Abstract
We evaluated the use of a novel multiple-locus variable-number tandem-repeat analysis (MLVA) method for typing of human Staphylococcus aureus. For a total of 150 clinical isolates, MLVA demonstrated the highest discriminatory power. MLVA correctly assigned isolates to outbreaks or identified isolates as unlinked. MLVA is a rapid and simple method for the epidemiological typing of S. aureus.
Pulsed-field gel electrophoresis (PFGE) is considered the gold standard method for the typing of Staphylococcus aureus isolates (1). Other commonly used typing schemes include multilocus sequence typing (MLST) and spa typing (www.mlst.net and www.spaserver.ridom.de) (2, 7). Recently, a method based on the unique lengths of the intergenic regions containing repetitive DNA loci (5, 8, 9, 12, 13), known as multiple-locus variable-number tandem-repeat analysis (MLVA), was introduced and described. Using this method, Hardy et al. described an MLVA scheme based on seven variable-number tandem repeats in S. aureus, termed staphylococcal interspersed repeat units (SIRUs) (5, 6). All of these methods have a number of drawbacks; most importantly, they are either fingerprinting methods, which are difficult to compare, or library typing methods lacking discriminatory power (3, 9, 10, 12, 13). We therefore developed a character-based MLVA scheme for human S. aureus that is based on the number of repeats of each locus, i.e., the allelic profile, which can then be used in combination with spa typing. A similar approach has already been reported for bovine S. aureus (4).
Our scheme was first tested using 100 European clinical human S. aureus isolates: 25 methicillin-susceptible and 75 methicillin-resistant isolates obtained between 1997 and 2004 were selected from the ENARE collection at the University Medical Centre Utrecht (UMCU), The Netherlands. These 100 specimens represented 35 MLST types and were well distributed throughout the S. aureus population (data not shown). MLVA typing was performed by using SIRU01, -05, -07, -13, -15, -16, and -21 (representing the spa gene) and sspA (5, 10). Each amplification was performed separately. The PCR products of SIRU01, -05, -07, -13, -15, -16, and sspA were analyzed on 2% agarose gel, while the SIRU21 PCR product was run on a 3% agarose gel. The number of repeat units (RUs) in each locus was determined by subtracting the sizes of the flanking regions from the size of the amplicon and then dividing the difference by the size of the repeat (Table 1). The result was then rounded to the nearest integer value.
TABLE 1.
Characteristics of the MLVA scheme
| Locus (RU size [bp]) | Formulaa | No. of RUs (% PCR negative) |
|---|---|---|
| SIRU01 (55) | (n − 157 − 30)/55 | 0-5 (4.5) |
| SIRU05 (60) | (n − 76 − 78)/60 | 1-22 (21.5) |
| SIRU07 (56) | (n − 27 − 160)/56 | 1-4 (2.2) |
| SIRU13 (64) | (n − 76 − 78)/64 | 0-26 (2.2) |
| SIRU15 (131) | (n − 48 − 174)/131 | 0-5 (0) |
| SIRU21 (24) | [(n − 12 − 81) − 16)]/24 | 1-16 (0) |
The formula for calculating the number of RUs per locus is as follows: (amplicon size [n] − size of left flanking region − size of right flanking region)/size of RU.
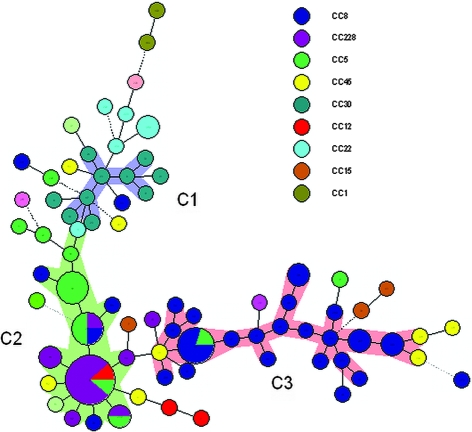
The MLVA showed the numbers of RUs to range from 0 to 26 (Table 1). SIRU16 and sspA were excluded from further study due to a lack of variation in RUs. Seventy-three isolates yielded complete MLVA profiles, and 27 isolates yielded partial profiles. The unamplified loci of the latter isolates were assigned the number 999. Combinations of RUs from SIRU01, -05, -07, -13, -15, and -21 yielded number strings that were considered to be the allelic profiles. An MLVA type (MT) was assigned to each of these profiles. The 76 MTs observed (Table 2) were clustered into three dominant MT genogroups, C1, C2, and C3 (Fig. 1). Most of the isolates belonged to clonal complexes 1, 5, 8, 12, 15, 22, 30, and 228 and were generally linked to genogroups. Isolates in clonal complex 45 were an exception. This difference may be explained either by unexpected evolutionary pressure or by the horizontal transfer of genetic material containing one or more SIRUs among the clonal complex 45 isolates.
TABLE 2.
MLVA allelic profiles, MTs, and spa types obtained from 100 S. aureus isolates with known MLST types
| Isolate no. | Sample | MLST type | Clonal complex | spa typea | MT | No. of RUs in locusb
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SIRU01 | SIRU05 | SIRU07 | SIRU13 | SIRU15 | SIRU21 | ||||||
| 1 | S0041 | 7 | 5 | 91 | 1 | 0 | 3 | 3 | 3 | 3 | 10 |
| 2 | S0033 | 12 | 12 | 160 | 2 | 1 | 7 | 1 | 3 | 1 | 7 |
| 3 | S0117 | 717 | 12 | 160 | 3 | 1 | 8 | 1 | 4 | 1 | 7 |
| 4 | S0061 | 45 | 45 | 3 | 4 | 1 | 2 | 1 | 4 | 1 | 8 |
| 5 | S0055 | 22 | 22 | 223 | 5 | 2 | 3 | 1 | 3 | 1 | 11 |
| 6 | S0134 | 34 | 30 | 2285* | 6 | 2 | 3 | 1 | 1 | 2 | 15 |
| 7 | S0141 | 15 | 15 | 346 | 7 | 2 | 5 | 2 | 2 | 1 | 10 |
| 8 | S0032 | 30 | 30 | 1945 | 8 | 2 | 3 | 2 | 1 | 2 | 12 |
| 9 | S0296 | 36 | 30 | 18 | 9 | 2 | 3 | 2 | 1 | 2 | 11 |
| 10 | S0084 | 149 | 5 | 2 | 10 | 2 | 2 | 1 | 4 | 1 | 10 |
| 11 | S0168 | 228 | 228 | 1 | 10 | 2 | 2 | 1 | 4 | 1 | 10 |
| 12 | S0072 | 228 | 228 | 1 | 10 | 2 | 2 | 1 | 4 | 1 | 10 |
| 13 | S0077 | 228 | 228 | 1 | 10 | 2 | 2 | 1 | 4 | 1 | 10 |
| 14 | S0085 | 228 | 228 | 1 | 10 | 2 | 2 | 1 | 4 | 1 | 10 |
| 15 | S0088 | 228 | 228 | 1 | 10 | 2 | 2 | 1 | 4 | 1 | 10 |
| 16 | S0065 | 228 | 228 | 1 | 10 | 2 | 2 | 1 | 4 | 1 | 10 |
| 17 | S0071 | 736 | 5 | 1 | 10 | 2 | 2 | 1 | 4 | 1 | 10 |
| 18 | S0056 | 228 | 228 | 1 | 10 | 2 | 2 | 1 | 4 | 1 | 10 |
| 19 | S0108 | 228 | 228 | 41 | 11 | 2 | 2 | 1 | 4 | 1 | 14 |
| 20 | S0074 | 228 | 228 | 41 | 11 | 2 | 2 | 1 | 4 | 1 | 14 |
| 21 | S0087 | 228 | 228 | 1 | 12 | 2 | 2 | 1 | 2 | 1 | 10 |
| 22 | S0062 | 225 | 5 | 3 | 13 | 2 | 2 | 2 | 3 | 2 | 8 |
| 23 | S0063 | 228 | 228 | 1 | 14 | 2 | 2 | 2 | 4 | 1 | 10 |
| 24 | S0070 | 228 | 228 | 1 | 15 | 2 | 2 | 2 | 1 | 0 | 10 |
| 25 | S0031 | 5 | 5 | 3 | 16 | 2 | 3 | 1 | 3 | 1 | 8 |
| 26 | S0054 | 34 | 30 | 369 | 17 | 2 | 3 | 1 | 2 | 2 | 11 |
| 27 | S0118 | 225 | 5 | 3 | 18 | 2 | 3 | 1 | 4 | 1 | 8 |
| 28 | S0075 | 225 | 5 | 3 | 18 | 2 | 3 | 1 | 4 | 1 | 8 |
| 29 | S0079 | 225 | 5 | 3 | 18 | 2 | 3 | 1 | 4 | 1 | 8 |
| 30 | S0081 | 225 | 5 | 3 | 18 | 2 | 3 | 1 | 4 | 1 | 8 |
| 31 | S0022 | 714 | 30 | 136 | 19 | 2 | 3 | 1 | 2 | 2 | 13 |
| 32 | S0047 | 715 | 30 | 166 | 20 | 2 | 3 | 1 | 2 | 2 | 12 |
| 33 | S0050 | 30 | 30 | 122 | 21 | 2 | 3 | 2 | 1 | 0 | 8 |
| 34 | S0057 | 225 | 5 | 3 | 22 | 2 | 3 | 2 | 3 | 2 | 8 |
| 35 | S0053 | 30 | 30 | 18 | 23 | 2 | 3 | 2 | 2 | 2 | 11 |
| 36 | S0021 | 713 | 30 | 18 | 24 | 2 | 4 | 2 | 2 | 2 | 11 |
| 37 | S0043 | 26 | 25 | 81 | 25 | 2 | 5 | 2 | 10 | 5 | 8 |
| 38 | S0049 | 239 | 8 | 275 | 26 | 2 | 3 | 1 | 8 | 1 | 8 |
| 39 | S0112 | 5 | 5 | 2 | 27 | 2 | 3 | 1 | 4 | 1 | 10 |
| 40 | S0128 | 5 | 5 | 2 | 27 | 2 | 3 | 1 | 4 | 1 | 10 |
| 41 | S0042 | 228 | 228 | 1 | 27 | 2 | 3 | 1 | 4 | 1 | 10 |
| 42 | S0045 | 247 | 8 | 52 | 27 | 2 | 3 | 1 | 4 | 1 | 10 |
| 43 | S0132 | 739 | 101 | 56 | 28 | 2 | 4 | 2 | 6 | 2 | 9 |
| 44 | S0111 | 247 | 8 | 2 | 29 | 2 | 1 | 1 | 4 | 1 | 10 |
| 45 | S0113 | 36 | 30 | 12 | 30 | 2 | 3 | 2 | 1 | 2 | 10 |
| 46 | S0060 | 45 | 45 | 15 | 31 | 3 | 2 | 2 | 1 | 1 | 10 |
| 47 | S0059 | 5 | 5 | 2 | 32 | 3 | 2 | 3 | 1 | 1 | 10 |
| 48 | S0027 | 8 | 8 | 9 | 32 | 3 | 2 | 3 | 1 | 1 | 10 |
| 49 | S0029 | 8 | 8 | 8 | 32 | 3 | 2 | 3 | 1 | 1 | 10 |
| 50 | S0044 | 8 | 8 | 8 | 32 | 3 | 2 | 3 | 1 | 1 | 10 |
| 51 | S0083 | 738 | 8 | 8 | 32 | 3 | 2 | 3 | 1 | 1 | 10 |
| 52 | S0068 | 8 | 8 | 8 | 33 | 3 | 3 | 2 | 2 | 2 | 10 |
| 53 | S0028 | 239 | 8 | 37 | 34 | 3 | 2 | 2 | 26 | 1 | 7 |
| 54 | S0120 | 684 | 8 | 37 | 35 | 3 | 2 | 2 | 1 | 1 | 7 |
| 55 | S0178 | 239 | 8 | 30 | 36 | 3 | 2 | 2 | 0 | 1 | 6 |
| 56 | S0177 | 239 | 8 | 30 | 36 | 3 | 2 | 2 | 0 | 1 | 6 |
| 57 | S0058 | 225 | 5 | 3 | 37 | 4 | 9 | 2 | 1 | 1 | 8 |
| 58 | S0030 | 15 | 15 | 84 | 38 | 4 | 7 | 2 | 2 | 5 | 11 |
| 59 | S0107 | 239 | 8 | 37 | 39 | 4 | 2 | 2 | 1 | 1 | 7 |
| 60 | S0109 | 8 | 8 | 8 | 40 | 4 | 2 | 3 | 0 | 1 | 10 |
| 61 | S0025 | 247 | 8 | 51 | 41 | 4 | 22 | 2 | 1 | 1 | 11 |
| 62 | S0035 | 247 | 8 | 51 | 42 | 4 | 4 | 2 | 0 | 1 | 11 |
| 63 | S0034 | 15 | 15 | 254 | 43 | 4 | 7 | 2 | 4 | 5 | 8 |
| 64 | S0297 | 8 | 8 | 8 | 44 | 4 | 1 | 3 | 0 | 1 | 10 |
| 65 | S0298 | 8 | 8 | 64 | 45 | 4 | 2 | 2 | 0 | 1 | 10 |
| 66 | S0127 | 239 | 8 | 138 | 46 | 4 | 2 | 2 | 0 | 1 | 6 |
| 67 | S0121 | 685 | 8 | 64 | 47 | 4 | 2 | 3 | 1 | 1 | 10 |
| 68 | S0129 | 247 | 8 | 75 | 48 | 4 | 4 | 2 | 1 | 1 | 12 |
| 69 | S0069 | 239 | 8 | 30 | 49 | 5 | 2 | 2 | 1 | 1 | 6 |
| 70 | S0066 | 735 | 45 | 305 | 50 | 5 | 3 | 2 | 4 | 2 | 11 |
| 71 | S0051 | 239 | 8 | 421 | 51 | 2 | 3 | 1 | 4 | 1 | 6 |
| 72 | S0052 | 45 | 45 | 693 | 52 | 2 | 2 | 1 | 4 | 1 | 1 |
| 73 | S0039 | 101 | 101 | 56 | 53 | 2 | 2 | 1 | 4 | 1 | 9 |
| 74 | S0067 | 254 | 8 | 9 | 74 | 999 | 2 | 2 | 1 | 1 | 10 |
| 75 | S0037 | 9 | Singletonc | 2176* | 77 | 4 | 2 | 999 | 0 | 1 | 4 |
| 76 | S0300 | 5 | 5 | 88 | 80 | 3 | 10 | 1 | 999 | 2 | 11 |
| 77 | S0299 | 72 | 8 | 126 | 83 | 3 | 10 | 999 | 999 | 2 | 8 |
| 78 | S0295 | 5 | 5 | 2 | 85 | 2 | 999 | 1 | 4 | 1 | 10 |
| 79 | S0046 | 228 | 228 | 1 | 85 | 2 | 999 | 1 | 4 | 1 | 10 |
| 80 | S0076 | 22 | 22 | 32 | 87 | 2 | 999 | 2 | 4 | 0 | 16 |
| 81 | S0078 | 22 | 22 | 32 | 87 | 2 | 999 | 2 | 4 | 0 | 16 |
| 82 | S0089 | 22 | 22 | 476 | 89 | 2 | 999 | 2 | 2 | 0 | 11 |
| 83 | S0116 | 97 | 97 | 527 | 91 | 2 | 999 | 3 | 3 | 1 | 12 |
| 84 | S0024 | 22 | 22 | 192 | 93 | 2 | 999 | 2 | 7 | 3 | 13 |
| 85 | S0036 | 45 | 45 | 1081 | 95 | 999 | 999 | 1 | 2 | 2 | 7 |
| 86 | S0086 | 45 | 45 | 4 | 97 | 999 | 999 | 2 | 0 | 1 | 9 |
| 87 | S0130 | 45 | 45 | 26 | 99 | 999 | 999 | 2 | 1 | 0 | 3 |
| 88 | S0023 | 45 | 45 | 1933 | 101 | 999 | 999 | 2 | 1 | 1 | 7 |
| 89 | S0166 | 239 | 8 | 37 | 103 | 999 | 999 | 2 | 0 | 1 | 7 |
| 90 | S0176 | 239 | 8 | 37 | 103 | 999 | 999 | 2 | 0 | 1 | 7 |
| 91 | S0167 | 239 | 8 | 37 | 105 | 4 | 999 | 2 | 0 | 1 | 7 |
| 92 | S0175 | 239 | 8 | 37 | 105 | 4 | 999 | 2 | 0 | 1 | 7 |
| 93 | S0026 | 247 | 8 | 844 | 107 | 4 | 999 | 2 | 0 | 1 | 8 |
| 94 | S0169 | 247 | 8 | 51 | 109 | 4 | 999 | 2 | 0 | 1 | 11 |
| 95 | S0110 | 247 | 8 | 52 | 111 | 4 | 999 | 2 | 0 | 1 | 10 |
| 96 | S0126 | 572 | 8 | 51 | 113 | 4 | 999 | 2 | 1 | 1 | 11 |
| 97 | S0119 | 1 | 1 | 922 | 115 | 1 | 999 | 3 | 1 | 2 | 6 |
| 98 | S0040 | 188 | 1 | 189 | 117 | 1 | 999 | 3 | 3 | 5 | 6 |
| 99 | S0115 | 239 | 8 | 359 | 119 | 5 | 999 | 4 | 3 | 2 | 9 |
| 100 | S0082 | 737 | 22 | 5 | 122 | 2 | 999 | 2 | 3 | 0 | 12 |
Asterisks indicate two novel spa types observed in this study.
999, no SIRU PCR amplification.
This MLST type did not group into any of the clonal complexes.
FIG. 1.
Population structure of S. aureus isolates based on MLVA. Each circle represents a different MLVA profile. Three clusters (C1, C2, and C3) were identified as shown by the three colors (blue, green, and pink). Heavy lines connecting two MTs denote a single-locus variant, thin lines denote MTs with a double-locus variant, and dotted lines connect MTs that differ by more than two loci.
spa typing was performed as described by Harmsen et al. (7) and yielded 50 sequence variations, or spa types, including 2 new spa types, 2285 and 2176. The numbers of repeats found in SIRU21 by spa typing and MLVA agreed completely.
The discriminatory power of MLVA, MLST, and spa typing showed that MLVA had a higher discriminatory power than both MLST and spa typing (for MLVA, 0.987 [95% confidence interval {CI}, 0.977 to 0.997]; for MLST, 0.941 [95% CI, 0.922 to 0.960]; and for spa typing, 0.963 [95% CI, 0.946 to 0.979]), even though the 95% CI for MLVA overlapped somewhat with that for spa typing. The adjusted Rand index for MLVA and spa typing and that for MLVA and MLST were 0.341 and 0.184, respectively. The Wallace coefficients (Table 3) indicated that MLVA was reasonably predictive of both MLST and spa type, whereas the reverse was not true.
TABLE 3.
Wallace coefficients for the methods used to characterize the 100 human S. aureus isolates
| Typing method | Wallace coefficient for:
|
||
|---|---|---|---|
| MLST | spa typing | MLVA | |
| MLST | 0.433 | 0.123 | |
| spa typing | 0.690 | 0.239 | |
| MLVA | 0.554 | 0.677 | |
A confirmation study was performed to validate our MLVA scheme. This study compared the MTs with the PFGE and spa types of 50 S. aureus isolates from the Department of Infection Control and Infection Prevention of UMCU. PFGE was performed using SmaI as previously described by Tenover et al. (11). The isolates were considered to be either epidemiologically related or devoid of any known epidemiological link, based on either their phage or their PFGE types (depending on the method used by the National Institute of Public Health and the Environment, The Netherlands, at the time of the initial isolation) and on their times of isolation. Two PCR fragments detected for SIRU13 in two isolates were assigned the value 888.
Totals of 31 MTs, 24 spa types, and 30 PFGE profiles were obtained (Table 4). The Simpson indices for MLVA, spa typing, and PFGE were 0.971, 0.908, and 0.970, respectively. The concordance between MLVA and spa typing was 93.5%, and that between MLVA and PFGE was 97.7%. The adjusted Rand indices for MLVA compared with PFGE and spa typing were 0.599 and 0.435, respectively. The Wallace coefficients showed that MLVA and spa typing were mutually predictive (data not shown).
TABLE 4.
MTs, spa types, and PFGE profiles of S. aureus isolatesa
| Isolate | Date of isolation | Phage type | PFGE clusterb | UMCU classificationc | PFGE profile | spa typed | MT | No. of RUs in locuse
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SIRU01 | SIRU05 | SIRU07 | SIRU13 | SIRU15 | SIRU21 | ||||||||
| 199 | 13 November 1997 | Z136 | O1 | A | 2175* | 178 | 1 | 999 | 3 | 1 | 2 | 9 | |
| 215 | 14 November 1997 | Z136 | O1 | A | 2175* | 178 | 1 | 999 | 3 | 1 | 2 | 9 | |
| 265 | 09 October 1998 | Z136 | O1 | A | 2175* | 178 | 1 | 999 | 3 | 1 | 2 | 9 | |
| 266 | 21 October 1998 | Z136 | O1 | A | 2175* | 178 | 1 | 999 | 3 | 1 | 2 | 9 | |
| 341 | 31 May 2000 | Z136 | U | F | 67 | 89 | 2 | 1 | 1 | 4 | 1 | 9 | |
| 593 | 01 August 2006 | 35 | U | U | 2 | 29 | 2 | 1 | 1 | 4 | 1 | 10 | |
| 130 | 31 May 1996 | III205 | O2 | B | 2 | 29 | 2 | 1 | 1 | 4 | 1 | 10 | |
| 132 | 10 June 1996 | III205 | O2 | B | 2 | 29 | 2 | 1 | 1 | 4 | 1 | 10 | |
| 158 | 04 January 1997 | III205 | O2 | B | 2 | 100 | 2 | 1 | 1 | 5 | 1 | 10 | |
| 241 | 23 June 1998 | III205 | U | E | 447 | 53 | 2 | 2 | 1 | 4 | 1 | 9 | |
| 478 | 01 March 2004 | 60a | U | E | 447 | 53 | 2 | 2 | 1 | 4 | 1 | 9 | |
| 298 | 18 June 1999 | III283 | U | C | 2 | 101 | 2 | 5 | 1 | 4 | 1 | 10 | |
| 319 | 25 November 1999 | III311 | U | C | 2 | 101 | 2 | 5 | 1 | 4 | 1 | 10 | |
| 519 | 16 December 2004 | 55 | O3 | C | 2 | 27 | 2 | 3 | 1 | 4 | 1 | 10 | |
| 524 | 05 January 2005 | 55 | O3 | C | 2 | 27 | 2 | 3 | 1 | 4 | 1 | 10 | |
| 530 | 07 January 2005 | 55 | O3 | C | 2 | 27 | 2 | 3 | 1 | 4 | 1 | 10 | |
| 529 | 05 January 2005 | 55 | O3 | C1 | 2173* | 102 | 2 | 3 | 2 | 4 | 1 | 10 | |
| 144 | 07 November 1996 | Z115 | O4 | G | 8 | 32 | 3 | 2 | 3 | 1 | 1 | 10 | |
| 145 | 21 February 1996 | Z115 | O4 | G | 8 | 47 | 4 | 2 | 3 | 1 | 1 | 10 | |
| 162 | 01 February 1997 | Z115 | O4 | G | 8 | 32 | 3 | 2 | 3 | 1 | 1 | 10 | |
| 255 | 15 September 1998 | Z115 | O4 | G | 8 | 32 | 3 | 2 | 3 | 1 | 1 | 10 | |
| 343 | 13 November 2000 | Z115 | U | G1 | 8 | 32 | 3 | 2 | 3 | 1 | 1 | 10 | |
| 506 | 29 June 2004 | 18 | U | H | 8 | 32 | 3 | 2 | 3 | 1 | 1 | 10 | |
| 516 | 06 December 2004 | 148 | O5 | D | 8 | 137 | 3 | 3 | 3 | 1 | 1 | 10 | |
| 518 | 11 December 2004 | 148 | O5 | D | 8 | 136 | 3 | 3 | 3 | 0 | 1 | 10 | |
| 520 | 11 December 2004 | 148 | O5 | D | 8 | 136 | 3 | 3 | 3 | 0 | 1 | 10 | |
| 347 | 27 March 2001 | Z231 | O6 | I | 52 | 412 | 4 | 25 | 2 | 1 | 1 | 10 | |
| 348 | 27 March 2001 | Z231 | O6 | I | 52 | 412 | 4 | 25 | 2 | 1 | 1 | 10 | |
| 363 | 07 April 2001 | Z231 | O6 | I | 52 | 412 | 4 | 25 | 2 | 1 | 1 | 10 | |
| 371 | 13 April 2001 | Z231 | O6 | I | 52 | 412 | 4 | 25 | 2 | 1 | 1 | 10 | |
| 395 | 06 December 2001 | Z231 | U | BB | 1257 | 169 | 4 | 2 | 2 | 1 | 1 | 10 | |
| 90 | 01 October 1995 | III84 | U | Z | 138 | 167 | 4 | 2 | 2 | 1 | 1 | 6 | |
| 125 | 15 February 1996 | III29 | U | M | 31 | 308 | 999 | 999 | 2 | 1 | 0 | 9 | |
| 129 | 21 May 1996 | V2 | U | U | 1 | 10 | 2 | 2 | 1 | 4 | 1 | 10 | |
| 284 | 10 November 1998 | III70 | U | K | 24 | 170 | 4 | 1 | 3 | 0 | 1 | 9 | |
| 331 | 18 February 2000 | III322 | U | Y | 2 | 27 | 2 | 3 | 1 | 4 | 1 | 10 | |
| 393 | 03 December 2001 | NT | U | W | 1932 | 90 | 2 | 3 | 2 | 2 | 2 | 10 | |
| 401 | 05 February 2002 | 147 | U | L | 44 | 275 | 4 | 999 | 1 | 3 | 6 | 14 | |
| 450 | 09 October 2003 | 28 | U | N | 728 | 308 | 999 | 999 | 2 | 0 | 1 | 5 | |
| 459 | 14 November 2003 | NT | U | P | 567 | 416 | 3 | 999 | 3 | 888 | 2 | 5 | |
| 466 | 04 December 2003 | 118a | U | T | 311 | 88 | 2 | 13 | 1 | 4 | 1 | 9 | |
| 482 | 16 June 2004 | 430/426 | U | X | 223 | 235 | 2 | 999 | 2 | 3 | 0 | 11 | |
| 502 | 23 October 2004 | 1293 | U | R | 1293 | 179 | 1 | 999 | 2 | 2 | 5 | 8 | |
| 503 | 28 October 2004 | 25a | U | S | 437 | 190 | 2 | 999 | 4 | 2 | 5 | 7 | |
| 559 | 13 August 2005 | 93 | U | F2 | 729 | 237 | 2 | 999 | 2 | 3 | 2 | 10 | |
| 561 | 26 October 2005 | 218 | U | AA | 8 | 168 | 4 | 1 | 3 | 1 | 1 | 10 | |
| Z548 | 04 March 2006 | NT | U | O | 11 | 402 | 3 | 999 | 3 | 888 | 2 | 7 | |
| 570 | 11 March 2006 | 99a | U | Y | 37 | 401 | 4 | 2 | 2 | 999 | 1 | 7 | |
| 578 | 24 April 2006 | 613 | U | Q | 316 | 177 | 1 | 999 | 3 | 1 | 5 | 6 | |
| 602 | 25 August 2006 | 46 | U | K1 | 8 | 44 | 4 | 1 | 3 | 0 | 1 | 10 | |
These classifications are compared to those previously given by the Department of Infection Control and Infection Prevention, UMCU, The Netherlands, on the basis of phage type or PFGE clustering.
NT, not typeable.
O1 to O6, outbreaks 1 to 6, respectively; U, unlinked epidemiological isolate.
Asterisks indicate two new spa types observed in this study.
999, no SIRU amplification found; 888, double fragments on SIRU PCR detected.
MLVA typing distinguished 10 MTs from 22 isolates belonging to six outbreaks and 24 MTs from 28 unlinked isolates (Table 4). MLVA, spa type, and PFGE profile completely agreed for two of the outbreaks (O1 and O6). Four other outbreaks (O2, O3, O4, and O5) were correctly assigned when single-locus differences in MLVA, long periods between isolate collection (isolates 506, 593, 298, and 319 and isolates from O4 and O5), and a foreign origin (isolate 395) were taken into account. The 28 unrelated isolates all exhibited unique MTs.
MLVA was able to distinguish between the different outbreaks when single-locus variants were considered to belong to the same outbreak and when date of isolation was taken into consideration.
Data in Table 5 indicate that MLVA profiles are relatively stable over time. Identical MTs and spa types were detected in the isolates from patients 3 and 4 during a 5-year period, while variations in only two loci (SIRU01 and -07) (spa types remained identical) were seen in isolates from patient 1 recovered 7 years after the first isolate was taken. Distinct genotypes, MTs, and spa types were detected in isolates from patient 2, which may be explained by a later infection with a different S. aureus strain. Thus, the long-term stability of the MLVA profiles is not a major concern for outbreak detection.
TABLE 5.
Evolution of S. aureus carriership based on MTs for four patients over timea
| Patient no. | Yr of isolation | Isolateb | spa type | MT | No. of RUs in locusc
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| SIRU01 | SIRU05 | SIRU07 | SIRU13 | SIRU15 | SIRU21 | |||||
| 1 | 1996 | 96-121 | 8 | 63 | 4 | 2 | 4 | 1 | 1 | 10 |
| 1999 | 99-307 | 8 | 63 | 4 | 2 | 4 | 1 | 1 | 10 | |
| 2001 | 01-386 | 8 | 63 | 4 | 2 | 4 | 1 | 1 | 10 | |
| 2003 | 03-438 | 8 | 31 | 3* | 2 | 2* | 1 | 1 | 10 | |
| 2 | 1999 | 99-288 | 37 | 35 | 3 | 2 | 2 | 1 | 1 | 7 |
| 2001 | 01-346 | 121 | 56 | 2* | 7* | 2 | 1 | 1 | 9* | |
| 3 | 1987 | 87-A117 | 75 | 48 | 4 | 4 | 2 | 1 | 1 | 12 |
| 1988 | 88-A208 | 75 | 48 | 4 | 4 | 2 | 1 | 1 | 12 | |
| 1989 | 89-A313 | 75 | 48 | 4 | 4 | 2 | 1 | 1 | 12 | |
| 1990 | 90-A358 | 75 | 48 | 4 | 4 | 2 | 1 | 1 | 12 | |
| 1992 | 92-A405 | 75 | 48 | 4 | 4 | 2 | 1 | 1 | 12 | |
| 4 | 1996 | 96-129 | 1 | 10 | 2 | 2 | 1 | 4 | 1 | 10 |
| 1996 | 96-139 | 1 | 10 | 2 | 2 | 1 | 4 | 1 | 10 | |
| 2001 | 01-384 | 1 | 10 | 2 | 2 | 1 | 4 | 1 | 10 | |
An isolate collected in 2003 from patient 2 showed two locus variations (SIRU01 and -07) compared to the isolates collected during the previous year, even though identical spa types were observed. Two isolates taken from patient 3 revealed three locus variations (SIRU01, -05, and -21) indicating that these two isolates were unrelated. The different spa types strengthen this finding.
The isolates are numbered according to year of isolation and strain number, e.g., 96-121 means the isolate collected in 1996 with strain number 121.
Asterisks indicate locus variation.
In summary, our MLVA scheme showed good typeability and excellent discriminatory power for the major clonal complexes and singletons of S. aureus. Moreover, the reliability of the method is very good: the number of RUs determined by MLVA for SIRU21, representing the spa gene, completely agreed with the number of repeats obtained when spa typing was performed on all isolates.
Footnotes
Published ahead of print on 23 July 2008.
REFERENCES
- 1.Bannerman, T. L., G. A. Hancock, F. C. Tenover, and J. M. Miller. 1995. Pulsed-field gel electrophoresis as a replacement for bacteriophage typing of Staphylococcus aureus. J. Clin. Microbiol. 33551-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Enright, M. C., N. P. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 381008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Francois, P., A. Huyghe, Y. Charbonnier, M. Bento, S. Herzig, I. Topolski, B. Fleury, D. Lew, P. Vaudaux, S. Harbarth, W. van Leeuwen, A. van Belkum, D. S. Blanc, D. Pittet, and J. Schrenzel. 2005. Use of an automated multiple-locus, variable-number tandem repeat-based method for rapid and high-throughput genotyping of Staphylococcus aureus isolates. J. Clin. Microbiol. 433346-3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gilbert, F. B., A. Fromageau, L. Gélineau, and B. Poutrel. 2006. Differentiation of bovine Staphylococcus aureus isolates by use of polymorphic tandem repeat typing. Vet. Microbiol. 117297-303. [DOI] [PubMed] [Google Scholar]
- 5.Hardy, K. J., B. A. Oppenheim, S. Gossain, F. Gao, and P. M. Hawkey. 2006. Use of variations in staphylococcal interspersed repeat units for molecular typing of methicillin-resistant Staphylococcus aureus strains. J. Clin. Microbiol. 44271-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hardy, K. J., D. W. Ussery, B. A. Oppenheim, and P. M. Hawkey. 2004. Distribution and characterization of staphylococcal interspersed repeat units (SIRUs) and potential use for strain differentiation. Microbiology 1504045-4052. [DOI] [PubMed] [Google Scholar]
- 7.Harmsen, D., H. Claus, W. Witte, J. Rothganger, H. Claus, D. Turnwald, and U. Vogel. 2003. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J. Clin. Microbiol. 415442-5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lindstedt, B. A. 2005. Multiple-locus variable number tandem repeats analysis for genetic fingerprinting of pathogenic bacteria. Electrophoresis 262567-2582. [DOI] [PubMed] [Google Scholar]
- 9.Malachowa, N., A. Sabat, M. Gniadkowski, J. Krzyston-Russjan, J. Empel, J. Miedzobrodzki, K. Kosowska-Shick, P. C. Appelbaum, and W. Hryniewicz. 2005. Comparison of multiple-locus variable-number tandem-repeat assay analysis with pulsed-field gel electrophoresis, spa typing, and multilocus sequence typing for clonal characterization of Staphylococcus aureus isolates. J. Clin. Microbiol. 433095-3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sabat, A., J. Krzyszton-Russjan, W. Strzalka, R. Filipek, K. Kosowska, W. Hryniewicz, J. Travis, and J. Potempa. 2003. New method for typing Staphylococcus aureus strains: multiple-locus variable-number tandem repeat analysis of polymorphism and genetic relationships of clinical isolates. J. Clin. Microbiol. 411801-1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 332233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tenover, F. C., R. R. Vaughn, L. K. McDougal, G. E. Fosheim, and J. E. McGowan, Jr. 2007. Multiple-locus variable-number tandem-repeat assay analysis of methicillin-resistant Staphylococcus aureus strains. J. Clin. Microbiol. 452215-2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Belkum, A. 2007. Tracing isolates of bacterial species by multilocus variable number of tandem repeat analysis (MLVA). FEMS Immunol. Med. Microbiol. 4922-27. [DOI] [PubMed] [Google Scholar]