Abstract
Morphogens are secreted signaling molecules that form concentration gradients and control cell fate in developing tissues. During development, it is essential that morphogen range is strictly regulated in order for correct cell type specification to occur. One of the best characterized morphogens is Drosophila Decapentaplegic (Dpp), a BMP signaling molecule that patterns the dorsal ectoderm of the embryo by activating the Mad and Medea (Med) transcription factors. We demonstrate that there is a spatial and temporal expansion of the expression patterns of Dpp target genes in SUMO pathway mutant embryos. We identify Med as the primary SUMOylation target in the Dpp pathway, and show that failure to SUMOylate Med leads to the increased Dpp signaling range observed in the SUMO pathway mutant embryos. Med is SUMO modified in the nucleus, and we provide evidence that SUMOylation triggers Med nuclear export. Hence, Med SUMOylation provides a mechanism by which nuclei can continue to monitor the presence of extracellular Dpp signal to activate target gene expression for an appropriate duration. Overall, our results identify an unusual strategy for regulating morphogen range that, rather than impacting on the morphogen itself, targets an intracellular transducer.
Keywords: SUMO, morphogen, Medea, Dpp, Drosophila embryo
The Drosophila TGF-β/BMP signaling molecule Dpp functions as a morphogen to pattern many different tissues throughout development (Ashe and Briscoe 2006). In the early embryo, evidence suggests that the potent signaling molecule is a heterodimer between Dpp and the related TGF-β signaling molecule Screw (Scw) (Shimmi et al. 2005). A Dpp/Scw gradient is established that subdivides the dorsal ectoderm of the embryo into amnioserosa and dorsal epidermis cell fates. Dpp/Scw signals through a heteromeric Thickveins (Tkv), Punt, and Saxophone (Sax) receptor complex leading to the phosphorylation of the Mothers against Dpp (Mad) transcription factor. Phosphorylated Mad (pMad) interacts with a second transcription factor, Med (Affolter et al. 2001). Together, these Smad transcription factors enter the nucleus where they activate at least three different thresholds of gene activity in response to the Dpp/Scw gradient (Ashe et al. 2000).
Med and its vertebrate ortholog Smad4 have been found to constitutively shuttle between the nucleus and cytoplasm in a signal-independent manner (Pierreux et al. 2000; Yao et al. 2008). Smad4 contains both a nuclear localization signal (NLS) and a CRM-1-dependent nuclear export signal (NES), and it has been proposed that the relative strengths of these two signals within a tissue regulate the amount of shuttling. Smad4, which shuttles into the nucleus in the absence of signal, is unable to activate transcription (Pierreux et al. 2000), possibly due to recruitment of a repressive complex harboring the SnoN oncoprotein (Stroschein et al. 1999).
The small ubiquitin-like modifier protein SUMO is conjugated to its substrate through the sequential activities of E1, E2, and E3 enzymes. Ubc-9, the essential E2 enzyme, catalyzes the conjugation of SUMO to the target lysine, typically located in a ΨKxE consensus motif (where Ψ is a large hydrophobic residue and x is any residue) (Hay 2005). An extended SUMO motif comprising negatively charged residues downstream from the SUMO site has also been proposed (Yang et al. 2006). However, some substrates are modified at sites that deviate from the consensus (Johnson 2004). Although four SUMO paralogs exist in mammals (Zhao 2007), only a single SUMO protein has been identified in flies, which is most similar to vertebrate SUMO-2/3 (Long and Griffith 2000). The nature of the proteins shown to be SUMOylated suggests that SUMOylation is predominantly a nuclear process (Hay 2005). Consistent with this, the SUMOylation machinery is localized mainly to the nucleus in both mammalian and fly cells (Rodriguez et al. 2001; Smith et al. 2004). Moreover, addition of both a consensus SUMO site and a NLS are necessary to confer SUMOylation upon an artificial substrate (Rodriguez et al. 2001). SUMOylation is a reversible modification, and a family of SUMO-specific proteases has been identified that catalyze deSUMOylation (Smith et al. 2004; Hay 2007).
Different types of proteins have been shown to be modified by SUMO, resulting in a range of biological effects including regulation of protein–protein interactions, subcellular localization, or stability (Hay 2005). As such, the SUMO pathway regulates different types of cellular events and processes including cell cycle progression, subcellular transport, gene expression, maintenance of genome integrity and subnuclear organization, circadian clock rhythmicity, and cellular differentiation. In terms of the latter, SUMO has been linked to development of the mammalian palate (Alkuraya et al. 2006), zebrafish neural tissue (Bakkers et al. 2005), Caenorhabditis elegans reproductive system (Broday et al. 2004; Leight et al. 2005; Poulin et al. 2005), and Drosophila anterior segments (Epps and Tanda 1998).
Many SUMO substrates for which developmental roles have been identified are proteins involved in the regulation of transcription. For example, SUMO modification of SOX transcriptional activators may provide a mechanism for modulating their activity in different developmental contexts (Savare et al. 2005). During C. elegans development, SUMOylation of the Polycomb group repressor protein SOP-2 is necessary for it to repress Hox genes (Zhang et al. 2004), whereas modification of the Drosophila gypsy insulator may influence the establishment of chromatin domains (Capelson and Corces 2006). Smad4, the vertebrate ortholog of Med (Wisotzkey et al. 1998), is SUMO modified in tissue culture cells, resulting in either activation (Lin et al. 2003a) or repression (Long and Griffith 2000) of its activity depending on the reporter analyzed. Although SUMOylation activates some transcription factors, it is more commonly associated with transcriptional repression. SUMO can mediate repression through the recruitment of corepressors, such as histone deacetylases, or through sequestration of the modified transcription factor in subnuclear compartments; e.g., PML bodies, which are unfavorable for transcription (Hay 2005). The type I TGF-β receptor is also SUMO modified, leading to enhanced recruitment and phosphorylation of Smad3. However, neither the type I or II BMP receptors are SUMOylated (Kang et al. 2008).
Here, we investigate the role of the SUMO pathway in modulating morphogen signaling in the context of the developing Drosophila embryo. We show that SUMOylation of Med negatively regulates its activity, thereby restricting the signaling range of the key Dpp morphogen in the Drosophila embryo. Mechanistic insight is provided by our data, which show that SUMO modification of Med promotes its nuclear export. Together, our data suggest that SUMO modification is an important elaboration on the French flag model of positional information with respect to Dpp signaling in the Drosophila embryo.
Results
The SUMO pathway negatively regulates Dpp signaling
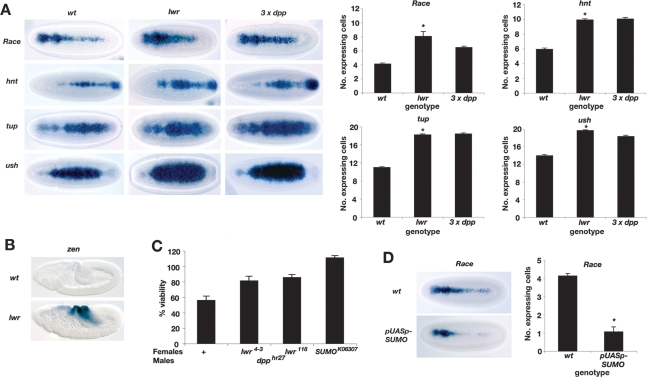
All the major components of the SUMO pathway are maternally expressed in the Drosophila embryo (data not shown), including the essential E2-conjugating enzyme Ubc9. Ubc9 is encoded by the lesswright (lwr, also called semushi) gene in flies, and embryos from females carrying homozygous lwr118 mutant germline clones have greatly reduced lwr mRNA (Epps and Tanda 1998). Therefore, we analyzed Dpp target gene expression in lwr118 mutant embryos with reduced maternal and zygotic lwr activity (hereafter referred to as lwr mutant embryos). The characteristic expression patterns of different Dpp threshold responses (Ashe et al. 2000) are expanded compared with wild-type embryos (Fig. 1A). The average number of expressing cells for the different target genes was quantified and compared with that of wild-type embryos and those with an extra copy of dpp (Fig. 1A). The expansion of Dpp target gene expression is similar to that of embryos carrying an extra copy of dpp, demonstrating that failure to SUMOylate one or more substrates in the embryo increases the range of the Dpp signal.
Figure 1.
The SUMO pathway negatively regulates Dpp signaling. (A) RNA in situ hybridization of Dpp target genes in embryos at the onset of gastrulation that are either wild-type, lwr mutant, or carrying three copies of dpp, as labeled. Dorsal views of the embryos are shown with anterior to the left. lwr mutant embryos were collected from females that were induced to produce lwr118 homozygous mutant germline clones using the FLP-DFS technique. Embryos generated using this method are smaller and more rounded in size than wild-type embryos. The graphs show quantification of the number of expressing cells in the middle of the embryo for each gene; n = 15; error bars are SEM; (*) P < 0.05 compared with wild type (Student’s t-test). (B) Stage 8 wild-type and lwr mutant embryos showing zen expression as visualized by RNA in situ hybridization. (C) Graph showing percent viability of dpphr27 mutant progeny from females that are either wild-type or heterozygous for the lwr118, lwr4-3, or sumok06307 mutations. Viability is expressed as a percentage of the number of dpphr27 mutant progeny compared with those that inherit the wild-type chromosome paternally. No genetic interaction is observed when the lwr mutations are transmitted paternally (data not shown). (D) RNA in situ hybridization of Race expression in embryos from tub-GAL4; pUASp-sumo females, which results in sumo overexpression. Quantification is as described in A.
Given this effect on the spatial range of the Dpp signal, we also tested for any temporal perturbation of signaling. Previously, zerknüllt (zen) expression has been used as a marker to demonstrate an increased duration of the Dpp signal, as zen normally ceases to be expressed in stage 8 wild-type embryos (Podos et al. 2001). Visualization of zen expression in embryos at the onset of gastrulation reveals that, like other Dpp target genes, it is expanded (data not shown). Moreover, in gastrulating embryos, zen expression persists in the lwr mutant embryos in contrast to wild-type embryos (Fig. 1B). Analysis of the cuticle phenotype of the lwr mutant embryos reveals that, in addition to the pair-rule phenotype described previously (Epps and Tanda 1998), a proportion have a posterior hole or a mild dorsal closure defect (Supplemental Fig. S1). A posterior hole has been described previously in dSmurf mutants, which lack the dSmurf ubiquitin ligase that degrades Mad, and attributed to hindgut defects due to an inappropriate perdurance of Dpp signaling (Podos et al. 2001). The puckered dorsal closure defect observed is also characteristic of ectopic Dpp signaling in late stage embryos (Martin-Blanco et al. 2000). Therefore, in the absence of SUMOylation there is temporal misregulation of Dpp signaling in addition to the effect on the spatial range.
As Dpp signaling is increased in the lwr mutant embryos, we analyzed the expression patterns of dpp and short gastrulation (sog) in the mutant embryos. The Sog protein has a pivotal role in formation of the Dpp gradient in the embryo (Ashe and Levine 1999). As shown in Supplemental Figure S2, the expression patterns of sog and dpp are similar to the wild-type patterns, suggesting that the lwr mutant phenotype is not due to alteration of the Dpp gradient, but instead affects a pathway component downstream from Dpp.
To confirm the link between SUMO and Dpp signaling, we also tested for genetic interactions between either the lwr118, lwr4-3, or sumok06307 mutations and the dpphr27 allele. lwr118 and lwr4-3 are independently isolated recessive lethal lwr alleles (Epps and Tanda 1998; Apionishev et al. 2001). dpphr27 is a weak hypomorphic allele (Wharton et al. 1993) that results in ∼50% lethality of mutant progeny when crossed with wild-type females (Fig. 1C). However, there is increased survival of dpp mutant progeny from females heterozygous for the lwr or sumo alleles, providing further support that Dpp signaling is increased in embryos with reduced SUMOylation capacity.
Given that compromising the SUMO pathway augments Dpp signaling, we tested the effect of SUMO overexpression in embryos using the GAL4/UAS system. Increased SUMO expression leads to a reduction in expression of the peak Dpp target gene Race in the presumptive amnioserosa (Fig. 1D), consistent with increased SUMOylation reducing Dpp signaling. Together, these data demonstrate that the SUMO pathway negatively regulates Dpp signaling in the embryo.
Med is SUMO modified in vitro
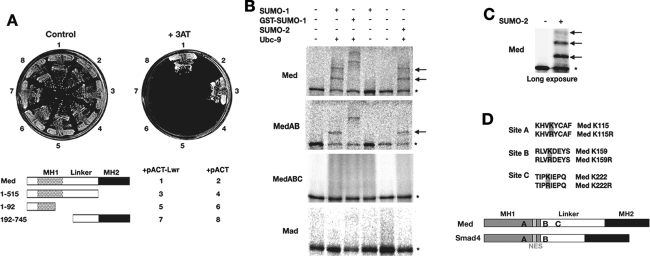
As Smad4 has been shown to be SUMOylated (Lin et al. 2003a), we tested whether Drosophila Med is also a target. First, we used the yeast two-hybrid assay to investigate a possible interaction between Lwr and Med. These two proteins do interact, and this interaction requires sequences flanking the junction between the MH1 and linker domains (Fig. 2A). We next used the in vitro SUMOylation assay (Desterro et al. 1998) to investigate Med as a SUMOylation target. Med is SUMO modified in vitro, yet mutation of the two amino acids—K113, K159 (MedAB)—equivalent to those shown to be necessary for SUMOylation of Smad4 (Lin et al. 2003a) does not abolish Med SUMOylation (Fig. 2B,D). Instead the K113R, K159R mutant Med contains one additional major site that can be SUMO modified. In fact, on a long exposure of a Med in vitro SUMO modification assay, in addition to two strong shifts equivalent to addition of SUMO, a third weak shift can be seen (Fig. 2C). Mutational analysis of lysine residues in various putative SUMO sites identified PKIE (K222) as the remaining SUMO site in Med. Mutation of all three SUMO sites in the MedABC mutant abolishes its SUMOylation in vitro (Fig. 2B). Analysis of Mad as a potential substrate using this in vitro assay revealed that it is not SUMO modified (Fig. 2B), consistent with the lack of a consensus SUMO site in the protein.
Figure 2.
Med is SUMOylated on three sites. (A) Yeast two-hybrid analysis showing an interaction between Med and Lwr proteins. The key shows the regions of Med protein that were introduced into yeast as GAL4 DNA-binding domain fusions, along with Lwr or control activation domain plasmids. Control growth and that on selection medium is shown. (B) In vitro SUMO assays for Mad and Med. In vitro translated, 35S-labeled wild-type Mad, Med, and mutant Med proteins were incubated in the presence and absence of different components as indicated. SUMO-1 and SUMO-2 are mammalian SUMO paralogs; GST-SUMO-1 is an additional control, as this larger protein gives a bigger size shift. Asterisks denote unmodified protein and arrows point to the positions of SUMOylated Med. Mad protein is not a substrate for SUMO modification in vitro. (C) A long exposure of a Med in vitro SUMOylation assay shows a weak third retarded band, consistent with the addition of three SUMOs. (D) The sites of SUMO modification are shown, with the introduced mutations. Their locations within the Med MH1 and Linker domains are indicated, with the SUMO sites that are conserved in Smad4 for comparison. The Med/Smad4 NES is also shown as a gray box.
Med is SUMOylated in vivo
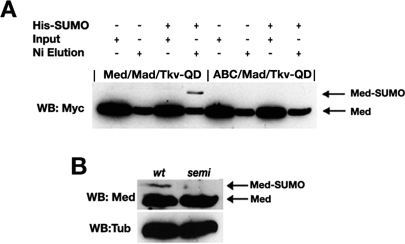
Next, we tested whether Med is SUMOylated in Drosophila S2 tissue culture cells by transfecting Myc epitope-tagged wild type or the triple Lys mutant of Med (denoted MedABC) along with activated Thickveins receptor (Tkv-QD), to activate the pathway in the absence of Dpp ligand, Mad, and His6-tagged Drosophila SUMO. The latter allowed nickel purification of SUMO-conjugated proteins (Rodriguez et al. 1999), and Western blot analysis with anti-Myc antibody reveals that Med is SUMOylated whereas the triple mutant is not (Fig. 3A). As Med protein binds to nickel, its presence serves as a control for the nickel purification. Using a Med antibody, we were also able to detect a more slowly migrating protein in extract prepared from wild-type but not lwr mutant embryos (Fig. 3B). This observation is consistent with the retarded protein being SUMOylated Med in wild-type embryos, whereas lwr mutant embryos are unable to SUMO modify Med. Overall, our SUMOylation data reveal that Med, but not Mad, is a SUMO target in Drosophila, and that Med has three possible sites for SUMO modification.
Figure 3.
Med is SUMO modified in vivo. (A) In vivo SUMOylation of Med. Myc epitope-tagged wild-type or mutant Med was transfected into S2 cells with Mad, activated Tkv receptor, and His6-SUMO as shown. His6-SUMO-conjugated proteins were purified by nickel affinity chromatography, and SUMO-modified Med was detected with an anti-Myc antibody. Inputs representing a proportion of the transfected cells, and the elutions from the nickel beads are shown. Med protein binds to nickel, and controls for the purification. (B) Med is SUMO modified in the embryo. Western blot analysis of Med in 2- to 4-h embryonic extracts detects a more slowly migrating form of the protein. This form is not detected in extracts from lwr mutant embryos, consistent with it representing SUMO-modified Med. A Tubulin Western blot is included as a loading control.
Expression of non-SUMOylatable Med phenocopies the lwr mutant phenotype
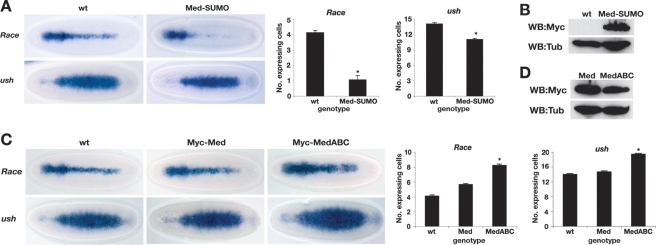
Although we identified Med as a SUMO target, many proteins have been suggested to be SUMOylated (Hay 2005). Therefore, to focus on the effect of Med SUMOylation in vivo, we introduced a Med-SUMO fusion protein into the embryo. We fused the SUMO cDNA sequence in-frame to the start of the Med coding sequence, and expressed this Myc epitope-tagged fusion protein directly under the control of the ubiquitin promoter, which drives maternal and zygotic expression (Fig. 4A,B; Lee et al. 1988). The Race and u-shaped (ush) type I and II Dpp threshold responses were analyzed and quantified in transgenic embryos. Overexpression of Med-SUMO leads to a loss of Race expression in the presumptive amnioserosa and a narrowing of the ush expression pattern (Fig. 4A), similar to that observed in dpp heterozygous embryos (Ashe et al. 2000). To control for the SUMO fusion inhibiting target gene expression by acting in a dominant-negative manner, we compared the effects of GAL4-driven overexpression of UAS-Med and UAS-Med-GFP transgenes on the number of amnioserosa Krüppel (Kr)-positive cells in later-stage embryos. The UAS-Med and UAS-Med-GFP transgenes both increase the number of Kr-positive cells (Supplemental Fig. S3A). We also tested our Ub:Med transgene that does not alter the number of amnioserosa cells in heterozygotes, presumably due to lower expression from the Ubiquitin promoter than that achieved with GAL4-UAS amplification. However, our Ub:Med-SUMO transgene reduces the number of amnioserosa cells (Supplemental Fig. S3B), consistent with a reduction in Dpp signaling. Together, these data argue against a nonspecific effect from an N-terminal fusion to Med. Consistent with this, recruitment of Med with N-terminally fused GFP into Smad complexes (Gao et al. 2005) has been described, as has transcriptionally active GFP-Smad4 (Nicolas et al. 2004). We also ruled out any effect being due to the integration sites of our transgenes, by verifying the reduction in target gene expression in two additional Med-SUMO transgenic lines (Supplemental Fig. S4A). Therefore, the Med-SUMO data suggest that SUMOylation of Med is sufficient to down-regulate Dpp target gene expression.
Figure 4.
Failure to SUMOylate Med increases Dpp signaling range. (A) Tethering SUMO to Med represses Dpp target genes. Dorsal views of gastrulating embryos showing Race and ush staining. Embryos are either wild type or express a Med-SUMO fusion protein. Graphs show quantification of the number of Race or ush expressing cells in the middle of the embryo; n = 15; error bars are SEM; (*) P < 0.05 (Student’s t-test). (B) Western blot analysis of 2- to 4-h extracts from the embryos shown in A confirms expression of the Myc epitope-tagged Med-SUMO fusion protein. The Tubulin signal serves as a loading control. (C) Med that cannot be SUMOylated has higher transcriptional activity. RNA in situ hybridization of gastrulating wild-type embryos and those overexpressing wild-type or mutant Med proteins, with Race and ush probes. Embryos are oriented with dorsal up. Graphs and quantification are as in A. (D) As in B except that extracts from embryos overexpressing wild-type or mutant Med are analyzed.
As a more critical test of the role of SUMO modification of Med, we investigated whether the Dpp signaling phenotype observed in the lwr mutant embryos (Fig. 1A) could be explained by a failure to SUMOylate Med. We introduced wild-type Med or the triple MedABC mutant, which cannot be SUMOylated, into the Drosophila embryo, again under the control of the ubiquitin promoter. As shown in Figure 4C, introduction of the wild-type Med transgene leads to a small expansion in the Race and ush expression patterns. However, introduction of the non-SUMOylatable ABC mutant of Med leads to a more substantial increase in the Race and ush expression patterns (Fig. 4C), despite the slightly lower accumulation of this mutant Med compared with wild-type Med (Fig. 4D). Quantification of the alterations in expression patterns is shown in Figure 4C. These effects are reproduced in a further two independent lines for each transgene (Supplemental Fig. S4B), ruling out position effects. Therefore, introduction of non-SUMOylatable Med phenocopies the lwr mutant phenotype in terms of Dpp target gene expression (cf. Figs. 4C and 1A). This result demonstrates that Med is the major SUMO target in the Dpp pathway, and that Med SUMOylation down-regulates Dpp-responsive transcription in the early embryo. We also used a luciferase reporter assay in tissue culture cells to show that the difference in the transcriptional activity of MedABC versus Med is alleviated when the SUMO pathway is inhibited using Gam1 (Supplemental Fig. S5; Boggio et al. 2004). This result suggests that the MedABC mutation primarily affects SUMOylation.
Med is SUMO modified in the nucleus
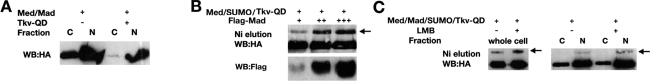
We next addressed whether Med SUMOylation is a nuclear or cytoplasmic event. As we used 293 cells for these experiments, we first analyzed the nuclear–cytoplasmic partitioning of transfected Med in this cell line both in the presence and absence of pathway activation. Successful separation of nuclear and cytoplasmic fractions was confirmed by the expected distribution of the cytoplasmic Tubulin and nuclear NBS proteins (data not shown). In the absence of Tkv-QD, we detect Med in both the cytoplasmic and nuclear fractions (Fig. 5A). Here, the nuclear Med presumably represents the pool that has shuttled into the nucleus in the absence of signal, as described for Smad4 (Pierreux et al. 2000). In the presence of signal, more transfected Med enters the nucleus (Fig. 5A), again consistent with the Smad4 data (Pierreux et al. 2000).
Figure 5.
Med is SUMO modified in the nucleus. (A) The relative proportion of Med protein in the cytoplasmic (C) and nuclear (N) fractions of 293 cells transfected with the indicated plasmids is detected by Western blotting with an anti-HA antibody. (B) The indicated plasmids were transfected into 293 cells, followed by nickel affinity chromatography of His6-SUMO conjugates. (Top panel) SUMO modification of HA-tagged wild-type Med protein was determined by Western blot analysis of the nickel eluates. The arrow points to SUMO-modified Med. (Bottom panel) Western blot showing the increase in Mad following its titration. (C) 293 cells were transfected with Mad, Med, His6-SUMO, and Tkv-QD plasmids, and where labeled (+) treated with Leptomycin B (LMB), an inhibitor of Smad4/Med export. SUMO conjugates were purified, and the amount of SUMO-modified Med in whole-cell extracts (left panel) or nuclear and cytoplasmic fractions (right panel) was visualized by Western blot analysis. Arrow indicates SUMOylated Med.
To address whether Med SUMOylation occurs in the nucleus or cytoplasm, we tested the effect on Med SUMOylation of increasing nuclear Med levels by titrating in Mad in the presence of Tkv-QD (Podos and Ferguson 1999). Adding more Mad leads to an increase in the amount of SUMO-modified Med (Fig. 5B), consistent with Med SUMOylation occurring in the nucleus.
If SUMOylation of Med occurs in the nucleus, then increasing the level of nuclear Med should increase the amount that is modified. To test this prediction, we treated cells with Leptomycin B (LMB), which blocks Smad4/Med nuclear export (Pierreux et al. 2000). LMB treatment leads to an increase in the amount of SUMO-modified Med (Fig. 5C), indicating that prolonged nuclear localization of Med promotes its SUMOylation, consistent with modification occurring in the nucleus. To test this more directly, we analyzed SUMO-modified Med in nuclear and cytoplasmic fractions isolated from the LMB-treated transfected cells. The data reveal that SUMOylated Med is detected in the nuclear fraction, and that this amount increases upon LMB treatment (Fig. 5C). Together, the data presented in Figure 5, B and C, strongly support the conclusion that Med is SUMO modified in the nucleus. This interpretation is also compatible with the existing data that SUMOylation is predominantly a nuclear process both in mammalian and Drosophila cells (Rodriguez et al. 2001; Smith et al. 2004).
SUMOylation of constitutively shuttling Med
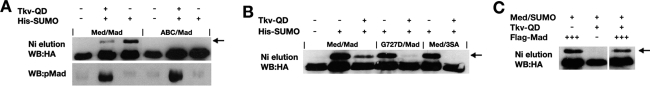
Med/Smad4 also enters the nucleus in the basal state due to signal-independent constitutive shuttling (Fig. 5A; Pierreux et al. 2000). As we provided evidence for nuclear Med SUMOylation, it seems likely that Med would also be modified in the absence of signal due to its shuttling. To test this, we compared the amount of SUMO-modified Med in the presence and absence of the activated Tkv-QD receptor. Non-SUMOylatable MedABC was included as a negative control. As shown in Figure 6A, Med is SUMOylated in the absence of Tkv-QD, consistent with the protein shuttling into the nucleus. However, more SUMO-modified Med is detected in the absence of signal than in its presence (Fig. 6A) despite the lower amount of nuclear Med under these conditions (Fig. 5A). This decrease suggests that pMad protects Med from SUMO modification in the presence of signal (see Discussion). We also detected pMad and, consistent with pathway activation being required for accumulation of pMad, we detected significant levels of pMad only when Tkv-QD is present. Slightly more pMad is associated with MedABC compared with Med in the presence of Tkv-QD (Fig. 6A, see Discussion).
Figure 6.
SUMO modification of shuttling Med. (A) 293 cells were transfected with the indicated plasmids and His6-SUMO conjugates were purified by nickel affinity chromatography. Western blots show the levels of SUMOylated HA-tagged Med (top panel) and associated pMad protein (bottom panel). Arrow indicates SUMO-modified Med. (B) SUMO assay as described in A with the indicated transfected plasmids. MedG727D is compromised with respect to its ability to interact with Mad and enter the nucleus, whereas the Mad3SA mutant cannot be phosphorylated or enter the nucleus following pathway activation. (C) Detection of SUMO-modified Med as described in A. Lanes 1–3 are all taken from the same exposure of the same Western blot; the gap reflects only the removal of irrelevant lanes.
We next made use of previously characterized Mad and Med mutants to compare the relative amounts of SUMO-modified Med in the presence and absence of signal. MedG727D is compromised in terms of its ability to interact with Mad and accumulate in the nucleus in a signal-dependent manner. Similarly, Mad3SA cannot be phosphorylated upon pathway activation and fails to enter the nucleus, thereby blocking nuclear accumulation of Med also (Wisotzkey et al. 1998). As these mutations only affect Mad–Med interaction, they should have no effect on constitutively shuttling Med and therefore the amount of SUMO-modified Med when signal is absent. As shown in Figure 6B, in the absence of Tkv-QD, the G727D mutation or presence of Mad3SA do not affect the level of Med SUMOylation.
In the presence of Tkv-QD, SUMOylation of MedG727D, which is disrupted in its ability to enter the nucleus, is greatly reduced (Fig. 6B). Visualization of pMad levels confirms that in the presence of MedG727D versus wild-type Med there is less associated pMad when the pathway is activated (data not shown), consistent with a reduced interaction of MedG727D with Mad as described previously (Wisotzkey et al. 1998). Similarly, in the presence of Tkv-QD and Mad3SA, no SUMO-modified Med is detected (Fig. 6B). As reported previously for the mutant Mad protein (Wisotzkey et al. 1998), no pMad is detected when Mad3SA, Med, and Tkv-QD are transfected (data not shown). These data, which show that Med is poorly SUMOylated when mutant Smad proteins defective in nuclear entry are tested, support our earlier conclusion that SUMO modification of Med occurs in the nucleus. However, they also suggest that Med cannot constitutively shuttle under these conditions as the strong levels of SUMOylated Med observed in the absence of signal when the mutant proteins are tested are not detected upon pathway activation.
To test whether Med constitutively shuttles in the presence of signal, we determined the effect on Med SUMOylation of transfecting cells with only Med and Tkv-QD. Control transfections of Mad and Med only or Mad, Med, and Tkv-QD were also included. Although SUMOylated Med is detected in these latter two control transfections, no SUMO-modified Med is detected in cells with Tkv-QD but no exogenous Mad (Fig. 6C). This result supports our conclusion that Med constitutive shuttling is inhibited upon pathway activation, and that Med nuclear entry under this condition is entirely dependent on Mad interaction.
Med SUMOylation triggers nuclear export
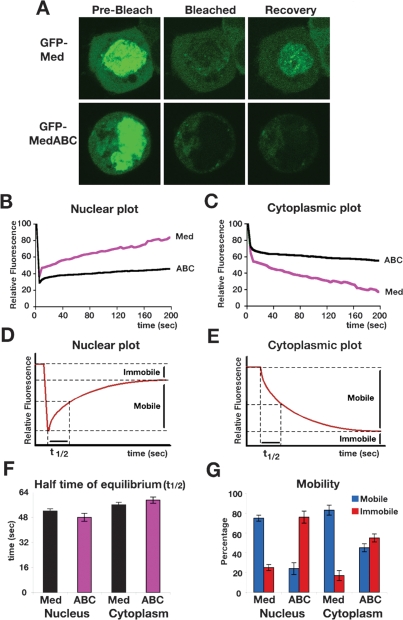
As Med SUMO modification is a nuclear event, we investigated whether SUMOylation affects Med’s nuclear–cytoplasmic partitioning by using Fluorescence Recovery After Photobleaching (FRAP) experiments. We used GFP-Med, which has been shown previously to be recruited into Smad complexes (Gao et al. 2005), and which is functional in the Drosophila embryo (Supplemental Fig. S3). We focused on the effect of Med SUMOylation under signaling conditions, by transfecting GFP-Med or GFP-MedABC, Mad, and Tkv-QD into 293 cells. Nuclei were photobleached and the rate of GFP-Med reappearance in the nucleus as well as the rate of disappearance of GFP-Med from the cytoplasm was measured. The raw FRAP data were subjected to background subtraction, correction for intensity changes, and normalization to the first prebleach time point (set at 100%). From these data, kinetic plots were generated showing fluorescence changes in the nucleus and cytoplasm over time (Fig. 7B,C). After photobleaching nuclear GFP-Med, there is a rapid recovery of nuclear fluorescence (Fig. 7A,B) and a concomitant loss of fluorescence from the cytoplasm (Fig. 7A,C). Detailed images of representative FRAP experiments are shown in Supplemental Figure S6. These data are consistent with bleached GFP-Med protein exiting the nucleus and being replaced with unbleached fluorescent protein from the cytoplasm. Such dynamic shuttling behavior in the presence of signal has been described previously for Smad4 (Nicolas et al. 2004). In contrast, analysis of the non-SUMOylatable GFP-MedABC mutant reveals that there is poorer recovery of fluorescent protein in the nucleus (Fig. 7A,B) and slower loss of MedABC fluorescence in the cytoplasm (Fig. 7A,C). These data suggest there is a deficiency in the transit of MedABC between the nucleus and the cytoplasm. We showed that Med is SUMOylated in the nucleus (Fig. 5); therefore, Med and MedABC are functionally equivalent until they enter the nucleus and SUMOylation of Med occurs. Thus, the simplest interpretation of the MedABC FRAP data is that the non-SUMOylatable MedABC protein cannot be exported, which has a secondary effect on the level of MedABC import.
Figure 7.
SUMOylation promotes Med nuclear export. (A) Panels show representative prebleach, nuclear bleach, and recovery images from experiments on cells transfected with Tkv-QD, Mad, and GFP-Med or GFP-MedABC. The recovery images correspond to the 200-sec time point. (B,C) Graphs show quantification of Med and MedABC nuclear (B) and cytoplasmic (C) recovery, each averaged from eight cells. (D,E) Example nuclear (D) and cytoplasmic (E) kinetic plots with the Mobile, Immobile fractions, and t1/2 shown. (F) Graph shows half time of equilibration (t1/2) for the Med and MedABC FRAP data. (G) Graph shows the percentage of Mobile and Immobile Med or MedABC as calculated from the nuclear and cytoplasmic recovery experiments. Error bars are SEM throughout.
The kinetic plots allow t1/2 to be calculated, which is defined as the time for exchange of half of the mobile fraction between the bleached and unbleached compartments, as indicated on example nuclear or cytoplasmic kinetic plots shown in Figure 7, D and E. They also reveal the proportion of mobile or immobile fluorescent protein in either the nucleus or cytoplasm (Fig. 7D,E). Surprisingly, analysis of the GFP-Med and GFP-MedABC data in this way reveals that the kinetics associated with the limited FRAP signals for MedABC are almost identical to those for wild-type Med, with equivalent half time of recoveries calculated for both (Fig. 7F). The key difference lies in the degree of fluorescence recovery or loss, which relates directly to the proportion of protein that is immobile (Fig. 7D,E). Our data demonstrate that a large proportion of the nuclear GFP-Med protein is mobile, whereas GFP-MedABC is largely immobile (Fig. 7G). Therefore, together our data suggest that there is nothing inherently defective with respect to the ability of MedABC to enter or exit the nucleus. Instead, there is a large pool of immobile MedABC protein, implying that the SUMO modification functions to convert immobile nuclear Med into protein that is competent for nuclear export.
Med SUMOylation limits the level of nuclear Smads in vivo
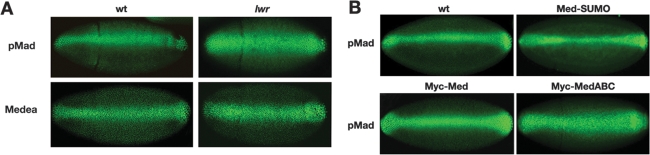
Our data reveal that SUMOylation of Med occurs in the nucleus and promotes nuclear export. Therefore, we used specific antibodies and confocal microscopy to visualize the nuclear distribution of pMad and Med in wild-type and lwr mutant embryos. Both pMad and Med have a broader nuclear distribution in the mutant embryos (Fig. 8A), with typical expansions of both Smads from five to six nuclei wide in wild-type embryos to eight or nine in lwr mutant embryos. The increase in nuclear Med in the absence of SUMOylation is consistent with SUMO promoting Med nuclear export. However, the effect on pMad is surprising, given that pMad can accumulate in the nucleus without stable binding to Med (Wisotzkey et al. 1998; see the Discussion).
Figure 8.
Med SUMOylation reduces nuclear Smad levels. (A) Confocal images of gastrulating wild-type and lwr mutant embryos showing pMad and Med distribution. Embryos are oriented with dorsal up. (B) pMad protein distribution in embryos that are either wild-type or carrying a Ub:Med-SUMO, Ub:Med, or Ub:MedABC transgene.
To further investigate the effect of Med SUMOylation on nuclear pMad distribution, we also visualized the distribution of pMad in our transgenic embryos. The nuclear pMad stripe is thinner in the Med-SUMO embryos (typically three to four stained nuclei at the narrowest point) compared with wild-type embryos (Fig. 8B). In embryos overexpressing wild-type Med, there is an increase in nuclear pMad so that the stripe encompasses seven to eight nuclei (Fig. 8B). As we also see an increase in Dpp target gene expression in these embryos (Fig. 4C), the increase in pMad here likely reflects more efficient pathway activation. However, introduction of the non-SUMOylatable MedABC transgene leads to an even broader distribution of pMad to a stripe of nine to 10 nuclei (Fig. 8B), demonstrating that pMad accumulates in the nucleus in the absence of Med SUMOylation. Together, the data shown in Figure 8, A and B, provide evidence that Med SUMOylation reduces the accumulation of nuclear Med and pMad in the Drosophila embryo.
Discussion
SUMO modification restricts Dpp signaling range and duration
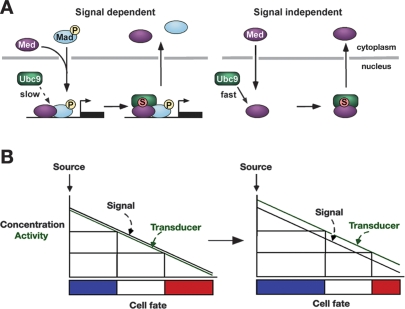
In this study we show that SUMOylation negatively regulates the spatial range and duration of the Dpp signal in the Drosophila embryo. The intracellular transducer Med is identified as the primary SUMO substrate, and we provide evidence that SUMOylation of Med occurs in the nucleus. Our data further demonstrate that Med SUMOylation promotes its nuclear export, leading to more restricted domains of activated Smads in wild-type embryos than those with reduced Lwr levels.
Together, our data suggest a model whereby Med enters the nucleus either by shuttling in a signalindependent manner or through pathway activation, leading to its SUMOylation (Fig. 9A). As we detect less SUMOylated Med in the presence of signal, we propose that pMad slows the rate of Med SUMOylation, possibly via an effect on Ubc9 recruitment. Our FRAP data and imaging of Med in lwr mutant embryos suggest that SUMO modification of Med acts as a trigger to promote its mobility and nuclear export. This finding could explain the necessity for pMad to delay SUMO modification of Med, in order that active Smad complexes have sufficient time to activate transcription. It has been reported previously that TGF-β signaling decreases the nuclear mobility of vertebrate Smad4 (Schmierer and Hill 2005). We propose that this decrease may reflect a slower rate of Smad4 SUMOylation in the presence of phosphorylated R-Smad, which in turn retains Smad4 in an unmodified immobile form.
Figure 9.
Med SUMOylation and gradient interpretation. (A) Summary of the effect of Med SUMOylation on its subcellular distribution. (B) SUMO modification of the Med intracellular transducer modulates the transcriptional output with respect to the positional information (see the text for details).
Like Med, the pMad domains are also expanded in lwr mutant embryos and those with non-SUMOylatable Med (Fig. 8). We also detect more pMad associated with the non-SUMOylatable MedABC mutant than with wild-type Med (Fig. 6A). Therefore, the loss of nuclear Med upon SUMOylation appears to promote loss of pMad, even though pMad can accumulate in the nucleus without Med interaction (Wisotzkey et al. 1998). Recently, pyruvate dehydrogenase phosphatase (PDP) has been shown to terminate Dpp signaling through dephosphorylation of pMad (Chen et al. 2006). Although it is presently unclear if PDP dephosphorylates pMad in the nucleus or cytoplasm, the Smad2/3 phosphatase PPM1A acts in the nucleus, resulting in Smad2/3 nuclear export (Lin et al. 2006). Therefore, it is possible that SUMO and PDP function together in the nucleus to terminate Dpp signaling. The expanded pMad domains observed when Med SUMOylation is prevented suggest a model in which Med SUMO modification in a wild-type embryo precedes pMad dephosphorylation. This model is consistent with the evidence that dephosphorylation of the receptor-activated Smad promotes complex dissociation and export (Inman et al. 2002; Lin et al. 2006).
SUMO-dependent export of Med from the nucleus following signal activation provides a mechanism to ensure that cells activate Dpp-dependent transcription only in response to the continual receipt of an extracellular Dpp signal. Removal of this sensing mechanism in lwr mutant embryos leads to an inappropriate signaling duration as detected by prolonged zen expression and the cuticle phenotypes.
The fate of SUMOylated Med is currently unknown. However, as Ulp1, one of the major SUMO deconjugating enzymes in Drosophila, is localized to the nuclear pore complex (Smith et al. 2004), it is likely that Med is deSUMOylated upon export. We suggest that ultimately SUMOylated Med is either recycled following deSUMOylation or degraded. Despite the apparently large cytoplasmic pool of Med (Sutherland et al. 2003), overexpression of wild-type Med expands Dpp target gene expression and the number of amnioserosa cells in early and late stage embryos, respectively (Fig. 4C; Supplemental Fig. S3A; Sutherland et al. 2003). These observations suggest that Med is limiting for signaling, in which case failure to recycle SUMO-modified Med would have a significant impact on the Med pool.
Med, which constitutively shuttles between the nucleus and cytoplasm in the absence of signal, is also SUMO modified in the nucleus. There is evidence that in the absence of signal, the Sno corepressor is recruited to nuclear Smad4 to prevent signal-independent transcriptional activation (Stroschein et al. 1999). By limiting Med’s time in the nucleus, SUMO-mediated nuclear export may be an additional strategy deployed to further protect against inappropriate transcriptional responses (Fig. 9A). Interestingly, our results suggest that activation of the Dpp pathway inhibits Med constitutive shuttling. This scenario is different from that described for vertebrate Smad4, which can shuttle independently of an R-Smad upon active TGF-β signaling (Schmierer and Hill 2005). Recently, basal shuttling of Smad4 has been shown to require Importin7/8, whereas the mechanism of nuclear import of constitutively shuttling Med is independent of Moleskin, the Drosophila ortholog of Importin7/8 (Yao et al. 2008). These findings provide further support to our conclusion that there are inherent differences between the constitutive shuttling properties of Med and Smad4.
SUMOylation promotes Med nuclear export
Our data identify a central role for SUMO in modulating the nuclear–cytoplasmic partitioning of the Smad transcription factors. Precedents already exist for SUMO in regulating both the import and export of proteins. For example, SUMO has been implicated in promoting the nuclear retention of the Elk-1 transcription factor (Salinas et al. 2004), adenoviral E1B-55K protein (Kindsmuller et al. 2007), and CtBP1 corepressor (Lin et al. 2003b). In terms of SUMO promoting nuclear export, as our data suggest for Med, examples include the TEL repressor protein (Wood et al. 2003), MEK1 kinase (Sobko et al. 2002), ribosome biogenesis factors (Panse et al. 2006), and p53 transcription factor (Carter et al. 2007).
Following genotoxic stress, SUMOylation of the IκB kinase regulator NEMO triggers a cascade of additional modifications including phosphorylation and ubiquitination that ultimately promote NEMO’s nuclear export (Huang et al. 2003). We note that Ectodermin, a nuclear ubiquitin ligase, constrains BMP signaling by promoting nuclear clearance of Smad4 (Dupont et al. 2005). Whether the fly ortholog of Ectodermin has a similar role, and indeed if there is any interplay between Ectoderminmediated ubiquitination and SUMOylation of Med in its nuclear export, remains to be determined. An alternative mechanism by which SUMO promotes Med export is based on that described for p53. p53 is monoubiquitinated by MDM2, which exposes the NES and allows recruitment of the PIASy E3 ligase leading to p53 SUMOylation. As a result, MDM2 dissociates and p53 nuclear export occurs (Carter et al. 2007). SUMOylation may re-expose the Med NES that has been inactivated upon signaling (Watanabe et al. 2000), promoting nuclear export. The location of the Med NES in between SUMO sites A and B (Fig. 2D) may lend itself to this type of regulation. Interestingly, SUMO sites A and B are the two that are conserved in vertebrate Smad4, as is the position of the NES. We speculate that SUMOylation will also direct nuclear export of vertebrate Smad4.
Although SUMO modification of Smad4 has been postulated to have both positive (Lin et al. 2003a) and negative (Long et al. 2004) effects on gene expression, Med SUMOylation leads to a reduction in its transcriptional activity in the context of Dpp signaling in the Drosophila embryo. These differences may reflect promoter-specific effects or particular characteristics of the transcription factor complex that depend on which receptor-activated Smad is associated with Med/Smad4.
Med SUMOylation and morphogen gradient interpretation
Studies of extracellular signals such as Dpp and Hedgehog support the generation of different gene activity thresholds by a “French flag” model of positional information (Wolpert 1996). Signal concentration provides positional information so that cells located nearest the source activate a peak threshold of gene activity and adopt a specific cell fate, whereas cells located further from the source express different threshold responses and assume distinct fates. Morphogen concentration at the source and sink is therefore crucial, and mechanisms that have been characterized for regulating patterning by morphogens have intuitively focused on the morphogen itself (Tabata and Takei 2004). However, our results identify a twist on the French flag model whereby the positional information provided by a specific concentration of morphogen can be refined by modulating the activity of an intracellular transducer. In this way the French flag floats in relation to Dpp activity (Fig. 9B), as the absolute amount of Dpp required for each fate is influenced by the activity of the SUMOylation pathway. Although our study has concentrated on the SUMO post-translational modification, any mechanism that hones the activity or distribution of an intracellular transducer will affect the interpretation of positional information and pattern formation in a similar way. Moreover, we predict that SUMO itself will be used to modulate the signaling outputs by other morphogens in different developmental contexts. A good candidate appears to be the Wnt morphogen, as links between SUMO and the Wnt pathway during Xenopus development been suggested (Yukita et al. 2004).
Smad post-translational modifications
The spatial and temporal range of the Dpp/BMP signal is controlled not only by Med SUMOylation but also by PDP dephosphorylation of pMad (Chen et al. 2006) and dSmurf-dependent ubiquitination of cytoplasmic Mad (Podos et al. 2001). Therefore, multiple mechanisms exist for constraining the activity of the Smad transcription factors, all of which are wasteful in terms of signal. Although wasteful, having a dedicated dampener in the form of SUMO modification may be tolerated so that the Dpp signaling pathway can be controlled somewhat in the event of inappropriate activation. This may be essential given the potency of Dpp signaling in inducing cell fates (Ferguson and Anderson 1992). Another possibility is that the disadvantage of losing signal through this built-in dampener is far outweighed by its use as a mechanism through which the presence of an extracellular signal can constantly be sensed.
In addition to the central role of Med/Smad4 in mediating the appropriate transcriptional outputs in response to signaling by all TGF-β ligands (Ashe and Briscoe 2006), the function of Smad4 as an essential tumor suppressor protein in humans has been well documented (Arteaga 2006). As well as SUMOylation, ubiquitination of the Med/Smad4 transcription factor has been described (Dupont et al. 2005). Therefore, it appears that multiple mechanisms are deployed during development to harness the activity of this pivotal signal-responsive transcription factor.
Materials and methods
Fly stocks, P-element mediated transformation, RNA in situ hybridization, and immunostaining
The following stocks were used: dppHin37/GlaDp(2;2)DTD48, dpphr27/CyO; tub-GAL4-VP16; FRT lwr118, FRT lwr4-3 (Epps and Tanda 1998; Apionishev et al. 2001); sumok06307/CyO; and yw67c23 flies were used as wild-type. lwr germline clone embryos were generated using the FLP-DFS technique (Chou and Perrimon 1996). In these embryos there is paternal rescue; therefore, the embryos shown are maternally and zygotically lwr mutant. Embryo collection, P-element-mediated transformation, and RNA in situ hybridizations using digoxigenin-labeled RNA probes and immunostaining with anti-pMad (Tanimoto et al. 2000) (1:500), anti-Med (Sutherland et al. 2003) (1:500), and anti-Kr antibodies (1:200) were performed using standard methods. For each expression pattern visualized by RNA in situ hybridization, the number of expressing cells in the middle of the embryo was counted in at least 15 embryos of the appropriate genotypes, which were stained in parallel with control embryos.
Plasmids
pAC-Myc Med, pAC-Flag Mad, pAC-TkvQD (Muller et al. 2003), pCMV-HA Med, pCMV-HA MedG727D, pCMV-Flag Mad, pCMV-Flag Mad3SA, pCMV TkvQD (Wisotzkey et al. 1998), pCT-GFPMed (Gao et al. 2005), and pCMV-His6 SUMO (Rodriguez et al. 1999) have been described previously. An EcoRI-digested PCR fragment of the Drosophila sumo cDNA with sequences encoding His6 inserted at the N terminus was inserted into pAC5.1 (Invitrogen) and pUASp (Rorth 1998). pWUM-Med was constructed by insertion of cDNA sequences into the pWUM plasmid that contains the Ubiqutin promoter followed by a Myc-epitope tag (Lee et al. 1988). The ABC mutations were engineered using the QuikChange site-directed mutagenesis kit (Stratagene). The Med-SUMO fusion was generated by PCR and contains the sumo cDNA encoding amino acids 1–80 inserted upstream of the Med AUG. For pUAS-GFP-Med, the open reading frame of the Med A isoform (AF027729, Wisotzkey et al. 1998) was PCR amplified from pBS-Med (Sutherland et al. 2003) and then cloned into the BglII–XhoI sites of the vector pP{UAS-EGFP} (Parker et al. 2001). In-frame fusion and full Med sequence were confirmed by sequencing, and these transgenic flies were generated by the CBRC Transgenic Drosophila Core. For the yeast two-hybrid experiments, fragments of the Med cDNA encoding the regions indicated in Figure 2A were isolated by PCR and cloned into the plasmid pGB (James et al. 1996). The entire lwr coding sequence was cloned into pACT2 (Clontech). The yeast two-hybrid assay was performed using standard methods.
Genetic interactions
+, lwr4-3/CyO, lwr118/CyO, or sumok06307/CyO females were crossed to dpphr27/Tft males. Viability was calculated by comparing adult progeny with wild-type bristles (mutant allele/ dpphr27) to Tufted individuals (mutant allele/Tft).
SUMO assays
In vitro transcription/translation was conducted using 1 μg of DNA and the rabbit reticulocyte lysate system (Invitrogen) according to the manufacturer’s instructions. In vitro SUMOylation assays were performed as described (Desterro et al. 1998). For the in vivo SUMO assays, 2 μg of Med, Mad, Tkv-QD, and SUMO were transfected into Drosophila S2 cells or HEK 293 cells using Effectene or Polyfect reagents (both Qiagen), respectively. Two days post-transfection, cells were harvested, and His6-SUMO conjugates were purified as described (Rodriguez et al. 1999) and analyzed using anti-Myc (Santa Cruz Biotechnologies), anti-HA (Abcam), or anti-phosphoSmad1 (Cell Signaling) antibodies. Cell fractionations were performed as described (Wong et al. 1999; Pierreux et al. 2000). Cells were treated with Leptomycin B (LMB) at a final concentration of 20 ng/mL for 2 h before harvesting.
FRAP
MDA-MB468 cells were transiently transfected using CaPO4 in glass-bottom microwell dishes (World Precision Instruments), and used for live imaging 48 h post-transfection. Imaging and photobleaching were conducted on a Zeiss LSM 510 using an argon laser (488 nm) at 55% capacity. The nucleus was photobleached at 100% laser transmission for 50 iterations. Prebleach and post-bleach images of the GFP-tagged Med were visualized using a 63× Plan Apochromat oil objective (NA 1.4) for up to 200 sec after the bleach at 4% laser transmission. Images were collected using the Zeiss LSM software (Carl Zeiss Micro Imaging Inc.). Fluorescence for the nucleus and whole cell was quantified using the National Institutes of Health Image J software. Background subtraction, data correction, and normalization were carried out as previously described (Rabut and Ellenberg 2005). The FRAP values were plotted as the recovery of relative nuclear fluorescence over time, whereas the relative cytoplasmic fluorescence levels were calculated by subtracting the nuclear fluorescence from whole-cell fluorescence, and plotted relative to time. Plots from greater than eight cells for each experiment were analyzed using nonlinear regression in PRISM version 5 software (GraphPad). τ1/2 values (the time required to reach half of the final intensity after photobleaching) and the Mobile (Mf)/Immobile (If) fractions were calculated as described previously (Rabut and Ellenberg 2005).
Acknowledgments
We thank David J. Sutherland, Svetlana Korochkina, and Viet Le for technical support; G. Pyrowolakis, M. Affolter, L. Alphey, L. Goldstein, A. Laughon, M. Peifer, and P. Rorth for plasmids; S. Tanda and K. Wharton for flies; P. ten Dijke and H. Jackle for the pMad and Kr antibodies, respectively; C. Hill for MDA-MB468 cells; S. Brown for advice; and R. Harris and A. Sawala for comments on the manuscript. This work was supported by a Wellcome grants to H.L.A. (077341/Z/05/Z) and M.P.A. (080349/Z/06/Z). The Raftery laboratory was supported by NIGMS.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.494808.
References
- Affolter M., Marty T., Vigano M.A., Jazwinska A. Nuclear interpretation of Dpp signaling in Drosophila. EMBO J. 2001;20:3298–3305. doi: 10.1093/emboj/20.13.3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkuraya F.S., Saadi I., Lund J.J., Turbe-Doan A., Morton C.C., Maas R.L. SUMO1 haploinsufficiency leads to cleft lip and palate. Science. 2006;313:1751. doi: 10.1126/science.1128406. [DOI] [PubMed] [Google Scholar]
- Apionishev S., Malhotra D., Raghavachari S., Tanda S., Rasooly R.S. The Drosophila UBC9 homologue lesswright mediates the disjunction of homologues in meiosis I. Genes Cells. 2001;6:215–224. doi: 10.1046/j.1365-2443.2001.00413.x. [DOI] [PubMed] [Google Scholar]
- Arteaga C.L. Inhibition of TGFβ signaling in cancer therapy. Curr. Opin. Genet. Dev. 2006;16:30–37. doi: 10.1016/j.gde.2005.12.009. [DOI] [PubMed] [Google Scholar]
- Ashe H.L., Briscoe J. The interpretation of morphogen gradients. Development. 2006;133:385–394. doi: 10.1242/dev.02238. [DOI] [PubMed] [Google Scholar]
- Ashe H.L., Levine M. Local inhibition and long-range enhancement of Dpp signal transduction by Sog. Nature. 1999;398:427–431. doi: 10.1038/18892. [DOI] [PubMed] [Google Scholar]
- Ashe H.L., Mannervik M., Levine M. Dpp signaling thresholds in the dorsal ectoderm of the Drosophila embryo. Development. 2000;127:3305–3312. doi: 10.1242/dev.127.15.3305. [DOI] [PubMed] [Google Scholar]
- Bakkers J., Camacho-Carvajal M., Nowak M., Kramer C., Danger B., Hammerschmidt M. Destabilization of ΔNp63α by Nedd4-mediated ubiquitination and Ubc9-mediated sumoylation, and its implications on dorsoventral patterning of the zebrafish embryo. Cell Cycle. 2005;4:790–800. doi: 10.4161/cc.4.6.1694. [DOI] [PubMed] [Google Scholar]
- Boggio R., Colombo R., Hay R.T., Draetta G.F., Chiocca S. A mechanism for inhibiting the SUMO pathway. Mol. Cell. 2004;16:549–561. doi: 10.1016/j.molcel.2004.11.007. [DOI] [PubMed] [Google Scholar]
- Broday L., Kolotuev I., Didier C., Bhoumik A., Gupta B.P., Sternberg P.W., Podbilewicz B., Ronai Z. The small ubiquitin-like modifier (SUMO) is required for gonadal and uterine-vulval morphogenesis in Caenorhabditis elegans. Genes & Dev. 2004;18:2380–2391. doi: 10.1101/gad.1227104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capelson M., Corces V.G. SUMO conjugation attenuates the activity of the gypsy chromatin insulator. EMBO J. 2006;25:1906–1914. doi: 10.1038/sj.emboj.7601068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter S., Bischof O., Dejean A., Vousden K.H. C-terminal modifications regulate MDM2 dissociation and nuclear export of p53. Nat. Cell Biol. 2007;9:428–435. doi: 10.1038/ncb1562. [DOI] [PubMed] [Google Scholar]
- Chen H.B., Shen J., Ip Y.T., Xu L. Identification of phosphatases for Smad in the BMP/DPP pathway. Genes & Dev. 2006;20:648–653. doi: 10.1101/gad.1384706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou T.B., Perrimon N. The autosomal FLP-DFS technique for generating germline mosaics in Drosophila melanogaster. Genetics. 1996;144:1673–1679. doi: 10.1093/genetics/144.4.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desterro J.M., Rodriguez M.S., Hay R.T. SUMO-1 modification of IκBα inhibits NF-κB activation. Mol. Cell. 1998;2:233–239. doi: 10.1016/s1097-2765(00)80133-1. [DOI] [PubMed] [Google Scholar]
- Dupont S., Zacchigna L., Cordenonsi M., Soligo S., Adorno M., Rugge M., Piccolo S. Germ-layer specification and control of cell growth by Ectodermin, a Smad4 ubiquitin ligase. Cell. 2005;121:87–99. doi: 10.1016/j.cell.2005.01.033. [DOI] [PubMed] [Google Scholar]
- Epps J.L., Tanda S. The Drosophila semushi mutation blocks nuclear import of bicoid during embryogenesis. Curr. Biol. 1998;8:1277–1280. doi: 10.1016/s0960-9822(07)00538-6. [DOI] [PubMed] [Google Scholar]
- Ferguson E.L., Anderson K.V. Decapentaplegic acts as a morphogen to organize dorsal-ventral pattern in the Drosophila embryo. Cell. 1992;71:451–461. doi: 10.1016/0092-8674(92)90514-d. [DOI] [PubMed] [Google Scholar]
- Gao S., Steffen J., Laughon A. Dpp-responsive silencers are bound by a trimeric Mad-Medea complex. J. Biol. Chem. 2005;280:36158–36164. doi: 10.1074/jbc.M506882200. [DOI] [PubMed] [Google Scholar]
- Hay R.T. SUMO: A history of modification. Mol. Cell. 2005;18:1–12. doi: 10.1016/j.molcel.2005.03.012. [DOI] [PubMed] [Google Scholar]
- Hay R.T. SUMO-specific proteases: A twist in the tail. Trends Cell Biol. 2007;17:370–376. doi: 10.1016/j.tcb.2007.08.002. [DOI] [PubMed] [Google Scholar]
- Huang T.T., Wuerzberger-Davis S.M., Wu Z.H., Miyamoto S. Sequential modification of NEMO/IKKγ by SUMO-1 and ubiquitin mediates NF-κB activation by genotoxic stress. Cell. 2003;115:565–576. doi: 10.1016/s0092-8674(03)00895-x. [DOI] [PubMed] [Google Scholar]
- Inman G.J., Nicolas F.J., Hill C.S. Nucleocytoplasmic shuttling of Smads 2, 3, and 4 permits sensing of TGF-β receptor activity. Mol. Cell. 2002;10:283–294. doi: 10.1016/s1097-2765(02)00585-3. [DOI] [PubMed] [Google Scholar]
- James P., Halladay J., Craig E.A. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson E.S. Protein modification by SUMO. Annu. Rev. Biochem. 2004;73:355–382. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- Kang J.S., Saunier E.F., Akhurst R.J., Derynck R. The type I TGF-β receptor is covalently modified and regulated by sumoylation. Nat. Cell Biol. 2008;10:654–664. doi: 10.1038/ncb1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindsmuller K., Groitl P., Hartl B., Blanchette P., Hauber J., Dobner T. Intranuclear targeting and nuclear export of the adenovirus E1B-55K protein are regulated by SUMO1 conjugation. Proc. Natl. Acad. Sci. 2007;104:6684–6689. doi: 10.1073/pnas.0702158104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.S., Simon J.A., Lis J.T. Structure and expression of ubiquitin genes of Drosophila melanogaster. Mol. Cell. Biol. 1988;8:4727–4735. doi: 10.1128/mcb.8.11.4727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leight E.R., Glossip D., Kornfeld K. Sumoylation of LIN-1 promotes transcriptional repression and inhibition of vulval cell fates. Development. 2005;132:1047–1056. doi: 10.1242/dev.01664. [DOI] [PubMed] [Google Scholar]
- Lin X., Liang M., Liang Y.Y., Brunicardi F.C., Melchior F., Feng X.H. Activation of transforming growth factor-β signaling by SUMO-1 modification of tumor suppressor Smad4/DPC4. J. Biol. Chem. 2003a;278:18714–18719. doi: 10.1074/jbc.M302243200. [DOI] [PubMed] [Google Scholar]
- Lin X., Sun B., Liang M., Liang Y.Y., Gast A., Hildebrand J., Brunicardi F.C., Melchior F., Feng X.H. Opposed regulation of corepressor CtBP by SUMOylation and PDZ binding. Mol. Cell. 2003b;11:1389–1396. doi: 10.1016/s1097-2765(03)00175-8. [DOI] [PubMed] [Google Scholar]
- Lin X., Duan X., Liang Y.Y., Su Y., Wrighton K.H., Long J., Hu M., Davis C.M., Wang J., Brunicardi F.C., et al. PPM1A functions as a Smad phosphatase to terminate TGFβ signaling. Cell. 2006;125:915–928. doi: 10.1016/j.cell.2006.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long X., Griffith L.C. Identification and characterization of a SUMO-1 conjugation system that modifies neuronal calcium/calmodulin-dependent protein kinase II in Drosophila melanogaster. J. Biol. Chem. 2000;275:40765–40776. doi: 10.1074/jbc.M003949200. [DOI] [PubMed] [Google Scholar]
- Long J., Wang G., He D., Liu F. Repression of Smad4 transcriptional activity by SUMO modification. Biochem. J. 2004;379:23–29. doi: 10.1042/BJ20031867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Blanco E., Pastor-Pareja J.C., Garcia-Bellido A. JNK and decapentaplegic signaling control adhesiveness and cytoskeleton dynamics during thorax closure in Drosophila. Proc. Natl. Acad. Sci. 2000;97:7888–7893. doi: 10.1073/pnas.97.14.7888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller B., Hartmann B., Pyrowolakis G., Affolter M., Basler K. Conversion of an extracellular Dpp/BMP morphogen gradient into an inverse transcriptional gradient. Cell. 2003;113:221–233. doi: 10.1016/s0092-8674(03)00241-1. [DOI] [PubMed] [Google Scholar]
- Nicolas F.J., De Bosscher K., Schmierer B., Hill C.S. Analysis of Smad nucleocytoplasmic shuttling in living cells. J. Cell Sci. 2004;117:4113–4125. doi: 10.1242/jcs.01289. [DOI] [PubMed] [Google Scholar]
- Panse V.G., Kressler D., Pauli A., Petfalski E., Gnadig M., Tollervey D., Hurt E. Formation and nuclear export of preribosomes are functionally linked to the small-ubiquitin-related modifier pathway. Traffic. 2006;7:1311–1321. doi: 10.1111/j.1600-0854.2006.00471.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker L., Gross S., Alphey L. Vectors for the expression of tagged proteins in Drosophila. Biotechniques. 2001;31:1280–1282, 1284, 1286. doi: 10.2144/01316st01. [DOI] [PubMed] [Google Scholar]
- Pierreux C.E., Nicolas F.J., Hill C.S. Transforming growth factor β-independent shuttling of Smad4 between the cytoplasm and nucleus. Mol. Cell. Biol. 2000;20:9041–9054. doi: 10.1128/mcb.20.23.9041-9054.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podos S.D., Ferguson E.L. Morphogen gradients: New insights from Dpp. Trends Genet. 1999;15:396–402. doi: 10.1016/s0168-9525(99)01854-5. [DOI] [PubMed] [Google Scholar]
- Podos S.D., Hanson K.K., Wang Y.C., Ferguson E.L. The DSmurf ubiquitin–protein ligase restricts BMP signaling spatially and temporally during Drosophila embryogenesis. Dev. Cell. 2001;1:567–578. doi: 10.1016/s1534-5807(01)00057-0. [DOI] [PubMed] [Google Scholar]
- Poulin G., Dong Y., Fraser A.G., Hopper N.A., Ahringer J. Chromatin regulation and sumoylation in the inhibition of Ras-induced vulval development in Caenorhabditis elegans. EMBO J. 2005;24:2613–2623. doi: 10.1038/sj.emboj.7600726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabut G., Ellenberg J. Photobleaching techniques to study mobility and molecular dynamics of proteins in live cells: FRAP, iFRAP, and FLIP. In: Goldman R.D., Spector D.L., editors. Live cell imaging: A laboratory manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2005. pp. 101–126. [DOI] [PubMed] [Google Scholar]
- Rodriguez M.S., Desterro J.M., Lain S., Midgley C.A., Lane D.P., Hay R.T. SUMO-1 modification activates the transcriptional response of p53. EMBO J. 1999;18:6455–6461. doi: 10.1093/emboj/18.22.6455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez M.S., Dargemont C., Hay R.T. SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J. Biol. Chem. 2001;276:12654– 12659. doi: 10.1074/jbc.M009476200. [DOI] [PubMed] [Google Scholar]
- Rorth P. Gal4 in the Drosophila female germline. Mech. Dev. 1998;78:113–118. doi: 10.1016/s0925-4773(98)00157-9. [DOI] [PubMed] [Google Scholar]
- Salinas S., Briancon-Marjollet A., Bossis G., Lopez M.A., Piechaczyk M., Jariel-Encontre I., Debant A., Hipskind R.A. SUMOylation regulates nucleo-cytoplasmic shuttling of Elk-1. J. Cell Biol. 2004;165:767–773. doi: 10.1083/jcb.200310136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savare J., Bonneaud N., Girard F. SUMO represses transcriptional activity of the Drosophila SoxNeuro and human Sox3 central nervous system-specific transcription factors. Mol. Biol. Cell. 2005;16:2660–2669. doi: 10.1091/mbc.E04-12-1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmierer B., Hill C.S. Kinetic analysis of Smad nucleocytoplasmic shuttling reveals a mechanism for transforming growth factor β-dependent nuclear accumulation of Smads. Mol. Cell. Biol. 2005;25:9845–9858. doi: 10.1128/MCB.25.22.9845-9858.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimmi O., Umulis D., Othmer H., O'Connor M.B. Facilitated transport of a Dpp/Scw heterodimer by Sog/Tsg leads to robust patterning of the Drosophila blastoderm embryo. Cell. 2005;120:873–886. doi: 10.1016/j.cell.2005.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M., Bhaskar V., Fernandez J., Courey A.J. Drosophila Ulp1, a nuclear pore-associated SUMO protease, prevents accumulation of cytoplasmic SUMO conjugates. J. Biol. Chem. 2004;279:43805–43814. doi: 10.1074/jbc.M404942200. [DOI] [PubMed] [Google Scholar]
- Sobko A., Ma H., Firtel R.A. Regulated SUMOylation and ubiquitination of DdMEK1 is required for proper chemotaxis. Dev. Cell. 2002;2:745–756. doi: 10.1016/s1534-5807(02)00186-7. [DOI] [PubMed] [Google Scholar]
- Stroschein S.L., Wang W., Zhou S., Zhou Q., Luo K. Negative feedback regulation of TGF-βsignaling by the SnoN oncoprotein. Science. 1999;286:771–774. doi: 10.1126/science.286.5440.771. [DOI] [PubMed] [Google Scholar]
- Sutherland D.J., Li M., Liu X.Q., Stefancsik R., Raftery L.A. Stepwise formation of a SMAD activity gradient during dorsal-ventral patterning of the Drosophila embryo. Development. 2003;130:5705–5716. doi: 10.1242/dev.00801. [DOI] [PubMed] [Google Scholar]
- Tabata T., Takei Y. Morphogens, their identification and regulation. Development. 2004;131:703–712. doi: 10.1242/dev.01043. [DOI] [PubMed] [Google Scholar]
- Tanimoto H., Itoh S., ten Dijke P., Tabata T. Hedgehog creates a gradient of DPP activity in Drosophila wing imaginal discs. Mol. Cell. 2000;5:59–71. doi: 10.1016/s1097-2765(00)80403-7. [DOI] [PubMed] [Google Scholar]
- Watanabe M., Masuyama N., Fukuda M., Nishida E. Regulation of intracellular dynamics of Smad4 by its leucine-rich nuclear export signal. EMBO Rep. 2000;1:176–182. doi: 10.1093/embo-reports/kvd029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wharton K.A., Ray R.P., Gelbart W.M. An activity gradient of decapentaplegic is necessary for the specification of dorsal pattern elements in the Drosophila embryo. Development. 1993;117:807–822. doi: 10.1242/dev.117.2.807. [DOI] [PubMed] [Google Scholar]
- Wisotzkey R.G., Mehra A., Sutherland D.J., Dobens L.L., Liu X., Dohrmann C., Attisano L., Raftery L.A. Medea is a Drosophila Smad4 homolog that is differentially required to potentiate DPP responses. Development. 1998;125:1433–1445. doi: 10.1242/dev.125.8.1433. [DOI] [PubMed] [Google Scholar]
- Wolpert L. One hundred years of positional information. Trends Genet. 1996;12:359–364. doi: 10.1016/s0168-9525(96)80019-9. [DOI] [PubMed] [Google Scholar]
- Wong C., Rougier-Chapman E.M., Frederick J.P., Datto M.B., Liberati N.T., Li J.M., Wang X.F. Smad3-Smad4 and AP-1 complexes synergize in transcriptional activation of the c-Jun promoter by transforming growth factor β. Mol. Cell. Biol. 1999;19:1821–1830. doi: 10.1128/mcb.19.3.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood L.D., Irvin B.J., Nucifora G., Luce K.S., Hiebert S.W. Small ubiquitin-like modifier conjugation regulates nuclear export of TEL, a putative tumor suppressor. Proc. Natl. Acad. Sci. 2003;100:3257–3262. doi: 10.1073/pnas.0637114100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S.H., Galanis A., Witty J., Sharrocks A.D. An extended consensus motif enhances the specificity of substrate modification by SUMO. EMBO J. 2006;25:5083–5093. doi: 10.1038/sj.emboj.7601383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X., Chen X., Cottonham C., Xu L. Preferential utilization of IMP7/8 in nuclear import of Smads. J. Biol. Chem. 2008 doi: 10.1074/jbc.M801320200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yukita A., Michiue T., Fukui A., Sakurai K., Yamamoto H., Ihara M., Kikuchi A., Asashima M. XSENP1, a novel sumo-specific protease in Xenopus, inhibits normal head formation by down-regulation of Wnt/β-catenin signalling. Genes Cells. 2004;9:723–736. doi: 10.1111/j.1356-9597.2004.00757.x. [DOI] [PubMed] [Google Scholar]
- Zhang H., Smolen G.A., Palmer R., Christoforou A., van den Heuvel S., Haber D.A. SUMO modification is required for in vivo Hox gene regulation by the Caenorhabditis elegans Polycomb group protein SOP-2. Nat. Genet. 2004;36:507–511. doi: 10.1038/ng1336. [DOI] [PubMed] [Google Scholar]
- Zhao J. Sumoylation regulates diverse biological processes. Cell. Mol. Life Sci. 2007;64:3017–3033. doi: 10.1007/s00018-007-7137-4. [DOI] [PMC free article] [PubMed] [Google Scholar]