Abstract
MicroRNAs (miRNAs) are small noncoding RNAs that regulate gene expression post-transcriptionally via antisense base-pairing. Although miRNAs are involved in a variety of important biological functions, little is known about their transcriptional regulation. Using yeast one-hybrid assays, we identified transcription factors with a FLYWCH Zn-finger DNA-binding domain that bind to the promoters of several Caenorhabditis elegans miRNA genes. The products of the flh-1 and flh-2 genes function redundantly to repress embryonic expression of lin-4, mir-48, and mir-241, miRNA genes that are normally expressed only post-embryonically. Although single mutations in either flh-1 or flh-2 genes result in a viable phenotype, double mutation of flh-1 and flh-2 results in early larval lethality and an enhanced derepression of their target miRNAs in embryos. Double mutations in flh-2 and a third FLYWCH Zn-finger-containing transcription factor, flh-3, also result in enhanced precocious expression of target miRNAs. Mutations of lin-4 or mir-48&mir-241 do not rescue the lethal flh-1; flh-2 double-mutant phenotype, suggesting that the inviability is not solely the result of precocious expression of these miRNAs. Therefore, the FLH-1 and FLH-2 proteins likely play a more general role in regulating gene expression in embryos.
Keywords: lin-4, microRNA transcription, FLYWCH
MicroRNAs (miRNAs) are an abundant class of small (21–22 nucleotides [nt]) noncoding regulatory RNAs found in plants and animals. This ancient class of regulatory RNAs modulates a variety of biological processes including developmental timing, metabolism, and cell fate through base-pairing with the 3′ untranslated region (UTR) of their target mRNAs (Ambros 2004; Bushati and Cohen 2007). Most animal miRNAs are transcribed by RNA polymerase II as part of longer primary transcripts (pri-miRNAs). These pri-miRNAs are then processed in a stepwise manner by protein complexes that include the RNase III enzymes Drosha and Dicer to produce the mature 21–22-nt miRNAs (for review, see Kim 2005).
Studies in mammals have shown that the biogenesis of some miRNAs can be regulated at the transcriptional level. For example, the proto-oncogene c-Myc directly activates the transcription of the mir-17-92 cluster, and c-Myc-induced overexpression of mir-17-92 induces tumor angiogenesis (O’Donnell et al. 2005; Coller et al. 2007). In contrast, c-Myc expression in lymphoma cells results in the transcriptional repression of a broad repertoire of miRNAs (Chang et al. 2008). Repression of mir-124a transcription by the RE1 silencing transcription factor (REST) contributes to the maintenance of neuronal identity (Conaco et al. 2006). Also, the myogenic transcription factors myogenin and myogenic differentiation 1 (MyoD) have been implicated in regulating the expression of two muscle-specific miRNAs, mir-1 and mir-133, during myogenesis (Rao et al. 2006).
The expression of some miRNAs can also be regulated post-transcriptionally. The down-regulation of several miRNAs in cancer cells are attributable to reduced processing by Drosha of pri-miRNAs (Thomson et al. 2006). In addition, the post-transcriptional regulation of the let-7 miRNA is associated with the neuronal differentiation of embryonic stem cells, and embryocarcinoma cells are an important feature in neuronal specification (Wulczyn et al. 2007).
The founding member of the miRNA class of small RNAs is the product of the Caenorhabditis elegans lin-4 gene (Lee et al. 1993). Expression of lin-4 miRNA is first detected in the middle of the first larval stage (L1) (Feinbaum and Ambros 1999), and its up-regulation results in the down-regulation of one of its target mRNAs, lin-14, through complementary sequences in the lin-14 3′ UTR (Wightman et al. 1993). Down-regulation of the LIN-14 protein then allows the transition from expression of the L1 stage to the expression of L2 stage developmental events to occur (Ambros and Horvitz 1987).
Two lines of evidence suggest that the temporal regulation of lin-4 occurs at the transcriptional level. First, Northern blotting analysis of the lin-4 miRNA in wild-type animals reveals the presence of two transcripts, an ∼65-nt and a 22-nt species. The longer transcript is a precursor of the mature 22-nt lin-4 (Lee et al. 1993). Both RNAs are up-regulated coordinately during the mid-L1 stage (R. Lee and V. Ambros, unpubl.), suggesting that the lin-4 precursor is activated transcriptionally during the L1 stage and then the mature lin-4 is rapidly processed from its precursor. Second, lin-4∷gfp transcriptional reporters containing only DNA sequences upstream of the miRNA recapitulate its temporal expression, indicating that these upstream sequences contain all the transcriptional regulatory elements required for the temporal regulation of lin-4 (Esquela-Kerscher et al. 2005; Baugh and Sternberg 2006; this study).
In this study, we identify a class of Zn-finger FLYWCH transcription factors that includes FLH-1, FLH-2, and FLH-3 (FLYWCH transcription factor-1, FLYWCH transcription factor-2, and FLYWCH transcription factor-3) that act redundantly during embryogenesis to repress the transcription of lin-4 and other miRNAs that are normally up-regulated postembryonically.
Results
FLH-1 binds to an upstream region of lin-4
To identify candidate proteins that could be direct regulators of lin-4 expression, we conducted yeast one-hybrid (Y1H) screens using an 87-bp fragment from the phylogenetically conserved upstream region of the lin-4 gene as bait (Lee et al. 1993). This DNA fragment (fragment 365–451) consists of nucleotides 365–451 (as measured 5′ from the start of the mature lin-4) of a 693-bp SalI lin-4 rescuing construct (Lee et al. 1993). Fragment 365–451 had been identified previously in gel mobility shift assays to contain sequences capable of developmentally regulated binding to a component contained in nuclear extracts from L1 larvae (R. Feinbaum and V. Ambros, unpubl.). As preys in the Y1H screens, we used a random-primed and an oligo dT-primed C. elegans cDNA library. We screened 2.1 × 106 yeast transformants and found several candidates exhibiting fragment 365–451 binding activity. Among these candidates was a 485-bp sequence encoding a portion (residues 105–265) of an uncharacterized ORF, y11d7a.12, which encodes a predicted transcription factor with a FLYWCH Zn-finger DNA-binding domain (Dorn and Krauss 2003; Reece-Hoyes et al. 2005). Based on the presence of the FLYWCH motif in the Y11D7A.12 protein, the gene name flh-1 was assigned to y11d7a.12.
The FLH-1-binding fragment in the lin-4 promoter is essential for repression of lin-4 in the embryo
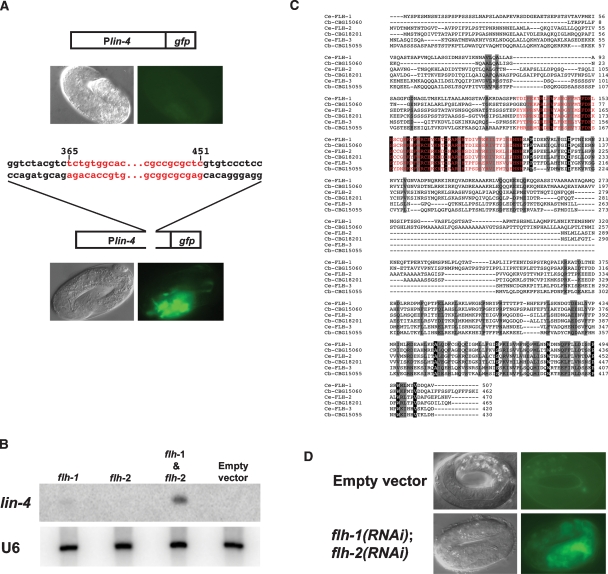
To determine whether sequences contained within fragment 365–451 are necessary for the proper temporal expression of lin-4 in vivo, we used a Plin-4∷gfp reporter, consisting of 2.4 kb of DNA sequences upstream of the mature lin-4 fused to GFP. As expected, animals with the Plin-4∷gfp (maIs134) transgene exhibited no GFP during embryogenesis (Fig. 1A). Consistent with previous reports, GFP expression from maIs134 is first detected at the mid-L1 stage and persists into adulthood in various cell types, including the hypodermis, vulva, pharynx, ventral nerve cord, and muscles (Esquela-Kerscher et al. 2005; Baugh and Sternberg 2006; this study). A modified Plin-4∷gfp reporter construct containing a deletion of nucleotides corresponding to the sequence of fragment 365–451 exhibited GFP expression in late-stage embryos (Fig. 1A), suggesting that this region in the lin-4 promoter contains cis-acting regulatory sequences necessary for the repression of lin-4 expression in the embryo, presumably through the binding of FLH-1.
Figure 1.
(A) Deletion of the FLH-1-binding site in Plin-4∷gfp results in precocious embryonic expression. In wild-type N2 embryos, the expression of Plin-4∷gfp (maIs134) consisting of 2.4 kb of the upstream region of the mature lin-4 fused to GFP recapitulates the endogenous expression pattern of lin-4. (Top panel) Expression of maIs134 is absent in embryos. (Bottom panel) Animals with a modified Plin-4∷gfp transgene with a deletion of the FLH-1-binding site (fragment 365–451) show aberrant fluorescent expression in late-stage embryos. (B) RNAi of FLH transcription factors results in increased levels of lin-4 during embryogenesis. flh-1(RNAi), but not flh-2(RNAi), results in detectable levels of lin-4 in RNA extracted from embryos. lin-4 levels are further elevated in flh-1(RNAi); flh-2(RNAi) embryos. No lin-4 was detectable in the control RNAi using the empty RNAi vector. The U6 snRNA was used as the loading control. (C) Protein sequence alignment of FLH transcription factors in C. elegans and C. briggsae. ClustalW alignment of the amino acid sequences of C. elegans FLH-1, FLH-2, and FLH-3 and their respective C. briggsae orthologs—CBG15060, CBG18201, and CBG15055—shows conservation of the FLYWCH motif and the C terminus. The flh-1 locus encodes two isoforms, FLH-1a and FLH-1b, that differ by three amino acids; residues 339–341 (PLQ) in FLH-1a are absent from FLH-1b. Black highlight indicates identical amino acids and gray boxes indicate similar amino acids. The FLYWCH motif is shown in red. (D) RNAi of FLH transcription factors leads to precocious embryonic expression of Plin-4∷gfp. Double RNAi-by-feeding of flh-1 and flh-2 in animals carrying the Plin-4∷gfp (maIs134) transgene results in the precocious expression of GFP in late-stage embryos. RNAi in animals using the empty RNAi vector exhibited no GFP in embryos.
RNAi of flh genes results in precocious embryonic expression of lin-4
To test whether FLH-1 is required for the repression of lin-4 expression in the embryo, we assayed lin-4 RNA levels in embryos produced by hermaphrodites treated with flh-1(RNAi). We used rrf-3(pk1426) animals that are hypersensitive to RNAi (Simmer et al. 2002). Northern blot analysis of total RNA extracted from flh-1(RNAi) embryos detected weak precocious expression of lin-4 in embryos (Fig. 1B). This precocious expression was enhanced by simultaneous RNAi of both flh-1 and flh-2 (c26e6.2), which encodes another of the C. elegans FLYWCH family of proteins (Fig. 1B,C). RNAi of flh-2 alone did not result in any detectable precocious lin-4 (Fig. 1B). Similarly, RNAi of flh-1 or flh-2 alone did not lead to appreciable precocious expression of GFP from our Plin-4∷gfp reporter (data not shown). However, Plin-4∷gfp was precociously active in double-RNAi, flh-1(RNAi); flh-2(RNAi) embryos (Fig. 1D). These results demonstrate that FLH-1 represses lin-4 expression by binding to a negative cis-regulatory promoter element and suggests a functional redundancy between FLH-1 and FLH-2.
Isolation and phenotype of deletion mutations in the flh genes
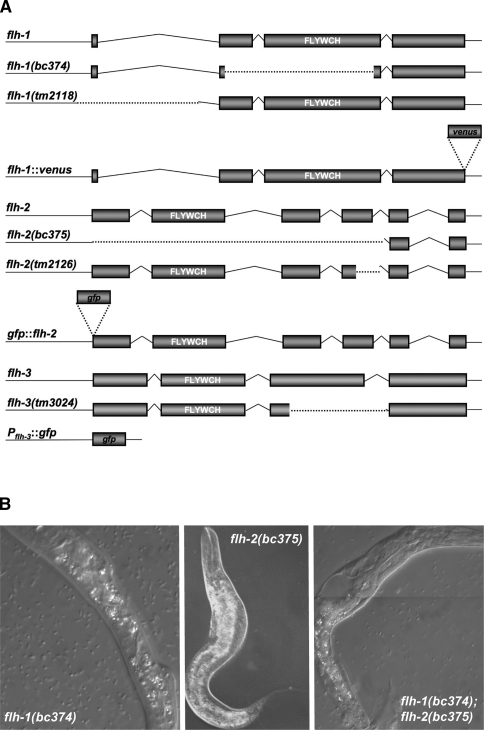
To further test the role of the FLYWCH family of proteins in the regulation of miRNA gene expression and C. elegans development, we obtained deletion mutations of flh-1 and flh-2 by screening a library of ethyl methanesulphonate (EMS)-mutagenized worms using gene-specific PCR primers. The y11d7a.12 deletion mutation, flh-1(bc374), is an 894-bp deletion that deletes most of exons 2 and 3 and results in the loss of the FLYWCH domain (Fig. 2A). The c26e6.2 mutation, flh-2(bc375), is a 2023-bp deletion that extends from the predicted translation start site to all of exon 4 (Fig. 2A), and it also contains an insertion/duplication of tttttatcagaccgcctgt at the deletion junction.
Figure 2.
(A) Schematic of the flh-1, flh-2, and flh-3 loci, mutants, and reporter transgenes. The flh-1 gene encodes two isoforms—FLH-1a and FLH-1b—that differ by three amino acids (Fig. 1C). In this study, we refer to the product of flh-1a as “FLH-1.” The nature of the flh-1(bc374) and flh-2(bc375) mutations is described under Results. The flh-1(tm2118) is a 707-bp deletion, from 287 bp upstream to 419 bp downstream of the translational start codon. The flh-2(tm2126) lesion is a 348-bp deletion. The flh-3 locus is immediately upstream of flh-1 and is transcribed in the opposite orientation. The flh-3(tm3024) mutant allele is a 337-bp deletion of most of exon 3. A description of the fluorescent transgenes can be found in the Materials and Methods. The white letters indicate the location of the FLYWCH domain. Dotted lines delineate deleted regions. The figure is not drawn to scale. (B) Phenotype of FLH-1 and FLH-2 mutants. Animals were observed using Nomarski DIC microscopy. (Left and middle panels) Single mutants of flh-1(bc374) and flh-2(bc375) exhibit a nearly wild-type phenotype with a low penetrance of larvae with morphological abnormalities. (Right panel) The double flh-1(bc374); flh-2(bc375) mutation also results in young larvae with morphological aberrations as well as a complete penetrance of early larval lethality.
Animals homozygous for either flh-1(bc374) or flh-2(bc375) exhibited a nearly wild-type phenotype with a low penetrance of young larvae with morphological abnormalities (2.8% for bc374, n = 502) (Fig. 2B). While the single FLYWCH mutants had an almost wild-type phenotype, loss of both flh-1 and flh-2 resulted in the complete penetrance of early larval lethality (Fig. 2B). L1 larvae homozygous for flh-1(bc374) and flh-2(bc375), examined following hatching, had either severe morphological abnormalities and/or appeared necrotic, and 100% (n = 92) of these double-mutant L1 larvae died before reaching the L2 larval stage.
Unlike the flh-1(bc374); flh-2(bc375) double mutant, animals double mutant for two other alleles of flh-1 and flh-2—flh-1(tm2118); flh-2(tm2126)—were viable. The viability of the flh-1(tm2118); flh-2(tm2126) double mutant is consistent with the less severe molecular lesions in these alleles as compared with flh-1(bc374) and flh-2(bc375) and indicates that flh-1(tm2118) and flh-1(tm2126) express residual protein and are probably not nulls (Fig. 2A; Supplemental Fig. 1). Although flh-1(tm2118); flh-2(tm2126) larvae appeared superficially normal, adults are uncoordinated and retain more embryos than N2 adults, suggesting defects in egg-laying.
miRNA expression profile of flh mutants
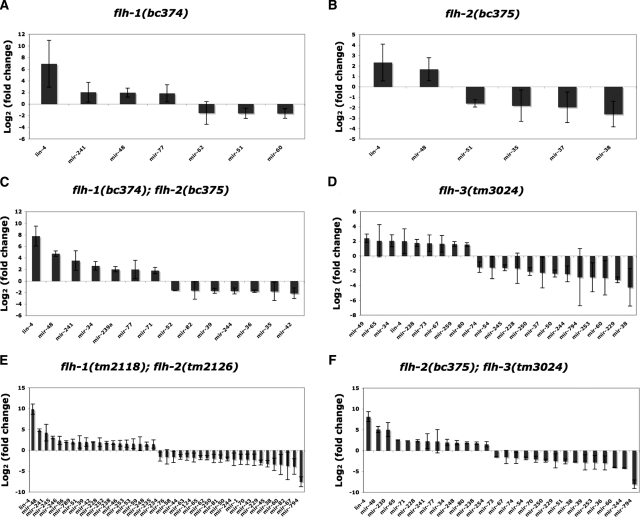
To determine whether the levels of other miRNAs besides lin-4 changed in flh mutants, we performed real-time RT–PCR (miRTaqMan) assays on 107 miRNAs using total RNA isolated from embryos from various flh mutants. For flh-1(bc374), we found that in addition to lin-4, the miRNAs mir-241 and mir-48 were also increased at least twofold in mutant embryos compared with wild-type (Fig. 3A; Supplemental Fig. 3a; Supplemental Table 1). Decreases of less than twofold were detected for mir-51 and mir-60 (Fig. 3A; Supplemental Fig. 3a; Supplemental Table 1). In flh-2(bc375), there was no significant increase in the levels of any of the miRNAs we tested (Fig. 3B; Supplemental Fig. 3b; Supplemental Table 2). The most marked changes were observed in double flh-1(bc374) and flh-2(bc375) embryos, where lin-4, mir-48, mir-241, and mir-34 were significantly overexpressed compared with wild type (Fig. 3C; Supplemental Fig. 3d; Supplemental Table 4). Embryos double mutant for flh-1(tm2118) and flh-2(tm2126) displayed similar increases in the levels of lin-4 and mir-241 as well as several other miRNAs, including mir-245, mir-246, and mir-56. flh-1(tm2118); flh-2(tm2126) embryos also exhibited a significant decrease in mir-794 (Fig. 3E; Supplemental Fig. 3e; Supplemental Table 5). miRTaqMan analysis of animals mutant for flh-3 (y11d7a.13), which encodes the third C. elegans FLYWCH motif-containing protein (Figs. 1C, 2A), showed a twofold increase in the levels of only mir-34 and mir-49 (Fig. 3D; Supplemental Fig. 3c; Supplemental Table 3). Animals with a combination of the flh-2(bc375) and flh-3(tm3024) mutations exhibited elevated levels of several miRNAs—lin-4, mir-48, mir-230, mir-65, mir-71, and mir-228 (Fig. 3F; Supplemental Fig. 3f; Supplemental Table 6). Our TaqMan results indicate that while single-gene flh mutations have only moderate or undetectable effects on miRNA levels, double flh gene mutations do have significant effects on miRNA levels, further confirming a redundant relationship between FLH-1, FLH-2, and FLH-3.
Figure 3.
MiRNA levels in flh mutants. Total RNA isolated from 100 late-stage embryos (see the Materials and Methods) of wild type (N2) and flh-1(bc374) (A), flh-2(bc375) (B), flh-1(bc374); flh-2(bc375) (C), flh-3(tm3024) (D), flh-1(tm2118); flh-2(tm2126) (E), and flh-2(bc375); flh-3(tm3024) (F) were subjected to miRNA TaqMan real-time PCR assays. miRNA levels in mutants compared with the N2 control were determined using the ΔΔC method (Materials and Methods), and are expressed here as log2 (fold change). Lists and graphs of all the analyzed ΔΔCt values are found in Supplemental Figure 3 and Supplemental Tables 1–6.
Genome-scale Y1H screens reveal additional interactions between miRNA promoters and FLH proteins
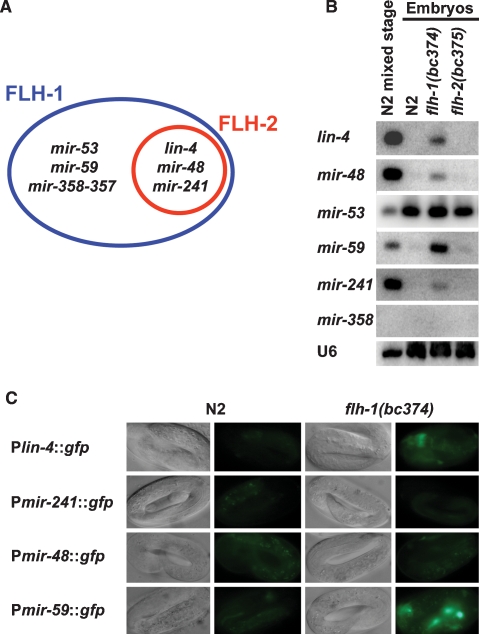
To date, 115 miRNA genes have been identified in C. elegans (miRBase version 4.0). Of these 115 miRNA genes, 82 are intergenic. Sixty-six of these 82 are located in intergenic regions and are likely to be transcribed as independent transcriptional units. The other 16 of the intergenic miRNAs are expressed from operons, with a single promoter for each operon. The remaining 33 miRNAs are situated within introns of protein-coding genes and therefore may be under the transcriptional control of their host genes. We cloned upstream promoter sequences for 71 out of the 82 intergenic miRNA genes and tested whether other miRNA promoters besides lin-4 could be directly bound by FLH-1 and FLH-2. In our genome-scale screen, we detected binding of FLH-1 to the promoters of lin-4, mir-241, mir-48, mir-53, mir-59, and mir-358-357, and binding of FLH-2 to the promoters of lin-4, mir-241, and mir-48 (Fig. 4A; Martinez et al. 2008). Transcriptional GFP reporters and Northern blots using RNA from embryos of flh-1(bc374) showed precocious expression of lin-4, mir-48, mir-59, and mir-241 (Fig. 4B) but not in flh-2(bc375) or N2 (Fig. 4B,C; data not shown). No change in expression levels was detected for mir-53 or mir-358 in either flh-1(bc374) or flh-2(bc375) (Fig. 4B; data not shown). Northern blot analysis of RNA from various developmental stages of flh-1(bc374) and flh-2(bc375) revealed little or no change in the temporal expression of their miRNA targets during post-embryogenesis (Supplemental Fig. 2), indicating a primary role of FLH-1 and FLH-2 in embryogenesis.
Figure 4.
(A) Venn diagram of miRNA targets identified by genome-scale Y1H screens. The large oval depicts miRNA promoter targets for FLH-1: lin-4, mir-241, mir-48, mir-53, mir-59, and mir-358-357. The inner circle denotes the miRNA promoter targets for FLH-2: lin-4, mir-241, and mir-48. (B) FLH-1 represses the expression of its miRNA targets during embryogenesis. Total RNA from N2, flh-1(bc374), or flh-2(bc375) embryos was extracted and analyzed by Northern blots as described in Materials and Methods. RNA from a mixed-stage population of N2 was loaded as reference. mir-358 was undetectable even after prolonged exposures. All blots were stripped and reprobed for the U6 snRNA as the loading control. (C) Transcriptional reporters for lin-4, mir-241, mir-48, and mir-59 show precocious expression during embryogenesis in an FLH-1 mutant. Late-stage embryos from N2 and flh-1(bc374) expressing the Plin-4∷gfp, Pmir-241∷gfp, Pmir-48∷gfp, or Pmir-59∷gfp transgenes were examined by fluorescence and DIC optics. All images were of identical exposure time and were processed in parallel.
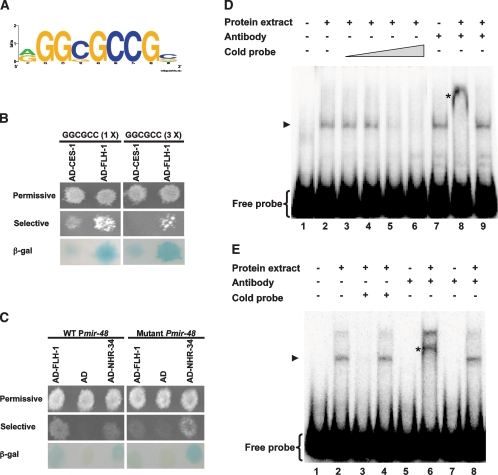
Identification of an FLH-1 consensus binding site
We used the sequences of the miRNA promoters that were found to bind FLH-1 in Y1H assays (Plin-4, Pmir-241, Pmir-48, Pmir-53, Pmir-59, and Pmir-358-357) to derive a putative FLH-1 consensus binding site using the Improbizer algorithm (Ao et al. 2004). We found that the predicted FLH-1-binding site contains an a/gGGCGCCG sequence (Fig. 5A) that tends to be located in the first 1 kb upstream of the annotated miRNA. miRNA promoters that bind FLH-1 by Y1H assays (“Y1H positives”) had higher Improbizer scores than a set of promoters that did not bind FLH-1 (“Y1H negatives”). Of the six promoters found to be positive for Y1H interaction with FLH-1, four (lin-4, mir-241, mir-48, and mir-59) contain the a/gGGCGCCG, and these four correspond to miRNAs that changed in expression in flh mutants as assessed by Northern blots and TaqMan assays. Only one Y1H negative promoter had an Improbizer score comparable with the Y1H positives. Interestingly, this miRNA (mir-34) did change in expression in flh mutants, suggesting that we failed to detect FLH binding to this promoter by Y1H assays. A second Y1H negative (mir-60) that corresponds to a miRNA that changes in flh mutants did not have a site with a good Improbizer score. This may represent a Y1H false negative or a case in which FLH proteins regulate this miRNA indirectly.
Figure 5.
(A) A predicted FLH-1 consensus binding site. The miRNA targets found for FLH-1 were used to predict a consensus binding site of a/gGGCGCCG using the Improbizer algorithm as described in Materials and Methods. (B) FLH-1 binds to the predicted consensus binding site in a Y1H assay. Y1H assays show the interaction between AD-FLH-1 and DNA baits containing either one (left panels) or three (right panels) binding sites. AD-CES-1 was used as the negative control. (C) Deletion of the predicted consensus binding site in Pmir-48 abolishes binding by FLH-1 in Y1H assays. (Left panels) Y1H assays show the interaction between AD-FLH-1 and DNA baits consisting of a wild-type Pmir-48. (Right panels) Deletion of the consensus sequence disrupts binding by AD-FLH-1. AD-NHR-34 was used as a positive control. We find AD-NHR-34 binding to wild-type Pmir-48, and deletion of the FLH-1-binding site does not disrupt AD-NHR-34 binding. (β-Gal) β-Galactosidase assay. (D) In vitro binding of FLH-1 to the consensus binding site in Plin-4. Gel mobility shift assays were performed using a [γ-32P] 5′-end-labeled 87-bp fragment (fragment 365–451) from Plin-4 incubated with total protein extract from N2 embryos. Competition assays used (lanes 3–6) increasing quantities of unlabeled fragment 365–451. (Lane 8) Antiserum against FLH-1 was added to show that the shifted material contains FLH-1. (Lane 9) Addition of a control rabbit IgG failed to form a supershifted complex. (Lane 1) Free probe only. (Lane 2) Probe incubated with protein extract. (Lanes 3–6) Probe incubated with protein extract and increasing amounts of unlabeled probe (0.1-, 1-, 10-, and 100-fold excess, respectively). (Lane 7) Probe with FLH-1 antibody but without protein extract. (Lane 8) Probe with protein extract and FLH-1 antiserum. (Lane 9) Probe with protein extract and control rabbit IgG. (Lanes 2–5,7,9) Arrowhead points to the shifted probe-FLH-1 complex. (Lane 8) Asterisk denotes the probe-FLH-1-FLH-1 antibody supershifted complex. (E) In vitro binding of FLH-1 to the consensus binding site in Pmir-48. Gel shift assays were done using a [γ-32P] 5′-end-labeled 51-bp fragment (fragment 200–251) from Pmir-48 and total protein extract from N2 embryos. (Lane 2) A shifted complex is seen in the sample incubated with the protein extract. Competition assays were done using both a 100-fold excess of unlabeled fragment 200-251 (lane 3) or unlabeled fragment 200–251 with a deletion of the consensus site (lane 4). (Lanes 6,8) Supershifted complexes were detected upon the addition of FLH-1 antiserum but not with control rabbit IgG. (Lane 1) Free probe only. (Lane 2) Probe incubated with protein extract. (Lane 3) Probe with protein extract and 100-fold excess of cold fragment 200–251. (Lane 4) Probe with protein extract and 100-fold excess of fragment 200–251 with a deletion of the consensus binding site. (Lane 5) Probe with FLH-1 antibody and no extract. (Lane 6) Probe and extract with FLH-1 antibody. (Lane 7) Probe with control rabbit IgG and no extract. (Lane 8) Probe and extract with control rabbit IgG. (Lanes 2,4,8) Arrowhead indicates the shifted probe–FLH-1 complex. (Lane 6) Asterisk shows the probe–FLH-1–FLH-1 antibody supershifted complex.
We found that DNA with the binding site consensus sequence can, indeed, interact with FLH-1 in Y1H assays (Fig. 5B). Moreover, deletion of the a/gGGCGCCG sequence from the mir-48 promoter abolishes FLH-1 binding (Fig. 5C).
To further verify the interaction between FLH-1 and the consensus sequence, we performed gel mobility shift assays using total protein extract from N2 embryos with the 87-bp fragment 365–451 from Plin-4 (Fig. 5D) and a 51-bp fragment (fragment 200–251; consisting of nucleotides 200–251 upstream of the mature mir-48) from Pmir-48 that also contains a consensus FLH-1-binding sequence (Fig. 5E). Addition of radiolabeled fragment 365–451 to the embryo extract resulted in a shifted complex that was competed away by unlabeled fragment 365–451 (Fig. 5D, lanes 3–6). A supershifted complex was formed upon the addition of anti-FLH-1 serum but not with a control antibody (Fig. 5D, lanes 8,9). Likewise, incubation of the embryo extract with a radiolabeled fragment from Pmir-48 (fragment 200–251) resulted in the formation of a shifted complex (Fig. 5E, lane 2). A supershifted complex was detected with the addition of anti-FLH-1 serum (Fig. 5E, lane 6) but not with a control antibody (Fig. 5E, lane 8). The shifted complex can be outcompeted upon the addition of unlabeled cold fragment 200–251 (Fig. 5E, lane 3). Addition of 100-fold excess of unlabeled fragment 200–251 deleted for the consensus site was ineffective in preventing FLH-1 binding to a wild-type radiolabeled fragment 200–251 (Fig. 5E, lane 4), consistent with the sequence-specific binding of FLH-1 to the Pmir-48 200–251 fragment via the consensus sequence. Interestingly, addition of 100-fold excess of mutated fragment 365–451 containing a deletion of the consensus site was still able to compete with wild-type fragment 365–451 (data not shown), indicating that FLH-1 can also bind to sequences outside the consensus site in fragment 365–451.
Expression pattern of FLH transcription factors
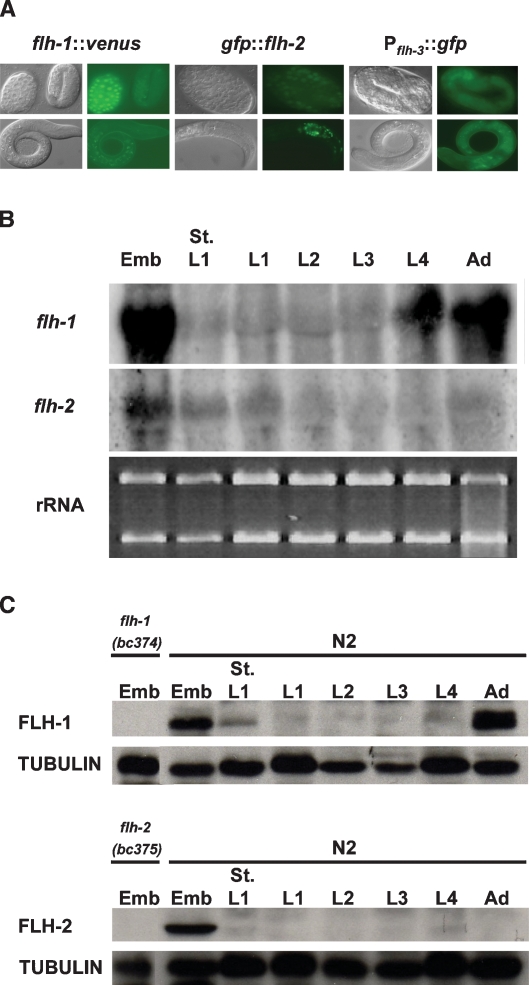
To visualize the expression pattern of FLH-1, FLH-2, and FLH-3, we made fluorescent translational or transcriptional fusions and examined their expression in transgenic worms. Expression of VENUS (Nagai et al. 2002) from the translational fusion flh-1∷venus localizes to most cells starting at the gastrulation stage, with its expression diminishing by the L1 stage (Fig. 6A). The expression pattern of the rescuing translational gfp∷flh-2 transgene (Fig. 6A; Supplemental Table 8) was somewhat different from that of flh-1∷venus. In embryogenesis, GFP was detected starting at the gastrulation stage. However, expression in head and tail cells persisted during the larval and adult stages (Fig. 6A). Fluorescence from the transcriptional fusion Pflh-3∷gfp was detected in late-stage embryos and L1 larvae (Fig. 6A).
Figure 6.
(A) Expression pattern of FLH-1, FLH-2, and FLH-3. A translational fusion consisting of a VENUS reporter fused to the C terminus of flh-1 (Pflh-1∷flh-1∷venus∷flh-1 3′ UTR) displays expression starting from mid-embryogenesis. (Left panel) VENUS expression is detected in most cells during the gastrulation stage but is down-regulated during late embryogenesis and is undetectable by L1. (Middle panel) GFP expressed from the rescuing translational fusion gfp∷flh-2 (Pflh-2∷gfp∷flh-2∷flh-2 3′ UTR) is detected in most cells during the gastrulation stage. (Middle panel) Unlike flh-1∷venus, the gfp∷flh-2 reporter shows detectable expression in head and tail cells of larvae and adults. (Right panel) The expression of a GFP transcriptional reporter for flh-3 (Pflh-3∷gfp) displayed the most intense expression during late stages of embryogenesis and little GFP in mid-stage larvae. (Right panel) Expression from Pflh-3∷gfp was also detected in L1 larvae. (B) Northern analysis of flh-1 and flh-2 mRNAs. Total RNA from N2 synchronized animals was analyzed by Northern blotting. Equivalent amounts of the RNA used for the Northern blots were run separately in parallel, and the levels of the rRNAs served as the loading control. The flh-1 and flh-2 mRNAs are similar in size to the ribosomal RNAs, and some cross-reactivity may have occurred between the flh-1 and flh-2 probes and the rRNAs. (C) Western analysis of FLH-1 and FLH-2. Protein lysates from synchronized animals were analyzed by Western blots with antisera to FLH-1, FLH-2, and tubulin. Protein extracts from the deletion mutants, flh-1(bc374) and flh-2(bc375), show that the antibodies are specific to their corresponding antigen. Emb, St. L1, and Ad indicate embryos, starved L1 larvae, and adults, respectively.
Northern blot analysis of total RNA extracted from populations of staged animals shows that the flh-1, flh-2, and flh-3 mRNAs are detected in embryos and reduced significantly after hatching (Fig. 6B; data not shown). While we were unable to observe VENUS from FLH-1∷VENUS, we detected the flh-1 mRNA in L4 animals and adults (Fig. 6B), suggesting that additional regulatory elements may be involved in the regulation of flh-1 expression that are not present in our flh-1∷venus transgene.
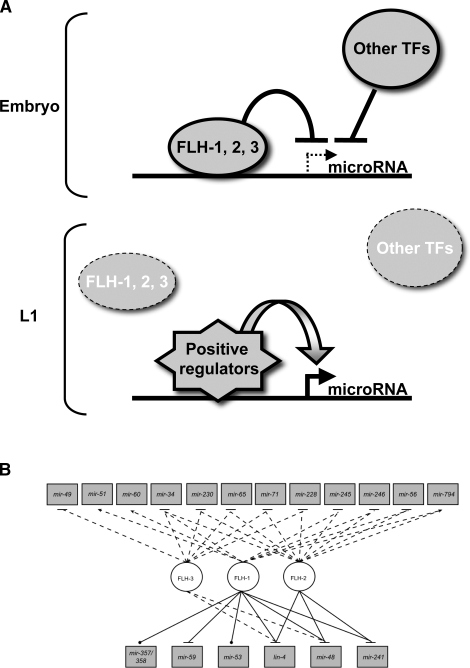
Western blot analyses using polyclonal antibodies against FLH-1 show that FLH-1 is present during embryogenesis and adulthood, while FLH-2 could be detected only in embryos (Fig. 6C). This protein expression, together with the results of the Northern blots showing reduction of flh-1 mRNA after embryogenesis, and with the temporal expression of the fluorescent transgenes, indicate that FLH-1 and FLH-2 function during embryogenesis to repress their target genes and then are down-regulated soon after hatching, consistent with the post-embryonic up-regulation of FLH targets (Fig. 8A, below).
Figure 8.
(A) A working model for FLH-1, FLH-2, and FLH-3 transcription factors. FLH-1, and/or FLH-2, in association with FLH-3, are proposed to directly bind to sequences containing the consensus a/gGGCGCCG in the promoters of lin-4, mir-241, and mir-48 during embryogenesis. This binding prevents the premature transcription of these miRNAs. Upon hatching, the FLH proteins are down-regulated, permitting the up-regulation of transcription of these miRNAs during larval development by the action of other factors, including putative transcriptional activators. (B) FLYWCH transcription factors regulate miRNA expression. Network depicting regulatory effects on miRNAs (rectangles) exerted by FLYWCH transcription factors (TFs) (circles). Solid lines are TF–Pmir interactions found by Y1H assays, dashed lines are miRNAs that change in expression in flh mutants, dotted lines are TF–Pmir physical interactions (no regulatory interaction determined), arrows are transcriptional activation, and blunted arrows are transcriptional repression.
Precocious expression of lin-4 in flh mutants reduces LIN-14 levels in embryos
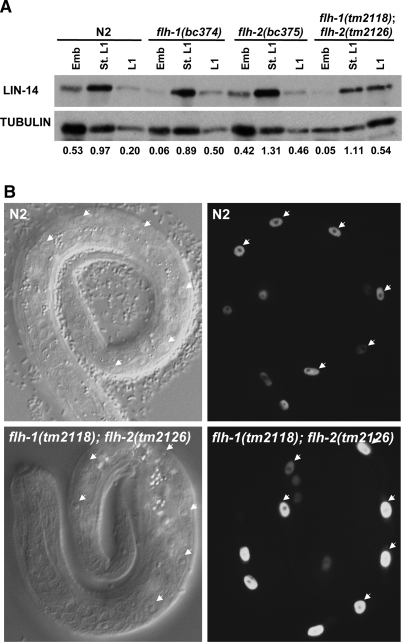
To determine if the elevated expression of lin-4 in embryos of flh mutants could lead to precocious down-regulation of LIN-14, the principal target of lin-4, we used Western blots to estimate LIN-14 levels in single and double flh-1 and flh-2 mutant embryos (Fig. 7A). Single-mutant flh-1(bc374) embryos or double-mutant flh-1(tm2118); flh-2(tm2126) embryos exhibited a ninefold to 10-fold decrease in LIN-14 levels compared with N2 or flh-2(bc375) embryos (Fig. 7A). Interestingly, although LIN-14 levels were significantly lower during embryogenesis in the single flh-1 and the double flh-1; flh-2 mutants, the level of LIN-14 in starved L1 larvae remained relatively unchanged compared with N2 (Fig. 7A). In L1 larvae, a slight increase in LIN-14 was detected in the single flh-1(bc374) and flh-2(bc375) mutants and in the double flh-1(tm2118); flh-2(tm2126) mutant (Fig. 7A). These results indicate that the precociously expressed lin-4 in embryos of the flh-1(bc374) and flh-1(tm2118); flh-2(tm2126) can lead to embryonic repression of LIN-14, but does not significantly affect LIN-14 levels post-embryonically.
Figure 7.
(A) LIN-14 levels in flh mutants. Western blots using protein extracts were first probed with anti-LIN-14 serum followed by stripping and reprobing with a tubulin antibody. Embryos hatched overnight in M9 medium were used as the source of starved L1s. The numerical values represent the ratio of LIN-14 to tubulin. (B) Seam cells number in L1 larvae. Embryos with a seam cell nuclei-specific transgene (scm∷gfp) were hatched overnight in sterile M9 followed by feeding on seeded NGM plates for 1–2 h. L1 larvae were then scored for the number of V lineage seam cells (V1–V6). Arrowheads point to the V1–V6 seam cells from one plane.
Elevated lin-4 levels in flh mutant embryos do not result in post-embryonic heterochronic defects
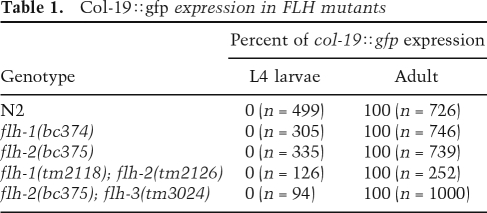
Because loss-of-function (lf) mutations of flh-1 and flh-1; flh-2 result in the up-regulation of lin-4 in embryos, and since it was shown previously that elevated lin-4 could result in a precocious expression of L2-adult developmental events (Feinbaum and Ambros 1999), we examined flh-1 and flh-2 mutant animals for defects in post-embryonic developmental timing. In particular, we used a transgenic col-19∷gfp reporter to monitor the timing of adult-specific developmental programs in the lateral hypodermis. The col-19 gene is an adult-specific collagen gene that is under the control of the heterochronic pathway (Liu et al. 1995). Heterochronic mutations that cause precocious development result in the premature expression of col-19∷gfp during larval stages, whereas mutations that cause retarded development result in the loss of col-19∷gfp expression in adults (Abrahante et al. 1998). We did not observe altered timing of col-19∷gfp expression in flh-1(bc374) or flh-2(bc375) single mutant in flh-1(tm2118); flh-2(tm2126) or flh-2(bc375); flh-3(tm3024) double mutants (Table 1). We were unable to examine flh-1(bc374); flh-2(bc375) double-mutant animals for post-embryonic heterochronic phenotypes because of the early larval lethality of the double-mutant combination.
Table 1.
Col-19∷gfp expression in FLH mutants
Animals with a high-copy transgene of the lin-4 gene display precocious phenotypes, including egg-laying defects, dumpy phenotype, and defects in tail and vulva morphology, that are reminiscent of lin-14(lf) animals (Feinbaum and Ambros 1999). Unlike worms that overexpress lin-4 from a transgene, single flh-1(bc374) animals, other than having a low penetrance of abnormally shaped larvae, do not show phenotypes similar to those of lin-14(lf) animals. flh-2(bc375) mutants, however, do exhibit a low penetrance of dumpy animals (Fig. 2B). In addition, the double flh-1(tm2118); flh-2(tm2126) mutants are uncoordinated and have egg-laying defects and an incomplete penetrance of dumpy animals.
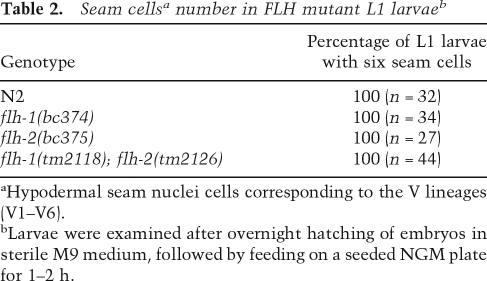
An additional phenotype of animals overexpressing lin-4 is the precocious expression of L2 larvae-specific cell division at the L1 larval stage (Feinbaum and Ambros 1999). To determine whether single or double mutants of flh-1 and flh-2 exhibit precocious L2-stage cell division, we examined the hypodermal cells (seam cells) of the V lineage (V1–V6). Wild-type L1 larvae hatch with six V-lineage seam cells. These six seam cells divide once in L1 to generate a daughter seam cell and a hypodermal cell. Because the hypodermal daughter cell does not divide, the number of V-lineage seam cells at the end of the L1 stage remains at six. At the L2 stage, five of the six seam cells generate two daughter cells, thus increasing the number of the V-lineage seam cells from six to 11 (Sulston and Horvitz 1977). In L1 larvae deficient for LIN-14, the seams cells undergo aberrant division programs characteristic of the L2 stage, thus resulting in the production of more than six seam cells (Ambros and Horvitz 1984). We used a seam cell nuclei-specific fluorescent marker (scm∷gfp) to examine whether the precocious expression of lin-4 in the flh mutants results in an abnormally high number of V-lineage seam cells in the L1 stage. As in wild-type L1 larvae, the number of seam cells remained at six in the single flh-1(bc374) and flh-2(bc375) mutants as well as in the double flh-1(tm2118); flh-2(tm2126) mutant (Table 2; Fig. 7B), indicating that the precocious expression of lin-4 during the embryonic stage is not sufficient to confer post-embryonic heterochronic defects.
Table 2.
Seam cellsa number in FLH mutant L1 larvaeb
aHypodermal seam nuclei cells corresponding to the V lineages (V1–V6).
bLarvae were examined after overnight hatching of embryos in sterile M9 medium, followed by feeding on a seeded NGM plate for 1–2 h.
FLH transcription factors likely regulate non-miRNA targets
To determine whether early larval lethality of the double flh-1(bc374); flh-2(bc375) mutation was caused primarily by overexpression of lin-4, mir-241, or mir-48 during embryogenesis, we built VT1589 {lin-4(e912) II/mnC1; flh-2(bc375) III; flh-1(bc374) IV/nT1 [qIs51] (IV;V)} and VT1645 {flh-2(bc375) III; flh-1(bc374) IV/nT1 [qIs51] (IV;V); nDf51 V} and asked whether the lethality of the flh-1; flh-2 double mutant could be rescued in animals deleted for lin-4 or mir-48 and mir-241. Animals that were homozygous for either lin-4(0) or nDf51 [mir-48(0) mir-241(0)] did not produce viable progeny that were homozygous for both flh-1(bc374) and flh-2(bc375). This implies that the inviability of flh-1(bc374); flh-2(bc375) animals is not primarily a consequence of excessive levels of lin-4 or of mir-48 and mir-241, but may be due to the collective up-regulation of additional miRNA targets of FLH-1 and FLH-2, and/or altered expression of yet to be identified protein-coding gene targets of FLH-1 and FLH-2.
Discussion
We report the identification of three transcription factors—FLH-1, FLH-2, and FLH-3—that act redundantly to repress the expression of several miRNAs in C. elegans during embryogenesis. FLH-1, FLH-2, and FLH-3 are three of four proteins in C. elegans that contain a FLYWCH DNA-binding domain (Reece-Hoyes et al. 2005). Proteins with FLYWCH domains have also been identified in insects and vertebrates (Dorn and Krauss 2003; Krauss and Dorn 2004; Babu et al. 2006). The FLYWCH motif is a Cys2His2-type zinc-finger domain characterized by the conserved sequence: F/Y-X(n)-L-X(n)-F/Y-X(n)-WXCX(6–12)CX(17–22)HXH (where X is any amino acid). It was first identified in isoforms of the putative chromatin-modulating protein modifier of (mdg4) locus [mod(mdg4)] in Drosophila melanogaster (Gerasimova et al. 1995; Buchner et al. 2000). Phenotypes of mod(mdg-4) mutant flies include effects on position effect variegation, chromatin insulation, nerve cell pathfinding, chromosomal meiotic pairing, and apoptosis (for review, see Dorn and Krauss 2003). It is not known whether mod(mdg4) mutations cause defects in miRNA expression in Drosophila. Drosophila Mod(mdg4) has not been shown to bind to DNA, but the Mod(mdg4) FLYWCH domain does seem to mediate protein–protein interactions, since it can directly interact with the DNA-binding protein Suppressor of Hairy-wing [Su(Hw)] as part of the gypsy insulator (Ghosh et al. 2001). Although we did not establish a direct in vivo interaction between the FLH-1, FLH-2, or FLH-3 proteins, Y2H assays detected a physical interaction between FLH-1 and FLH-3 (Walhout et al. 2002). The potential functional significance of interactions among the C. elegans FLH proteins in vivo or their interactions with other proteins remains to be determined. The functional redundancy between the flh-1, flh-2, and flh-3 genes that we show here is consistent with a range of possible molecular relationships among the FLH-1, FLH-2, and FLH-3 proteins in vivo, including binding common sets of DNA sites, common protein partners, and/or functioning separately in redundant pathways.
The fourth C. elegans protein with a FLYWCH domain is peb-1 (Thatcher et al. 2001). The function and DNA-binding activity of peb-1 requires the FLYWCH motif as a deletion in this region results in deficiencies in pharynx development and molting (Beaster-Jones and Okkema 2004; Fernandez et al. 2004). Y1H analysis showed that PEB-1 binds to the promoter of several miRNAs (Martinez et al. 2008). However, unlike flh-1, miRNA TaqMan analyses of peb-1 mutant embryos did not show any aberrance in lin-4 levels (data not shown).
We found that the loss of both FLH-1 and FLH-2 results in early larval lethality. It is apparent from our studies that this phenotype is not due simply to the overexpression of lin-4, mir-48, or mir-241 during embryogenesis since a lin-4(lf) mutation or deletion of mir-48 and mir-241 did not rescue the flh lethal phenotype. Also, the increases in lin-4, mir-241, and mir-48 levels observed in the viable flh mutants [flh-1(tm2118); flh-2(tm2126) or flh-2(bc375); flh-3(tm3024)] were approximately similar to the increases in those miRNAs observed in the inviable mutant [flh-1(bc374); flh-2(bc375)] (Fig. 3C,E,F; Supplemental Fig. 3d–f; Supplemental Tables 4–6). Lethality in the latter case could be attributable to a slight but concerted overexpression of other miRNAs. However, we do not favor that option since worms with a mutation in a gene whose product is involved in the general processing of all the miRNAs, alg-1, also fails to rescue the lethality of the double FLYWCH mutant (data not shown). It is likely, then, that the FLH proteins regulate the transcription of protein-coding genes, as well as miRNA genes, that collectively contribute to the flh mutant phenotypes.
We did not observe post-embryonic developmental timing defects in flh mutants that overexpress in embryos miRNAs known to be developmental timing regulators. For example, although transgenic overexpression in early larval stages of mir-48 or lin-4 has been shown to cause precocious expression of later larval cell fates (Feinbaum and Ambros 1999; Li et al. 2005), the viable flh mutants that we examined did not display such larval developmental timing phenotypes. This is consistent with our observation that mir-48 and lin-4 are primarily overexpressed during embryogenesis in flh mutants, and are relatively normal during development of mutant larvae. It appears that the FLH proteins are particularly involved in inhibiting miRNA expression in embryos, and other regulatory mechanisms, including transcriptional activators, govern up-regulation of these same miRNAs in larvae (Fig. 8).
It is evident that the temporal (and most likely also spatial) regulation of miRNAs involves a complex network of negative and positive transcription factors as well as post-transcriptional regulation. Indeed, a recent genome-wide analysis of C. elegans transcription factors and intergenic miRNA promoters has revealed nearly 350 transcription factor–miRNA interactions (Martinez et al. 2008). Future experiments are needed to fully understand the roles of transcription factors in governing the temporal and spatial expression of miRNAs, including how developmental and physiological signals modulate the transcription factors regulating miRNA gene expression.
Materials and methods
Y1H assays
Fragment 365–451 of Plin-4 was cloned into pBM2389, upstream of a GAL1 promoter that drives the expression of HIS3 (Liu et al. 1993), generating plasmids pBM2389.AF (forward orientation) and pBM2389.AR (reverse orientation). The DNA fragment was also cloned into pSE640, upstream of a CYC1 promoter driving the expression of LacZ (American Type Culture Collection), generating plasmids pSE640.AF (forward orientation) and pSE640.AR (reverse orientation). These reporter plasmids were integrated into Saccharomyces cerevisiae strain YM4271. The yeast strains harboring the integrated plasmids were then transformed with two mixed-staged C. elegans cDNA libraries, pACT-RB1 (oligo dT primed) and pACT-RB2 (random hexamer primed; gifts from Dr. Robert Barstead). Yeast transformants with pBM2389.AF or pBM2389.AR and an activator plasmid were selected for tryptophan, leucine, and histidine prototrophy and with 40 mM 3-amino-1,2,4-triazole. Plasmids from positive candidates from the histidine screen were extracted and transformed into YM4271 harboring pSE640.AF or pSE640.AR, where the interaction was confirmed using in situ β-galactosidase assays. All yeast manipulations were done following standard procedures (Adams et al. 1997).
Gateway-compatible Y1H assays were done as described before (Deplancke et al. 2006; Vermeirssen et al. 2007) using 2 kb upstream of the mature miRNA as baits and FLH-1 and FLH-2 as preys.
Plasmid construction
A 5.6-kb fragment containing a rescuing wild-type copy of the unc-119 gene was inserted into the promoter-less GFP vector pPD95.75 to generate pMO23. All PCR reactions were done using Platinum Pfx DNA polymerase (Invitrogen). The Plin-4∷gfp reporter plasmid was made by amplifying a 2.4-kb fragment immediately upstream of the mature lin-4 and inserting it into pMO23. Plasmids for Pmir-241∷gfp, Pmir-48∷gfp, Pmir-59∷gfp, and Pmir-358-357∷gfp were made by amplifying 1.5–2-kb fragments upstream of the mature miRNA and ligating them into pMO23. The Plin-4∷gfp plasmid with a deletion of fragment 365–451 was made by the overlap extension PCR method (Ho et al. 1989).
A VENUS translational reporter for flh-1 (Pflh-1 [5 kb]∷flh-1∷venus∷flh-1 3′ UTR [290 bp]) was constructed using a combination of overlap extension PCR and Gateway cloning (Walhout et al. 2000). A GFP translational reporter for flh-2 (Pflh-2 [4.8 kb]∷gfp∷flh-2∷flh-2 3′ UTR [411 bp]) was made using a pBluescript SK(+) vector with GFP and the unc-119 mini-gene transformation marker from pDP#MM051 (Maduro and Pilgrim 1995). The transcriptional fusion reporter for flh-3 (Pflh-3 [4.5 kb]∷gfp∷flh-3 3′ UTR [1 kb]) was made by Gateway cloning. Details of all plasmid constructions and primer sequences will be provided upon request.
C. elegans strains
Worms were grown using standard procedures at 20°C on Nematode Growth Medium (NGM) plates (Sulston and Hodgkin 1988). The wild-type strain was C. elegans var. Bristol strain N2 (Brenner 1974). Deletion alleles isolated from mutagenesis libraries were backcrossed to N2 at least six to eight times before characterization. All nematode strains used in this study are listed in Supplemental Table 8.
C. elegans transformation
Gold microparticle biolistic bombardment (for review, see Praitis 2006) of DP38 [unc-119(ed3)] was used to create transgenic worms carrying fluorescent reporters. We used a transformation procedure described by Berezikov et al. (2004) using a PDS-1000/He system with the Hepta adaptor (Bio-Rad). At least two independent lines were obtained per bombardment.
RNAi-by-feeding
Embryos obtained following hypochlorite treatment of gravid adults were placed on RNAi plates (NGM with 100 μg/mL ampicillin, 15 μg/mL tetracycline, and 1 mM IPTG; seeded with bacteria expressing dsRNA) (Kamath et al. 2001). Once they reached L4, they were transferred onto fresh RNAi plates.
Isolation of FLH-1 and FLH-2 deletions
Deletion alleles flh-1(bc374) and flh-2(bc375) were isolated from a population of worms mutagenized with EMS using the poison primer method (Edgley et al. 2002). DNA sequencing was performed to assess the nature of the lesions. Sequences of the screening primers will be made available upon request.
miRNA TaqMan real-time PCR assays
One-hundred late-stage embryos were collected into Worm Lysis Buffer (50 mM KCl, 10 mM Tris at pH 8.3, 2.5 mM MgCl2, 0.45% NP-40, 0.45% Tween 20, 0.01% gelatin, 30 μg/mL proteinase K), subjected to 10 cycles of freezing and thawing, followed by incubation for 1 h at 65°C and for 20 min at 95°C. A Trizol (Invitrogen) extraction was done, and the RNA template was coprecipitated with glycogen (Ambion). The RNA was used in TaqMan assays following the instructions of the manufacturer using an ABI 7900HT Fast-Real Time PCR System (Applied Biosystems) (Chen et al. 2005). The miRNA Ct values were analyzed in triplicate from three independent biological samples. The comparative Ct (2−ΔΔCt) method (Livak and Schmittgen 2001) was used to calculate the average ΔΔCt values using the small nucleolar RNA, sn2841 or U18, as the normalization standard. Only those values for which the three independent biological replicates exhibited the same trend (increase or decrease) were considered in our analysis. ΔΔCt values were then normalized by subtracting the average ΔΔCt value for all the miRNAs in the experiment. Normalized ΔΔCt values for each miRNA assay were averaged across all replicates to generate a ΔΔCt final value, and the standard error of the mean was determined. Z-scores were calculated as −ΔΔCtfinal/SD. Z-scores ≥2 or ≤−2 were considered significant.
Northern blot analysis
Total RNA was extracted and analyzed (5–20 μg) by Northern blotting as described by Ambros and Lee (2004) using Starfire probes (Integrated DNA Technologies) complementary to the miRNA or to U6 snRNA. Northern blots for flh-1, flh-2, and flh-3 were done as described by Burnett (1997). PCR fragment probes for flh-1 and flh-2 were radiolabeled with [α-32P]dATP using the Decaprime II Random Primed DNA Labeling Kit (Ambion), and hybridized probe was detected using PhosphorImager screens and ImageQuant (Molecular Dynamics). Northern blots used to detect flh-1 and flh-2 were reprobed for flh-3 using a PCR fragment specific for flh-3 following the instructions of the DIG High Prime DNA Labeling and Detection Starter Kit II (Roche).
Western blot analysis
Embryos were obtained from the hypochlorite treatment of staged gravid adults. Starved L1s were collected following the overnight hatching of embryos in M9 buffer at 20°C. Worm pellets were resuspended in an equal volume of Lysis Buffer (4% SDS, 100 mM Tris at pH 6.8, 20% glycerol) and boiled for 20 min. Protein concentration was assessed using the RC DC Protein Assay Reagent (Bio-Rad). Protein extracts were resolved in 10% SDS-PAGE gels and transferred to PVDF membranes. Westerns were done with rabbit antisera against LIN-14 (Hristova et al. 2005), γ-tubulin (Sigma T1450), FLH-1, or FLH-2. Quantification of proteins was done using ImageJ (NIH).
Preparation of anti-FLH-1 and anti-FLH-2
A histidine-tagged full-length FLH-1 protein was expressed in Escherichia coli from the plasmid pHIS.Parallel1 (Sheffield et al. 1999), and purified protein was used to raise polyclonal antibodies in rabbits (Pocono Rabbit Farm and Laboratory). Polyclonal antibodies for FLH-2 were raised in rabbits immunized with a KLH conjugated peptide consisting of the last 20 amino acids (Open Biosystems).
Prediction of the FLH-1 consensus binding site
The sequences of the promoters that tested positive for FLH-1 binding by Y1H were analyzed using Improbizer (Ao et al. 2004) to predict a consensus binding site. Two types of input sequences were analyzed, either up to 2 kb or 1 kb upstream of the annotated miRNAs (Martinez et al. 2008). We used three types of background sequences: (1) all promoters sequences from the promoterome (Dupuy et al. 2004) (∼20,000 sequences lengths 300 bp to 2.5 kb); (2) a subset of sequences from the promoterome containing only regions between 2 and 2.5 kb upstream of the translation start site; and (3) the same background as foreground (same set of positive sequences used as background model). The motif shown was the most redundant site among all six searches. Improbizer scores reflect how well a site present in a given promoter fits the position weight matrix (for details, see http://www.soe.ucsc.edu/∼kent/improbizer/improbizer.html). The sequence logo was created using WebLogo (http://weblogo.berkeley.edu).
Cloning the FLH-1 consensus binding site
We cloned the predicted FLH-1-binding site from the promoter of mir-358. The sequence tested contained four extra nucleotides on each side of the predicted hit to account for the possibility that flanking nucleotides important for binding may have been missed in the motif searches. Complementary DNA primers were designed to contain one (FLYWCH-1x and FLYWCH-1-y) or three (FLYWCH-3x and FLYWCH-3-y) tandem FLH-binding sites. A Gateway compatible entry vector, pMW#4, was ligated with the annealed primers, and the FLH-1-binding sites were subsequently cloned into pMW#2 and pMW#3 integrated into the genome of S. cerevisiae YM4271 and used in Y1H assays as described previously (Deplancke et al. 2006).
Deletion of the FLH-1-binding site in Pmir-48
The FLH-1 consensus binding site was deleted from Pmir-48 using the QuikChange II Site-Directed Mutagenesis kit (Stratagene) following the instructions of the manufacturer with an entry clone with 2 kb of Pmir-48 as template. This entry clone was subsequently used to generate a Y1H bait as described previously (Deplancke et al. 2006).
Electrophoretic mobility shift assay
Wild-type gravid hermaphrodites were collected and embryos were harvested by hypochlorite treatment (Sulston and Hodgkin 1988). Embryos (50 μL of packed pellet) were washed, resuspended in 650 μL of 10 mM HEPES (pH 7.2) supplemented with Halt protease inhibitor cocktail (Pierce) and disrupted by 30 strokes of an ice-cold 3-mL stainless steel dounce homogenizer. Fifty micrograms of the resulting extract were incubated at room temperature in Binding Buffer (10 mM HEPES at pH 7.2, 25 mM NaCl, 1 mM MgCl2, 50 μM ZnCl2, 5% glycerol) with 2 μg of BSA and 0.5 μg of poly (dI–dC). A 30-min preincubation was done for cold probe chases and supershift assays using anti-FLH-1 or rabbit IgG Ab-1 control antibody (Thermo Scientific). Following the addition of a [γ-32P]ATP 5′-end-labeled wild-type or mutant fragments 365–451 (from Plin-4) or 200–251 (from Pmir-48), samples were incubated for an additional 20 min, immediately loaded into a 5% native gel, and electrophoresed at room temperature.
Acknowledgments
We are grateful to C. Chen from Applied Biosystems for TaqMan assay reagents, S. Mitani from the National Bioresource Project for deletion strains, and C. Mello for providing the facility and supplies to do some of the radioactive experiments. Some nematode strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the NIH National Center for Research Resources (NCRR). We thank B. Deplancke for help with the FLH-binding site cloning, A. Fire for pPD95.75, and Z. Derewenda for pHIS.Parallel1. M.C.O. was funded in part by NIH post-doctoral fellowship GM070118-02. This work was supported by NIH grants GM348642 and DK068429 to V.A. and A.J.M.W., respectively.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1678808.
References
- Abrahante J.E., Miller E.A., Rougvie A.E. Identification of heterochronic mutants in Caenorhabditis elegans. Temporal misexpression of a collagen::green fluorescent protein fusion gene. Genetics. 1998;149:1335–1351. doi: 10.1093/genetics/149.3.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams A., Gottschling D.E., Kaiser C.A., Stearns T. Methods in yeast genetics. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1997. [Google Scholar]
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Ambros V., Horvitz H.R. Heterochronic mutants of the nematode Caenorhabditis elegans. Science. 1984;226:409–416. doi: 10.1126/science.6494891. [DOI] [PubMed] [Google Scholar]
- Ambros V., Horvitz H.R. The lin-14 locus of Caenorhabditis elegans controls the time of expression of specific postembryonic developmental events. Genes & Dev. 1987;1:398–414. doi: 10.1101/gad.1.4.398. [DOI] [PubMed] [Google Scholar]
- Ambros V., Lee R.C. Identification of microRNAs and other tiny noncoding RNAs by cDNA cloning. Methods Mol. Biol. 2004;265:131–158. doi: 10.1385/1-59259-775-0:131. [DOI] [PubMed] [Google Scholar]
- Ao W., Gaudet J., Kent W.J., Muttumu S., Mango S.E. Environmentally induced foregut remodeling by PHA-4/FoxA and DAF-12/NHR. Science. 2004;305:1743–1746. doi: 10.1126/science.1102216. [DOI] [PubMed] [Google Scholar]
- Babu M.M., Iyer L.M., Balaji S., Aravind L. The natural history of the WRKY-GCM1 zinc fingers and the relationship between transcription factors and transposons. Nucleic Acids Res. 2006;34:6505–6520. doi: 10.1093/nar/gkl888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baugh L.R., Sternberg P.W. DAF-16/FOXO regulates transcription of cki-1/Cip/Kip and repression of lin-4 during C. elegans L1 arrest. Curr. Biol. 2006;16:780–785. doi: 10.1016/j.cub.2006.03.021. [DOI] [PubMed] [Google Scholar]
- Beaster-Jones L., Okkema P.G. DNA binding and in vivo function of C. elegans PEB-1 require a conserved FLYWCH motif. J. Mol. Biol. 2004;339:695–706. doi: 10.1016/j.jmb.2004.04.030. [DOI] [PubMed] [Google Scholar]
- Berezikov E., Bargmann C.I., Plasterk R.H. Homologous gene targeting in Caenorhabditis elegans by biolistic transformation. Nucleic Acids Res. 2004;32:e40. doi: 10.1093/nar/gnh033. http://nar.oxfordjournals.org/cgi/content/full/32/4/e40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchner K., Roth P., Schotta G., Krauss V., Saumweber H., Reuter G., Dorn R. Genetic and molecular complexity of the position effect variegation modifier mod(mdg4) in Drosophila. Genetics. 2000;155:141–157. doi: 10.1093/genetics/155.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnett W.V. Northern blotting of RNA denatured in glyoxal without buffer recirculation. Biotechniques. 1997;22:668–671. doi: 10.2144/97224st01. [DOI] [PubMed] [Google Scholar]
- Bushati N., Cohen S.M. MicroRNA functions. Annu. Rev. Cell Dev. Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- Chang T.C., Yu D., Lee Y.S., Wentzel E.A., Arking D.E., West K.M., Dang C.V., Thomas-Tikhonenko A., Mendell J.T. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat. Genet. 2008;40:43–50. doi: 10.1038/ng.2007.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Ridzon D.A., Broomer A.J., Zhou Z., Lee D.H., Nguyen J.T., Barbisin M., Xu N.L., Mahuvakar V.R., Andersen M.R., et al. Real-time quantification of microRNAs by stem–loop RT–PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller H.A., Forman J.J., Legesse-Miller A. “Myc’ed messages”: myc induces transcription of E2F1 while inhibiting its translation via a microRNA polycistron. PLoS Genet. 2007;3:e146. doi: 10.1371/journal.pgen.0030146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conaco C., Otto S., Han J.J., Mandel G. Reciprocal actions of REST and a microRNA promote neuronal identity. Proc. Natl. Acad. Sci. 2006;103:2422–2427. doi: 10.1073/pnas.0511041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deplancke B., Mukhopadhyay A., Ao W., Elewa A.M., Grove C.A., Martinez N.J., Sequerra R., Doucette-Stamm L., Reece-Hoyes J.S., Hope I.A., et al. A gene-centered C. elegans protein–DNA interaction network. Cell. 2006;125:1193–1205. doi: 10.1016/j.cell.2006.04.038. [DOI] [PubMed] [Google Scholar]
- Dorn R., Krauss V. The modifier of mdg4 locus in Drosophila: Functional complexity is resolved by trans splicing. Genetica. 2003;117:165–177. doi: 10.1023/a:1022983810016. [DOI] [PubMed] [Google Scholar]
- Dupuy D., Li Q.R., Deplancke B., Boxem M., Hao T., Lamesch P., Sequerra R., Bosak S., Doucette-Stamm L., Hope I.A., et al. A first version of the Caenorhabditis elegans Promoterome. Genome Res. 2004;14:2169–2175. doi: 10.1101/gr.2497604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgley M., D’Souza A., Moulder G., McKay S., Shen B., Gilchrist E., Moerman D., Barstead R.2002Improved detection of small deletions in complex pools of DNA Nucleic Acids Res. 30e52 . http://nar.oxfordjournals.org/cgi/content/full/30/12/e52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esquela-Kerscher A., Johnson S.M., Bai L., Saito K., Partridge J., Reinert K.L., Slack F.J. Post-embryonic expression of C. elegans microRNAs belonging to the lin-4 and let-7 families in the hypodermis and the reproductive system. Dev. Dyn. 2005;234:868–877. doi: 10.1002/dvdy.20572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinbaum R., Ambros V. The timing of lin-4 RNA accumulation controls the timing of postembryonic developmental events in Caenorhabditis elegans. Dev. Biol. 1999;210:87–95. doi: 10.1006/dbio.1999.9272. [DOI] [PubMed] [Google Scholar]
- Fernandez A.P., Gibbons J., Okkema P.G. C. elegans peb-1 mutants exhibit pleiotropic defects in molting, feeding, and morphology. Dev. Biol. 2004;276:352–366. doi: 10.1016/j.ydbio.2004.08.040. [DOI] [PubMed] [Google Scholar]
- Gerasimova T.I., Gdula D.A., Gerasimov D.V., Simonova O., Corces V.G. A Drosophila protein that imparts directionality on a chromatin insulator is an enhancer of position-effect variegation. Cell. 1995;82:587–597. doi: 10.1016/0092-8674(95)90031-4. [DOI] [PubMed] [Google Scholar]
- Ghosh D., Gerasimova T.I., Corces V.G. Interactions between the Su(Hw) and Mod(mdg4) proteins required for gypsy insulator function. EMBO J. 2001;20:2518–2527. doi: 10.1093/emboj/20.10.2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho S.N., Hunt H.D., Horton R.M., Pullen J.K., Pease L.R. Site directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- Hristova M., Birse D., Hong Y., Ambros V. The Caenorhabditis elegans LIN-14 is a novel transcription factor that controls the developmental timing of transcription from the insulin/insulin-like growth factor gene ins-33 by direct DNA binding. Mol. Cell Biol. 2005;25:11059–11072. doi: 10.1128/MCB.25.24.11059-11072.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath R.S., Martinez-Campos M., Zipperlen P., Fraser A.G., Ahringer J. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in C. elegans. Genome Biol. 2001;2:1–10. doi: 10.1186/gb-2000-2-1-research0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim V.N. MicroRNA biogenesis: Coordinated cropping and dicing. Nat. Rev. Mol. Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- Krauss V., Dorn R. Evolution of the trans-splicing Drosophila locus mod(mdg4) in several species of Diptera and Lepidoptera. Gene. 2004;331:165–176. doi: 10.1016/j.gene.2004.02.019. [DOI] [PubMed] [Google Scholar]
- Lee R., Feinbaum R., Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Li M., Jones-Rhoades M.W., Lau N.C., Bartel D.P., Rougvie A.E. Regulatory mutations of mir-48, a C. elegans let-7 family microRNA, cause developmental timing defects. Dev. Cell. 2005;9:415–422. doi: 10.1016/j.devcel.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Liu J., Wilson T.E., Milbrandt J., Johnston M. Methods companion. Methods Enzymol. 1993;5:125–137. [Google Scholar]
- Liu Z., Kirch S., Ambros V. The Caenorhabditis elegans heterochronic gene pathway controls stage-specific transcription of collagen genes. Development. 1995;121:2471–2478. doi: 10.1242/dev.121.8.2471. [DOI] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2[−ΔΔC(T)] method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Maduro M., Pilgrim D. Identification and cloning of unc-119, a gene expressed in the Caenorhabditis elegans nervous system. Genetics. 1995;141:977–988. doi: 10.1093/genetics/141.3.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez N.J., Ow M.C., Barrasa M.I., Hammell M., Sequerra R., Doucette-Stamm L., Roth F.P., Ambros V.R., Walhout A.J.M. A C. elegans genome-scale microRNA network contains composite feedback motifs with high flux capacity. Genes & Dev. 2008 doi: 10.1101/gad.1678608. (this issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai T., Ibata K., Park E.S., Kubota M., Mikoshiba K., Miyawaki A. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 2002;20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- O’Donnell K.A., Wentzel E.A., Zeller K.I., Dang C.V., Mendell J.T. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- Praitis V. Creation of transgenic lines using microparticle bombardment methods. Methods Mol. Biol. 2006;351:93–107. doi: 10.1385/1-59745-151-7:93. [DOI] [PubMed] [Google Scholar]
- Rao P.K., Kumar R.M., Farkhondeh M., Baskerville S., Lodish H.F. Myogenic factors that regulate expression of muscle-specific microRNAs. Proc. Natl. Acad. Sci. 2006;103:8721–8726. doi: 10.1073/pnas.0602831103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reece-Hoyes J.S., Deplancke B., Shingles J., Grove C.A., Hope I.A., Walhout A.J. A compendium of Caenorhabditis elegans regulatory transcription factors: A resource for mapping transcription regulatory networks. Genome Biol. 2005;6:R110. doi: 10.1186/gb-2005-6-13-r110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheffield P., Garrard S., Derewenda Z. Overcoming expression and purification problems of RhoGDI using a family of “parallel” expression vectors. Protein Expr. Purif. 1999;15:34–39. doi: 10.1006/prep.1998.1003. [DOI] [PubMed] [Google Scholar]
- Simmer F., Tijsterman M., Parrish S., Koushika S.P., Nonet M.L., Fire A., Ahringer J., Plasterk R.H.A. Loss of the putative RNA-directed RNA polymerase RRF-3 makes C. elegans hypersensitive to RNAi. Curr. Biol. 2002;12:1317–1319. doi: 10.1016/s0960-9822(02)01041-2. [DOI] [PubMed] [Google Scholar]
- Sulston J.E., Hodgkin J. Methods. In: Wood W.B., et al., editors. The neamtode Caenorhabditis elegans. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1988. pp. 587–606. [Google Scholar]
- Sulston J.E., Horvitz H.R. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev. Biol. 1977;56:110–156. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- Thatcher J.D., Fernandez A.P., Beaster-Jones L., Haun C., Okkema P.G. The Caenorhabditis elegans peb-1 gene encodes a novel DNA-binding protein involved in morphogenesis of the pharynx, vulva, and hindgut. Dev. Biol. 2001;229:480–493. doi: 10.1006/dbio.2000.9978. [DOI] [PubMed] [Google Scholar]
- Thomson J.M., Newman M., Parker J.S., Morin-Kensicki E.M., Wright T., Hammond S.M. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes & Dev. 2006;20:2202–2207. doi: 10.1101/gad.1444406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeirssen V., Barrasa M.I., Hidalgo C.A., Babon J.A., Sequerra R., Doucette-Stamm L., Barabási A.L., Walhout A.J. Transcription factor modularity in a gene-centered C. elegans core neuronal protein–DNA interaction network. Genome Res. 2007;17:1061–1071. doi: 10.1101/gr.6148107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walhout A.J., Temple G.F., Brasch M.A., Hartley J.L., Lorson M.A., van den Heuvel S., Vidal M. GATEWAY recombinational cloning: Application to the cloning of large numbers of open reading frames or ORFeomes. Methods Enzymol. 2000;328:575–592. doi: 10.1016/s0076-6879(00)28419-x. [DOI] [PubMed] [Google Scholar]
- Walhout A.J., Reboul J., Shtanko O., Bertin N., Vaglio P., Ge H., Lee H., Doucette-Stamm L., Gunsalus K.C., Schetter A.J., et al. Integrating interactome, phenome, and transcriptome mapping data for the C. elegans germline. Curr. Biol. 2002;12:1952–1958. doi: 10.1016/s0960-9822(02)01279-4. [DOI] [PubMed] [Google Scholar]
- Wightman B., Ha I., Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- Wulczyn F.G., Smirnova L., Rybak A., Brandt C., Kwidzinski E., Ninnemann O., Strehle M., Seiler A., Schumacher S., Nitsch R. Post-transcriptional regulation of the let-7 microRNA during neural cell specification. FASEB J. 2007;21:415–426. doi: 10.1096/fj.06-6130com. [DOI] [PubMed] [Google Scholar]