Abstract
After the initial description of Arthrobacter spp. isolated from clinical specimens in the mid-1990s, very few further reports on Arthrobacter spp. have appeared in the clinical microbiology literature. The aim of the present study was to elucidate the distribution of Arthrobacter spp. and Arthrobacter-like bacteria encountered in clinical specimens by studying 50 consecutively isolated or received strains of large-colony-forming, whiteish-grayish, non-cheese-like-smelling, nonfermentative gram-positive rods by applying phenotypic methods as well as 16S rRNA gene sequencing. We observed a very heterogenous distribution, with the 50 strains belonging to 20 different taxa and each of 13 strains as a single representative of its particular taxon. Thirty-eight strains represented true Arthrobacter strains, 7 strains belonged to the genus Brevibacterium, 2 were Microbacterium species, and each of 3 single strains was a member of the rarely encountered genera Pseudoclavibacter, Leucobacter, and Brachybacterium, respectively. A. cumminsii (n = 14) and A. oxydans (n = 11) were the most frequently found species. The present report describes the first three A. aurescens strains isolated from human clinical specimens. Comprehensive antimicrobial susceptibility data are given for the 38 Arthrobacter isolates.
Arthrobacter spp. belong to the heterogenous group of coryneform bacteria and had not been reported to have been isolated from human clinical specimens until the mid-1990s (7). This was somewhat surprising, because Arthrobacter spp. are the most frequently isolated coryneform bacteria when soil specimens are incubated aerobically, indicating that humans are unceasingly exposed to these bacteria. After the initial descriptions (7, 9), only a few other studies have been published regarding the appearance of Arthrobacter spp. in clinical specimens (1, 10, 11, 12, 23). Therefore, we have continued to collect Arthrobacter isolates either from our routine clinical laboratory procedures or as reference cultures sent to us. The present report outlines the data pertaining to 50 consecutive Arthrobacter or Arthrobacter-like strains collected by one of the authors (G. Funke). Arthrobacter and Arthrobacter-like strains were defined for the purposes of the present study as large-colony-forming, whiteish-grayish, non-cheese-like-smelling, nonfermentative gram-positive rods. The aim of the study was to investigate the identity of Arthrobacter and Arthrobacter-like strains by use of phenotypic and molecular genetic methods in order to finally reveal the true distribution within clinical Arthrobacter isolates. In addition, we determined MICs of a variety of antimicrobials against these bacteria, since limited data exist in the relevant literature on the antimicrobial susceptibility patterns of Arthrobacter spp. As a byproduct of our investigations, we describe two new bacterial species, namely, Arthrobacter sanguinis sp. nov. and Brevibacterium ravenspurgense sp. nov.
MATERIALS AND METHODS
Strains.
The 50 strains examined in the present study were isolated in the routine clinical microbiology laboratories of Gärtner & Colleagues Laboratories, Ravensburg, Germany, or were referred to this institution by collaborating laboratories. None of the isolates had been included in any of our previous studies except strains 1369, NML 90-0364, NML 91-0435, NML 92-0385, NML 92-0394, NML 92-0600, and NML 93-0702 (7). The strains had been stored at −20°C in skim milk. For the investigations, strains were grown on Columbia sheep blood agar (SBA) plates (BD, Heidelberg, Germany) and passaged twice on SBA plates at 35°C in ambient air before use.
Phenotypic testing.
The techniques applied have been outlined in detail before (8, 23). For assimilation tests of the nonfermenting gram-positive rods, we applied commercial AUX medium (bioMérieux, Marcy l′Etoile, France) to the API 50CH kit (bioMérieux) (5). Reading of the assimilation reactions was done at 48 and 120 h of incubation at 35°C.
Chemotaxonomic investigations.
Analyses of cellular fatty acids and the diamino acid of the bacterial cell wall were performed as outlined before (8).
Molecular genetic investigations.
Analysis of the complete 16S rRNA gene sequences was performed according to a published protocol (6). Almost complete (>1,350 bp) 16S rRNA gene sequences were determined for each clinical strain by aligning multiple overlapping sequences by use of the Lasergene 5 package (DNASTAR Inc., Madison, WI). Phylogenetic trees were constructed using the neighbor-joining method, included in the MEGA4 suite software (22), based on a comparison of approximately 1,350 nucleotides. Bootstrap values, expressed as percentages of 1,000 replications, are given at each branching point in the figures.
Identification.
Strains were identified on the species level when the 16S rRNA gene sequence of the individual strain was >99.0% homologous to the type strain of a certain species (19) and when phenotypic testing did not indicate any aberrant reactions regarding the published data for this particular species.
Antimicrobial susceptibility testing.
The CLSI standard for determination and interpretation of antimicrobial MICs for Corynebacterium spp. (2) was applied. Briefly, by use of a broth microdilution method, bacterial cells representing an inoculum equivalent to a 0.5 McFarland standard were grown in cation-adjusted Mueller-Hinton broth with lysed horse blood and incubated for up to 48 h. Reading of MICs was done by two independent researchers.
Nucleotide accession numbers.
The GenBank accession numbers of the complete 16S rRNA gene sequences of all 50 clinical isolates included in the present study are given in Table 1. The GenBank accession number of the 16S rRNA gene sequence of the Arthrobacter sanguinis type strain is EU086805. The GenBank accession number of the 16S rRNA gene sequence of the Brevibacterium ravenspurgense type strain is EU086793.
TABLE 1.
Strains included in the present study
| Strain collection no. | Patient age (yr), sexa | Clinical source | Identification | GenBank accession no. |
|---|---|---|---|---|
| 20 (CCUG 56047)b | 45, m | Wound swab | Brevibacterium ravenspurgense sp. nov. | EU086793 |
| 27 | 59, f | Sinus aspirate | Brevibacterium otitidis | EU086795 |
| 28 | 27, f | Deep wound | Brevibacterium paucivorans | EU086796 |
| 60 | 53, m | Wound swab | Brevibacterium casei | EU086802 |
| 84 | 2, m | Wound swab | Arthrobacter aurescens | EU086809 |
| 102 | 39, m | Wound secretion | Microbacterium foliorum | EU086781 |
| 115 | 89, f | Wound swab | Arthrobacter oxydans | EU086782 |
| 120 | 24, f | Wound swab | Arthrobacter oxydans | EU086783 |
| 124 | 34, f | Urine | Arthrobacter cumminsii | EU086784 |
| 148 | 78, f | Urine | Arthrobacter aurescens | EU086789 |
| 219 | 78, f | Wound swab | Arthrobacter cumminsii | EU086794 |
| 300 | 51, f | Blood culture | Arthrobacter cumminsii | EU086797 |
| 352 | 53, m | Tracheal secretion | Arthrobacter cumminsii | EU086798 |
| 379 | 46, m | Otitis externa | Brevibacterium casei | EU086799 |
| 448 | 62, m | Nasal swab | Microbacterium oxydans | EU086800 |
| 486 | 45, f | Urine | Arthrobacter cumminsii | EU086827 |
| 511 | 62, m | Wound swab | Brachybacterium sp. | EU086801 |
| 664 | 65, m | Urine | Arthrobacter cumminsii | EU086819 |
| 665 | 74, m | Aortic valve | Pseudoclavibacter sp. | EU086820 |
| 690 | 25, m | Blood culture | Arthrobacter sp. (uncultured soil bacterium clone AKAU3746) | EU086821 |
| 696 | 66, m | Wound swab | Brevibacterium otitidis | EU086822 |
| 720 | 83, m | Wound swab | Arthrobacter cumminsii | EU086803 |
| 740 (CCUG 44254) | 45, f | Cervix | Arthrobacter cumminsii | EU086804 |
| 741 (CCUG 46407) | 63, m | Human blood | Arthrobacter sanguinis sp. nov. | EU086805 |
| 742 (CCUG 29118) | 82, f | Otitis externa | Arthrobacter cumminsii | EU086806 |
| 743 (CCUG 35230) | 27, m | Human tibia | Leucobacter sp. | EU086807 |
| 744 (CCUG 46391) | 88, f | Human blood | Brevibacterium ravenspurgense sp. nov. | EU086808 |
| 1361 | 36, f | Urine | Arthrobacter cumminsii | EU086785 |
| 1366 | NK | Urine | Arthrobacter albus | EU086786 |
| 1369 | NK | Vaginal swab | Arthrobacter oxydans | EU086823 |
| 1378 | NK | Urine | Arthrobacter cumminsii | EU086787 |
| 1391 | NK | Otitis externa | Arthrobacter cumminsii | EU086788 |
| 1515 (CCUG 33745) | 47, m | Human blood | Arthrobacter cumminsii | EU086790 |
| NMLc 90-0364 | NK | Eye | Arthrobacter oxydans | EU086810 |
| NML 91-0435 | NK | Eye | Arthrobacter oxydans | EU086824 |
| NML 92-0385 | NK | Blood culture | Arthrobacter sp. strain BS20 | EU086825 |
| NML 92-0394 | NK | Human blood | Arthrobacter oxydans | EU086826 |
| NML 92-0600 | NK | Blood culture | Arthrobacter oxydans | EU086811 |
| NML 93-0693 | NK | NK | Arthrobacter oxydans | EU086791 |
| NML 93-0702 | NK | NK | Arthrobacter oxydans | EU086792 |
| NML 93-0734 | NK | NK | Arthrobacter aurescens | EU086812 |
| NML 95-0018 | NK | Hand wound | Arthrobacter sp. strain An16 | EU086813 |
| NML 95-0188 | 28, f | Blood culture | Arthrobacter sp. strain 19B | EU086814 |
| NML 98-0077 | NK | Urine | Arthrobacter protophormiae | EU086815 |
| NML 99-0063 | NK | Blood culture | Arthrobacter albus | EU086816 |
| NML 99-0140 | NK | Blood culture | Arthrobacter cumminsii | EU086817 |
| NML 00-0248 | NK | Blood culture | Arthrobacter sp. strain W8 | EU086777 |
| NML 01-0266 | NK | Neck abscess | Arthrobacter oryzae | EU086778 |
| NML 02-0288 | NK | Blood culture | Arthrobacter oxydans | EU086779 |
| NML 03-0063 | NK | Lung swab at autopsy | Arthrobacter oxydans | EU086780 |
m, male; f, female; NK, not known.
CCUG, Culture Collection University of Gothenburg, Gothenburg, Sweden.
NML, National Microbiology Laboratory.
RESULTS
Table 1 lists all the data of the strains included in the present study. Patient data were available in 30 of 50 cases and showed that 16 patients were males and 14 females. The mean of the ages of the patients was 52.5 years (range, 2 to 89 years). Twelve strains each were isolated either from blood cultures or from wounds, and 8 strains came from urine samples. Five strains were isolated from primarily sterile tissues, and only one strain came from respiratory tract material.
We observed that the 50 strains included in the present study were representatives of 20 different taxa and that each of 13 strains was a single representative of a particular taxon. 16S rRNA gene homologies ranged from 99.03% to 99.93%, with a mean of 99.60%, excluding the two newly defined species (see below) and the members of the genera Pseudoclavibacter, Leucobacter, and Brachybacterium (see below). Thirty-eight strains represented true Arthrobacter species, 7 strains belonged to the genus Brevibacterium, 2 strains were microbacteria, and each of 3 single strains was a member of the genera Pseudoclavibacter, Leucobacter, or Brachybacterium, respectively. Within the arthrobacters, A. cumminsii (n = 14) and A. oxydans (n = 11) were the most frequently detected species and together represented half of the strains included in the present study. Three strains were identified as A. aurescens. Strain 690 was a representative of the so-far-noncultivated Arthrobacter bacterial clone AKAU3746, and strains NML 92-0385, NML 95-0018, and NML 95-0188 were representatives of Arthrobacter sp. strains BS20, An16, and 19B, respectively, none of which could be identified on the species level.
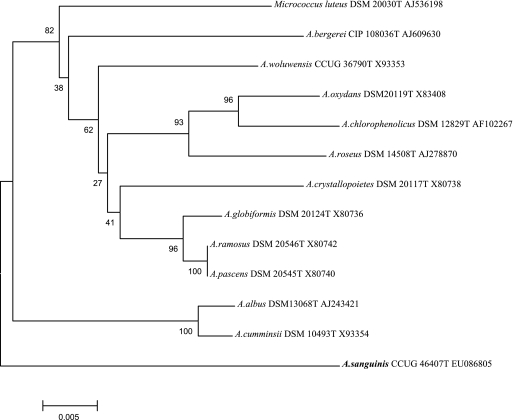
Strain 741 was phylogenetically distinct from all of the other valid Arthrobacter species and is, therefore, described as a new species, Arthrobacter sanguinis sp. nov. (see below). 16S rRNA gene homologies with all other 51 presently valid Arthrobacter spp. ranged from 91.8 to 94.7%, with A. crystallopoietes being its closest known phylogenetic neighbor. Table 2 gives the results of biochemical reactions differentiating A. sanguinis sp. nov. from its closest phylogenetic neighbors, with 16S rRNA gene homology values between 94.5 and 94.7%. Cellular fatty acids were of the branched type, with C15:0ai and C17:0ai predominating, and lysine was the diamino acid of the cell wall, which was compatible with an assignment of the unknown bacterium to the genus Arthrobacter. Figure 1 shows the phylogenetic position of A. sanguinis with respect to its neighbors, demonstrating that A. sanguinis represents a unique deep branch within the genus Arthrobacter.
TABLE 2.
Phenotypic features differentiating Arthrobacter sanguinis sp. nov. from its closest phylogenetic neighbors
| Feature | Reaction for:
|
|||
|---|---|---|---|---|
| A. sanguinis | A. crystallo- poietes | A. cumminsii | A. globiformis | |
| Activity of: | ||||
| α-Galactosidase | + | − | − | − |
| β-Galactosidase | + | − | − | + |
| N-Acetyl-β-glucosaminidase | + | − | − | − |
| α-Glucosidase | + | + | − | + |
| α-Mannosidase | + | − | − | + |
| Utilization ofa: | ||||
| Amygdaline | + | − | − | − |
| d-Arabitol | + | − | − | − |
| l-Arabinose | − | − | − | + |
| Cellobiose | + | + | − | − |
| Dulcitol | − | − | − | + |
| Galactose | + | − | − | + |
| N-Acetyl-glucosamine | + | − | − | + |
| Glycerol | + | − | − | + |
| Inositol | − | + | − | + |
| Maltose | + | − | − | + |
| Mannitol | + | + | − | + |
| Melezitose | − | − | − | + |
| Melibiose | + | − | − | + |
| Raffinose | + | − | − | + |
| Rhamnose | − | − | − | + |
| Sorbitol | + | − | − | + |
| Trehalose | + | − | − | + |
| Turanose | + | − | − | + |
Utilization reactions were tested as outlined in reference 5.
FIG. 1.
Phylogenetic tree based on 16S rRNA gene sequences, showing the position of Arthrobacter sanguinis sp. nov. within its closest phylogenetic neighbors. Micrococcus luteus was used as an outlier. The bar represents percent substitutions.
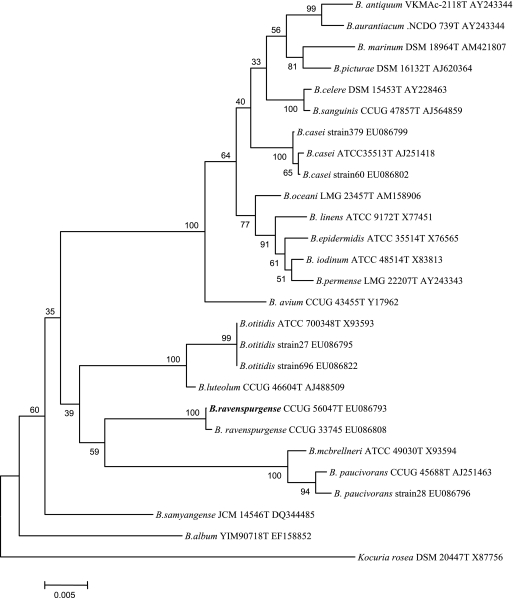
Of the seven Brevibacterium strains, two turned out to be B. casei, two were B. otitidis, and one was B. paucivorans. Two further strains could not be assigned to any of the established brevibacterial species. Phylogenetic analyses revealed that the 16S rRNA genes from these two isolates shared homology of only between 93.3 and 95.2% with those of the 19 other presently defined Brevibacterium species. Cellular fatty acids were of the branched type, with C15:0ai and C17:0ai predominating, and meso-diamino pimelic acid was detected as the diamino acid of the cell wall, which was compatible with an assignment to the genus Brevibacterium. The two unknown strains showed 99.7% (1,444 of 1,448 bp) 16S rRNA gene homology and were defined as a new species, Brevibacterium ravenspurgense sp. nov. Their phylogenetic relationship with all other presently defined brevibacteria is depicted in Fig. 2.
FIG. 2.
Phylogenetic tree of the genus Brevibacterium based on 16S rRNA gene sequences showing the position of Brevibacterium ravenspurgense sp. nov. as well as of the other Brevibacterium strains from the present study. Kocuria rosea was used as an outlier. The bar represents percent substitutions.
Two strains turned out to be Microbacterium oxydans and M. foliorum, respectively, and did not, in these two instances, exhibit yellow pigmentation. One strain each was a member of the genera Pseudoclavibacter, Leucobacter, and Brachybacterium. Those three isolates could not be identified on the species level, since the phenotypic and genetic data of other strains belonging to these genera are presently not comprehensive enough to allow unambiguous assignment to a particular species.
Antimicrobial susceptibility testing of the 38 Arthrobacter strains showed that 90.5% of the results fell into the susceptible category, 3.9% into the intermediate category, and 5.5% into the resistant category (Table 3). All antimicrobials tested, except ciprofloxacin and erythromycin, demonstrated good activities against Arthrobacter spp.
TABLE 3.
Antimicrobial susceptibility patterns of Arthrobacter strains (n = 38)a
| Antimicrobial agent | MIC (μg/ml)
|
No. (%) of isolates in indicated susceptibility category
|
||||
|---|---|---|---|---|---|---|
| Range | 50% | 90% | Susceptible | Intermediate | Resistant | |
| Cefotaxime | ≤0.03 to 8 | 0.12 | 1 | 36 (95) | 1 (3) | 1 (3) |
| Ciprofloxacin | 0.12 to 16 | 2 | 4 | 16 (42) | 13 (34) | 9 (24) |
| Doxycycline | ≤0.06 to 16 | 0.12 | 1 | 37 (97) | 0 (0) | 1 (3) |
| Erythromycin | 0.03 to >32 | 0.12 | 32 | 32 (84) | 0 (0) | 6 (16) |
| Gentamicin | 0.25 to 16 | 1 | 4 | 36 (95) | 1 (3) | 1 (3) |
| Linezolid | 0.12 to 2 | 1 | 2 | 38 (100) | 0 (0) | 0 (0) |
| Meropenem | ≤0.03 to 2 | 0.25 | 1 | 38 (100) | 0 (0) | 0 (0) |
| Penicillin | ≤0.03 to >64 | 0.12 | 0.5 | 36 (95) | 0 (0) | 2 (5) |
| Rifampin | ≤0.015 to 4 | ≤0.015 | 0.25 | 37 (97) | 0 (0) | 1 (3) |
| Vancomycin | 0.25 to 2 | 0.5 | 1 | 38 (100) | 0 (0) | 0 (0) |
The Arthrobacter strains listed in Table 1 were tested.
DISCUSSION
The present report represents the most comprehensive study of Arthrobacter and Arthrobacter-like strains isolated from clinical specimens published to date. Previous studies had included a maximum of 15 Arthrobacter strains (7, 9, 23). That the previous reports examined a relatively small number of strains is not surprising, since, in our experience, arthrobacters are two to three times less frequently isolated from clinical specimens than other nonfermenting coryneforms such as microbacteria or brevibacteria (G. Funke, unpublished observation). Despite the large number of Arthrobacter isolates, one limitation of our present study is that the clinical data of the patients were limited, but this lack is very often present in studies coming from reference centers.
The most frequently found Arthrobacter species in the present series was A. cumminsii. That same result had been published in another report a decade ago (9), and other authors later confirmed this observation (23). A. oxydans represented more than 20% of the clinical strains in the present study. In the study of Wauters et al. (23), A. oxydans represented 2 of 5 clinical Arthrobacter strains. A. oxydans has been, at least in some studies, the most frequently found Arthrobacter species in specimens from soil (17), which might have been the source from which our clinical strains originated.
The third most frequently found Arthrobacter species was A. aurescens, which has not been reported before as being isolated from human clinical specimens. A. aurescens can be differentiated from the closely related A. nitroguajacolicus (99.7% 16S rRNA gene homology) by a negative sucrose utilization reaction (13); such a result was seen for all three clinical strains included in the present study.
We observed an enormous heterogeneity within the 50 studied large-colony-forming, whitish-grayish, non-cheese-like-smelling, nonfermentative gram-positive rods, with each of 13 strains representing a single strain belonging to a particular taxon. Strain 690 represented an isolate of the previously noncultured Arthrobacter clone AKAU3746; whether this strain is a representative of a new Arthrobacter species remains to be elucidated by extensive quantitative DNA-DNA hybridization studies, since strain 690 did not have a 16S rRNA gene divergence percentage of greater than 3% compared to that of its most closely defined phylogenetic neighbor in the present study. In contrast, the 16S rRNA gene divergence value for strain 741 was greater than 5% for all of the Arthrobacter spp., which clearly demonstrated that this strain deserves recognition as an individual species (19, 20) (for species description, see below).
Our two A. albus strains are only the third and fourth strains of this species that have appeared in the literature (23). We can confirm that the susceptibility to desferrioxamine concentrations of 1,000 μg per disk may allow the separation between A. cumminsii (susceptible) and A. albus (resistant) (23); of the 14 A. cumminsii strains tested in the present study, only strain 219 was resistant to desferrioxamine. Although it is acknowledged that our number of A. albus strains (plus the 16S rRNA gene data from the type strain of A. albus) was limited, the following 16S rRNA gene signature nucleotides seem to allow a differentiation between the closely related A. cumminsii and A. albus (based on A. cumminsii numbering using the type strain sequence X93354): at position 157, a C in A. cumminsii versus a nucleotide deletion in A. albus; between positions 161 and 162, a deletion in A. cumminsii versus a T in A. albus; at position 163, G versus A; at position 504, G versus A; at position 988, C versus T; and at position 1096, T versus G.
The susceptibility patterns of the 38 Arthrobacter strains were similar to the data reported before (7, 9), with nearly all isolates exhibiting susceptibility to β-lactams, doxycycline, gentamicin, linezolid, rifampin, and vancomycin. Only the MICs of gentamicin tended to be lower with the broth microdilution method used in the present investigations compared to the results seen with the agar dilution method used in a previous study (7). We did not detect multiresistant Arthrobacter isolates, which contrasts with results reported before for A. woluwensis (1, 7).
The two B. casei strains found in the present study did not exhibit the distinctive cheese-like smell usually detected in B. casei (5). So far, only four B. otitidis strains have been described in the literature (3, 16, 25) and only seven strains have been described for B. paucivorans (24). The two strains representing an unknown Brevibacterium (i.e., B. ravenspurgense) had a distinctive sticky colony consistency which has not been observed for other true brevibacteria except for some B. paucivorans strains. B. ravenspurgense can be differentiated from B. paucivorans by the following reactions: strains of B. ravenspurgense are positive for pyrazinamidase and esterase (C4) whereas B. paucivorans strains are not; in contrast, B. paucivorans strains are variable with respect to N-acetyl-β-glucosaminidase reactivity whereas the two strains of B. ravenspurgense described in the present report were negative for this particular reaction. Like B. mcbrellneri and B. paucivorans (16, 24), the two unknown Brevibacterium strains did not utilize any of the carbohydrates in the test system used whereas the majority of the other brevibacteria are quite reactive in this test system (5). Interestingly, we did not observe gelatinase activity for the unknown Brevibacterium strains whereas nearly all other brevibacteria express this enzyme activity (5, 24).
When nonfermenting microbacteria do not exhibit yellow pigmentation, they can be easily confused with Arthrobacter spp. In the present series, 4% of the strains represented nonfermenting Microbacterium strains. It is noteworthy that two former Arthrobacter species (“Arthrobacter flavescens” and “Arthrobacter terregens”) have been reclassified as Microbacterium species (see http://www.bacterio.cict.fr/a/arthrobacter.html), indicating the close phenotypic relationship between these two genera. Furthermore, it should be mentioned that Arthrobacter and Microbacterium are the two largest genera within the nonfermenting coryneform bacteria, with each presently comprising more than 50 valid species.
This paper reports only the fourth Pseudoclavibacter strain isolated from humans. The type species of the genus had been previously designated “Brevibacterium helvolum,” and the genus Zimmermannella is a later synonym of Pseudoclavibacter (14, 15). Up to now, Leucobacter strains have not been reported to have been isolated from human clinical material (14, 18, 21). In the relevant literature, only two Brachybacterium strains isolated from humans have appeared so far, both of which exhibited fermentative metabolism (4).
For the routine clinical laboratory, we recommend the use of molecular identification techniques (e.g., full-length 16S rRNA gene sequencing) in order to identify clinically relevant large-colony-forming, whitish-grayish, non-cheese-like-smelling, nonfermentative gram-positive rods because of the great degree of heterogeneity within this group of bacteria as shown in the present study.
Arthrobacter sanguinis sp. nov.
Arthrobacter sanguinis (san′gui.nis. L. masc. gen. n. sanguinis of blood, indicating that the bacterium was isolated from a blood culture).
The cells are coryneform bacteria without irregular branching, and spores are not formed. The organism is obligately aerobic. The colonies are whitish-grayish, slightly convex, of creamy texture, and up to 2 mm in diameter after 24 h of incubation at 35°C on Columbia SBA plates. Activities of the following enzymes are detected: catalase, acid phosphatase, alkaline phosphatase, esterase (C4), esterase lipase (C8), α-galactosidase, β-galactosidase, gelatinase, N-acetyl-β-glucosaminidase, α-glucosidase, leucine arylamidase, α-mannosidase, pyrazinamidase, and trypsin. Activities of α-chymotrypsin, cystine arylamidase, α-fucosidase, β-glucosidase, β-glucuronidase, lipase (C14), nitrate reductase, naphthol-AS-BI-phosphohydrolase, urease, and valine arylamidase are not observed. The bacterium is capable of utilizing N-acetylglucosamine, amygdalin, d-arabitol, cellobiose, fructose, galactose, gentiobiose, glucose, glycerol, maltose, mannitol, mannose, melibiose, potassium gluconate, potassium 2-ketogluconate, raffinose, sucrose, sorbitol, trehalose, and turanose as carbon sources. The type strain did not utilize adonitol, d-arabinose, l-arabinose, l-arabitol, arbutin, dulcitol, erythritol, fucose, methyl-α-d-glucopyranoside, glycogen, inositol, inulin, potassium 5-ketogluconate, lactose, lyxose, methyl-α-d-mannopyranoside, melezitose, rhamnose, ribose, salicin, sorbose, starch, tagatose, methyl-β-d-xylopyranoside, xylitol, or xylose. Lysine is the diamino acid of the peptidoglycan, and C15:0ai and C17:0ai are the predominant cellular acid acids. The type strain is CCUG 46407 and has been deposited in the Culture of the University of Gothenburg, Sweden, and as strain DSM 21259 in the German Collection of Microorganisms and Cell Cultures.
Brevibacterium ravenspurgense sp. nov.
Brevibacterium ravenspurgense (ra.vens.pur.gen′se. N.L. adj. from Ravenspurgum, Latin name of the town of Ravensburg, Germany, where the type strain of this species was isolated).
Cells are coryneform bacteria without irregular branching, and spores are not formed. The organism is obligately aerobic. The colonies are whitish-grayish, slightly convex, have a sticky consistency, and are up to 2 mm in diameter after 24 h of incubation at 35°C on Columbia SBA plates. Activities of the following enzymes are detected: catalase, esterase (C4), esterase lipase (C8), leucine arylamidase, naphthol-AS-BI-phosphohydrolase, and pyrazinamidase. Activities of the following enzymes could not be observed: acid phosphatase, α-chymotrypsin, α-fucosidase, α-galactosidase, β-galactosidase, gelatinase, N-acetyl-β-glucosaminidase, α-glucosidase, β-glucosidase, β-glucuronidase, lipase (C14), α-mannosidase, nitrate reductase, pyrrolidonyl arylamidase, urease, and valine arylamidase. Activities of alkaline phosphatase and trypsin are variable. The organism does not utilize carbohydrates in the system described by Funke and Carlotti (5). meso-Diamino pimelic acid is the diamino acid of the peptidoglycan, and C15:0ai and C17:0ai are the predominant cellular fatty acids. The type strain is CCUG 56047 and has been deposited in the Culture of the University of Gothenburg, Sweden, and as strain DSM 21258 in the German Collection of Microorganisms and Cell Cultures.
Acknowledgments
This paper is part of the medical doctoral thesis of one of the authors (I.M.) at the medical faculty of the University of Ulm, Germany. We thank E. Falsen, Culture Collection University of Gothenburg, Sweden, for kindly providing strains.
Footnotes
Published ahead of print on 23 July 2008.
REFERENCES
- 1.Bernasconi, E., C. Valsangiacomo, R. Peduzzi, A. Carota, T. Moccetti, and G. Funke. 2004. Arthrobacter woluwensis subacute infective endocarditis: case report and review of the literature. Clin. Infect. Dis. 38e27-e31. [DOI] [PubMed] [Google Scholar]
- 2.Clinical and Laboratory Standards Institute. 2006. Methods for antimicrobial dilution and disk susceptibility testing of infrequently isolated or fastidious bacteria. Document M45-A. CLSI, Wayne, PA. [DOI] [PubMed]
- 3.Dass, K. N., M. A. Smith, V. J. Gill, S. A. Goldstein, and D. R. Lucey. 2002. Brevibacterium endocarditis: a first report. Clin. Infect. Dis. 35e20-e21. [DOI] [PubMed] [Google Scholar]
- 4.Funke, G., and K. A. Bernard. 2007. Coryneform gram-positive rods, p. 485-514. In P. R. Murray, E. J. Baron, J. H. Jorgensen, M. L. Landry, and M. A. Pfaller (ed.), Manual of clinical microbiology, 9th ed. ASM Press, Washington, DC.
- 5.Funke, G., and A. Carlotti. 1994. Differentiation of Brevibacterium spp. encountered in clinical specimens. J. Clin. Microbiol. 321729-1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Funke, G., R. Frodl, and H. Sommer. 2004. First comprehensively documented case of Paracoccus yeei infection in a human. J. Clin. Microbiol. 423366-3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Funke, G., R. A. Hutson, K. A. Bernard, G. E. Pfyffer, G. Wauters, and M. D. Collins. 1996. Isolation of Arthrobacter spp. from clinical specimens and description of Arthrobacter cumminsii sp. nov. and Arthrobacter woluwensis sp. nov. J. Clin. Microbiol. 342356-2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Funke, G., G. Martinetti Lucchini, G. E. Pfyffer, M. Marchiani, and A. von Graevenitz. 1993. Characteristics of CDC group 1 and group 1-like coryneform bacteria isolated from clinical specimens. J. Clin. Microbiol. 312907-2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Funke, G., M. Pagano-Niederer, B. Sjödén, and E. Falsen. 1998. Characteristics of Arthrobacter cumminsii, the most frequently encountered Arthrobacter species in human clinical specimens. J. Clin. Microbiol. 361539-1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hou, X. G., Y. Kawamura, F. Sultana, S. Shu, K. Hirose, K. Goto, and T. Ezaki. 1998. Description of Arthrobacter creatinolyticus sp. nov., isolated from human urine. Int. J. Syst. Bacteriol. 48423-429. [DOI] [PubMed] [Google Scholar]
- 11.Hsu, C. L., L. Y. Shih, H. S. Leu, C. L. Wu, and G. Funke. 1998. Septicemia due to Arthrobacter species in a neutropenic patient with acute lymphoblastic leukemia. Clin. Infect. Dis. 271334-1335. [DOI] [PubMed] [Google Scholar]
- 12.Huang, Y., N. Zhao, L. He, L. Wang, Z. Liu, M. You, and F. Guan. 2005. Arthrobacter scleromae sp. nov. isolated from human clinical specimens. J. Clin. Microbiol. 431451-1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kotoucková, L., P. Schumann, E. Durnová, C. Spröer, I. Sedlácek, J. Neca, Z. Zdráhal, and M. Nemec. 2004. Arthrobacter nitroguajacolicus sp. nov., a novel 4-nitroguaiacol-degrading actinobacterium. Int. J. Syst. Evol. Microbiol. 54773-777. [DOI] [PubMed] [Google Scholar]
- 14.Lin, Y.-C., K. Uemori, D. A. de Briel, V. Arunpairojana, and A. Yokota. 2004. Zimmermannella helvola gen. nov., sp. nov., Zimmermannella alba sp. nov., Zimmermanella bifida sp. nov., Zimmermannella faecalis sp. nov. and Leucobacter albus sp. nov., novel members of the family Microbacteriaceae. Int. J. Syst. Evol. Microbiol. 541669-1676. [DOI] [PubMed] [Google Scholar]
- 15.Manaia, C. M., B. Nogales, N. Weiss, and O. C. Nunes. 2004. Gulosibacter molinativorax gen. nov., sp. nov., a molinate-degrading bacterium, and classification of ‘Brevibacterium helvolum’ DSM 20419 as Pseudoclavibacter helvolus gen. nov., sp. nov. Int. J. Syst. Evol. Microbiol. 54783-789. [DOI] [PubMed] [Google Scholar]
- 16.Pascual, C., M. D. Collins, G. Funke, and D. G. Pitcher. 1996. Phenotypic and genotypic characterization of two Brevibacterium strains from the human ear: description of Brevibacterium otitidis sp. nov. Med. Microbiol. Lett. 5113-123. [Google Scholar]
- 17.Smit, E., P. Leeflang, S. Gommans, J. van den Broek, S. van Mil, and K. Wernars. 2001. Diversity and seasonal fluctuations of the dominant members of the bacterial soil community in a wheat field as determined by cultivation and molecular methods. Appl. Environ. Microbiol. 672284-2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Somvanshi, V. S., E. Lang, P. Schumann, R. Pukall, R. M. Kroppenstedt, S. Ganguly, and E. Stackebrandt. 2007. Leucobacter iarius sp. nov., in the family Microbacteriaceae. Int. J. Syst. Evol. Microbiol. 57682-686. [DOI] [PubMed] [Google Scholar]
- 19.Stackebrandt, E., and J. Ebers. 2006. Taxonomic parameters revisited: tarnished gold standards. Microbiol. Today 33152-155. [Google Scholar]
- 20.Stackebrandt, E., and B. M. Goebel. 1994. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 44846-849. [Google Scholar]
- 21.Takeuchi, M., N. Weiss, P. Schumann, and A. Yokota. 1996. Leucobacter komagatae gen. nov., sp. nov., a new aerobic gram-positive, nonsporulating rod with 2,4-diaminobutyric acid in the cell wall. Int. J. Syst. Bacteriol. 46967-971. [DOI] [PubMed] [Google Scholar]
- 22.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 241596-1599. [DOI] [PubMed] [Google Scholar]
- 23.Wauters, G., J. Charlier, M. Janssens, and M. Delmée. 2000. Identification of Arthrobacter oxydans, Arthrobacter luteolus sp. nov., and Arthrobacter albus sp. nov., isolated from human clinical specimens. J. Clin. Microbiol. 382412-2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wauters, G., J. Charlier, M. Janssens, and M. Delmée. 2001. Brevibacterium paucivorans sp. nov., from human clinical specimens. Int. J. Syst. Evol. Microbiol. 511703-1707. [DOI] [PubMed] [Google Scholar]
- 25.Wauters, G., B. van Bosterhaut, V. Avesani, R. Cuvelier, J. Charlier, M. Janssens, and M. Delmée. 2000. Peritonitis due to Brevibacterium otitidis in a patient undergoing continuous ambulatory peritoneal dialysis. J. Clin. Microbiol. 384292-4293. [DOI] [PMC free article] [PubMed] [Google Scholar]