Abstract
The Foodborne Viruses in Europe network has developed integrated epidemiological and virological outbreak reporting with aggregation and sharing of data through a joint database. We analyzed data from reported outbreaks of norovirus (NoV)-caused gastroenteritis from 13 European countries (July 2001 to July 2006) for trends in time and indications of different epidemiology of genotypes and variants. Of the 13 countries participating in this surveillance network, 11 were capable of collecting integrated epidemiological and virological surveillance data and 10 countries reported outbreaks throughout the entire period. Large differences in the numbers and rates of reported outbreaks per country were observed, reflecting the differences in the focus and coverage of national surveillance systems. GII.4 strains predominated throughout the 5-year surveillance period, but the proportion of outbreaks associated with GII.4 rose remarkably during years in which NoV activity was particularly high. Spring and summer peaks indicated the emergence of genetically distinct variants within GII.4 across Europe and were followed by increased NoV activity during the 2002-2003 and 2004-2005 winter seasons. GII.4 viruses predominated in health care settings and in person-to-person transmission. The consecutive emergence of new GII.4 variants is highly indicative of immune-driven selection. Their predominance in health care settings suggests properties that facilitate transmission in settings with a high concentration of people such as higher virus loads in excreta or a higher incidence of vomiting. Understanding the mechanisms driving the changes in epidemiology and clinical impact of these rapidly evolving RNA viruses is essential to design effective intervention and prevention measures.
Noroviruses (NoVs) are small, nonenveloped RNA viruses that are increasingly reported as causes of gastroenteritis across the world. Data from population-based studies suggest that NoVs are the most common cause of infectious gastroenteritis at the community level in developed countries. Little information is available about the role of NoV in gastroenteritis in developing countries (7, 12, 31). While illness associated with NoVs is typically mild and self-limiting, their high incidence and transmissibility result in large numbers of outbreaks, for which NoV has become notorious. Outbreaks occur in people of all ages and are particularly common in health care settings or other institutions where transmission may be facilitated by crowding and possibly lower standards of hygiene (2, 13, 28). Here, the impact of NoV may be more severe (30), and costs of controlling outbreaks may be high.
Recent events have suggested changes in the epidemiology of NoVs, when an unexpectedly high number of outbreaks on cruise ships signaled the start of a major epidemic in 2002 (27). Similarly, the spring of 2006 witnessed numerous outbreaks on cruise liners across Europe (22), triggering a joint outbreak investigation by the European Centers for Disease Control and the Foodborne Viruses in Europe (FBVE) network to identify possible sources for these outbreaks and provide clues from detailed investigations on the basis of the joint data set (43). Reports from other parts of the world have also suggested increased incidence of NoV outbreaks (8, 37).
NoVs are genetically highly variable and have been divided into five genogroups, which can differ as much as 40% with regard to the amino acid composition of the major capsid protein (VP1) (49).
Genogroups are further divided into genotypes, defined by strains with a higher level of homology across the VP1 (80%). An increasing number of genotypes are recognized as well as additional sublineages within genotypes (14, 17, 25, 39, 42). GII strains, particularly GII.4, are found most commonly all over the world (2, 13, 39).
We analyzed the data submitted to a joint database for 24 institutes in 13 countries in Europe, to study the trends in NoV outbreak reporting and the distribution of circulating strains of NoVs identified between July 2001 and July 2006. We show that rapid evolution of NoV occurs all over Europe and speculate that emergence of new variants at regular intervals is associated with high levels of outbreak reporting.
MATERIALS AND METHODS
The network.
The FBVE network was established in 1999 and is a collaboration between epidemiologists and virologists in 13 countries. A web-based database was established in 2001 by the network to provide a systematic collection of data on outbreaks of viral gastroenteritis in Europe (20). A data set with epidemiological information was reported, along with data on the diagnostic methods used, as well as sequence information from outbreak strains. A description of the projects and an example of the outbreak questionnaire and the database design can be found via www.fbve.nl.
Definitions used.
A NoV outbreak was defined as an outbreak (two or more cases linked in place and time) of gastroenteritis with laboratory-confirmed NoV infection. Gastroenteritis was defined as two or more episodes of vomiting in a 12-h period and lasting at least 12 h and/or two or more loose stools in a 12-h period and lasting at least 12 h. Laboratory confirmation of a NoV outbreak means two or more of a minimum of five stool specimens obtained from persons in the acute phase of the illness testing positive.
The NoV high season was defined as date of onset of the outbreak between 1 July year x and 30 June year x + 1.
Strain characterization.
The genotype of NoV was determined based on partial sequence analysis of the polymerase gene and/or capsid gene (15, 44). Definitive assignment of a genotype in the database entries was performed by one molecular virologist from the coordinating team according to our publicly available typing system (www.rivm.nl/bnwww). The GII.4 strains were then subdivided into five variants, based on distinct phylogenetic clustering and unique motifs (40). The variants were assigned as GII.4-year, based on the first year in which this clustering became apparent. Due to a lag in reporting and the need to have a number of sequences before a cluster can be recognized, this is often a year later than the actual first outbreak with this new variant.
Initially, all genotyping within the network was based on partial polymerase gene sequencing, but following developments in the diagnostics of NoV some countries have switched to genotyping based on partial capsid gene sequences. For this study, the polymerase and capsid genotypes were combined and grouped into three classes: GII.4, all other genogroup II strains, and all non-genogroup II strains. For part of the analyses the GII.4 group was subdivided into the separate variants. If available, a strain was assigned to one of these classes or variants based on phylogenetic clustering of the polymerase sequence. When only the capsid sequence was determined, this was used for classification.
Recombination is common in NoV, and as a rule the recombination point is located at the open reading frame 1 (ORF1)/ORF2 overlap region, which is between the polymerase and capsid regions used in this study (5, 35). This implies that results from polymerase-based genotyping and capsid-based genotyping cannot be combined without specifically addressing this issue. We concluded that combining the two typing methods using the above-described division into three classes was valid on the basis of an analysis of the subset of 264 entries in the data set used in this study which contained both polymerase and capsid sequences. This showed that only 2% (n = 5) of the strains would be assigned to another class when using capsid genotype.
Data analysis.
For this study, we selected all outbreaks with a diagnosis of NoV and date of onset between 1 July 2001 and 31 June 2006 from a download of the FBVE outbreak reporting database of 4 June 2007. The number of outbreaks each month and in each country was determined. Per analysis a subset of outbreaks was selected based on the availability of data. For these selections, the number of outbreaks by genotype and variant was determined in each month, by suspected mode of transmission and by setting. GII.4 strains were compared to GII non-4 and GI strains. Potential risk factors were determined from the minimum data set including month and year of onset of the outbreak, setting, and suspected mode of transmission, using multinomial logistic regression models. To limit the number of dummy variables in the model, potential risk factors were converted to binary variables. Variables significant during univariate analyses (P < 0.05) were included in a multivariate model. The variables remained in the multivariate model if P values were below 0.05 while using the backward selection procedure, when they were found to be confounding factors for other variables in the model (β changing at least 10%) or when they were effect modifiers (χ2 of the interaction term significant at P < 0.05).
RESULTS
Representativeness of data.
For the period of 1 July 2001 to 30 June 2006, 7,637 NoV outbreaks were reported to the FBVE reporting database (Table 1). The rates (number of outbreaks as a proportion of the population size of each country) differed by country, ranging from 0.0/million in 2001-2002 and 2003-2004 in Italy, where only one outbreak was reported in both years, to 30.4/million in 2005-2006 for Ireland.
TABLE 1.
Outbreaks per season per country
| Countryb | Value by seasona:
|
||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2001-2002
|
2002-2003
|
2003-2004
|
2004-2005
|
2005-2006
|
Whole period, 2001-2006
|
||||||||||||||||||
| N all (%)c | Rated | N type (%)e | N type + epi (%)f | N all (%) | Rate | N type (%) | N type + epi (%) | N all (%) | Rate | N type (%) | N type + epi (%) | N all (%) | Rate | N type (%) | N type + epi (%) | N all (%) | Rate | N type (%) | N type + epi (%) | N all (%) | N type (%) | N type + epi (%) | |
| DE | 74 (8.7) | 0.9 | 74 (15.7) | 74 (21.8) | 99 (8.8) | 1.2 | 99 (22.1) | 98 (27.4) | 0 | 0 | 0 | 1,500 (64.1) | 18.2 | 0 | 0 | 2,135 (71.5) | 25.9 | 0 | 0 | 3,808 (49.9) | 173 (9.4) | 172 (13.2) | |
| DK | 21 (2.5) | 3.9 | 1 (0.2) | 1 (0.3) | 5 (0.4) | 0.9 | 2 (0.4) | 1 (0.3) | 4 (1.2) | 0.7 | 0 | 0 | 7 (0.3) | 1.3 | 3 (0.7) | 3 (1.1) | 18 (0.6) | 3.3 | 17 (4.3) | 16 (5.8) | 55 (0.7) | 23 (1.2) | 21 (1.6) |
| ES | 43 (5.0) | 1.0 | 34 (7.2) | 19 (5.6) | 36 (3.2) | 0.9 | 24 (5.4) | 15 (4.2) | 4 (1.2) | 0.1 | 3 (2.6) | 0 | 14 (0.6) | 0.3 | 13 (3.1) | 3 (1.1) | 30 (1.0) | 0.7 | 27 (6.8) | 23 (8.4) | 127 (1.7) | 101 (5.5) | 60 (4.6) |
| FI | 55 (6.5) | 10.6 | 45 (9.6) | 9 (2.6) | 118 (10.5) | 22.7 | 22 (4.9) | 3 (0.8) | 22 (6.6) | 4.2 | 18 (15.4) | 3 (4.5) | 54 (2.3) | 10.3 | 31 (7.5) | 5 (1.9) | 44 (1.5) | 8.4 | 16 (4.0) | 9 (3.3) | 293 (3.8) | 132 (7.1) | 29 (2.2) |
| FR | 9 (1.1) | 0.1 | 7 (1.5) | 5 (1.5) | 13 (1.2) | 0.2 | 11 (2.5) | 11 (3.1) | 9 (2.7) | 0.1 | 8 (6.8) | 4 (8.1) | 26 (1.1) | 0.4 | 26 (6.3) | 18 (6.8) | 31 (1.0) | 0.5 | 31 (7.8) | 24 (8.7) | 88 (1.2) | 83 (4.5) | 62 (4.8) |
| EW | 485 (56.9) | 8.2 | 191 (40.6) | 133 (39.1) | 563 (50.0) | 9.5 | 60 (13.4) | 42 (11.7) | 192 (57.8) | 3.2 | 11 (9.4) | 0 | 357 (15.3) | 5.9 | 41 (9.9) | 0 | 341 (11.4) | 5.6 | 81 (20.4) | 0 | 1,938 (25.4) | 384 (20.8) | 175 (13.4) |
| HU | 92 (10.8) | 9.0 | 61 (13.0) | 43 (12.6) | 112 (9.9) | 11.0 | 97 (21.7) | 63 (17.6) | 51 (15.4) | 5.0 | 39 (33.3) | 22 (33.3) | 81 (3.5) | 8.0 | 71 (17.1) | 41 (15.4) | 62 (2.1) | 6.2 | 41 (10.3) | 34 (12.4) | 398 (5.2) | 309 (16.7) | 203 (15.6) |
| IE | 0 | 0 | 0 | 81 (3.5) | 19.7 | 68 (16.4) | 67 (25.2) | 128 (4.3) | 30.4 | 44 (11.1) | 44 (16.0) | 209 (2.7) | 112 (6.1) | 111 (8.5) | |||||||||
| IT | 1 (0.1) | 0.0 | 1 (0.2) | 0 | 2 (0.2) | 0.0 | 2 (0.4) | 1 (0.3) | 1 (0.3) | 0.0 | 0 | 0 | 9 (0.4) | 0.2 | 5 (1.2) | 1 (0.4) | 3 (0.1) | 0.1 | 2 (0.5) | 1 (0.4) | 16 (0.2) | 10 (0.5) | 3 (0.2) |
| NL | 59 (6.9) | 3.7 | 55 (11.7) | 54 (15.9) | 148 (13.1) | 9.1 | 107 (23.9) | 101 (28.2) | 27 (8.1) | 1.7 | 25 (21.4) | 25 (37.9) | 168 (7.2) | 10.3 | 132 (31.9) | 104 (39.1) | 120 (4.0) | 7.3 | 101 (25.4) | 89 (32.4) | 522 (6.8) | 420 (22.7) | 373 (28.6) |
| NO | 0 | 0 | 0 | 16 (0.7) | 3.5 | 0 | 0 | 22 (0.7) | 4.7 | 0 | 0 | 38 (0.5) | 0 | 0 | |||||||||
| SE | 7 (0.8) | 0.8 | 1 (0.2) | 2 (0.6) | 12 (1.1) | 1.3 | 10 (2.2) | 9 (2.5) | 15 (4.5) | 1.7 | 13 (11.1) | 12 (18.2) | 11 (0.5) | 1.2 | 10 (2.4) | 10 (3.8) | 26 (0.9) | 2.9 | 20 (5.0) | 20 (7.3) | 71 (0.9) | 54 (2.9) | 53 (4.1) |
| SL | 6 (0.7) | 3.0 | 0 | 0 | 19 (1.7) | 9.5 | 14 (3.1) | 14 (3.9) | 7 (2.1) | 3.5 | 0 | 0 | 16 (0.7) | 8.0 | 14 (3.4) | 14 (5.3) | 25 (0.8) | 12.5 | 18 (4.5) | 15 (5.5) | 73 (1.0) | 46 (2.5) | 43 (3.3) |
| Total | 852 (100) | 2.4 | 470 (100) | 340 (100) | 1,127 (100) | 3.5 | 448 (100) | 358 (100) | 332 (100) | 1.6 | 117 (100) | 66 (100) | 2,340 (100) | 6.4 | 414 (100) | 266 (100) | 2,985 (100) | 8.2 | 398 (100) | 275 (100) | 7,636 (100) | 1,847 (100) | 1,305 (100) |
Date of onset between 1 July year x and 30 June year x + 1.
DE, Germany; DK, Denmark; ES, Spain; FI, Finland; FR, France; EW, England and Wales; HU, Hungary; IE, Ireland; IT, Italy; NL, The Netherlands; NO, Norway; SE, Sweden; SL, Slovenia.
N all (%), number of outbreaks per country and % of all outbreaks of that season.
Rate, number of outbreaks per million inhabitants per country (population on 1 January of 2002 to 2006; source, Eurostat [http://epp.eurostat.ec.europa.eu]). Total rate per season is calculated using only the contributing countries within that season in the denominator.
N type (%), number of outbreaks with typing data and % of all outbreaks with typing data.
N type + epi (%), number of outbreaks with typing data and known mode of transmission and setting and % of all outbreaks with typing data and known mode of transmission and setting.
Two countries, Germany and England and Wales (these latter two being considered one country for reporting purposes), supplied 3,808 (49.9%) and 1,938 (25.4%) of all outbreaks in the data set, respectively. Ten countries—Denmark, Spain, Finland, France, England and Wales, Hungary, Italy, The Netherlands, Sweden, and Slovenia—reported outbreaks throughout the 5-year period. Twenty-four percent (1,847/7,637) of the reported outbreaks contained data on genotype (based on submitted sequences). Within this subset, 50% of the data were provided by three countries, England and Wales, Hungary, and The Netherlands, with 20.8%, 16.7% and 22.7%, respectively. Seven countries—Spain, Finland, France, England and Wales, Hungary, The Netherlands, and Sweden—submitted outbreak reports including sequences throughout the complete period.
The subset of reports containing both genotyping and epidemiological background information on setting and mode of transmission was 1,305 records (17.1%). The data from large contributors were analyzed separately to look for possible biases in the global analyses in order to differentiate between national and international trends. These data are presented separately where relevant.
Trends in overall outbreak reporting, 1 July 2001 to 30 June 2006.
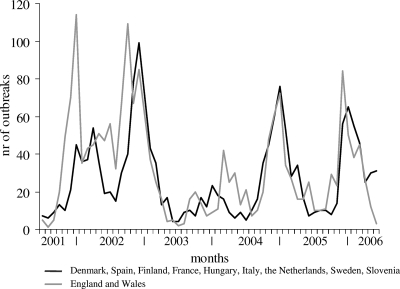
Monthly trends in outbreak reporting for the 10 countries which reported throughout the complete period showed a clear winter seasonality (Fig. 1). The size and month of the peak differed annually, with the largest number of outbreaks reported in the 2002-2003 period and remarkably low levels of reporting in the 2003-2004 period. The outbreaks from England and Wales are shown separately from those of the other countries because they show a slightly different pattern. In the years 2001-2002, 2002-2003, and 2005-2006 the peak in England and Wales was observed somewhat earlier than in the other countries, and increased reporting was observed in the spring of 2004 unlike in other countries. Off-seasonal outbreak activity was observed across Europe in the spring of 2002. The seasonal peak month in each country was determined for each season. No direction of spread across Europe could be identified visually this way (data not shown).
FIG. 1.
Number of reported outbreaks per month of onset for all countries reporting throughout the complete period.
Trends in NoV outbreaks by genotype, 1 July 2001 to 30 June 2006.
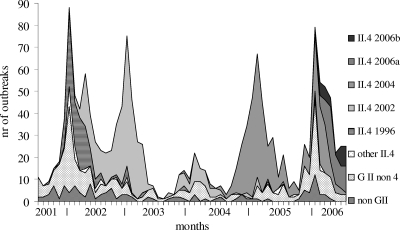
Outbreaks were grouped by genotype for the countries that submitted these data throughout the reporting period (Fig. 2), showing that GII.4 strains were the predominant genotype within the 5 years under surveillance. In England and Wales the proportion of GII.4 outbreaks was lower throughout the entire period. The winter seasonality was clearest for outbreaks caused by GII.4 strains, and a weaker seasonal pattern was visible for other GII viruses. Within the GII non-4 strains mostly II.2, II.7, and II.b strains were found (data not shown; typing based only on polymerase sequences) with the 2001-2002 peak of GII non-4 outbreaks mainly caused by II.b. II.b is the preliminary name for a distinct strain with a polymerase sequence that did not cluster with any known sequence globally. It was initially associated with oyster-associated outbreaks all linked to the same source and subsequently found in other settings (4). The 2005-2006 peak was dominated by II.7 and II.b (based on polymerase typing; data not shown).
FIG. 2.
Number of outbreaks with genotypes and variants per month from Spain, Finland, France, England and Wales, Hungary, The Netherlands, and Sweden (the subset of countries which have outbreak reports with sequence data throughout the complete period).
No seasonal pattern was observed for non-GII outbreaks. Univariate logistic regression analysis confirmed that GII.4 strains were more common between October and March than were non-GII strains (odds ratio [OR], 1.9; 95% confidence interval [CI], 1.2 to 3.1; P = 0.006). For other GII outbreaks this was also significant (OR, 1.8; 95% CI, 1.7 to 3.0; P = 0.0322), whereas non-GII outbreaks occurred throughout the year. A similar pattern was observed for individual countries, although this was not statistically significant due to low numbers. The 2002-2003 and the 2004-2005 winter peaks were almost exclusively caused by GII.4 NoV.
Trends for individual GII.4 variants, 1 July 2001 to 30 June 2006.
Following initial observations on emergence of new variants within GII.4, we decided to study the trends in individual GII.4 variants. Plotting trends for individual GII.4 variants showed consecutive emergence and disappearance of new variants across Europe (Fig. 2). In the 2001-2002 season the predominant II.4 variant was GII.4 1996, which had also been circulating in years prior to this survey (40). In 2002-2003 and 2004-2005 the peaks were almost exclusively caused by a new variant which was first reported a few months earlier. In 2006, two distinct variants were seen cocirculating. The first report of variant GII.4 2002 in the database was from two outbreaks in January 2002 in residential institutions in The Netherlands, with person-to-person transmission. Subsequently this variant rapidly became predominant in the course of the year. The following season, 2003-2004, showed very low NoV activity, but the first outbreaks with the new II.4 2004 variant were seen cocirculating with the 2002 epidemic strain. In the 2004-2005 season this new variant had become predominant, with only a few outbreaks of II.4 2002. In the 2005-2006 season the 2002 variant was not detected anymore.
Variant II.4 2004 was reported for the first time in an outbreak which took place in November 2003 in Finland with an unknown setting and mode of transmission. The first reports of variant II.4 2006a in the database were in two outbreaks in February 2006 in France and The Netherlands in residential institutions, with person-to-person transmission. Variant II.4 2006b was firstly reported in an outbreak in December 2005 in two residential institutions in Spain. Patterns for 2002 and 2004 variants were similar across the network. For the 2006 variants, some geographic differences were observed. In Hungary, only the 2006b variant was found during the study period. No geographical direction of spread could be deduced visually from the first outbreak or the peak month for the individual variants in the different countries (data not shown).
Modes of transmission overall and by genotype.
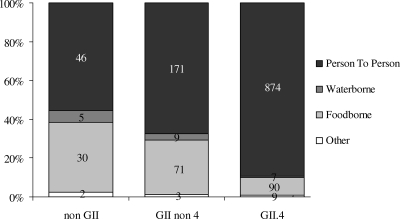
Of all outbreaks with a reported mode of transmission (n = 5,036), 88% (n = 4,429) were suspected to be person-to-person outbreaks, 10% (n = 506) were food-borne outbreaks, and 2% (n = 76) were waterborne outbreaks. For the subset of outbreaks with genotyping data (n = 1,317), again the main (suspected) mode of transmission (83%) was person to person (Fig. 3). As Fig. 3 shows, person-to-person outbreaks were relatively more often caused by GII.4 viruses and food-borne outbreaks were relatively more often caused by non-II genotypes. Univariate logistic regression analysis showed that person-to-person transmission was an independent risk factor for GII.4 outbreaks (OR, 6.3; 95% CI, 3.8 to 46.6; P < 0.001) but not other GII outbreaks (OR, 1.6; 95% CI, 1.0 to 2.6; P = 0.0736), compared to genogroup non-II outbreaks.
FIG. 3.
Suspected mode of transmission for all genotyped outbreaks with reported mode of transmission (n = 1,317).
Setting of outbreaks, overall and by genotype.
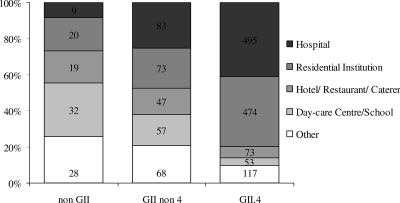
Of all outbreaks with a reported setting (n = 6,579), 72% (n = 4,710) took place in a health care setting, i.e., residential institutions (36%, n = 2,383) and hospitals (35%, n = 2,327).
This was similar to the proportion found in the 1,641 outbreaks for which genotyping information was available (70%, n = 1,154). There was a significant difference between genotypes: 80% of GII.4 outbreaks were in health care settings, compared with 43% of the outbreaks caused by other genotypes (Fig. 4). In day care centers and schools GI strains were found relatively more often. Univariate logistic regression analysis of outbreaks in health care settings showed significantly higher risk for both GII.4 (OR, 11.8; 95% CI, 7.0 to 1,086.6; P < 0.001) and other GII (OR, 2.8; 95% CI, 1.6 to 4.9; P = 0.0004) outbreaks.
FIG. 4.
Setting per genotype for all genotyped outbreaks with a reported setting (n = 1,648).
Multivariate analysis.
Setting, transmission mode, and seasonality were included in a multivariate logistic regression model comparing GII.4 to other GII outbreaks. During the backward selection procedure setting and mode remained in the model as significant risk factors. No confounding from season was observed. The analysis resulted in ORs as shown in Table 2. Outbreaks in health care settings were almost nine times more likely to be caused by GII.4 strains than by NoVs belonging to genotypes other than GII. Outbreaks listed as person-to-person outbreaks were almost twice as likely to be GII.4 outbreaks.
TABLE 2.
ORs of multinomial logistic regression, NoV strain groups for person-to-person transmission, high season, and health care outbreaksa
| Variable | GII.4
|
GII non-4
|
||
|---|---|---|---|---|
| OR | P | OR | P | |
| Person to person | 1.9 | 0.026 | 1.0 | 0.8981 |
| Health careb | 8.8 | <0.0001 | 2.6 | 0.003 |
| nc | 976 | 250 | ||
Reference category for the equation is non-genogroup II (n = 79).
Hospital or residential institution.
Total n = 1,305.
DISCUSSION
Our outbreak surveillance found large differences in numbers and rates of outbreaks in each country (Table 1), which are most likely a result of the differences in national surveillance systems rather than true differences in prevalence of NoV outbreaks (24, 26, 29). This is not a specific problem of NoV surveillance, as almost all international surveillance networks cope with lack of standardization, reporting delay, and missing values in health event reporting (1, 9, 33). In addition, NoVs are not on the list of priority diseases for surveillance in the European Union. Therefore, such differences will remain in the near future. France, Denmark, and Sweden report only suspected food-borne NoV outbreaks, and Italy and Spain do not have a national NoV surveillance system and report regional data. Norway and Ireland are new members of the network and started reporting outbreaks in 2004; Slovenia started in 2002. In Germany the system of NoV reporting to the FBVE database changed considerably during the period described in this paper. Cases of NoV infection have been notifiable since 2001 in Germany. In 2005 Germany started reporting outbreaks to the FBVE database collected through a new surveillance system. Cases are registered, and subsequently outbreaks are established by linking individual cases into groups (10). For some of the outbreaks the virus is characterized, but due to stringent privacy laws these characterized viruses cannot be linked to individual outbreaks. Thus, although Germany has reported sequences to the FBVE database during the entire study period, these cannot be used in the overviews in this paper. Equally the structure of outbreak reporting in England and Wales, Finland, and Norway precludes systematic provision of integrated laboratory and epidemiology data. Several countries submitted additional sequences without outbreak information to the FBVE database, of which Germany and Denmark did so in large quantities (n > 200). These data cannot be used in this epidemiological overview but will be included in a phylogenetic overview elsewhere.
Taking into account the differences in the scope and system of national surveillance is necessary when doing comparative analysis of data as presented in this study (21). In every analysis, depending on the availability and validity of the required data, a different set of outbreaks is selected.
We looked at trends in outbreak reporting, which confirmed the clear winter seasonality of NoV outbreaks (32). The variation in the size of the seasonal peaks has been described earlier for individual countries, which reported increased activity during the 2002-2003 and 2004-2005 winter seasons (14, 19, 23, 27, 39). However, it is difficult to draw conclusions on these trends based on surveillance data alone. Combining these data with molecular virological information, it becomes clear that the GII strains cause the seasonality and striking changes in the epidemiology have been observed: the proportion of all outbreaks caused by GII.4 rose remarkably during the observed peak years of 2002-2003 and 2004-2005 (2, 13, 39), and further analysis showed that this was in fact a succession of distinguishable variants emerging and disappearing rapidly. Molecular analysis has shown that these viruses have evolved from circulating strains by accumulation of amino acid mutations at surface-exposed regions of the viral particle, which is highly indicative of immune-driven selection, although other explanations may exist (14, 25, 39). The period under surveillance for the whole group is short, but data from the countries with stable surveillance over longer periods of time suggest that the high GII.4 presence and rate of change is a relatively recent phenomenon. The first observed GII.4 variant (GII.4 1996) that appeared globally was associated with high rates of outbreak reporting in 1995-1996 (46). These viruses continued to circulate with less severe impact, until the 2002 new variant was identified through our network (27). Again, these viruses emerged globally, and increased levels of outbreaks were reported across the world (45, 47).
New GII.4 variants have emerged every other year and disappeared a few years later, which can be seen as extremely fast evolution (6, 14, 34, 39). This pattern is similar to observations of influenza viruses, where new variants are known antigenic drift variants, but changes in NoVs in recent years have occurred faster than those in influenza viruses (41). Work is ongoing on global comparisons of circulating NoV variants, and preliminary results confirm that the 2004 and 2006 variants are now seen globally (38). Although during the study period variant II.4 2006a was not found in Hungary, this variant has been reported from that country starting from October 2006. It is not yet possible to establish any direction in geographical spread, but this may be revealed in a later stage by using scan statistics (24).
The observed changes in circulating genotypes may provide part of the explanation for the apparently increased problems with NoVs in health care settings in recent years (18, 48). Viruses of the GII.4 genotype are predominant in health care settings where people are at risk of complications of gastroenteritis (2, 3, 11, 13, 16, 28, 30). This highly significant association, combined with the predominance of GII.4 in person-to-person transmission, suggests that GII.4 strains have properties that facilitate transmission in settings with a high concentration of people, such as higher virus loads in excreta or a higher incidence of vomiting. Quantitative data on virus shedding and on clinical symptoms are needed to test this hypothesis. A study in The Netherlands suggested increased mortality due to gastroenteritis associated with the winter seasonal peaks of 2002-2003 and 2004-2005, and similar studies are ongoing in England and Wales and France (L. van Asten, J. Siebenga, C. van den Wijngaard, R. Verheij, H. Vliet, W. van Pelt, and M. Koopmans, submitted for publication). Data from the work of Verhoef et al. (43) suggested that unusual spring and summer activity can be seen as a predictor for severe winter seasons, reflecting the impact of new variant GII.4 NoVs on cruise ships as well. The unpredictable nature of this rapidly evolving RNA virus with striking changes in epidemiology is reason for concern. Understanding the mechanisms driving this process is essential to design effective intervention and prevention measures. For this a transnational approach is necessary, and thus, the need for a more standardized and systematic approach is clear, both for surveillance and for the molecular virological data collection and analysis. The global NoV initiative aims to work toward this standardized nomenclature, including recombinant strains (25, 36).
Acknowledgments
This work was supported by the European Commission in three projects: (i) FBVE (Foodborne Viruses in Europe), rapid detection of transnational food-borne viral infections and elucidation of transmission routes through molecular tracing and development of a common database, contract no. QLK1-1999-00594; (ii) DIVINE, prevention of emerging enteric viral infections: diagnoses, viability testing, networking and epidemiology, contract no. 2003213; and (iii) EVENT (Enteric Virus Emergence, New Tools), providing tools to prevent emergence of enteric viruses, contract no. 502571.
Footnotes
Published ahead of print on 23 July 2008.
REFERENCES
- 1.Amato-Gauci, A., and A. Ammon. 2007. ECDC to launch first report on communicable diseases epidemiology in the European Union. Euro Surveill. 12E070607.2. [DOI] [PubMed] [Google Scholar]
- 2.Blanton, L. H., S. M. Adams, R. S. Beard, G. Wei, S. N. Bulens, M. A. Widdowson, R. I. Glass, and S. S. Monroe. 2006. Molecular and epidemiologic trends of caliciviruses associated with outbreaks of acute gastroenteritis in the United States, 2000-2004. J. Infect. Dis. 193413-421. [DOI] [PubMed] [Google Scholar]
- 3.Bon, F., K. Ambert-Balay, H. Giraudon, J. Kaplon, S. Le Guyader, M. Pommepuy, A. Gallay, V. Vaillant, H. de Valk, R. Chikhi-Brachet, A. Flahaut, P. Pothier, and E. Kohli. 2005. Molecular epidemiology of caliciviruses detected in sporadic and outbreak cases of gastroenteritis in France from December 1998 to February 2004. J. Clin. Microbiol. 434659-4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buesa, J., B. Collado, P. Lopez-Andujar, R. Abu-Mallouh, J. Rodriguez Diaz, A. Garcia Diaz, J. Prat, S. Guix, T. Llovet, G. Prats, and A. Bosch. 2002. Molecular epidemiology of caliciviruses causing outbreaks and sporadic cases of acute gastroenteritis in Spain. J. Clin. Microbiol. 402854-2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bull, R. A., M. M. Tanaka, and P. A. White. 2007. Norovirus recombination. J. Gen. Virol. 883347-3359. [DOI] [PubMed] [Google Scholar]
- 6.Bull, R. A., E. T. Tu, C. J. McIver, W. D. Rawlinson, and P. A. White. 2006. Emergence of a new norovirus genotype II.4 variant associated with global outbreaks of gastroenteritis. J. Clin. Microbiol. 44327-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Castilho, J. G., V. Munford, H. R. Resque, U. Fagundes-Neto, J. Vinje, and M. L. Racz. 2006. Genetic diversity of norovirus among children with gastroenteritis in Sao Paulo State, Brazil. J. Clin. Microbiol. 443947-3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Centers for Disease Control and Prevention. 2007. Norovirus activity—United States, 2006-2007. MMWR Morb. Mortal. Wkly. Rep. 56842-846. [PubMed] [Google Scholar]
- 9.Doyle, T. J., M. K. Glynn, and S. L. Groseclose. 2002. Completeness of notifiable infectious disease reporting in the United States: an analytical literature review. Am. J. Epidemiol. 155866-874. [DOI] [PubMed] [Google Scholar]
- 10.Faensen, D., H. Claus, J. Benzler, A. Ammon, T. Pfoch, T. Breuer, and G. Krause. 2006. SurvNet@RKI—a multistate electronic reporting system for communicable diseases. Euro Surveill. 11100-103. [PubMed] [Google Scholar]
- 11.Fankhauser, R. L., J. S. Noel, S. S. Monroe, T. Ando, and R. I. Glass. 1998. Molecular epidemiology of “Norwalk-like viruses” in outbreaks of gastroenteritis in the United States. J. Infect. Dis. 1781571-1578. [DOI] [PubMed] [Google Scholar]
- 12.Gabbay, Y. B., R. I. Glass, S. S. Monroe, C. Carcamo, M. K. Estes, J. D. Mascarenhas, and A. C. Linhares. 1994. Prevalence of antibodies to Norwalk virus among Amerindians in isolated Amazonian communities. Am. J. Epidemiol. 139728-733. [DOI] [PubMed] [Google Scholar]
- 13.Gallimore, C. I., J. Green, D. Lewis, A. F. Richards, B. A. Lopman, A. D. Hale, R. Eglin, J. J. Gray, and D. W. Brown. 2004. Diversity of noroviruses cocirculating in the north of England from 1998 to 2001. J. Clin. Microbiol. 421396-1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gallimore, C. I., M. Iturriza-Gomara, J. Xerry, J. Adigwe, and J. J. Gray. 2007. Inter-seasonal diversity of norovirus genotypes: emergence and selection of virus variants. Arch. Virol. 1521295-1303. [DOI] [PubMed] [Google Scholar]
- 15.Green, K. Y., A. Z. Kapikian, and R. M. Chanock. 2001. Human caliciviruses, p. 841-874. In D. M. Knipe, P. M. Howley, and D. E. Griffin (ed.), Fields virology. Lippincott-Raven, Philadelphia, PA.
- 16.Hale, A., K. Mattick, D. Lewis, M. Estes, X. Jiang, J. Green, R. Eglin, and D. Brown. 2000. Distinct epidemiological patterns of Norwalk-like virus infection. J. Med. Virol. 6299-103. [DOI] [PubMed] [Google Scholar]
- 17.Ho, E. C., P. K. Cheng, A. W. Lau, A. H. Wong, and W. W. Lim. 2007. Atypical norovirus epidemic in Hong Kong during summer of 2006 was caused by a new genogroup II/4 variant. J. Clin. Microbiol. 452205-2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hutson, A. M., R. L. Atmar, and M. K. Estes. 2004. Norovirus disease: changing epidemiology and host susceptibility factors. Trends Microbiol. 12279-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koch, J. 2004. Norovirus-infections: high incidence expected in the 2004/2005 season. Epidemiol. Bull. 50439-440. [Google Scholar]
- 20.Koopmans, M., H. Vennema, H. Heersma, E. van Strien, Y. van Duynhoven, D. Brown, M. Reacher, and B. Lopman. 2003. Early identification of common-source foodborne virus outbreaks in Europe. Emerg. Infect. Dis. 91136-1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kroneman, A., J. Harris, H. Vennema, E. Duizer, Y. van Duynhoven, J. Gray, M. Iturriza, B. Bottiger, G. Falkenhorst, C. Johnsen, C. H. von Bonsdorff, L. Maunula, M. Kuusi, P. Pothier, A. Gallay, E. Schreier, J. Koch, G. Szucs, G. Reuter, K. Krisztalovics, M. Lynch, P. McKeown, B. Foley, S. Coughlan, F. M. Ruggeri, I. Di Bartolo, K. Vainio, E. Isakbaeva, M. Poljsak-Prijatelj, A. H. Grom, A. Bosch, J. Buesa, A. S. Fauquier, G. Hernandez-Pezzi, K. O. Hedlund, and M. Koopmans. 2008. Data quality of 5 years of central norovirus outbreak reporting in the European Network for food-borne viruses. J. Public Health 3082-90. [DOI] [PubMed] [Google Scholar]
- 22.Kroneman, A., H. Vennema, J. Harris, G. Reuter, C. H. von Bonsdorff, K. O. Hedlund, K. Vainio, V. Jackson, P. Pothier, J. Koch, E. Schreier, B. E. Bottiger, and M. Koopmans. 2006. Increase in norovirus activity reported in Europe. Euro Surveill. 11E061214.1. [DOI] [PubMed] [Google Scholar]
- 23.Kroneman, A., H. Vennema, Y. van Duijnhoven, E. Duizer, and M. Koopmans. 23 December 2004, posting date. High number of norovirus outbreaks associated with a GGII.4 variant in the Netherlands and elsewhere: does this herald a worldwide increase? Euro Surveill. http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=2606.
- 24.Kulldorff, M. A. 1997. A spatial scan statistic. Commun. Stat. Theor. Methods 261481-1496. [Google Scholar]
- 25.Lindesmith, L. C., E. F. Donaldson, A. D. Lobue, J. L. Cannon, D. P. Zheng, J. Vinje, and R. S. Baric. 2008. Mechanisms of GII.4 norovirus persistence in human populations. PLoS Med. 5e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lopman, B., Y. van Duynhoven, F. X. Hanon, M. Reacher, M. Koopmans, and D. Brown. 2002. Laboratory capability in Europe for foodborne viruses. Euro Surveill. 761-65. [DOI] [PubMed] [Google Scholar]
- 27.Lopman, B., H. Vennema, E. Kohli, P. Pothier, A. Sanchez, A. Negredo, J. Buesa, E. Schreier, M. Reacher, D. Brown, J. Gray, M. Iturriza, C. Gallimore, B. Bottiger, K. O. Hedlund, M. Torven, C. H. von Bonsdorff, L. Maunula, M. Poljsak-Prijatelj, J. Zimsek, G. Reuter, G. Szucs, B. Melegh, L. Svennson, Y. van Duijnhoven, and M. Koopmans. 2004. Increase in viral gastroenteritis outbreaks in Europe and epidemic spread of new norovirus variant. Lancet 363682-688. [DOI] [PubMed] [Google Scholar]
- 28.Lopman, B. A., D. W. Brown, and M. Koopmans. 2002. Human caliciviruses in Europe. J. Clin. Virol. 24137-160. [DOI] [PubMed] [Google Scholar]
- 29.Lopman, B. A., M. H. Reacher, Y. Van Duijnhoven, F. X. Hanon, D. Brown, and M. Koopmans. 2003. Viral gastroenteritis outbreaks in Europe, 1995-2000. Emerg. Infect. Dis. 990-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mattner, F., D. Sohr, A. Heim, P. Gastmeier, H. Vennema, and M. Koopmans. 2006. Risk groups for clinical complications of norovirus infections: an outbreak investigation. Clin. Microbiol. Infect. 1269-74. [DOI] [PubMed] [Google Scholar]
- 31.Monica, B., S. Ramani, I. Banerjee, B. Primrose, M. Iturriza-Gomara, C. I. Gallimore, D. W. Brown, M. Fathima, P. D. Moses, J. J. Gray, and G. Kang. 2007. Human caliciviruses in symptomatic and asymptomatic infections in children in Vellore, South India. J. Med. Virol. 79544-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mounts, A. W., T. Ando, M. Koopmans, J. S. Bresee, J. Noel, and R. I. Glass. 2000. Cold weather seasonality of gastroenteritis associated with Norwalk-like viruses. J. Infect. Dis. 181(Suppl. 2)S284-S287. [DOI] [PubMed] [Google Scholar]
- 33.Reintjes, R., M. Thelen, R. Reiche, and A. Csohan. 2007. Benchmarking national surveillance systems: a new tool for the comparison of communicable disease surveillance and control in Europe. Eur. J. Public Health 17375-380. [DOI] [PubMed] [Google Scholar]
- 34.Reuter, G., K. Krisztalovics, H. Vennema, M. Koopmans, and G. Szucs. 2005. Evidence of the etiological predominance of norovirus in gastroenteritis outbreaks—emerging new-variant and recombinant noroviruses in Hungary. J. Med. Virol. 76598-607. [DOI] [PubMed] [Google Scholar]
- 35.Reuter, G., H. Vennema, M. Koopmans, and G. Szucs. 2006. Epidemic spread of recombinant noroviruses with four capsid types in Hungary. J. Clin. Virol. 3584-88. [DOI] [PubMed] [Google Scholar]
- 36.Rohayem, J., J. Munch, and A. Rethwilm. 2005. Evidence of recombination in the norovirus capsid gene. J. Virol. 794977-4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sakon, N., K. Yamazaki, T. Yoda, T. Tsukamoto, T. Kase, K. Taniguchi, K. Takahashi, and T. Otake. 2007. Norovirus storm in Osaka, Japan, last winter (2006/2007). Jpn. J. Infect. Dis. 60409-410. [PubMed] [Google Scholar]
- 38.Siebenga, J. J., S. Bidawid, S. Broor, Z. Dua, Z. Fang, C. I. Gallimore, G. Greening, and J. Hewitt. 2007. Global molecular epidemiology of subsequent emerging variants of the dominant GGII.4 genotype of noroviruses between 2001 and 2007, p. S3-S6. In Proceedings of the Third International Calicivirus Conference, vol. 15. [Google Scholar]
- 39.Siebenga, J. J., H. Vennema, E. Duizer, and M. P. Koopmans. 2007. Gastroenteritis caused by norovirus GGII.4, The Netherlands, 1994-2005. Emerg. Infect. Dis. 13144-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Siebenga, J. J., H. Vennema, B. Renckens, E. de Bruin, B. van der Veer, R. J. Siezen, and M. Koopmans. 2007. Epochal evolution of GGII.4 norovirus capsid proteins from 1995 to 2006. J. Virol. 819932-9941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith, D. J., A. S. Lapedes, J. C. de Jong, T. M. Bestebroer, G. F. Rimmelzwaan, A. D. Osterhaus, and R. A. Fouchier. 2004. Mapping the antigenic and genetic evolution of influenza virus. Science 305371-376. [DOI] [PubMed] [Google Scholar]
- 42.Vainio, K., and M. Myrmel. 2006. Molecular epidemiology of norovirus outbreaks in Norway during 2000 to 2005 and comparison of four norovirus real-time reverse transcriptase PCR assays. J. Clin. Microbiol. 443695-3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Verhoef, L., E. Depoortere, I. Boxman, E. Duizer, Y. van Duynhoven, J. Harris, C. Johnsen, A. Kroneman, S. Le Guyader, W. Lim, L. Maunula, H. Melda, R. Ratcliff, G. Reuter, E. Schreier, J. Siebenga, K. Vainio, C. Varela, H. Vennema, and M. Koopmans. 2008. Emergence of new norovirus variants on spring cruise ships and prediction of winter epidemics. Emerg. Infect. Dis. 14238-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vinje, J., J. Green, D. C. Lewis, C. I. Gallimore, D. W. Brown, and M. P. Koopmans. 2000. Genetic polymorphism across regions of the three open reading frames of “Norwalk-like viruses.” Arch. Virol. 145223-241. [DOI] [PubMed] [Google Scholar]
- 45.Vipond, I. B., E. O. Caul, D. Hirst, B. Carmen, A. Curry, B. A. Lopman, P. Pead, M. A. Pickett, P. R. Lambden, and I. N. Clarke. 2004. National epidemic of Lordsdale norovirus in the UK. J. Clin. Virol. 30243-247. [DOI] [PubMed] [Google Scholar]
- 46.White, P. A., G. S. Hansman, A. Li, J. Dable, M. Isaacs, M. Ferson, C. J. McIver, and W. D. Rawlinson. 2002. Norwalk-like virus 95/96-US strain is a major cause of gastroenteritis outbreaks in Australia. J. Med. Virol. 68113-118. [DOI] [PubMed] [Google Scholar]
- 47.Widdowson, M. A., E. H. Cramer, L. Hadley, J. S. Bresee, R. S. Beard, S. N. Bulens, M. Charles, W. Chege, E. Isakbaeva, J. G. Wright, E. Mintz, D. Forney, J. Massey, R. I. Glass, and S. S. Monroe. 2004. Outbreaks of acute gastroenteritis on cruise ships and on land: identification of a predominant circulating strain of norovirus—United States, 2002. J. Infect. Dis. 19027-36. [DOI] [PubMed] [Google Scholar]
- 48.Widdowson, M. A., S. S. Monroe, and R. I. Glass. 2005. Are noroviruses emerging? Emerg. Infect. Dis. 11735-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zheng, D. P., T. Ando, R. L. Fankhauser, R. S. Beard, R. I. Glass, and S. S. Monroe. 2006. Norovirus classification and proposed strain nomenclature. Virology 346312-323. [DOI] [PubMed] [Google Scholar]