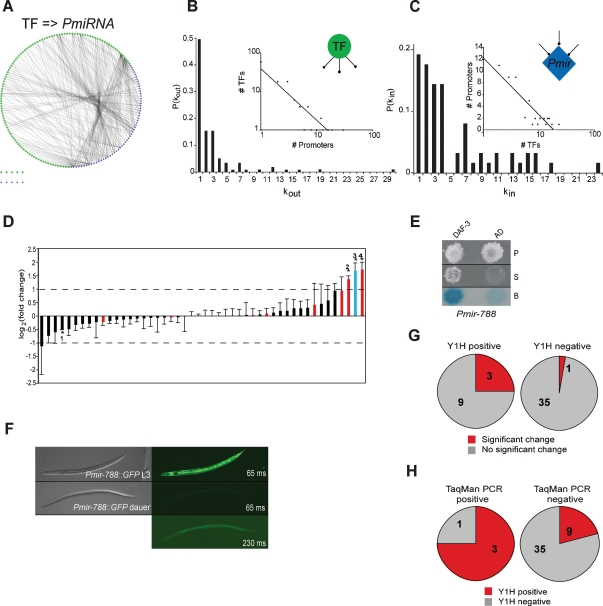
All high-confidence TF → miRNA interactions were visualized into a network model (Fig. 1A). The distribution of both the outgoing connectivity (“out-degree,” or number of miRNA promoters bound by a given TF) (Fig. 1B) and the incoming connectivity (“in-degree,” or number of TFs bound to a given miRNA promoter) (Fig. 1C) of this network is similar to those of C. elegans protein-coding gene networks (Deplancke et al. 2006a; Vermeirssen et al. 2007a). This indicates that the overall architecture of miRNA transcription regulatory networks is similar to that of protein-coding gene networks. Hence, at least based on these two network properties, miRNA expression overall is regulated in a similar manner as protein-coding genes.
Figure 1.
A genome-scale C. elegans TF → miRNA transcription regulatory network. (A) TF → miRNA interactions identified by high-throughput Y1H assays were visualized into a transcription regulatory network using Cytoscape (Shannon et al. 2003). (Blue diamonds) miRNA promoters; (green circles) TFs. (B) Out-degree; P(kout) is the proportion of miRNA promoters per TF. (Inset) The out-degree distribution best fits a power law (R2 = 0.82). (C) In-degree; P(kin) is the proportion of TFs per miRNA promoter. (Inset) The in-degree best fits an exponential distribution (R2 = 0.84). (D) TaqMan PCR assays of 48 miRNAs in N2 and daf-3(mgDf90) mutant dauer larvae. The average log2(fold change) of five experiments is shown. Error bars indicate standard error of the mean. Asterisks indicate significant changes. (1) mir-85. (2) mir-48. (3) mir-788. (4) mir-241. (Red bars) miRNAs bound by DAF-3 in Y1H assays; (blue bar) mir-788. The dashed line indicates a twofold difference. (E) Y1H assay confirming the interaction between DAF-3 and Pmir-788. (P) Permissive media; (S) selective media; (B) β-galactosidase assay; (AD) empty vector. (F) mir-788 is repressed in dauer larvae. (Left) Nomarski image. (Right) GFP fluorescence. The top right and right middle panels are 65-msec exposures, whereas the bottom right panel is a 230-msec exposure of the same field as in the right middle panel to visualize the presence of the animal. (G,H) Correlation between Y1H and TaqMan PCR data. Both the proportion and the actual numbers are depicted.
DAF-3 is a TF that interacted with multiple miRNA promoters (Supplemental Table S4). It is involved in dauer formation, a developmentally arrested, alternative third larval stage that occurs under adverse environmental conditions (Patterson et al. 1997). DAF-3 expression increases when worms enter the dauer stage (Wang and Kim 2003). To examine the regulatory effect of DAF-3 on miRNA expression, we compared the levels of 48 miRNAs (see the Materials and Methods) in wild-type and daf-3(mgDf90) mutant dauer larvae by TaqMan PCR assays, and ranked miRNAs according to their change in expression (Fig. 1D). Four miRNAs changed significantly in expression levels in daf-3(mgDf90) mutants. Three of these were increased in the mutant, and one was slightly decreased (∼1.4-fold). One of the miRNAs that increased in the daf-3(mgDf90) mutant, mir-788 (Fig. 1D, blue bar) was only recently identified (Ruby et al. 2006) and had therefore not been included in our Y1H experiments. We cloned the mir-788 promoter and tested it for binding to all 755 available C. elegans TFs (Vermeirssen et al. 2007b). Pmir-788 interacted with eight TFs, including DAF-3 (Fig. 1E; Supplemental Table S5). The promoters of the other two miRNAs that showed a significant increase in expression also bound to DAF-3. Therefore, all three miRNAs with significantly increased expression in the daf-3(mgDf90) mutant correspond to Y1H positives. This indicates that DAF-3 can function as a transcriptional repressor, which is in agreement with previous observations (Thatcher et al. 1999; Deplancke et al. 2006a). In addition, mir-788 promoter activity was repressed upon dauer formation, which is in agreement with the increase of DAF-3 during this stage (Fig. 1F; Wang and Kim 2003). Overall, 25% (3/12) of the miRNAs that interact with DAF-3 in Y1H assays are significantly increased in daf-3(mgDf90) dauer animals, while only one out of the 36 miRNAs (3%) that do not interact with DAF-3 showed a small, but significant change in expression (Fig. 1G). Conversely, the promoters of 75% of the miRNAs that change significantly in daf-3(mgDf90) mutants were bound by DAF-3, while only ∼20% of the miRNAs that do not change are controlled by a promoter bound by DAF-3 (Fig. 1H). Together, these results demonstrate that Y1H and TaqMan PCR data correlate (Fisher exact test, P-value = 0.04), and provide insights into the transcriptional consequences of physical TF–promoter interactions within the context of an intact animal. It is important to note that those miRNAs that interacted with DAF-3 but that did not change in expression in daf-3(mgDf90) animals may be regulated under different developmental or physiological conditions during the lifetime of the animal. Another possibility is that they may change in expression in one or a few cells within the animal, and thus fall below the detection limit of whole animal TaqMan PCR assays.
A C. elegans miRNA → TF post-transcription regulatory network
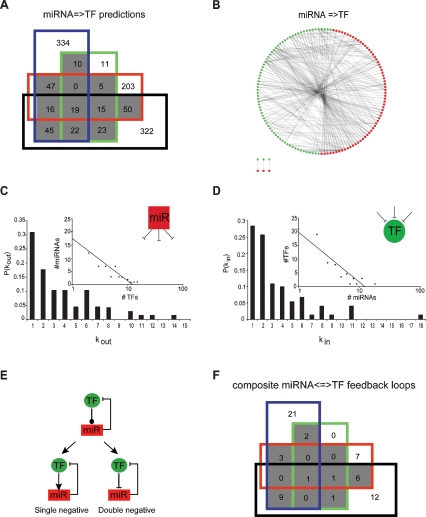
We generated a post-transcriptional miRNA → TF network by identifying which of the TFs found in Y1H assays are predicted miRNA targets. Since target prediction algorithms can be noisy (Sethupathy et al. 2006), we only used targets predicted by two or more, from a total of four miRNA target prediction algorithms, including Targetscan, Pictar, miRanda, and RNAhybrid (Fig. 2A; Supplemental Table S6; see the Materials and Methods). We identified 252 high-confidence miRNA → TF interactions involving 67 miRNAs and 73 TFs. The most highly connected miRNAs are members from the let-7, mir-80, and mir-2 families. The most highly connected TFs are ZAG-1, ZTF-10, and LIN-26, all of which belong to the ZF-C2H2 family; and ELT-3 and NHR-14, which belong to the ZF-GATA and ZF-NHR families, respectively.
Figure 2.
A C. elegans miRNA → TF post-transcription regulatory network. (A) Four-way Venn diagram showing the number of miRNA → TF predictions for TFs found in the transcriptional network. (Blue) RNA–hybrid; (green) Pictar; (red) miRanda; (black) TargetScanS; (gray) predictions common in two or more algorithms. (B) The predicted miRNA → TF post-transcription regulatory network. (Red squares) miRNAs; (green circles) TFs. (C) Out-degree; P(kout) is the proportion of TF targets per miRNA. (Inset) The out-degree best fits an exponential distribution (R2 = 0.90). (D) In-degree; P(kin) is the proportion of miRNA targeting a TF. (Inset) The in-degree best fits an exponential distribution (R2 = 0.84). (E, top) Cartoon of the two types of composite miRNA ↔ TF feedback loops: single negative (bottom left) and double negative (bottom right). A line with a dot indicates physical interaction; the blunt arrow indicates repression. (F) Four-way Venn diagram showing the number of composite miRNA ↔ TF feedback loops obtained after network integration. (Gray) Loops common in two or more algorithms.
All miRNA → TF interactions were visualized into a network model (Fig. 2B). The out-degree of this network (the number of TFs targeted by a given miRNA) is best fit by an exponential distribution (Fig. 2C). Most biological networks characterized to date exhibit a different, power-law degree distribution in which a small number of nodes (network hubs) are extremely highly connected compared with the rest of the nodes in the network (Jeong et al. 2000). The biological significance of this observation became apparent when it was found that hubs in protein–protein interaction networks are often essential for survival or development of the organism (Jeong et al. 2001). For instance, the out-degree distribution of TFs in transcriptional networks follows a power law, and the TF hubs in these networks tend to be essential for viability (Fig. 1B; Deplancke et al. 2006a; Vermeirssen et al. 2007a). The exponential out-degree distribution of the miRNA → TF post-transcriptional network indicates that no clear miRNA hubs can be identified. Interestingly, C. elegans can tolerate removal of most individual miRNAs without obvious developmental defects (Miska et al. 2007). The exponential out-degree distribution of miRNAs and lack of essentiality for most of them both agree with the hypothesis that miRNAs predominantly function to fine-tune gene expression instead of establishing crucial developmental gene expression programs (Bartel and Chen 2004; Hornstein and Shomron 2006).
The in-degree distribution of all miRNA target genes (the number of miRNAs that regulate a target) follows a power law (data not shown). However, the in-degree of the miRNA → TF post-transcriptional network is best fit by an exponential distribution (Fig. 2D). When all genes are considered, we find that the target hubs are enriched for TFs (Fisher exact test, P-value < 0.001), which is in agreement with previous observations in other organisms (Enright et al. 2003; Shalgi et al. 2007). Thus, the exponential in-degree distribution of the miRNA → TF network is likely best explained by the exclusive sampling of TF-encoding target genes.
miRNAs and TFs function together in composite feedback loops
We define a “type I” miRNA ↔ TF composite feedback loop as a miRNA and a TF that mutually regulate each other (Fig. 2E). To systematically identify such loops, we integrated the transcriptional (TF → miRNA) and post-transcriptional (miRNA → TF) networks into a directed, bipartite miRNA gene regulatory network and counted the number of composite miRNA ↔ TF loops. We found 23 type I composite miRNA ↔ TF loops in the integrated network, involving 14 miRNAs and 16 TFs (Fig. 2F; Supplemental Table S7). The 16 TFs represent a variety of families indicating that loops are not biased toward particular types of TFs.
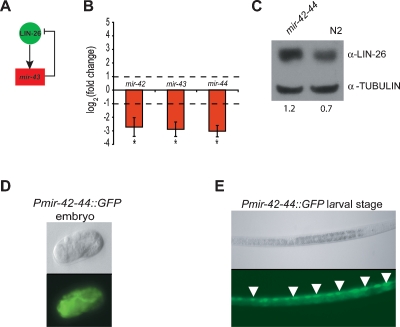
Approaches to confirm the in vivo relevance of composite miRNA ↔ TF feedback loops require assays to determine the regulatory consequences of Y1H interactions, and to assay TF levels in miRNA mutants. There are two possible type I composite miRNA ↔ TF feedback loop subtypes: “single-negative” feedback loops in which the TF activates the miRNA, and “double-negative” feedback loops in which the TF represses the miRNA (Fig. 2E). Here, we focused on the mir-43 ↔ LIN-26 composite feedback loop, which we found to belong to the single-negative class (Fig. 3A). To determine whether LIN-26 activates or represses miRNA expression, we performed TaqMan PCR assays in wild-type and lin-26(ok939) mutant embryos. We found that mir-43 levels decrease approximately eightfold in lin-26(ok939) mutants, compared with wild-type embryos. Two other miRNAs, mir-42 and mir-44, are coexpressed with mir-43 in an operon, and, as expected, they also decrease in lin-26(ok939) mutant embryos (Fig. 3B). These data demonstrate that LIN-26 is an activator of mir-43 expression. One miRNA (mir-63) whose promoter was bound by LIN-26 in Y1H assays increases in lin-26(ok939) animals, suggesting that LIN-26 may also function as a transcriptional repressor (Supplemental Fig. S1A). In total, the expression of four out of six miRNAs targeted by LIN-26 in Y1H assays (67%) changed significantly in lin-26(ok939) mutant embryos. After confirming the transcriptional LIN-26 → mir-43 interaction, we used Western blotting to show that LIN-26 protein levels are increased in mir-42-44(mgDf49) mutant embryos (Fig. 3C) and larvae (Supplemental Fig. S1B), which also confirms the post-transcriptional mir-43 → LIN-26 interaction. Together, these data demonstrate that mir-43 and LIN-26 function in a single-negative composite feedback loop.
Figure 3.
The mir-43 ↔ LIN-26 composite feedback loop. (A) LIN-26 and mir-43 function in a single-negative composite feedback loop. (B) TaqMan PCR analysis shows that mir-43 and the two miRNAs with which it is transcribed from an operon (mir-42 and mir-44) are down-regulated in lin-26(ok939) mutants compared with wild-type animals. The average log2(fold change) of three experiments with standard error of the mean is shown. The dashed lines indicate a twofold difference. Asterisks indicate significant changes. (C) Western blotting shows that LIN-26 is up-regulated in mir-42-44(nDf49) mutant embryos compared with N2 wild-type embryos. α-Tubulin antibody was used as a loading control. Numerical values represent LIN-26 levels after normalization to tubulin. (D) Pmir-42-44 drives expression in the developing embryo. (E) Pmir-42-44 drives expression in seam cells (a subset of seam cells is indicated by white arrows) in C. elegans larvae.
Single-negative type I feedback loops can direct stable coexpression of both components (see the Discussion). Thus, we hypothesized that mir-43 and LIN-26 are coexpressed at least in some tissues. LIN-26 is expressed throughout the lifetime of C. elegans, starting in the early embryo, and is involved in epithelial differentiation (Labouesse et al. 1994). It is expressed in various epithelial tissues, including the hypodermis and seam cells (Landmann et al. 2004). We created transgenic animals that harbor a Pmir-42-44∷GFP fusion and found that Pmir-42-44 drives GFP expression in embryos and throughout development (Fig. 3D,E). In larval stages, expression was detected in hypodermal seam cells, suggesting that LIN-26 and mir-43 are indeed coexpressed (Fig. 3E).
Most single miRNA mutants do not confer a detectable phenotype (Miska et al. 2007), and we were not able to detect a phenotype of a deletion encompassing the mir-42-44 locus. Thus, mir-43 could be involved in epithelial differentiation (as suggested by its expression pattern) but may act redundantly with other (miRNA) genes. Comprehensive analysis of gene expression in C. elegans will likely help to identify additional genes that may function in this process.
miRNA ↔ TF composite feedback loops form higher-order network structures
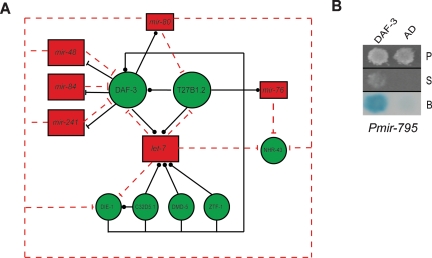
Several miRNAs and TFs are involved in higher-order network subgraphs that include several loops. For instance, we identified higher-order composite feedback loops that contain one miRNA and two TFs (“type II” loops), or one TF and two miRNAs (“type III” loops) (Supplemental Table S7). An example of an even more complex subgraph involving multiple miRNAs, TFs, and composite feedback loops is shown in Figure 4A. The promoters of all members of the let-7 family of miRNAs (which includes let-7, mir-48, mir-84, and mir-241) are bound by DAF-3, and these miRNAs are also predicted to target DAF-3. DAF-3 regulates the expression of mir-241 and mir-48 in dauer formation and may also regulate mir-84 (Fig. 2D). Regulation of let-7 by DAF-3 in vivo could not be examined because let-7 was undetectable in dauer animals using TaqMan PCR assays (data not shown). An additional conserved let-7 family member, mir-795, has only recently been discovered (Ruby et al. 2006). We cloned the promoter of mir-795 and found that it can also interact with DAF-3 in Y1H assays (Fig. 4B). This highly interconnected subgraph suggests that the let-7 family collectively plays a role in dauer formation. We also incorporated available protein–DNA interactions for protein-coding genes (Deplancke et al. 2006a; Vermeirssen et al. 2007a, b). By doing so, we identified several feed-forward loops, for instance, between DAF-3, T27B1.2, and let-7. Since we do not yet have comprehensive protein–DNA interaction data for protein-coding genes, we cannot examine whether in C. elegans, as has been proposed for other systems, feed-forward loops involving miRNAs constitute a network motif (Shalgi et al. 2007; Tsang et al. 2007).
Figure 4.
(A) Example of a higher-order network subgraph containing multiple composite miRNA ↔ TF feedback loops. Black arrows indicate transcriptional interactions; dashed red arrows indicate post-transcriptional interactions. Red rectangles indicate miRNAs, and green circles indicate TFs. Repressive interactions are indicated by blunt arrows, and interactions for which the functional consequence is unknown are indicated by dotted arrows. (B) Y1H assay demonstrating the interaction between Pmir-795 and DAF-3. (P) Permissive media; (S) selective media; (B) β-galactosidase assay; (AD) empty vector.
Composite miRNA ↔ TF feedback loops constitute a network motif
To test whether composite miRNA ↔ TF feedback loops constitute a genuine network motif, we examined if they are enriched in the integrated miRNA gene regulatory network compared with randomized networks. We used three different methods to generate randomized networks. “Edge switching” (ES) is the most stringent method that maintains the individual degree of each node in the network, and changes only the interaction partners (Milo et al. 2002). “Node Replacement I” (NR-I) changes the individual degree of the nodes, the identities of the nodes as well as the interaction partners, but keeps the overall degree distribution of the network constant. Finally, “Node Replacement II” (NR-II) randomizes everything: the identities of the nodes, the interaction partners, and the individual and overall degrees. The use of these three methods not only allows us to determine whether miRNA ↔ TF feedback loops constitute a network motif, but also to investigate potential effects of network architecture (see below).
We found that the integrated miRNA gene regulatory network contains approximately twice as many composite miRNA ↔ TF feedback loops as the average number of loops found in randomized networks (P-value = 0.004 for ES, 0.004 for NR-I, and 0.0002 for NR-II) (Table 1). This demonstrates that composite miRNA ↔ TF feedback loops constitute a genuine network motif.
Table 1.
Network motif analysis
miRNAs and TFs in composite feedback loops provide a high information flow
Interestingly, NR-I and NR-II yielded on average the same number of composite miRNA ↔ TF feedback loops (10.6) (Supplemental Table S7). Randomized networks generated by NR-II possess a more random degree distribution than randomized networks generated by NR-I. Since both methods produce the same average number of composite feedback loops, this suggests that the overall distribution of in-degrees and out-degrees of either miRNAs or TFs does not contribute to the propensity of forming composite miRNA ↔ TF feedback loops. This is in agreement with previously reported mathematical models that examined the expected number of feedback loops in different types of networks with random, scale-free or condensed degree distributions (Itzkovitz et al. 2003).
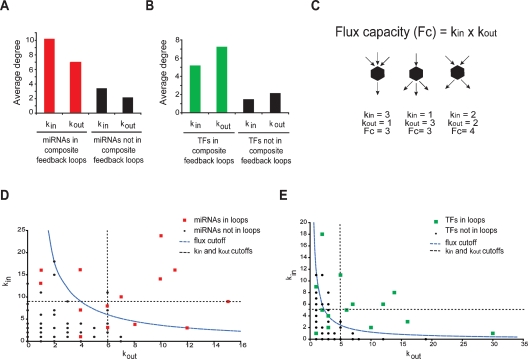
We investigated the individual degrees of nodes that participate in loops. We found that miRNAs and TFs in composite miRNA ↔ TF feedback loops have a higher in-degree and out-degree than nodes that do not participate in loops (Fig. 5A,B). In other words, miRNAs in composite feedback loops regulate more TFs and are regulated by more TFs, and vice versa. We ranked miRNAs and TFs according to their degree, and annotated whether they participate in a loop or not, and found a significant association between loop participation and a high in-degree or out-degree (Supplemental Table S8). We found the same association when nodes with kin = 0 or kout = 0 are removed (Supplemental Table S8). These observations show that high in-degrees and out-degrees are indicators of loop participation.
Figure 5.
TFs and miRNAs in composite feedback loops are characterized by a high flux capacity (Fc). (A) Average in-degree and out-degree of miRNAs that participate in loops (red) or that do not (black). (B) Average in-degree and out-degree of TFs that participate in loops (green) or that do not (black). (C) The Fc of a node is defined by the product of the in- and out-degree. As the example indicates, nodes with the same total number of edges can have a different flux. (D,E) Plot of in-degree (kin) versus out-degree (kout) for each miRNA (D) and TF (E) in the integrated network. Red squares indicate miRNAs involved in composite feedback loops; green squares indicate TFs involved in feedback loops; black circles indicate miRNAs (D) and TFs (E) not involved in composite feedback loops. Dashed lines represent cut-offs for kin, kout, and Fc for the 15% most highly connected nodes.
To better capture the combined high in-degree and out-degree properties of a node, we introduce a new network parameter, referred to as “flux capacity” (Fc = kin × kout) (Fig. 5C). By plotting the out-degree versus the in-degree of each node in the network, we found that a high Fc better describes the difference between nodes that participate in loops and those that do not (Fig. 5D,E; Supplemental Table S8). The association between a high Fc and loop participation suggests that this type of local architecture in a network may predispose loop formation. Indeed, in randomized networks, nodes with a high Fc participate more frequently in loops than nodes with a low Fc (Supplemental Fig. S2A, B). However, this association is less prominent than the enrichment in the real network (Supplemental Fig. S2C). It is important to note that the integrated miRNA network contains twice as many miRNA ↔ TF feedback loops than randomized networks, even when the individual and overall degrees remain unaltered (Table 1, Edge switching). This indicates that, while Fc is a good indicator for feedback loop participation, there are other determinants involved as well.
Discussion
In this study, we present the first experimentally mapped genome-scale TF → miRNA transcription regulatory network in any organism. The integration of this network with a computationally predicted miRNA → TF post-transcriptional network revealed 23 composite miRNA ↔ TF feedback loops in which the TF that binds a miRNA promoter is itself regulated by that same miRNA. This dramatically extends the number of miRNA ↔ TF feedback loops identified to date in any organism. The overall number of miRNA ↔ TF composite feedback loops is likely even higher because both Y1H assays and computational miRNA target prediction algorithms miss interactions. For instance, miRNA target predictions currently mostly include only those that are conserved in related organisms. However, it is likely that several miRNA → target interactions may be species-specific.
There are several explanations for missed interactions in the transcriptional TF → miRNA network. For instance, we did not retrieve any interactions for lsy-6 or mir-1; lsy-6 is a neuronal miRNA, and mir-1 is expressed in muscle, so one could expect to retrieve neuronal and muscle TFs, respectively (Chang et al. 2004; Simon et al. 2008). There are several explanations for false negatives in our data set. First, our library screens are not saturated. For example, when we rescreened Plsy-6, we retrieved CEH-27 and ODR-7, both of which are neuronal TFs (Vermeirssen et al. 2007a). Interestingly, both TargetScan and RNAhybrid predict putative lsy-6-binding sites in the 3′UTR of ceh-27, suggesting that they may constitute another composite miRNA ↔ TF feedback loop (data not shown; these data have been added to EDGEdb, but are not included in the network). Another composite miRNA ↔ TF feedback loop that we did not include in the integrated miRNA network involves mir-788 and IRX-1. This loop was not included because mir-788 is one of several miRNAs identified after our initial analyses (Ruby et al. 2006). We found that IRX-1 interacts with the promoter of mir-788 by Y1H assays (Supplemental Table S5); and Targetscan and RNAhybrid both predict that mir-788 targets the irx-1 3′UTR (Supplemental Table S7). Interestingly, mir-788 and irx-1 are both expressed in the hypodermis, suggesting that they may function in a single-negative composite feedback loop (Reece-Hoyes et al. 2007; this study). Thus, the total number of composite miRNA ↔ TF feedback loops identified in this study is actually 25. These loops provide a framework for further functional analysis, both in terms of the underlying biology and the effects they have on gene expression programs.
The second explanation for the presence of false negatives is that some TFs may not function in the context of Y1H assays. For instance, binding that requires heterodimerization or post-translational modification of TFs is missed in Y1H assays (Deplancke et al. 2006a). Finally, transcription regulation of miRNAs may be controlled by cis-regulatory elements that reside outside of the promoter fragment used in Y1H assays. In the future, it will be important to map the transcription start site of pri-miRNA transcripts to better delineate their promoters and to further improve Y1H assays to enable the retrieval of heterodimers. To test the effect of false negatives, we generated 10 networks in which we randomly removed 10% of the TF → miRNA interactions, and found that feedback loops are still enriched compared with randomized networks (Supplemental Table S9). Thus, we conclude that the presence of false negatives does not affect our overall findings.
Both the transcriptional TF → miRNA and post-transcriptional miRNA → TF networks may also contain false-positive interactions. For instance, many genes do not have an annotated or experimentally determined 3′UTR, and for those genes, the algorithms predict sites in the genomic sequence downstream from the stop codon. Since target prediction algorithms are noisy, we did not include any interactions that were identified by only a single miRNA target prediction algorithm.
Y1H assays may also result in false-positive TF → miRNA interactions. For instance, the DNA fragments used may contain regulatory elements that do not regulate the transcription of miRNAs but that of neighboring genes. In addition, although Y1H assays identify reproducible interactions, it may be difficult to detect their regulatory consequence in vivo (see below). We aimed to minimize the inclusion of technical false positives in the TF → miRNA network by applying a stringent Y1H scoring system that takes the quality of the bait, the prey, and the interaction into account (for a detailed description of the scoring system, see Vermeirssen et al. 2007a). After applying this system, we retained 347 interactions, out of 483 that were present in the raw data (data not shown). The quality of the transcriptional miRNA network is demonstrated by the in vivo regulatory confirmation of many physical TF → miRNA interactions. We found that DAF-3 represses miRNA expression in dauer animals and LIN-26 activates some of its Y1H targets and represses others in embryos. In addition, we found that the FLYWCH TFs FLH-1 and FLH-2 that interact with multiple miRNA promoters repress miRNA expression in the embryo (Ow et al. 2008). Interactions for which we did not detect a regulatory consequence by TaqMan PCR assays should be regarded as inconclusive because they could fall below the detection limit of TaqMan PCR or occur in other developmental or environmental conditions. Indeed, we previously observed that some interactions that occur in a particular cell type or tissue can be detected only as a modest effect by quantitative PCR when whole animals are assayed (Deplancke et al. 2006a).
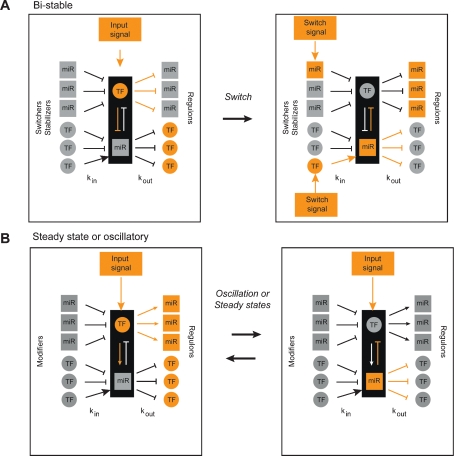
Composite miRNA ↔ TF feedback loops likely participate in specific gene regulatory circuits that precisely control gene expression programs in development or homeostasis. For instance, double-negative feedback loops can generate mutually exclusive or bistable expression of the miRNA or TF, and, hence their downstream targets (Fig. 6A; Johnston et al. 2005). A bistable system can switch between two states, depending on which of multiple potential input signals are active (Gardner et al. 2000). Once a state is established, the input signal is no longer necessary. As a result, bistable systems provide robust and noise-free gene expression programs. Single-negative feedback loops (Fig. 6B) can result in stable expression of both components by reducing stochastic fluctuations in gene expression (Tsang et al. 2007). Alternatively, such a loop can result in oscillatory expression of both components, which depends on additional input signals (Hirata et al. 2002). This could be important in processes such as the cell cycle, molting at different larval stages, or other cyclic processes.
Figure 6.
Model for the function of composite feedback loops in gene expression programs. (A) Bistable systems are generated by double-negative feedback loops. (B) Steady state or oscillatory systems can be generated by single-negative feedback loops. For each type of loop an example is shown in which orange indicates nodes or edges that are “on,” and gray indicates nodes or edges that are “off.”
Many miRNAs and TFs that participate in composite feedback loops are characterized by a high Fc. The high out-degree reflects that both the miRNA and the TF have many downstream targets, or regulons, and the loop ensures that the expression of these regulons is tightly correlated. For instance, in bistable systems (Fig. 6A), expression of the miRNA and TF regulons is mutually exclusive. In steady-state or oscillatory systems, however, the regulons can be coexpressed, either at steady-state levels or in oscillation (Fig. 6B). The high in-degree of both the miRNAs and the TFs that participate in loops suggests that regulon control is highly adaptable: The systems can be subjected to different stabilizers, switches, or modifiers, for instance, in different tissues or under different developmental or environmental conditions.
Two recent bioinformatics studies proposed that miRNAs and their targets are involved in feed-forward as well as feedback loops (Shalgi et al. 2007; Tsang et al. 2007). Shalgi and colleagues searched for pairs of miRNAs and TFs that coregulate target genes by identifying putative miRNA sites and TF-binding sites that co-occur in individual genes. They observed that such miRNA–TF pairs are predicted to regulate each other more frequently than randomly picked pairs, suggesting the existence of feedback loops. Tsang et al. (2007) proposed that correlation or anti-correlation between miRNAs and their targets can result from various types of feed-forward and feedback loops involving miRNAs, their predicted target genes, and upstream regulators (e.g., TFs, kinases), but did not predict any actual loops. We now provide 23 (25 when lsy-6 ↔ CEH-27 and mir-788 ↔ IRX-1 are included) novel miRNA ↔ TF feedback loops and demonstrate that these correspond to a genuine network motif.
Feedback motifs are rare in pure transcriptional networks (Milo et al. 2002; Shen-Orr et al. 2002). We demonstrate that miRNAs are a post-transcriptional missing link to form feedback motifs. It is likely that other post-transcriptional interactions are also involved in feedback regulation. Previously, Margalit and colleagues had shown that protein–protein interactions play a role in generating composite feedback loops in the transcriptional network of Saccharomyces cerevisiae (Yeger-Lotem et al. 2004). It is likely that protein–protein interactions also contribute to the formation of loops involving miRNAs (or other regulators) in C. elegans networks. In the future, it will be important to integrate miRNA gene regulatory networks with genome-scale protein–protein interaction networks and other functional networks as well.
Taken together, we propose that composite miRNA ↔ TF feedback loops provide a common mechanism of gene regulation at a systems level in C. elegans. Similar system-level analyses will reveal whether the individual composite miRNA ↔ TF feedback loops found in other organisms are also examples of a network motif, and whether this mechanism is evolutionarily conserved.
Materials and methods
miRNA promoter definition
We used the 115 miRNA gene predictions available in WormBase WS130 (http://www.wormbase.org) and miRNA registry version 4.0 (http://microrna.sanger.ac.uk). A miRNA promoter is defined here as the intergenic region upstream of the predicted stem–loop sequence annotated in miRBase version4.0 (Supplemental Table S1). We used a minimal length of 300 bp and a maximal length of 2 kb. In total, 79 promoters (for a total of 95 miRNAs) were selected as DNA baits for Y1H assays (Supplemental Table S2). Seventy-one promoters (controlling 84 miRNAs) were successfully cloned into pMW#2 and pMW#3 by Gateway cloning (Walhout et al. 2000b) and integrated into the genome of S. cerevisiae YM4271 (Deplancke et al. 2006b).
Gateway-compatible Y1H assays
Detailed Y1H protocols are described elsewhere (Deplancke et al. 2006b). Y1H screens were performed with each miRNA promoter bait strain versus both AD-wrmcDNA (>106 colonies screened) (Walhout et al. 2000b) and AD-TF (>3 × 105 colonies screened) (Deplancke et al. 2004) prey libraries. For Pmir-61-250, both reporters were highly self-active and, therefore, this bait could not be used. All interactions were retested by PCR/gap repair (Deplancke et al. 2006b). PCR products corresponding to preys that retested were sequenced by Agencourt Bioscience Corporation. Interactors were identified by BLASTX. In total, 669 Interaction Sequence Tags (ISTs) were obtained (Walhout et al. 2000a). Y1H matrix experiments were performed by transforming all interactors obtained in the screens (for which a clone was available), and several TFs found in previous studies (Deplancke et al. 2006a; Vermeirssen et al. 2007a) into each promoter bait strain (130 preys were used in total) (Supplemental Table S3). In addition, six baits and Pmir-788 were screened versus our AD-TF yeast array (Supplemental Table S4; Vermeirssen et al. 2007b). Ninety-eight percent of the ∼10,500 transformations were successful. All interactions obtained were subjected to a stringent standardized scoring and filtering system (Vermeirssen et al. 2007a). Only interactions with a score ≥5 were retained (Supplemental Table S4). All interactions are available in the EDGEdb database (Barrasa et al. 2007).
C. elegans strains
C. elegans N2 wild-type, GR1311 [daf-3(mgDf90)], VC663 [lin-26(ok939)], and MT13372 [mir-42-44(nDf49)] strains were cultured on OP50 seeded NGM plates at 20°C unless otherwise noted.
TaqMan PCR assays
Templates for miRNA TaqMan PCR assays were obtained by collecting 50 N2 and daf-3(mgDf90) dauer animals, or 100 N2 and homozygous lin-26(ok939) segregant mid- to late-stage embryos into lysis buffer (50 mM KCl, 10 mM Tris at pH 8.3, 2.5 mM MgCl2, 0.45% NP-40, 0.45% Tween 20, and 0.01% gelatin). The samples were subjected to 10 cycles of freezing and thawing and incubated for 1 h at 65°C and for 20 min at 95°C. After Trizol Reagent (Invitrogen 15596-026) extraction, the RNA was coprecipitated with glycogen. MicroRNA TaqMan PCR assays were performed following the recommendations of the manufacturer (Applied Biosystems). A TaqMan PCR assay for the small nuclear RNA sn2343 was used as normalization standard.
Induction of dauer larvae formation
Dauer pheromone was prepared as described (Vowels and Thomas 1994). N2 and daf-3(mgDf90) embryos were hatched on 5-mm pheromone plates (NGM without peptone, supplemented with 100 mg/mL streptomycin and seeded with 6× OP50) and incubated for 3 d at 25°C.
Normalization and analysis of TaqMan PCR data
daf-3(mgDf90) versus N2
A total of five independent biological experiments were performed using TaqMan probes for 107 miRNAs in daf-3(mgDf90) and N2 wild-type dauer animals, and each experiment was done in triplicate. In each experiment, a Ctmean value of the three technical replicates was calculated. The standard deviations (SD) within technical repeats were very low (∼0.1–0.4) (data not shown). miRNAs with Ctmean values ≥35 in either wild-type or daf-3(mf90) animals were discarded (21 miRNAs in total). A ΔΔCt value was calculated using the following formula:
Experiments were normalized by first calculating the average ΔΔCt value of all miRNAs within each experiment and then subtracting this value from each individual ΔΔCt value. We only used miRNAs that had ΔΔCt values in four or five experiments (nine miRNAs were discarded). Normalized ΔΔCt values for each miRNA were averaged across all experiments to calculate ΔΔCtfinal, and the standard error of the mean was determined. Only the 48 miRNAs for which both Y1H and TaqMan PCR data were available are visualized in Figure 1D (the fold difference in expression is defined as 2−ΔΔCt). Z-scores were calculated as −ΔΔCtfinal/SD. Z-scores ≥2 were considered significant.
lin-26(ok939) versus N2
A total of three independent biological experiments were performed using TaqMan probes for the eight miRNAs whose promoters were bound by LIN-26 in Y1H assays. In each experiment, a Ctmean value of the three technical replicates was calculated. miRNAs with Ctmean ≥ 35 in either wild-type or lin-26(ok939) animals were discarded (two miRNAs). A ΔΔCt value was calculated using the following formula:
The average ΔΔCt of the three experiments and the standard error of the mean for all three experiments were calculated and are visualized in Figure 3B and Supplemental Figure S1A (the fold difference in expression is defined as 2−ΔΔCt).
Transgenesis
Transgenic promoter∷GFP animals were generated as described (Reece-Hoyes et al. 2007).
miRNA target predictions
Four programs were used to obtain miRNA target predictions: Pictar (Lall et al. 2006), miRanda Targets version 4 (Griffiths-Jones et al. 2006), TargetScan release 3.1 (Lewis et al. 2005), and RNA-hybrid (Rehmsmeier et al. 2004) (by running the algorithm locally). For RNA hybrid predictions, 3′UTR sequences were obtained from WormBase WS159. For genes that did not have an annotated 3′UTR, 300 nt downstream from the stop codon of the longest annotated transcript was taken. Only genes with annotated Caenorhabditis briggsae orthologs were used. Pairing of the seed region was performed allowing either GU pairs, or one bulge on the mRNA side within the seed region (but no G:U wobbles). Predictions were filtered for minimum free energies (MFE) less than −15. RNA-hybrid was run both for C. elegans and C. briggsae, and only predictions common in both were kept. The following modification was made to the RNA-hybrid code: The original program would find the best hybrid (smaller MFE) for a particular miRNA position, block out that entire site in the 3′UTR, and no longer consider any of those nucleotides for other possible matches with that miRNA. This resulted in sites with minimal MFE but that may contain seed imperfections, and the program would miss perfect seed match sites with a slightly higher MFE that could be found by just shifting one or two nucleotides. We modified the code to allow the selection of sites with a better seed match and post-processed the output to eliminate duplicate predictions (Hammell et al. 2008). For subsequent analyses, we only used targets predicted by multiple programs.
For miRNA → TF target predictions, we included all TFs found in Y1H assays, including novel putative TFs. TargetScan targets are defined with GenBank NM identifiers (IDs). Targets predicted by other algorithms are listed with WormBase IDs. We mapped WormBase IDs to NM IDs using a conversion data file kindly provided by G. Bell.
An “all genes” list was assembled by downloading “confirmed” and “partially confirmed” gene IDs from WormBase WS170 (we obtained 14,631 non-TF gene IDs). We were able to match 13,794 WormBase IDs to NM IDs. Therefore, the “all genes” list used to retrieve miRNA predictions had a total of 14,754 genes: 960 TF genes (including novel putative TFs) and 13,794 non-TF genes.
Western blotting
Wild-type and mir-42-44(nDf49) mutant worms were grown in 60-mm OP50 seeded NGM plates and bleached (25% commercial bleach/0.25 M KOH, 5–10 min) when most animals were gravid adults. Eggs were washed in M9 buffer and either collected for extract preparation or incubated for 18 h at 20°C on S medium to allow hatching. To obtain larval stages, worms were placed on OP50 seeded NGM plates and harvested after 15 h (L1) and 46 h (L4), respectively. Worms were washed four times in M9 buffer and resuspended in 2 mL of M9 buffer. Worms were centrifuged (2000 rpm, 1 min), transferred to a weighed 1.5-mL Eppendorf tube, and centrifuged again. Supernatant was removed, and the worm pellet was weighed. To estimate worm pellet volume, we assumed that 1 g = 1 mL. The worm pellet was resuspended in an equal volume of prewarmed 2× sample buffer (2% SDS, 10% glycerol, 0.1% bromophenol blue, 100 mM dithiothreitol, 50 mM Tris-HCL at pH 6.8) and boiled for 10 min. To reduce viscosity, samples were sheared using a 1-cc syringe. Insoluble debris was removed by centrifugation at 14,000 rpm for 1 min, and supernatant was transferred to a clean Eppendorf tube. Approximately 90,000 embryos, 30,000 L1 animals, and 10,000 L4 animals were used for each genotype. Equal volumes of mutant and wild-type extracts were run on NuPAGE 4%–12% bis-Tris Gel (Invitrogen NP0323) and electroblotted onto PVDF membranes. PVDF membranes were incubated overnight at 4°C with anti-LIN-26 antibody (a kind gift from J. Polanowska; 1:2,000 dilution in TBS-Tween with 5% dry milk) or for 1 h with murine anti-α-tubulin antibody (Sigma #T6074) for 1 h at room temperature. HRP-conjugated secondary antibody (goat anti-rabbit for LIN-26 and anti-mouse for α-tubulin, respectively) incubations were done for 1 or 2 h at room temperature.
Network randomizations
The integrated miRNA-TF gene regulatory network is a bipartite directed network that is composed of two types of nodes and two types of edges: TF → PmiRNA and miRNA → TF interactions. To avoid randomly generating meaningless interactions such as miRNA → PmiRNA or TF → TF, the transcriptional and post-transcriptional networks were randomized separately and then combined for motif analysis. Several miRNAs are transcribed from operons that contain two or more miRNAs (Supplemental Table S2). Whereas one miRNA within an operon may be part of a network motif, the others do not have to be part of the same motif. To enable correct motif analysis, we added a third type of edge between miRNA promoters and each of the miRNAs they control. For example, LIN-26 binds the promoter of Pmir-42-44 (that controls mir-42, mir-43, and mir-44) but only mir-43 targets LIN-26. The Pmir → miRNA edges were never randomized.
We used three randomization strategies, and only nodes present in the real networks were used (i.e., all miRNAs whose promoters were cloned, and TFs retrieved by Y1H assays): (1) ES (Milo et al. 2002). Two edges are randomly picked from the network, and the target nodes between them are exchanged (e.g., A-B and C-D will become A-D and C-B). The switch is only performed if the new edges are not already present in the newly created network. A random number of switches, between 100 and 200 times the number of edges, are performed to create randomized networks. With ES, the individual in-degrees and out-degrees of nodes are maintained, and therefore the overall distributions of in-degree, out-degree, and Fc are kept as well. (2) NR-I. This method maintains the overall in-degree and out-degree distributions but randomizes the in-degree and out-degree of individual nodes. The origin nodes (the first component of an edge) are first replaced (e.g., A is replaced with E in the A-B edge). All positions of the origin node are replaced with the same substitute node (e.g., A-B, A-C become E-B, E-C). Then, target nodes are randomized in the same way as the origin nodes (e.g., E-B, D-B become E-F, D-F). (3) NR-II. This method randomly replaces the nodes in the networks without preserving the degree distribution. Nodes were randomized one edge at a time and replaced with a randomly picked node of the same type. If a node substitution results in an edge that is already present, we randomly select a different node to replace it. Nodes can be picked multiple times, resulting in a more random degree distribution and a random individual node degree.
To assess the influence of Fc on loop participation in randomized networks, 400 randomized networks were made using the ES method. We counted the number of times each node, with a specific kout and kin, was part of a loop and the number of times it was not part of a loop in the randomized networks. We then calculated and plotted the ratio between the number of times a node was in a loop versus the number of times it was not in a loop.
Network motif analysis
We used Mfinder (Kashtan et al. 2004) to count the number of motifs in the original and randomized networks. Type I composite miRNA ↔ TF feedback loops are represented by three nodes in the integrated miRNA gene regulatory network: the TF, the miRNA promoter, and the miRNA itself (see above) (Mfinder motif ID98). The higher-order motifs in Supplemental Table S7 include type II (Mfinder motif ID 4546) and type III (Mfinder motif ID 1090054). P-values were calculated empirically, using the distribution of loop counts in the appropriate set of generated randomized networks. Specifically, the P-value is defined as the proportion of random networks that have the same or larger number of motifs as observed in the original network. A P-value ≤0.01 was considered significant.
Acknowledgments
We thank members of the Walhout laboratory and J. Dekker for discussions and critical reading of the manuscript. We thank B. Deplancke for help with the initial cloning of miRNA promoters, and the sequencing staff at Agencourt Bioscience for technical assistance. We thank J. Polanowska for the LIN-26 antibody, M. Green (University of Massachusetts Medical School) for the anti-αTUBULIN antibody, and G. Bell (Bioinformatics and Research Computing, Whitehead Institute) for the gene name conversion file. We are grateful to C. Chen from Applied Biosystems for providing reagents for the TaqMan PCR assays to V.A., and to X. Karp for the dauer pheromone. Finally, some nematode strains were provided by the Caenorhabditis Genetic Center (CGC), which is funded by the NCRR. This work was supported by NIH grant DK068429 to A.J.M.W. and GM348642 to V.A. F.P.R. is supported by NIH grants (HG003224, HG0017115, HL81341, and HG004233), and by the Keck Foundation. M.C.O. was funded in part by NIH post-doctoral fellowship GM070118-02.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1678608.
References
- Alon U. Network motifs: Theory and experimental approaches. Nat. Rev. Genet. 2007;8:450–461. doi: 10.1038/nrg2102. [DOI] [PubMed] [Google Scholar]
- Ambros V., Lee R.C., Lavanway A., Williams P.T., Jewell D. MicroRNAs and other tiny endogenous RNAs in C. elegans. Curr. Biol. 2003;13:807–818. doi: 10.1016/s0960-9822(03)00287-2. [DOI] [PubMed] [Google Scholar]
- Barrasa M.I., Vaglio P., Cavasino F., Jacotot L., Walhout A.J.M. EDGEdb: A transcription factor–DNA interaction database for the analysis of C. elegans differential gene expression. BMC Genomics. 2007;8:21. doi: 10.1186/1471-2164-8-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bartel D.P., Chen C.Z. Micromanagers of gene expression: The potentially widespread influence of metazoan microRNAs. Nat. Rev. Genet. 2004;5:396–400. doi: 10.1038/nrg1328. [DOI] [PubMed] [Google Scholar]
- Baskerville S., Bartel D.P. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA. 2005;11:241–247. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer L.A., Lee T.I., Cole M.F., Johnstone S.E., Levine S.S., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracht J., Hunter S., Eachus R., Weeks P., Pasquinelli A.E. Trans-splicing and polyadenylation of let-7 microRNA primary transcripts. RNA. 2004;10:1586–1594. doi: 10.1261/rna.7122604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S., Johnston R.J., Frokjaer-Jensen C., Lockery S., Hobert O. MicroRNAs act sequentially and asymmetrically to control chemosensory laterality in the nematode. Nature. 2004;430:785–789. doi: 10.1038/nature02752. [DOI] [PubMed] [Google Scholar]
- Deplancke B., Dupuy D., Vidal M., Walhout A.J.M. A Gateway-compatible yeast one-hybrid system. Genome Res. 2004;14:2093–2101. doi: 10.1101/gr.2445504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deplancke B., Mukhopadhyay A., Ao W., Elewa A.M., Grove C.A., Martinez N.J., Sequerra R., Doucette-Stam L., Reece-Hoyes J.S., Hope I.A., et al. A gene-centered C. elegans protein–DNA interaction network. Cell. 2006a;125:1193–1205. doi: 10.1016/j.cell.2006.04.038. [DOI] [PubMed] [Google Scholar]
- Deplancke B., Vermeirssen V., Arda H.E., Martinez N.J., Walhout A.J.M. Gateway-compatible yeast one-hybrid screens. Cold Spring Harb. Protoc. 2006b doi: 10.1101/pdb. prot4590. [DOI] [PubMed] [Google Scholar]
- Enright A.J., John B., Gaul U., Tuschl T., Sander C., Marks D.S. MicroRNA targets in Drosophila. Genome Biol. 2003;5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farh K.K., Grimson A., Jan C., Lewis B.P., Johnston W.K., Lim L.P., Burge C.B., Bartel D.P. The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science. 2005;310:1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- Fazi F., Rosa A., Fatica A., Gelmetti V., De Marchis M.L., Nervi C., Bozzoni I. A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and C/EBPα regulates human granulopoiesis. Cell. 2005;123:819–831. doi: 10.1016/j.cell.2005.09.023. [DOI] [PubMed] [Google Scholar]
- Gardner T.S., Cantor C.R., Collins J.J. Construction of a genetic toggle switch in Escherichia coli. Nature. 2000;403:339–342. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S., Grocock R.J., van Dongen S., Bateman A., Enright A.J. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammell M., Long D., Zhang L., Lee A., Carmack C.S., Han M., Ding Y., Ambros V. mirWIP: microRNA target prediction based on microRNA-containing ribonucleoprotein-enriched transcripts. Nat. Methods. 2008 doi: 10.1038/nmeth.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbison C.T., Gordon D.B., Lee T.I., Rinaldi N.J., Macisaac K.D., Danford T.W., Hannett N.M., Tagne J.B., Reynolds D.B., Yoo J., et al. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata H., Yoshiura S., Ohtsuka T., Bessho Y., Harada T., Yoshikawa K., Kageyama R. Oscillatory expression of the bHLH factor Hes1 regulated by a negative feedback loop. Science. 2002;298:840–843. doi: 10.1126/science.1074560. [DOI] [PubMed] [Google Scholar]
- Hollenhorst P.C., Shah A.A., Hopkins C., Graves B.J. Genome-wide analyses reveal properties of redundant and specific promoter occupancy within the ETS gene family. Genes & Dev. 2007;21:1882–1894. doi: 10.1101/gad.1561707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornstein E., Shomron N. Canalization of development by microRNAs. Nat. Genet. 2006;38:S20–S24. doi: 10.1038/ng1803. [DOI] [PubMed] [Google Scholar]
- Itzkovitz S., Milo R., Kashtan N., Ziv G., Alon U. Subgraphs in random networks. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2003;68:026127. doi: 10.1103/PhysRevE.68. 026127. [DOI] [PubMed] [Google Scholar]
- Jeong H., Tombor B., Albert R., Oltvai Z.N., Barabasi A.-L. The large-scale organization of metabolic networks. Nature. 2000;407:651–654. doi: 10.1038/35036627. [DOI] [PubMed] [Google Scholar]
- Jeong H., Mason S.P., Barabasi A.-L., Oltvai Z.N. Lethality and centrality in protein networks. Nature. 2001;411:41–42. doi: 10.1038/35075138. [DOI] [PubMed] [Google Scholar]
- Johnson S.M., Lin S.-Y., Slack F.J. The time of appearance of the C. elegans let-7 microRNA is transcriptionally controlled utilizing a temporal regulatory element in its promoter. Dev. Biol. 2003;259:364–379. doi: 10.1016/s0012-1606(03)00202-1. [DOI] [PubMed] [Google Scholar]
- Johnson S.M., Grosshans H., Shingara J., Byrom M., Jarvis R., Cheng A., Labourier E., Reinert K.L., Brown D., Slack F. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- Johnston R.J., Chang S., Etchberger J.F., Ortiz C.O., Hobert O. MicroRNAs acting in a double-negative feedback loop to control a neuronal cell fate decision. Proc. Natl. Acad. Sci. 2005;102:12449–12454. doi: 10.1073/pnas.0505530102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashtan N., Itzkovitz S., Milo R., Alon U. Efficient sampling algorithm for estimating subgraph concentrations and detecting network motifs. Bioinformatics. 2004;20:1746– 1758. doi: 10.1093/bioinformatics/bth163. [DOI] [PubMed] [Google Scholar]
- Kim V.N. MicroRNA biogenesis: Coordinated cropping and dicing. Nat. Rev. Mol. Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- Kim J., Inoue K., Ishii J., Vanti W.B., Voronov S.V., Murchison E., Hannon G., Abeliovich A. A microRNA feedback circuit in midbrain dopamine neurons. Science. 2007;317:1220–1224. doi: 10.1126/science.1140481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labouesse M., Sookhareea S., Horvitz H.R. The Caenorhabditis elegans gene lin-26 is required to specify the fates of hypodermal cells and encodes a presumptive zinc-finger transcription factor. Development. 1994;120:2359–2368. doi: 10.1242/dev.120.9.2359. [DOI] [PubMed] [Google Scholar]
- Lall S., Grun D., Krek A., Chen K., Wang Y.-L., Dewey C.N., Sood P., Colombo T., Bray N., MacMenamin P., et al. A genome-wide map of conserved microRNA targets in C. elegans. Curr. Biol. 2006;16:460–471. doi: 10.1016/j.cub.2006.01.050. [DOI] [PubMed] [Google Scholar]
- Landmann F., Quintin S., Labouesse M. Multiple regulatory elements with spatially and temporally distinct activities control the expression of the epithelial differentiation gene lin-26 in C. elegans. Dev. Biol. 2004;265:478–490. doi: 10.1016/j.ydbio.2003.09.009. [DOI] [PubMed] [Google Scholar]
- Lee R.C., Feinbaum R.L., Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Lee Y., Kim M., Han J., Yeom K.-H., Lee S., Baek S.H., Kim V.N. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis B.P., Burge C.B., Bartel D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Li M., Jones-Rhoades M.W., Lau N.C., Bartel D.P., Rougvie A.E. Regulatory mutations of mir-48, a C. elegans let-7 family microRNA, cause developmental timing defects. Dev. Cell. 2005;9:415–422. doi: 10.1016/j.devcel.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Lim L.P., Lau N.C., Weinstein E.G., Abdelhakim A., Yekta S., Rhoades M.W., Burge C.B., Bartel D.P. The microRNAs of Caenorhabditis elegans. Genes & Dev. 2003;17:991–1008. doi: 10.1101/gad.1074403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milo R., Shen-Orr S., Itzkovitz S., Kashtan N., Chklovskii D., Alon U. Network motifs: Simple building blocks of complex networks. Science. 2002;298:824–827. doi: 10.1126/science.298.5594.824. [DOI] [PubMed] [Google Scholar]
- Miska E.A., Alvarez-Saavedra E., Abbott A.L., Lau N.C., Hellman A.B., McGonagle S.M., Bartel D.P., Ambros V.R., Horvitz H.R. Most Caenorhabditis elegans microRNAs are individually not essential for development or viability. PLoS Genet. 2007;3:e215. doi: 10.1371/journal. pgen.0030215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ow M.C., Martinez N.J., Olsen P.H., Silverman S., Barrasa M.I., Conradt B., Walhout A.J.M., Ambros V. The FLYWCH transcription factors FLH-1, FLH-2, and FLH-3 repress embryonic expression of microRNA genes in C. elegans. Genes & Dev. 2008 doi: 10.1101/gad. 1678808. (this issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson G.I., Koweek A., Wong A., Liu Y., Ruvkun G. The DAF-3 Smad protein antagonizes TGF-Β-related receptor signaling in the Caenorhabditis elegans dauer pathway. Genes & Dev. 1997;11:2679–2690. doi: 10.1101/gad.11.20.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reece-Hoyes J.S., Deplancke B., Shingles J., Grove C.A., Hope I.A., Walhout A.J.M. A compendium of C. elegans regulatory transcription factors: A resource for mapping transcription regulatory networks. Genome Biol. 2005;6:R110. doi: 10.1186/gb-2005-6-13-r110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reece-Hoyes J.S., Shingles J., Dupuy D., Grove C.A., Walhout A.J., Vidal M., Hope I.A. Insight into transcription factor gene duplication from Caenorhabditis elegans Promoterome-driven expression patterns. BMC Genomics. 2007;8:27. doi: 10.1186/1471-2164-8-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehmsmeier M., Steffen P., Hochsmann M., Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby J.G., Jan C., Player C., Axtell M.J., Lee W., Nusbaum C., Ge H., Bartel D.P. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell. 2006;127:1193–1207. doi: 10.1016/j.cell.2006.10.040. [DOI] [PubMed] [Google Scholar]
- Sandmann T., Girardot C., Brehme M., Tongprasit W., Stolc V., Furlong E.E. A core transcriptional network for early mesoderm development in Drosophila melanogaster. Genes & Dev. 2007;21:436–449. doi: 10.1101/gad.1509007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethupathy P., Megraw M., Hatzigergiou A.G. A guide through present computational approaches for the identification of mammalian microRNA targets. Nat. Methods. 2006;3:881–886. doi: 10.1038/nmeth954. [DOI] [PubMed] [Google Scholar]
- Shalgi R., Lieber D., Oren M., Pilpel Y. Global and local architecture of the mammalian microRNA-transcription factor regulatory network. PLoS Comput. Biol. 2007;3:e131. doi: 10.1371/journal.pcbi.0030131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen-Orr S.S., Milo R., Mangan S., Alon U. Network motifs in the transcriptional regulation network of Escherichia coli. Nat. Genet. 2002;31:64–68. doi: 10.1038/ng881. [DOI] [PubMed] [Google Scholar]
- Simon D.J., Madison J.M., Conery A.L., Thompson-Peer K.L., Soskis M., Ruvkun G.B., Kaplan J.M., Kim J.K. The microRNA miR-1 regulates a MEF-2-dependent retrograde signal at neuromuscular junctions. Cell. 2008;133:891–902. doi: 10.1016/j.cell.2008.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sood P., Krek A., Zavolan M., Macino G., Rajewsky N. Cell-type-specific signatures of microRNAs on target mRNA expression. Proc. Natl. Acad. Sci. 2006;103:2746–2751. doi: 10.1073/pnas.0511045103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark A., Brennecke J., Bushati N., Russell R.B., Cohen S.M. Animal MicroRNAs confer robustness to gene expression and have a significant impact on 3′UTR evolution. Cell. 2005;123:1133–1146. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- Sylvestre Y., De Guire V., Querido E., Mukhopadhyay U.K., Bourdeau V., Major F., Ferbeyre G., Chartrand P. An E2F/miR-20a autoregulatory feedback loop. J. Biol. Chem. 2007;282:2135–2143. doi: 10.1074/jbc.M608939200. [DOI] [PubMed] [Google Scholar]
- Thatcher J.D., Haun C., Okkema P.G. The DAF-3 Smad binds DNA and represses gene expression in the Caenorhabditis elegans pharynx. Development. 1999;126:97–107. doi: 10.1242/dev.126.1.97. [DOI] [PubMed] [Google Scholar]
- Tsang J., Zhu J., van Oudenaarden A. MicroRNA-mediated feedback and feedforward loops are recurrent network motifs in mammals. Mol. Cell. 2007;26:753–767. doi: 10.1016/j.molcel.2007.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varghese J., Cohen S.M. microRNA miR-14 acts to modulate a positive autoregulatory loop controlling steroid hormone signaling in Drosophila. Genes & Dev. 2007;21:2277–2282. doi: 10.1101/gad.439807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeirssen V., Barrasa M.I., Hidalgo C., Babon J.A.B., Sequerra R., Doucette-Stam L., Barabasi A.L., Walhout A.J.M. Transcription factor modularity in a gene-centered C. elegans core neuronal protein–DNA interaction network. Genome Res. 2007a;17:1061–1071. doi: 10.1101/gr.6148107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeirssen V., Deplancke B., Barrasa M.I., Reece-Hoyes J.S., Arda H.E., Grove C.A., Martinez N.J., Sequerra R., Doucette-Stamm L., Brent M., et al. Matrix and Steiner-triple-system smart pooling assays for high-performance transcription regulatory network mapping. Nat. Methods. 2007b;4:659–664. doi: 10.1038/nmeth1063. [DOI] [PubMed] [Google Scholar]
- Vowels J.J., Thomas J.H. Multiple chemosensory defects in daf-11 and daf-21 mutants of Caenorhabditis elegans. Genetics. 1994;138:303–316. doi: 10.1093/genetics/138.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walhout A.J.M. Unraveling transcription regulatory networks by protein–DNA and protein–protein interaction mapping. Genome Res. 2006;16:1445–1454. doi: 10.1101/gr.5321506. [DOI] [PubMed] [Google Scholar]
- Walhout A.J.M., Sordella R., Lu X., Hartley J.L., Temple G.F., Brasch M.A., Thierry-Mieg N., Vidal M. Protein interaction mapping in C. elegans using proteins involved in vulval development. Science. 2000a;287:116–122. doi: 10.1126/science.287.5450.116. [DOI] [PubMed] [Google Scholar]
- Walhout A.J.M., Temple G.F., Brasch M.A., Hartley J.L., Lorson M.A., van den Heuvel S., Vidal M. GATEWAY recombinational cloning: Application to the cloning of large numbers of open reading frames or ORFeomes. Methods Enzymol. 2000b;328:575–592. doi: 10.1016/s0076-6879(00)28419-x. [DOI] [PubMed] [Google Scholar]
- Wang J., Kim S.K. Global analysis of dauer gene expression in C. elegans. Development. 2003;130:1621–1634. doi: 10.1242/dev.00363. [DOI] [PubMed] [Google Scholar]
- Wightman B., Ha I., Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- Yeger-Lotem E., Sattath S., Kashtan N., Itzkovitz S., Milo R., Pinter R.Y., Alon U., Margalit H. Network motifs in integrated cellular networks of transcription-regulation and protein–protein interaction. Proc. Natl. Acad. Sci. 2004;101:5934–5939. doi: 10.1073/pnas.0306752101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo A.S., Greenwald I. LIN-12/Notch activation leads to microRNA-mediated downregulation of Vav in C. elegans. Science. 2005;310:1330–1333. doi: 10.1126/science.1119481. [DOI] [PMC free article] [PubMed] [Google Scholar]