Abstract
The first case of cervical lymphadenitis due to infection by a new Erwinia-like organism is reported. The organism was identified initially as Pantoea sp. by a Vitek 2-based assessment but was finally identified as a member of the genus Erwinia by 16S rRNA gene sequence analysis. The isolate displayed 98.9% 16S rRNA gene sequence similarity to that of E. tasmaniensis and showed phenotypic characteristics that were different from other Erwinia species.
CASE REPORT
A 79-year-old woman visited our hospital for evaluation of left-side cervical lymphadenopathy and a headache that had begun 3 days previously. She had an unremarkable medical history and no underlying illness. On physical examination, the left side of her neck was found to have multiple 0.5- to 1.5-cm-sized tender, conglomerated lymph nodes, but other findings were unremarkable. Initial laboratory analyses revealed a white blood cell count of 9,300 cells/mm3 (neutrophil, 46.4%); a hemoglobin level of 15.9 g/dl; a platelet count of 277,000/mm3; alanine aminotransferase (AST)/aspartate aminotransferase (ALT) ratio of 139:119 IU/liter; a total bilirubin concentration of 0.4 mg/dl; a creatinine concentration of 0.8 mg/dl; a creatine kinase concentration of 90 IU/liter; and a lactate dehydrogenase concentration of 505 IU/liter. Neck ultrasonography showed multifocal, necrotic lymph nodes on the left side of the neck, with perinodal infiltration. An excisional biopsy was performed in the operating room, and gross examination revealed a 1.3- by 1.2- by 0.7-cm3 gray-white soft tissue lesion with surrounding tissue that was severely inflamed and necrotic. For histopathological assessment, samples of the biopsied lymph node tissue were fixed in 10% neutral buffered formalin and embedded in paraffin. Paraffin sections were stained with hematoxylin and eosin and periodic acid-Schiff, with and without diastase treatment. Stains used for detecting microorganisms included Gram's stain, Grocott's methanamine silver stain, and Ziehl-Neelsen's stain for mycobacteria. Pathological examination revealed central necrosis with peripheral hemorrhages, with no evidence of malignancy or tuberculosis. The tuberculosis PCR test and cultures for aerobic, anaerobic, mycobacterial, and fungal organisms were done using the biopsy specimens. A test of the fluid from the severely necrotic tissue with bleeding, which was similar to hematoma material, was tried with Gram staining. As a result, one to three gram-negative organisms per high-power field were seen. In addition, a few white blood cells were also observed in the materials. A bacterium isolated from tissue obtained from the biopsy was gram negative and was identified as Pantoea sp. by Vitek 2 (bioMerieux) in the clinical microbiology laboratory. For a more precise identification, 16S rRNA gene analysis was undertaken. In the interim, the patient was treated with oral ciprofloxacin (500 mg every 12 h) for 2 weeks. The patient's recovery was complete, and she was ultimately discharged.
The isolate, a gram-negative bacillus that grew on both sheep blood and MacConkey agar at 37°C, was named ABB-Jeju-1. On the same blood plate, two colony types were detected within 24 h; one colony was yellow, and the other was white. Their sizes were similar (1.0 to 1.5 mm), and their biochemical profiles and 16S rRNA gene sequences were the same. The colonies with similar shapes and sizes were found in the MacConkey agar plate. The Vitek 2 GNI+ card system (bioMérieux, Hazelwood, MO) identified them as colonies of Pantoea sp. with confidence levels of 99.00% and 91.23%. They were oxidase negative and catalase positive. For phenotypic characterization, the API 20NE, API 20E, and API 50CH systems (bioMérieux, Hazelwood, MO) were used according to the recommendations of the manufacturer. Also, using the API systems, the isolate could be identified as Pantoea spp. In the API 20NE and 20E results, these colonies were positive for the reduction of nitrates to nitrites, esculin hydrolysis, acetoin production, and β-galactosidase, glucose, arabinose, mannitol, N-acetyl-glucosamine, maltose, gluconate, malate, phenyl-acetate, inositol, rhamnose, sucrose, melibiose, and arabinose assimilation. Results were negative for indole production, glucose acidification, arginine dihydrolase, lysine decarboxylase, ornithine decarboxylase, urease, gelatin hydrolysis, citrate utilization, H2S production, and tryptophane deaminase and mannose, caprate, adipate, citrate, sorbitol, and amydalin assimilation. When the organisms were assayed with the API 50CH system, they were also positive for l-arabinose, ribose, d-xylose, galactose, fructose, cellobiose, trehalose, gentiobiose, and d-arabitol and were negative for the assimilation of glycerol, erythritol, d-arabinose, l-xylose, adonitol, β-methyl-d-xyloside, dulcitol, α-methyl-d-mannoside, α-methyl-d-glucoside, arbutin, salicin, lactose, melibiose, inulin, melezitose, raffinose, starch, glycogen, xylitol, d-turanose, d-lyxose, d-tagatose, d-fucose, l-fucose, l-arabitol, gluconate, 2-keto-gluconate, and 5-keto-gluconate.
For the 16S rRNA gene-based identification, genomic DNA was extracted using a G-Spin genomic DNA extraction kit (iNtRon, Seoul, Korea). The 16S rRNA gene was amplified using primer sets 16S-F3 (5′-CAG GCC TAA CAC ATG CAA GT-3′) and 16S-R3 (5′-GGG CGG WGT GTA CAA GGC-3′) (5). Template DNA and 20 pmol of each primer were added to a PCR mixture tube (AccuPower PCR PreMix; Bioneer, Daejeon, Korea). The reaction mixture was subjected to 30 cycles of PCR; each cycle consisted of 30 s at 95°C, 30 s at 60°C, and 1 min at 72°C, followed by a final extension at 72°C for 5 min. The PCR product was purified using a PCR purification kit (CoreOne, Seoul, Korea). The purified PCR product was sequenced directly in both directions with the same primers as those used for amplification. DNA sequences were edited by EditSeq and MegAlign programs (DNASTAR, Madison, WI). A 1,418-bp sequence of the 16S rRNA gene was obtained from the bacterium.
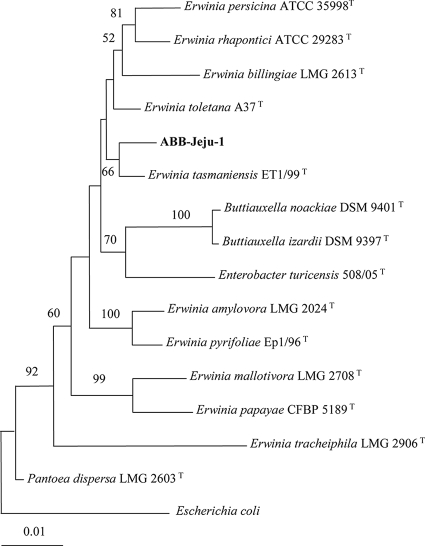
The isolate's 16S rRNA gene sequence was compared with those in the EzTaxon public database (http://www.eztaxon.org/) (2) and GenBank (http://www.ncbi.nlm.nih.gov/blast) using BLAST searches. Strains with the greatest pairwise similarity to strain ABB-Jeju-1 were Erwinia tasmaniensis Et1/99T (98.9%), E. toletana A37T (98.8%), and E. billingiae LMG 2613T (98.1%). The isolate was more than 97% similar in sequence to those of E. persicina ATCC 35998T, E. rhapontici ATCC 29283T, E. pyrifoliae Ep1/96T, E. amylovora LMG 2024T, Pantoea dispersa LMG 2603T, Enterobacter turicensis508/05T, Buttiauxella noackiae DSM 9401T, and Buttiauxella izardii DSM 9397T, based on pairwise comparisons. A phylogenetic tree based on 16S rRNA gene sequences also suggested that strain ABB-Jeju-1 clustered with E. tasmaniensis ET1/99T, and this association was moderately supported by a bootstrap value of 66% (Fig. 1). Very recently, Stackbrandt and Ebers (9) stated that a strain a with 16S rRNA gene sequence similarity lower than 99% with known species should be subjected to testing as a novel species by comparison of 16S rRNA gene sequence similarities and DNA-DNA reassociation values. Accordingly, strain ABB-Jeju-1 could not be regarded as E. tasmaniensis with confidence and was therefore designated as an Erwinia-like organism. When phenotypic characteristics were compared, the isolate differed from E. tasmaniensis in that it utilized rhamnose but not citrate, and it reduced nitrates into nitrites, while E. tasmaniensis did not (4).
FIG. 1.
Phylogenetic tree of strain ABB-Jeju-1 and type strains of other closely related species based on 16S rRNA gene sequences. This tree was reconstructed by the neighbor-joining method, and Escherichia coli was used as an outgroup. Numbers at branching nodes are percentages of 1,000 bootstrap replications. Only values greater than 50% are shown. The scale bar represents one substitution per 100 nucleotides.
In vitro susceptibility testing was performed by a broth microdilution method described in Clinical and Laboratory Standard Institute (CLSI) guidelines (3). As shown in Table 1, the isolate was susceptible or intermediate to most antimicrobial agents tested except for cefoxitin.
TABLE 1.
Results of antimicrobial susceptibility testing for strain ABB-Jeju-1
| Antimicrobial(s) | MIC(s) (mg/liter) | Susceptibilitya |
|---|---|---|
| Ampicillin | 16 | I |
| Amikacin | <2 | S |
| Penicillin | 16 | |
| Amoxicillin/clavulanate | 4/2 | S |
| Ceftazidime | <1 | S |
| Cefotaxime | <1 | S |
| Cefazolin | <1 | S |
| Cefepime | <1 | S |
| Cefoxitin | 32 | R |
| Cefuroxime | 8 | I |
| Ceftriaxone | 0.25 | S |
| Ciprofloxacin | <0.03 | S |
| Levofloxacin | <0.03 | S |
| Gentamicin | <1 | S |
| Nitrofurantoin | 64 | I |
| Rifampin | 16 | |
| Trimethoprim-sulfamethoxazole | 0.12-2.37 | S |
| Tetracycline | 1 | S |
S, susceptible; I, intermediate; R, resistant. MIC interpretative breakpoints of penicillin and rifampin are not shown by CLSI (3) for members of the family Enterobacteriaceae.
The genus Erwinia consists of 12 validated species (http://www.bacterio.cict.fr/), which are associated with or are pathogens of plants (7). Species in this genus are similar to Pantoea and Enterobacter species, and many Erwinia species have been reclassified into these groups. Clinical isolates of Erwinia sp. are exceedingly rare; only one species, Erwinia persicina, has been isolated from the urine of a woman with a urinary tract infection (6).
The isolation of a bacterium from the excisional biopsy tissue of a patient with cervical lymphadenitis is reported. This bacterium was found to be a new Erwinia-like species related most closely to Erwinia tasmaniensis. E. tasmaniensis is a recently described species (4) isolated from the flowers and bark of apple and pear trees in Australia but is not pathogenic to plants.
To date, Erwinia species, with the exception of E. persicina, are not associated with human disease. Although E. persicina was isolated from a human patient with a urinary tract infection (6), it is not clear if the organism actually caused the infection. In the present case, however, the strain ABB-Jeju-1 was isolated from a sterile site (a lymph node) during a surgical operation. In addition, tissue from the excisional biopsy showed a clear inflammatory reaction. Thus, this report describes the second case of an Erwinia strain found in clinical samples of humans and may be the first report of a human infection caused by Erwinia.
According to a study performed in Korea, Kikuchi's disease (34.7%), tuberculous lymphadenitis (22.4%), and nonspecific lymphadenitis (22.4%) were diagnosed histopathologically in patients with cervical lymphadenitis, which was confirmed by ultrasound-guided core needle gun biopsy (8). Cervical lymphadenitis is sometimes due to pyogenic infections. While the role of anaerobic bacteria in cervical lymphadenitis in children has been emphasized, mycobacteria, fungi, and aerobic bacteria such as Staphylococcus aureus and group A beta-hemolytic streptococci are important in adults (1). Although several gram-negative bacterial species such as Klebsiella pneumoniae, Escherichia coli, Serratia sp., Salmonella sp., and Pseudomonas aeruginosa have been isolated from patients with cervical lymphadenitis (1), gram-negative bacteria are a less-frequent cause of cervical lymphadenitis. Cervical lymphadenitis caused by Toxoplasma gondii infection has been reported recently in Korea (10), but cases caused by infection of Erwinia or even by Pantoea species, which are species closely related to those of Erwinia, have not been reported.
Erwinia species are not included in any commercially available identification system such as Vitek and MicroScan. For this reason, the E. tasmaniensis-like organism presently identified was initially identified as Pantoea sp. Our report of an E. tasmaniensis-like organism isolated from a patient with cervical lymphadenitis expands the number of Erwinia species associated with human infection.
Nucleotide sequence accession number.
The sequence of the 16S rRNA gene of strain ABB-Jeju-1 has been deposited in the GenBank database under accession number EU434702.
Acknowledgments
This study was partly supported by the Asian-Pacific Research Foundation for Infectious Diseases (ARFID) and the Samsung Biomedical Research Institute (SBRI).
Footnotes
Published ahead of print on 9 July 2008.
REFERENCES
- 1.Brook, I., and E. H. Frazier. 1998. Microbiology of cervical lymphadenitis in adults. Acta Otolaryngol. 118443-446. [DOI] [PubMed] [Google Scholar]
- 2.Chun, J., J.-H. Lee, Y. Jung, M. Kim, S. Kim, B. K. Kim, and Y. W. Lim. 2007. EzTaxon: a web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. Int. J. Syst. Evol. Microbiol. 572259-2261. [DOI] [PubMed] [Google Scholar]
- 3.Clinical and Laboratory Standards Institute/NCCLS. 2007. Performance standards for antimicrobial susceptibility testing; 17th informational supplement. CLSI/NCCLS M100-S17. Clinical and Laboratory Standards Institute, Wayne, PA.
- 4.Geider, K., G. Auling, Z. Du, V. Jakovljevic, S. Jock, and B. Volksch. 2006. Erwinia tasmaniensis sp. nov., a non-phytopathogenic bacterium from apple and pear trees. Int. J. Syst. Evol. Microbiol. 562937-2943. [DOI] [PubMed] [Google Scholar]
- 5.Ko, K. S., K. R. Peck, W. S. Oh, N. Y. Lee, J. H. Lee, and J.-H. Song. 2005. New species of Bordetella, Bordetella ansorpii sp. nov., isolated from the purulent exudates of an epidermal cyst. J. Clin. Microbiol. 432516-2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O'Hara, C. M., A. G. Steigerwalt, B. C. Hill, J. M. Miller, and D. J. Brenner. 1998. First report of a human isolate of Erwinia persicinus. J. Clin. Microbiol. 36248-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rojas, A. M., J. E. G. de los Rios, M. F.-L. Saux, P. Jimenez, P. Reche, S. Bonneau, L. Sutra, F. Mathieu-Daude, and M. McClelland. 2004. Erwinia toletana sp. nov., associated with Pseudomonas savastanoi-induced tree knots. Int. J. Syst. Evol. Microbiol. 542217-2222. [DOI] [PubMed] [Google Scholar]
- 8.Song, J. Y., H. J. Cheong, S. Y. Kee, J. Lee, J. W. Sohn, M. J. Kim, S. I. Seo, I. S. Kim, and W. J. Kim. 2007. Disease spectrum of cervical lymphadenitis: analysis based on ultrasound-guided core-needle gun biopsy. J. Infect. 55310-316. [DOI] [PubMed] [Google Scholar]
- 9.Stackbrandt, E., and J. Ebers. 2006. Taxonomic parameters revisited: tarnished gold standards. Microbiology Today 33152-155. [Google Scholar]
- 10.Suh, Y. J., W. Kim, W.-B. Park, and C.-S. Chun. 2002. Cervical lymphadenitis caused by Toxoplasma gondii. J. Kor. Surg. Soc. 62271-273. [Google Scholar]