Abstract
The etiopathogenesis of the skin disease digital dermatitis (DD), an important cause of lameness in cattle, remains uncertain. Microscopically, the disease appears to be polymicrobial, with spirochetes as the predominant bacteria. The objective of this study was to identify the main part of the bacteria involved in DD lesions of cattle by using culture-independent molecular methods. Ten different phylotypes of Treponema were identified either by 16S rRNA gene sequencing of bacteria from DD lesions or by fluorescence in situ hybridization (FISH) analysis using phylotype-specific 16S rRNA-directed oligonucleotide probes. Two phylotypes, phylotype 1 (PT1) and PT2, were not closely related to any characterized treponemal species. PT7 was 99.3% identical to Treponema denticola, while PT9 resembled T. vincentii by 96%. The remaining phylotypes, PT3, PT4, PT5, PT6, and PT8, and Treponema brennaborense had previously been isolated from DD lesions. Forty DD biopsy specimens were examined for Treponema by FISH. With one exception, all of the biopsy specimens revealed epidermotropic, intermingled infection with three or more different phylotypes (mean, 4.7). The most prevalent species were PT1 (95%), PT6 (93%), and PT3 (85%). While colonization by PT3 was confined to the surface of the epidermis, both PT1 and PT6 invaded deep into the stratum spinosum and were seen in ulcerated dermal papillae. In two cases, all 10 phylotypes were demonstrated. Furthermore, FISH with a Treponema group-specific probe showed that Treponema accounted for more than 90% of the total bacterial population in the biopsy specimens. These data strongly suggest that a group of apparently symbiotic Treponema species are involved as primary bacterial pathogens in DD.
Digital dermatitis (DD), an inflammatory skin disease of cattle, is characterized by focal, circumscribed, and papillomatous lesions localized to the lower limbs. The DD lesion is acute to chronic, is painful at palpation, and causes lameness and wasting. The disease was first described to occur in Italy in 1974 (7) and has since been found in many parts of the world, where it now constitutes a significant welfare and production problem (23, 27). A bacterial etiology of DD is supported by the observation that cattle infected with DD respond to antibiotic treatment and that virus isolation from affected tissues has been unsuccessful (13). The disease appears to be polymicrobial; a variety of bacteria has been cultivated from DD lesions (19, 32), and although spirochetes seem to be the predominating group, various rods and cocci can also be recognized in microscopic sections of DD specimens (10, 39). A recently discovered species, Guggenheimella bovis, has been suggested as a potential pathogen but has until now been isolated only from a few DD-infected animals (30, 35). Still, the high number of invasive spirochetes observed suggests an active contribution of this group to DD pathogenesis (8, 9, 12, 16, 24, 35). This hypothesis is further substantiated by the fact that serum samples from cattle infected with DD contain elevated levels of antibody to Treponema antigens (13, 25, 36). Also, it has been demonstrated that four Treponema strains previously isolated from DD-infected cows were able to induce abscess formation in mice (18).
Similarities between human periodontitis and DD in cattle have been observed, as both are tissue-destructive diseases with a multibacterial etiology where spirochetes appear to be the predominant species (2, 17). In human periodontal disease, however, at least 60 different, mostly not yet cultivated, phylotypes of spirochetes have been identified (26). For some species of Treponema, most notably Treponema denticola, an active role in disease pathogenesis has been documented (33).
The task of clarifying the etiology of DD is seriously hampered by the fact that spirochetes are notoriously difficult to cultivate (15). So far, only a limited number of treponemal species from DD lesions have been cultivated in vitro (11, 14, 19, 36, 37), and only one, Treponema brennaborense, has been formally classified (31). Consequently, culture-independent phylogenetic methods, such as comparative 16S rRNA gene sequence analysis and fluorescence in situ hybridization (FISH), are valuable tools in determining the diversity, phylogeny, prevalence, and spatial distribution of these unclassified DD spirochetes. Previous analyses of treponemal 16S rRNA gene sequences PCR amplified directly from DD lesions have identified at least five different phylotypes of bovine Treponema, two of which share a close phylogenetic relationship with the human oral treponemes T. denticola and T. vincentii, which are suspected to be involved in periodontitis (8, 9, 28). FISH experiments have revealed a stratification of a few phylotypes within the epidermis (24). Still, part of the bacterial population of DD infections remains unresolved.
The aim of this study was to identify the major part of the bacterial populations found in DD samples from Danish cattle, with a special emphasis on treponemes. This was achieved by comparative 16S rRNA gene analysis of 237 bacterial clones which were PCR amplified from DD biopsy specimens. Subsequently, by use of 16S rRNA oligonucleotide probes directed against newly identified as well as previously described treponemal phylotypes, FISH was applied to estimate the prevalence and spatial distribution of the bacterial phylotypes.
MATERIALS AND METHODS
Processing of tissue specimens.
From a slaughterhouse, 32 lower limbs of cattle (dairy cows as well as beef cattle) were collected on two different occasions (April and September), without any available clinical history, e.g., chronicity/acuteness of the lesions, use of antibiotic therapy, or age of the animals. The limbs exhibited different stages of DD. The limbs were washed with water, and lesions were photographed and recorded. A total of 41 lesions were identified and skin biopsy specimens sampled. The lesions were observed at dorsal (n = 15) or plantar (n = 26) localization in the proximal border of the interdigital space. For molecular analysis, one part of the biopsy specimens was immediately frozen and kept stored at −20°C. For histological examination, the tissues were fixed in 10% neutral buffered formalin, embedded in paraffin, sectioned 5 μm thick, and mounted on SuperFrost/plus slides (Menzel-Gläser, Braunschweig, Germany). All biopsy specimens were stained by hematoxylin and eosin (H&E) and evaluated histopathologically. Sections for in situ hybridization were deparaffinized in xylene and transferred to 100% alcohol before processing.
Classification of lesions.
Lesions were histopathologically classified as DD according to the method of Read and Walker (27) (with minor modifications) if they consisted of (i) a focal circumscribed acanthotic epidermis with or without parakeratotic papillomatous proliferation, (ii) the loss of the stratum granulosum, or (iii) the presence of an inflammatory infiltrate in the dermis. Furthermore, the lesions were graded according to the degree of keratinolysis in the stratum corneum and the stratum spinosum as (score of 1) focal, (score of 2) moderate, or (score of 3) extensive to diffuse and according to the inflammatory response in dermis as (score of 1) mild or absent, (score of 2) moderate, or (score of 3) severe. Table 1 provides an overview of the results.
TABLE 1.
Spatial distribution of treponemes
| Biopsy specimen no. | Limb no. and localization | Histo-pathology score (keratinolysis score/dermatitis score)a | Spatial distribution of treponemesb by FISH for phylotype or group (probe no.) |
Distribution (%) of Treponema group (probe no. 202) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PT1 (188) | PT6 (178) | PT3 (192) | PT5 (199) | PT2 (200) | PT7 (198) | PT4 (201) | T. brennaborense (179) | PT8 (189) | PT9 (214) | ||||
| 1 | 1, plantar | 2/2 | +++ | +++ | + | 90 | |||||||
| 2 | 2, dorsal | 3/3 | +++ | +++ | +++ | + | 90 | ||||||
| 3 | 3, plantar | 2/1 | +++ | +++ | +++ | + | 85 | ||||||
| 4 | 4, dorsal | 3/3 | +++ | +++ | +++ | + | ++ | 98 | |||||
| 5 | 5, dorsal | 3/2 | +++ | +++ | ++ | + | ++ | 85 | |||||
| 6 | 5, plantar | 2/1 | +++ | +++ | ++ | ++ | + | + | 85 | ||||
| 7 | 6, dorsal | 3/2 | +++ | +++ | ++ | 85 | |||||||
| 8 | 7, dorsal | 3/3 | +++ | +++ | ++ | 90 | |||||||
| 9 | 8, dorsal | 3/3 | +++ | +++ | ++ | + | 90 | ||||||
| 10 | 8, plantar | 1/1 | +++ | +++ | ++ | 85 | |||||||
| 11 | 9, dorsal | 3/3 | +++ | +++ | ++ | 90 | |||||||
| 12 | 9, plantar | 3/3 | +++ | +++ | ++ | 85 | |||||||
| 13 | 10, dorsal | 2/1 | +++ | +++ | ++ | ++ | 90 | ||||||
| 14 | 10, plantar | 3/3 | +++ | +++ | ++ | ++ | ++ | ++ | 85 | ||||
| 15 | 11, dorsal | 3/3 | +++ | +++ | ++ | 90 | |||||||
| 16 | 12, dorsal | 3/3 | +++ | +++ | ++ | 90 | |||||||
| 17 | 12, plantar | 1/1 | +++ | +++ | ++ | 90 | |||||||
| 18 | 13, dorsal | 3/3 | +++ | +++ | ++ | 90 | |||||||
| 19 | 14, dorsal | 1/3 | +++ | +++ | ++ | ++ | + | 90 | |||||
| 20 | 14, plantar | 2/2 | +++ | +++ | ++ | ++ | +++ | 90 | |||||
| 21 | 15, dorsal | 3/2 | +++ | +++ | ++ | ++ | +++ | 90 | |||||
| 22 | 15, plantar | 1/1 | +++ | +++ | ++ | ++ | +++ | ++ | 90 | ||||
| 23 | 16, dorsal | 3/3 | ++ | +++ | +++ | ++ | + | 90 | |||||
| 24 | 16, plantar | 1/1 | +++ | ++ | ++ | ++ | ++ | 90 | |||||
| 25 | 17, plantar | 3/3 | +++ | +++ | ++ | ++ | 90 | ||||||
| 26 | 18, plantar | 3/3 | +++ | +++ | +++ | +++ | + | +++ | 90 | ||||
| 27 | 19, dorsal | 2/2 | +++ | +++ | +++ | ++ | ++ | + | 90 | ||||
| 28 | 19, plantar | 3/2 | +++ | +++ | +++ | ++ | + | 90 | |||||
| 29 | 20, plantar | 3/3 | +++ | +++ | +++ | +++ | ++ | + | 90 | ||||
| 30 | 21, plantar | 1/1 | +++ | +++ | ++ | 90 | |||||||
| 31 | 22, plantar | 3/3c | 0 | ||||||||||
| 32 | 23, plantar | 1/3 | +++ | +++ | ++ | ++ | ++ | ++ | 90 | ||||
| 33 | 24, plantar | 1/1 | +++ | +++ | + | 85 | |||||||
| 34 | 25, plantar | 3/3 | +++ | +++ | +++ | + | ++ | ++ | 95 | ||||
| 35 | 26, plantar | 2/1 | +++ | +++ | ++ | +++ | 90 | ||||||
| 36 | 27, plantar | 3/2 | +++ | +++ | ++ | ++ | +++ | ++ | ++ | +++ | +++ | + | 90 |
| 37 | 28, plantar | 2/2 | +++ | +++ | +++ | +++ | +++ | + | 90 | ||||
| 38 | 29, plantar | 3/3 | +++ | +++ | +++ | +++ | + | +++ | +++ | +++ | +++ | ++ | 90 |
| 39 | 30, plantar | 3/2 | +++ | +++ | +++ | +++ | ++ | + | 90 | ||||
| 40 | 31, plantar | 1/1 | ++d | 100 | |||||||||
| 41 | 32, plantar | 3/3 | +++ | +++ | +++ | +++ | +++ | 85 | |||||
Pathology was scored as follows. Degree of keratinolysis: focal, 1; moderate, 2; extensive, 3. Degree of chronic perivascular dermatitis: mild or absent, 1; moderate, 2; severe, 3.
+, sparse (1 to 5% of the total number of bacteria) in the middle of the epidermis; ++, 5 to 10% of the total bacterial population; +++, 10 to 30% of the total bacterial population.
Gross lesions different from DD.
Superficial colonization, 100% of invasive bacteria.
DNA preparation.
DNA was extracted from nine randomly chosen biopsy specimens (specimens no. 4, 18, 24, 25, 27, 29, 32, 35, and 41). Samples of approximately 5 by 3 mm were crudely minced with a scalpel, each subsequently immersed in 400 μl lysis buffer and 40 μl of proteinase K provided with a DNeasy blood and tissue kit (Qiagen, Hilden, Germany), and incubated at 56°C for 16 h. The remaining part of the procedure was performed with the DNeasy blood and tissue kit according to the protocol provided by the manufacturer.
PCR amplification and cloning procedures.
The primers used for 16S rRNA gene amplification are listed in Table 2. The primer combinations 10FX/1509R and Trep-46F/1509R were used to generate DNA fragments of approximately 1.5 kb. PCR was carried out in a 50-μl reaction mix which contained 1× cloned Pfu polymerase reaction buffer (Stratagene, La Jolla, CA), 400 μM of each deoxynucleoside triphosphate (Amersham Biosciences, Piscataway, NJ), 0.4 μM of each primer, 2.5 U of cloned Pfu DNA polymerase (Stratagene), and 5 μl of template DNA. Thermal cycling using a T3 thermocycler (Biometra, Göttingen, Germany) was performed as follows: denaturation at 94°C for 4 min, followed by 30 cycles of denaturation at 94°C for 45 s, annealing at 55°C for 45 s, and extension at 72°C for 3 min, followed by a final elongation step of 20 min. The purity of the product was determined by electrophoresis in a 1.5% agarose gel. The PCR products from the different biopsy specimens were combined and then purified by using a MiniElute PCR purification kit (Qiagen, Hilden, Germany). The concentration and quality of the purified products were measured with an ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE).
TABLE 2.
Names and sequences of 16S rRNA-targeting oligonucleotide probes and 16S rRNA gene-targeting primers used in this study
| Primer/probe name (no.) | Sequence | Reference or source |
|---|---|---|
| Primers | ||
| Trep-46F | 5′-GTY TTA AGC ATG CAA GTC-3′ | 8 |
| 10FX | 5′-AGA GTT TGA TCC TGG CTN AG-3′ | 38 |
| 519F | 5′-CCA GCA GCC GCG GTA ATA C-3′ | 22 |
| 519R | 5′-GTA TTA CCG CGG CTG CTG G-3′ | 22 |
| 1054FX | 5′-CAT GGY YGT CGT CAG CTC GT-3′ | 38 |
| 1054RX | 5′-ACG AGC TGA CGA CRR CCA TG-3′ | 38 |
| 1509R | 5′-GTT ACC TTG TTA CGA CTT CAC-3′ | 21 |
| Oligonucleotide probes | ||
| S-D-eub-338-alexa (79) | 5′-GTC ATT CCA TCG AAA CAT A-3′ | 1 |
| S-S-Trep-HW-170 (178) | 5′-CAA GGC CGT AGC CTC CTT-3′ | This study |
| S-S-Trep-T16-432 (198) | 5′-CAT CTC ACA GGC ATT CCC-3′ | This study |
| S-S-Trep-DDK3-481 (189) | 5′-CCC TTA TTC ACA TGA TTA CCG T-3′ | 8 |
| S-S-Trep-I:B:C7-432 (199) | 5′-CAT CAG ATG AGC ATT CCC-3′ | This study |
| S-S-Trep-DDKL12-432 (201) | 5′-CAT CTC AAG GTC ATT CCC-3′ | This study |
| S-S-Trep-Dig3-432 (188) | 5′-CAT CCC AGT ATC ATT CCC-3′ | This study |
| S-S-Trep-Dig4-432 (192) | 5′-CAT CTC AGT GTC ATT CCC-3′ | This study |
| S-S-Trep-T5-432 (200) | 5′-CAT CTT TGC ATC ATT ACC-3′ | This study |
| S-S-T.brenna-133 (179) | 5′-CCT CAC AGC TCT CTA ACC TC-3′ | 31 |
| S-S-TrepGenus-725 (202) | 5′-CAG AAA CYC GCC TTC GCC-3′ | This study |
| S-S-Trep-DD7-T143-196 (214) | 5′-CTT TCC TTA CTA TCT CTT G-3′ | This study |
| S-S-F.necro-183-Cy3 (114) | 5′-GAT TCC TCC ATG CGA AAA-3′ | 5 |
Cloning was performed by use of a Zero Blunt TOPO PCR cloning kit for sequencing (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Clone libraries were created from 16S rRNA gene sequences amplified from extracted DNA. PCR amplicons, approximately 1.5 kb long, were ligated into the pCR4Blunt-TOPO plasmid vector and transformed into One Shot electrocompetent Escherichia coli cells supplied with the kit. The transformed cells were then plated onto Luria-Bertani agar plates supplemented with ampicillin (50 μg/ml) and incubated overnight at 37°C. Clones, randomly selected for further analysis, were placed into 50 μl of sterile water and boiled for 10 min. The size of inserts was determined by PCR with flanking vector primers T3 forward primer and T7 reverse primer, followed by electrophoresis on a 1.5% agarose gel.
Sequencing and phylogenetic analysis.
The PCR products were sequenced by cycle sequencing on an ABI 3130 genetic analyzer (Applied Biosystems, Foster City, CA) using a BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems) according to the manufacturer's instructions. The clone library was initially analyzed by partial sequencing of 237 randomly chosen clones. By use of the primer 519F, fragments of between 600 and 700 bases were obtained to determine the identity or approximate phylogeny. Almost full sequences were obtained for potential novel phylotypes (about 1.4 kb with vector primers and the primers listed in Table 2). Analysis and phylogenetic-tree construction were performed with BioNumerics, version 4.0 (Applied-Math, Sint-Martens-Latem, Belgium). For identification of the closest relatives, the sequences of the unidentified inserts were compared against sequences in GenBank (http://www.ncbi.nlm.nih.gov/BLAST/) and the Ribosomal Database Project (RDP) (http://rdp.cme.msu.edu/index.jsp). The software Pintail, version 1.0 (http://www.bioinformatics-toolkit.org/Pintail/), was used to identify chimeric sequences (3).
FISH.
The oligonucleotide probes used in this study are listed in Table 2. They were selected using the software ARB (http://www.arb-home.de) or the program Primrose, version 2.17 (http://www.bioinformatics-toolkit.org/Primrose/index.html) (4). The oligonucleotide probes were 5′ labeled with fluorescein isothiocyanate (FITC), Alexa Fluor 488, or the isothiocyanate derivative Cy3 (Eurofins MWG Operon, Ebersberg, Germany, or DNA Technology A/S, Risskov, Denmark). The hybridization was carried out at 45°C with 40 μl of hybridization buffer (100 mM Tris [pH 7.2], 0.9 M NaCl, 0.1% sodium dodecyl sulfate) and 200 ng of each probe for 16 h in a Sequenza slide rack (Thermo Shandon, Cheshire, United Kingdom). The sections were then washed three times in prewarmed (45°C) hybridization buffer for 15 min and subsequently three times in prewarmed (45°C) washing solution (100 mM Tris [pH 7.2], 0.9 M NaCl). The sections were rinsed in water, air dried, and mounted in Vectashield (Vector Laboratories Inc., Burlingame, CA) for epifluorescence microscopy.
An Axioimager M1 epifluorescence microscope equipped for epifluorescence with a 100-W HBO lamp and filter sets 43 and 38 were used to visualize Cy3 and Alexa Fluor 488, respectively. Images were obtained using an AxioCam MRm version 3 FireWiremonocrome camera and AxioVision software, version 4.5 (Carl Zeiss, Oberkochen, Germany).
Nucleotide sequence accession numbers.
The 16S rRNA gene sequences determined in this study are available from the EMBL/GenBank/DDBJ sequence databases under accession numbers AM942445 for PT1, AM942446 for PT2, AM942447 for PT3, AM942448 for PT4, AM942449 for PT5, AM942450 for PT6, AM942451 for PT7, AM980447 for PT8, and AM980448 for PT9.
RESULTS
Classification of lesions.
Histopathology revealed papillomatous proliferation of the stratum spinosum (acanthosis) with severely elongated rete ridge formation and long dermal papillae in all cases. Forty biopsy specimens displayed typical DD lesions characterized by focal, circumscribed (1 to 3 cm in diameter) plaques/warts of variable thickness and parakeratotic papillomatous proliferation. The localization (dorsal or plantar on the foot) and the pathology score of the individual lesions are listed in Table 1. Seven lesions were scored 1 in both keratinolysis of the stratum corneum/stratum spinosum and cellular inflammatory reaction in the dermis, and 17 lesions were scored 3 in both keratinolysis and dermatitis, while the remaining 16 lesions were scored in between. The diversity of gross and microscopic appearances among the DD lesions is illustrated in Fig. 1.
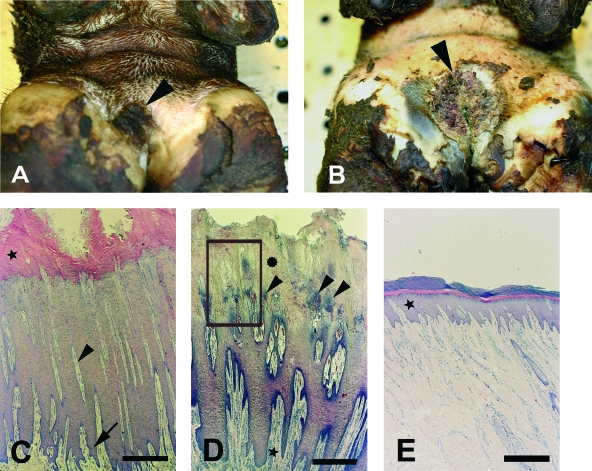
FIG. 1.
Gross histopathology of bovine DD in the interdigital area. (A) Biopsy specimen no. 40, showing circumscribed, focal, moist plaque with increased thickness of epidermis (arrowhead). (B) Biopsy specimen no. 41, showing circumscribed, ulcerated lesion with papillomatous proliferations (arrowhead). (C) Cross-section of biopsy specimen no. 30, revealing severe epidermal hyperplasia of the stratum spinosum with long, slender dermal papillae (arrowhead) and deep epidermal (arrow) and orthokeratotic (★) hyperkeratosis. Staining was done with H&E. Bar = 0.5 mm. (D) Cross-section of biopsy specimen no. 38, revealing severe epidermal hyperplasia of the stratum spinosum with bacterial colonization and exudation from dermal papillae (arrowhead). Keratinolysis of stratum corneum ( ) and perivascular dermatitis (★) are also shown. Staining was done with H&E the boxed area represents an area similar to that shown in Fig. 3). Bar = 0.5 mm. (E) Skin from the interdigital area, with a normally appearing epidermis (★). Staining was done with H&E. Bar = 0.5 mm.
A thickened epidermis with orthokeratotic hyperkeratosis of an otherwise intact stratum corneum and severe acanthosis, as illustrated in Fig. 1C, revealed only minimal bacterial colonization, whereas foci or larger areas with variable keratinolysis of the stratum corneum and stratum spinosum usually displayed extensive bacterial colonization as a weak bluish stain by H&E (Fig. 1D). In addition, erosion and ulceration of dermal papillae were correlated with the keratinolysis while the degree of cellular proliferation in the stratum spinosum appeared to be independent of the keratinolysis. The skin surrounding the DD lesions appeared normal, as shown in Fig. 1E.
In the single biopsy specimen (no. 31) which was not consistent with DD, a thick crust was observed diffusely in the proximal border of the plantar interdigital space. Histopathologically, the acanthotic and parakeratotic epidermis showed diffuse vacuolation of keratinocytes.
Sequence and phylogenetic analyses of 16S rRNA gene clone library.
Clone libraries were constructed by combining DNA from nine different DD infection biopsy specimens and using two different sets of primers, a general set and a Treponema-specific set. Fragments of approximately 1.5 kb from bacterial isolates were PCR amplified and cloned. From the resulting clone libraries, a total of 237 randomly chosen bacterial clones were partially sequenced (600 to 700 bases) to identify the predominant phylogenetic groups. Comparative sequence analysis revealed a number of different phylogenetic clades. Of the 237 clones, 122 were spirochetes, all belonging to the Treponema group. Almost 50% of the remaining clones were identified as Fusobacterium necrophorum (99% sequence identity). Also represented were Streptococcus dysgalactiae (99% sequence identity), Pasteurella sp. (95% sequence identity), and Klebsiella oxytoca (99% sequence identity).
Nearly complete 16S rRNA gene sequences (corresponding to positions 50 to 1485 in the E. coli 16S rRNA gene) were determined for 25 clones. These clones were primarily spirochetes representing putative new phylotypes. Six of the sequences were identified as chimeric by the program Pintail and were consequently excluded from further analysis.
Assuming that sequences more than 98% identical are considered to represent a single “phylotype” likely to be derived from a single species (34), nine different phylotypes of Treponema could be identified from the clonal library in this investigation. The phylotypes and the names of their closest cultivable relatives in GenBank are listed in Table 3. The sequences were aligned and trimmed to the length of the smallest sequence from GenBank. A phylogenetic tree based upon the comparison of 541 bases of the 16S rRNA gene sequences is presented in Fig. 2. The tree was constructed using the Jukes and Cantor correction and the neighbor-joining method (20, 29).
TABLE 3.
Phylotypes of Treponema spp. identified from clone library
| Clone no. (probe no.) | Name and GenBank accession no. of closest known cultivable relative (maximum percent identity) | No. of clones isolated (n = 122) |
|---|---|---|
| PT1 (188) | Treponema refringensAF426101 (93) | 34 |
| PT2 (200) | Treponema refringensAF426101 (89) | 35 |
| PT3 (192)/clone DDKL-20 | Treponema refringensAF426101 (94) | 1 |
| PT4 (201)/clone DDKL-12 | Treponema refringensAF426101 (92) | 5 |
| PT5 (199)/clone DDKL-13 | Treponema vincentiiAF033310 (98.6) | 6 |
| PT6 (178)/clone DDKL-4 | Treponema phagedenisM57739 (98) | 25 |
| PT7 (198) | Treponema denticolaAE017226 (99.3) | 12 |
| PT8 (189)/clone DDKL-3 | Treponema denticolaAE017226 (95) | 3 |
| PT9 (214) | Treponema vincentiiAY369250 (96) | 1 |
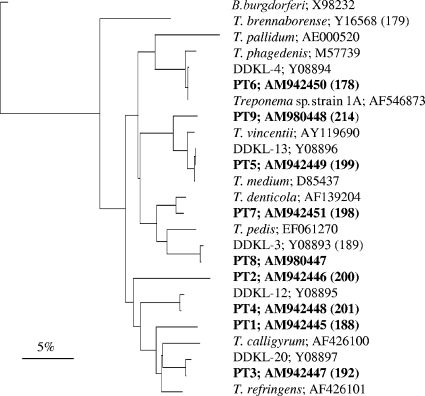
FIG. 2.
Phylogenetic tree based on 16S rRNA gene sequence comparison over 541 aligned bases between the Treponema phylotypes isolated from DD lesions in this (marked in bold) and previous studies and cultivable strains of Treponema. Borrelia burgdorferi was included as an outgroup. FISH probe numbers (in parentheses) and GenBank accession numbers are shown. The tree was constructed by the neighbor-joining method, and the similarity matrices were corrected for base changes using the Jukes and Cantor correction. The scale bar represents a 5% difference in nucleotide sequences.
Two phylotypes, PT1 and PT2, had no close relatives among previously identified treponemes. In both cases, the closest match in GenBank was with T. refringens, with sequence identities of 93% and 89%, respectively. The seven remaining 16S rRNA gene sequences shared close similarity to known species or phylotypes. The clones PT3, PT4, PT5, PT6, and PT8 were nearly identical (more than 99% similarity) to the phylotypes DDKL-20, DDKL-12, DDKL-13, DDKL-4, and DDKL-3, respectively, all described in a similar study (8). Mostly, the differences between these corresponding sequences were due to ambiguous nucleotides in the DDKL sequences. With respect to treponemes isolated from humans, PT5 and PT9 were both related to the oral treponeme T. vincentii, by 98.6% and 96% sequence similarity, respectively. PT6 exhibited 98% similarity with T. phagedenis, a normal commensal of the human urogenital tract. Finally, PT7 was nearly identical to T. denticola (99.3% sequence identity) while PT8 shared 95% sequence similarity with T. denticola.
FISH analysis of DD lesions.
In order to perform direct identification of the different treponemes, specific 16S rRNA-targeting fluorescence-labeled oligonucleotide probes were designed for the phylotypes identified from the 16S rRNA gene clone library. Also included in the FISH analysis were probes targeting T. brennaborense (31), F. necrophorum (5), the domain Bacteria (1), and finally a Treponema probe designed to detect all of the phylotypes found in the DD lesions (except for T. brennaborense). In addition, this probe covered a number of other Treponema species: T. refringens, T. calligyrum, T. denticola, T. vincentii, T. putidum, and T. pallidum. The names and sequences of the probes are listed in Table 2. By using two different fluorochrome-labeled probes simultaneously, double staining of bacteria was feasible.
Double in situ hybridization with the general probe for bacteria and the Treponema group probe revealed treponemes in 40 DD biopsy specimens. A massive and deep colonization of the epidermis by treponemes was common, as shown in Fig. 3A. The only biopsy specimen (no. 31) without DD, however, was completely negative for Treponema. This biopsy specimen was severely infected by bacteria morphologically different from spirochetes (various rods and cocci).
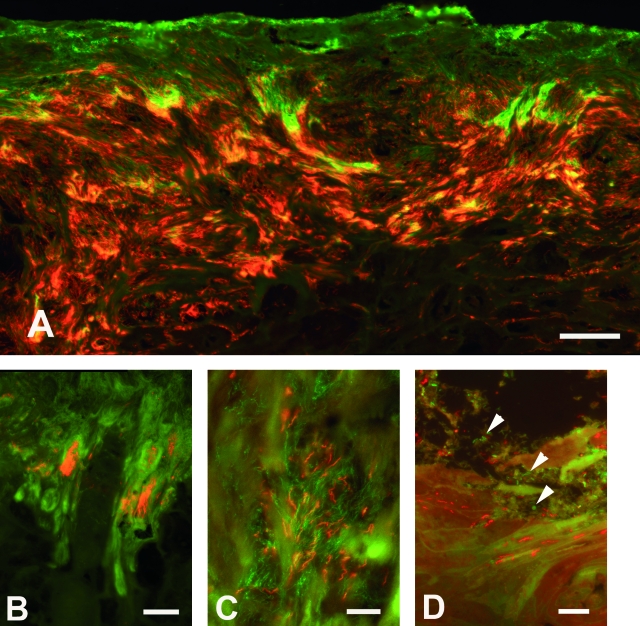
FIG. 3.
Demonstration of treponemas and other bacteria in DD lesions by FISH. (A) Massive and deep colonization of the epidermis by treponemas (red/orange) targeted by the Treponema group probe (no. 202, Cy3); other bacteria appear green (probe no. 79, FITC). Double in situ hybridization, biopsy specimen no. 36. Bar = 50 μm. (B) PT6 treponemas (red/orange) constituting approximately 10% of the total number of bacteria (green) colonizing the epidermis. Double in situ hybridization with the probes no. 178 (Cy3) and no. 79 (FITC), biopsy specimen no. 14. Bar = 75 μm. (C) Demonstration of two different treponemas intermingled deep in the stratum spinosum. PT1 (probe no. 188) labeled with Cy3 and PT6 (probe no. 178) labeled with Alexa Fluor 488, biopsy specimen no. 35. Bar = 15 μm. (D) Treponemas (PT3, red) are seen infiltrating the superficial layers of the stratum corneum, while other bacteria (green) (arrowheads) are seen on the surface. Double in situ hybridization with probes no. 192 (Cy3) and no. 79 (Alexa Fluor 488), biopsy specimen no. 40. Bar = 15 μm.
The results of FISH with the 10 phylotype/species-specific probes in the biopsy specimens are listed in Table 1 together with the pathology scores. The prevalence of the 10 phylotypes of Treponema found in the DD lesions is illustrated in Fig. 4. In general, the within-specimen prevalence and spatial distribution of the different treponemes showed three colonization patterns: (i) a sparse colonization of the middle part of the epidermis, contributing 1 to 5% of the bacterial population, (ii) predominant colonization of the surface and superficial layer of epidermis, usually contributing only 5 to 10% of the bacterial population, or (iii) a diffuse colonization of the entire thickness of the keratinolysed epidermis with the exception of the superficial layer and the surface. In addition, the treponemes infiltrated the dermis at the tip of the dermal papillae and usually constituted 10 to 30% of the total bacterial population, as illustrated in Fig. 3B. An estimation of the prevalence of the various phylotypes was based on a visual inspection of simultaneous FISH with the specific probes and the general bacterial probe.
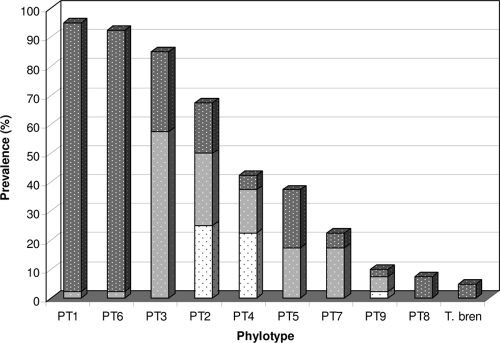
FIG. 4.
Prevalence of Treponema phylotypes in FISH analysis of 40 biopsy specimens from 32 Danish cows. The shading of the bars represents the contribution of each phylotype to the total bacterial population: dark gray signifies 10 to 30%, light gray 5 to 10%, and white 1 to 5%. T. bren, T. brennaborense.
FISH analysis of the 40 DD samples, using phylotype-specific 16S rRNA-targeting probes, demonstrated that PT1, PT6, and PT3 were the most frequently detected phylotypes in the DD lesions. PT1 and PT6 were observed in at least 93% of the samples and were found intermingled in the deep parts of the epidermis (Fig. 3C), both with high prevalence, each representing approximately 10 to 30% of the total bacterial population. PT3 was identified in 85% of the samples. This bacterium accounted for 5 to 10% of the total bacterial population and had a more superficial growth than the two phylotypes mentioned above, including colonization of the surface (Fig. 3D). In biopsy specimen no. 40, with a minimal pathology score (1/1), PT3 was the only Treponema phylotype present, representing 100% of the invasive bacteria identified, and its pattern was a sparse colonization of the surface. The treponemes, however, were seen invading only small, superficial epidermal clefts surrounded by slightly lysed keratinocytes.
PT2 and PT4 were present in 68% and 43% of the samples, respectively, both with a low prevalence (approximately 1 to 5%) and a scattered distribution in the middle of the lesions. PT5 was observed in 38% of the samples. This bacterium also displayed a superficial growth pattern. PT9 was present in various concentrations in approximately 10% of the samples. PT8 and T. brennaborense were seen in less than 10% of the biopsy specimens, but when present, both were found in large numbers (10 to 30% of the total number of Treponema bacteria) and deep within the epidermis. PT7, which shares 99.3% sequence identity with T. denticola, was found in moderate numbers (mostly 5 to 10% of the total bacterial population) in 23% of the DD tissue sections.
Colonizing the epidermis, the treponemes were found in the stratum spinosum with variable keratinolysis, e.g., they were seen in the intercellular space between degenerated keratinocytes as well as intracellularly in lysed cells. With the exception of biopsy specimen 31 and the seven biopsy specimens that scored 1/1 in pathology, treponemes were observed infiltrating the top of the dermal papillae. Similarly to the pattern observed in the stratum spinosum, the spirochetes were seen side by side tangentially to the surface. An indication of increased dermatropic growth of treponemes was not observed.
In general, the Treponema phylotypes were estimated to account for at least 90% of the total number of bacteria found in the DD lesions. Of the remaining 10% of the bacteria, a minor number had a spirochete-like morphology but were not caught by the Treponema group or T. brennaborense probes. The majority of the remaining bacteria consisted of various rods and cocci located in the upper and middle parts of the keratinolysed epidermis. Among these bacteria, F. necrophorum was observed in 22% of the biopsy specimens, usually situated in the superficial parts of the keratinolysed epidermis.
DISCUSSION
In this study, multiple different treponemes were identified from biopsy specimens taken above the interdigital clefts of cattle with variable stages of DD. By FISH, 1 to 10 different phylotypes (mean, 4.7) were identified and estimated to constitute more than 90% of the total number of bacteria in the lesions. Contrary to previous work (8, 24), it was here demonstrated that it is possible to perform FISH analysis in formalin-fixed sections with a satisfactory result by using the fluorescent markers Cy3 and Alexa Fluor 488 for probe labeling (Fig. 3).
Of the 10 phylotypes isolated from the DD lesions, 9 were found by comparative 16S rRNA gene sequence analysis, 5 of which, PT3, PT4, PT5, PT6, and PT8, had all previously been isolated from DD lesions and 3 of which, PT3, PT5, and PT6, have recently been cultivated (8, 19). With a sequence identity of 98% or more, PT5 and PT6 clustered with the species T. vincentii and T. phagedenis, respectively. T. phagedenis is a nonpathogenic human treponeme, while T. vincentii is commonly found in human periodontitis lesions (8). Four phylotypes, PT1, PT2, PT7, and PT9, had not previously been identified in DD lesions. The two first mentioned are not closely related to any cultivable species, while PT7 is identical to the periodontal pathogen T. denticola (sequence similarity of 99.3%) and PT9 shares 96% sequence similarity with T. vincentii. Also identified in two of the DD lesions by FISH analysis was T. brennaborense (31).
Compared to cultivable species of Treponema, the 10 phylotypes cluster in different phylogenetic groups (Fig. 2). One phylogenetic grouping is rather interesting, as it includes, besides the previously described PT3 and PT4 (8), the two new phylotypes PT1 and PT2. The closest known relatives of these new phylotypes are the cultivable, nonpathogenic human isolates T. refringens and T. calligyrum, which are both part of the normal flora of male and female genitals of humans and animals (6). What makes this group particularly interesting is the fact that the phylotypes belonging to this cluster are found in relatively high numbers in the DD lesions.
The two phylotypes with the highest prevalence and greatest spatial distribution were PT1, identified in this study, and PT6, previously identified in German cattle (8). These bacteria were found to constitute the frontier of invasive spirochetes in the deepest parts of the epidermis and in the dermis (Fig. 3C and Fig. 4). This location of PT6 has previously been observed by FISH analysis (24).
The third to fifth most prevalent phylotypes, PT3, PT2, and PT4, also belonged to the “T. refringens cluster” (Fig. 2). PT3 was almost as prevalent as the two phylotypes mentioned above, but this bacterium was in general found in the superficial parts of the lesion and constituted in more than 50% of the biopsy specimens only 5 to 10% of the total bacterial population. Whereas most of the Treponema species identified in this study appeared to be in bundles, PT2 and PT4 both displayed a scattered distribution in the middle parts of the lesion.
The two phylotypes PT5 and PT7, which are closely related to the human periodontal pathogens T. vincentii and T. denticola, respectively (Fig. 2), were both found in relatively low numbers in the superficial part of the DD lesions. For PT7, this result is in concordance with previous observations (24), where T. denticola was found only in detritus and superficial layers of the stratum spinosum. In the same investigation, however, a probe (TRE I) directed against PT5 showed this bacterium to be located deep in the stratum spinosum.
The three Treponema species with the lowest prevalence in biopsy specimens were PT9, PT8, and T. brennaborense. The two last mentioned were present in less than 10% of the biopsy specimens, but when present both were observed deep within the tissue and in large numbers constituting from 10 to 30% of the total bacterial population. In this study, PT8 and T. brennaborense were found in biopsy specimens with a high treponemal biodiversity, i.e., samples with between 6 and 10 different phylotypes were present (Table 1). Consequently, one may speculate that they constitute part of a secondary invasion in cases of advanced disease progression.
In a previous FISH analysis on smears of material from DD lesions (8), PT8 was the most frequently found phylotype, whereas the other four phylotypes identified were present only in small numbers. This contrasting result may be explained by the fact that the FISH analyses were performed on a relatively small sample size (four biopsy specimens) that may have been taken from lesions with advanced disease progression. A support for this view is the fact that the results of the present investigation are more in accordance with those of Moter et al. (24), who also performed FISH analysis on tissue sections.
Fusobacterium necrophorum was seen in 22% of the Danish biopsy specimens but always in the superficial keratinolysed layers of the epidermis and consequently represents a secondary invader. The fact that almost 50% of the clones sequenced from the clone library were identified as F. necrophorum suggests that DNA from the superficial parts of the DD biopsy specimens was overrepresented in the DNA extractions.
The study revealed the consistent presence of at least one of the three phylotypes PT1, PT6, and PT3 in the affected tissue. In fact, in 90% of the biopsy specimens, PT1 and PT6 were present simultaneously, and all three phylotypes were coidentified in 78% of the samples. Furthermore, in 38% of the biopsy specimens with the highest pathology score (3/3) only these three phylotypes were demonstrated. Moreover, an increased number of phylotypes present did not seem to aggravate the lesion, since the mean pathology score for the 11 biopsy specimens with six or more phylotypes was not higher than the overall pathology score (4.5 versus 4.6). The possibility that strains of Treponema differ in pathogenicity has further been substantiated by a study applying a mouse abscess model. In this study, four T. phagedenis-like spirochete strains showed evidence of possessing different pathogenic potentials (18). The observations by Elliott et al. are also in accordance with the fact that PT6, which is closely related to T. phagedenis, appears to be one of the most prevalent species in the Danish DD biopsy specimens. Furthermore, T. phagedenis-like spirochetes, residing deep within DD lesions, appear to facilitate DD lesion development by impairing various bovine macrophage functions, such as innate immunity and wound repair (40).
One possible hypothesis could be that a certain symbiosis or synergism exists between the most prevalent phylotypes and that coinfection is necessary for survival and for initiation of keratinolysis of the epidermis. Alternatively, the most prevalent phylotypes could each be capable of producing infection on their own or with other less frequent phylotypes. The observations from two biopsy specimens, no. 25 and no. 40 (Table 1), might be supportive of this second hypothesis. In biopsy specimen no. 25, which is in an advanced state of infection (histopathology score of 3/3), only PT6 of the three most prevalent phylotypes is present, along with three other less prevalent types. However, PT1 and PT3 might have been present at an earlier state of infection. In biopsy specimen no. 40, which appears to be in an early state of infection (histopathology score of 1/1), only one phylotype, PT3, was observed. Still, one might speculate that this bacterium might have been followed by others at a later state of infection or that the infection might never have developed seriously.
Pathogenetically, the marked damage of keratinocytes is believed to induce significant epidermal proliferation in DD (17). The seven lesions with only minimal keratinolytic damage and dermatitis (score of 1/1 in pathology), however, revealed a proliferation of the stratum spinosum similar to that of the other lesions with a higher pathology score, suggesting that the proliferation, possibly induced by toxins released by the surface-colonizing treponemes or other bacteria, precedes the keratinolytic damages.
Simultaneous hybridization with the probe for the Treponema group and the probe for the domain Bacteria showed that the majority of the bacteria in the lesions (90 to 95%) stained positive with the probe targeting most of the Treponema group. Deep in the stratum spinosum and the dermal papillae, only spirochetal morphotypes could be observed, providing more convincing evidence that treponemes are actively involved as primary pathogens.
As mentioned in the introduction, both DD and periodontal disease are considered mixed bacterial infections, with Treponema species among the most predominant organisms detected from the lesions. In periodontal disease, T. denticola is considered a key organism together with other proteolytic gram-negative bacteria, especially Porphyromonas gingivalis (33). In DD lesions, Treponema species are also suspected to be strongly implicated in disease progression, along with other bacterial morphotypes, including bacilli and cocci, which have been observed scattered between the spirochetes (10, 39). Until now, however, no specific species or phylotypes have been identified as the major pathogens in DD.
In this study, a number of possible key pathogenic phylotypes of treponemes were identified, including the novel PT1. Furthermore, in the Danish DD biopsy specimens, bacteria other than spirochetes were observed only in the debris covering the ulcer and in the superficial layers of the lesions, while spirochetal morphotypes appeared as the sole invaders of the deeper layers of the epidermis. As the biopsy specimens in this study originated from a slaughterhouse, no history of the animals was available. Future studies of DD may include within-herd prevalence of the different Treponema phylotypes as well as investigation of the treponemes present in the early stages of DD lesions and, if possible, a time course study of lesion development.
Interestingly, the four phylotypes which were closely related to the human periodontal pathogens T. denticola and T. vincentii appeared to be of less importance in the disease progression of DD. Meanwhile, PT6, which is almost identical to the human commensal T. phagedenis, was found in large numbers in 90% of the Danish DD biopsy specimens and is here considered a possible key pathogenic candidate. So, while it appears that species of human treponemes are able to infect cattle, some species-specific pathogenicity is indicated by this result. It could be interesting to investigate whether some of the other bovine phylotypes observed in the DD lesions in this and previous studies are also present in human periodontal lesions.
Acknowledgments
We thank Annie Ravn Pedersen and Joanna Zeitman Amenuvor for excellent technical assistance.
This work was supported by a grant from the Danish Dairy Board.
Footnotes
Published ahead of print on 18 June 2008.
REFERENCES
- 1.Amann, R. I., L. Krumholz, and D. A. Stahl. 1990. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J. Bacteriol. 172:762-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armitage, G. C., W. R. Dickinson, R. S. Jenderseck, S. M. Levine, and D. W. Chambers. 1982. Relationship between the percentage of subgingival spirochetes and the severity of periodontal disease. J. Periodontol. 53:550-556. [DOI] [PubMed] [Google Scholar]
- 3.Ashelford, K. E., N. A. Chuzhanova, J. C. Fry, A. J. Jones, and A. J. Weightman. 2005. At least 1 in 20 16S rRNA sequence records currently held in public repositories is estimated to contain substantial anomalies. Appl. Environ. Microbiol. 71:7724-7736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ashelford, K. E., A. J. Weightman, and J. C. Fry. 2002. PRIMROSE: a computer program for generating and estimating the phylogenetic range of 16S rRNA oligonucleotide probes and primers in conjunction with the RDP-II database. Nucleic Acids Res. 30:3481-3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boye, M., B. Aalbaek, and J. S. Agerholm. 2006. Fusobacterium necrophorum determined as abortifacient in sheep by laser capture microdissection and fluorescence in situ hybridization. Mol. Cell. Probes 20:330-336. [DOI] [PubMed] [Google Scholar]
- 6.Canale-Parola, E. 1984. The spirochetes, p. 38-70. In N. R. Krieg and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, 1st ed., vol. 1. Williams & Wilkins, Baltimore, MD. [Google Scholar]
- 7.Cheli, R., and C. Mortellaro. 1974. La dermatite digitale del bovino, p. 208-213. In P. Gallarati (ed.), Proceedings of the 8th International Conference on Diseases of Cattle. Piacenza, Milan, Italy.
- 8.Choi, B. K., H. Nattermann, S. Grund, W. Haider, and U. B. Göbel. 1997. Spirochetes from digital dermatitis lesions in cattle are closely related to treponemes associated with human periodontitis. Int. J. Syst. Bacteriol. 47:175-181. [DOI] [PubMed] [Google Scholar]
- 9.Collighan, R. J., and M. J. Woodward. 1997. Spirochaetes and other bacterial species associated with bovine digital dermatitis. FEMS Microbiol. Lett. 156:37-41. [DOI] [PubMed] [Google Scholar]
- 10.Cruz, C. E., C. A. Pescador, Y. Nakajima, and D. Driemeier. 2005. Immunopathological investigations on bovine digital epidermitis. Vet. Rec. 157:834-840. [DOI] [PubMed] [Google Scholar]
- 11.Demirkan, I., S. D. Carter, C. A. Hart, and M. J. Woodward. 1999. Isolation and cultivation of a spirochaete from bovine digital dermatitis. Vet. Rec. 145:497-498. [DOI] [PubMed] [Google Scholar]
- 12.Demirkan, I., S. D. Carter, R. D. Murray, R. W. Blowey, and M. J. Woodward. 1998. The frequent detection of a treponeme in bovine digital dermatitis by immunocytochemistry and polymerase chain reaction. Vet. Microbiol. 60:285-292. [DOI] [PubMed] [Google Scholar]
- 13.Demirkan, I., R. L. Walker, R. D. Murray, R. W. Blowey, and S. D. Carter. 1999. Serological evidence of spirochaetal infections associated with digital dermatitis in dairy cattle. Vet. J. 157:69-77. [DOI] [PubMed] [Google Scholar]
- 14.Demirkan, I., H. F. Williams, A. Dhawi, S. D. Carter, C. Winstanley, K. D. Bruce, and C. A. Hart. 2006. Characterization of a spirochaete isolated from a case of bovine digital dermatitis. J. Appl. Microbiol. 101:948-955. [DOI] [PubMed] [Google Scholar]
- 15.Dewhirst, F. E., M. A. Tamer, R. E. Ericson, C. N. Lau, V. A. Levanos, S. K. Boches, J. L. Galvin, and B. J. Paster. 2000. The diversity of periodontal spirochetes by 16S rRNA analysis. Oral Microbiol. Immunol. 15:196-202. [DOI] [PubMed] [Google Scholar]
- 16.Döpfer, D., A. Koopmans, F. A. Meijer, I. Szakall, Y. H. Schukken, W. Klee, R. B. Bosma, J. L. Cornelisse, A. J. van Asten, and A. A. ter Huurne. 1997. Histological and bacteriological evaluation of digital dermatitis in cattle, with special reference to spirochaetes and Campylobacter faecalis. Vet. Rec. 140:620-623. [DOI] [PubMed] [Google Scholar]
- 17.Edwards, A. M., D. Dymock, and H. F. Jenkinson. 2003. From tooth to hoof: treponemes in tissue-destructive diseases. J. Appl. Microbiol. 94:767-780. [DOI] [PubMed] [Google Scholar]
- 18.Elliott, M. K., D. P. Alt, and R. L. Zuerner. 2007. Lesion formation and antibody response induced by papillomatous digital dermatitis-associated spirochetes in a murine abscess model. Infect. Immun. 75:4400-4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Evans, N. J., J. M. Brown, I. Demirkan, R. D. Murray, W. D. Vink, R. W. Blowey, C. A. Hart, and S. D. Carter. 2008. Three unique groups of spirochetes isolated from digital dermatitis lesions in UK cattle. Vet. Microbiol. 130:141-150. [DOI] [PubMed] [Google Scholar]
- 20.Jukes, T. H., and C. R. Cantor. 1969. Evolution of protein molecules, p. 21-132. In H. N. Munroe (ed.), Mammalian protein metabolism. Academic Press, New York, NY.
- 21.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. Wiley & Sons, Chichester, United Kingdom.
- 22.Lane, D. J., B. Pace, G. J. Olsen, D. A. Stahl, M. L. Sogin, and N. R. Pace. 1985. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc. Natl. Acad. Sci. USA 82:6955-6959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Losinger, W. C. 2006. Economic impacts of reduced milk production associated with papillomatous digital dermatitis in dairy cows in the USA. J. Dairy Res. 73:244-256. [DOI] [PubMed] [Google Scholar]
- 24.Moter, A., G. Leist, R. Rudolph, K. Schrank, B. K. Choi, M. Wagner, and U. B. Göbel. 1998. Fluorescence in situ hybridization shows spatial distribution of as yet uncultured treponemes in biopsies from digital dermatitis lesions. Microbiology 144:2459-2467. [DOI] [PubMed] [Google Scholar]
- 25.Murray, R. D., D. Y. Downham, I. Demirkan, and S. D. Carter. 2002. Some relationships between spirochaete infections and digital dermatitis in four UK dairy herds. Res. Vet. Sci. 73:223-230. [DOI] [PubMed] [Google Scholar]
- 26.Paster, B. J., S. K. Boches, J. L. Galvin, R. E. Ericson, C. N. Lau, V. A. Levanos, A. Sahasrabudhe, and F. E. Dewhirst. 2001. Bacterial diversity in human subgingival plaque. J. Bacteriol. 183:3770-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Read, D. H., and R. L. Walker. 1998. Papillomatous digital dermatitis (footwarts) in California dairy cattle: clinical and gross pathologic findings. J. Vet. Diagn. Investig. 10:67-76. [DOI] [PubMed] [Google Scholar]
- 28.Rijpkema, S. G., G. P. David, S. L. Hughes, and M. J. Woodward. 1997. Partial identification of spirochaetes from two dairy cows with digital dermatitis by polymerase chain reaction analysis of the 16S ribosomal RNA gene. Vet. Rec. 140:257-259. [DOI] [PubMed] [Google Scholar]
- 29.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 30.Schlafer, S., M. Nordhoff, C. Wyss, S. Strub, J. Hubner, D. M. Gescher, A. Petrich, U. B. Göbel, and A. Moter. 2008. Involvement of Guggenheimella bovis in digital dermatitis lesions of dairy cows. Vet. Microbiol. 128:118-125. [DOI] [PubMed] [Google Scholar]
- 31.Schrank, K., B. K. Choi, S. Grund, A. Moter, K. Heuner, H. Nattermann, and U. B. Göbel. 1999. Treponema brennaborense sp. nov., a novel spirochaete isolated from a dairy cow suffering from digital dermatitis. Int. J. Syst. Bacteriol. 49:43-50. [DOI] [PubMed] [Google Scholar]
- 32.Schroeder, C. M., K. W. Parlor, T. L. Marsh, N. K. Ames, A. K. Goeman, and R. D. Walker. 2003. Characterization of the predominant anaerobic bacterium recovered from digital dermatitis lesions in three Michigan dairy cows. Anaerobe 9:151-155. [DOI] [PubMed] [Google Scholar]
- 33.Sela, M. N. 2001. Role of Treponema denticola in periodontal diseases. Crit. Rev. Oral Biol. Med. 12:399-413. [DOI] [PubMed] [Google Scholar]
- 34.Stephens, C. 2000. Microbiology: intimate strangers. Curr. Biol. 10:272-275. [DOI] [PubMed] [Google Scholar]
- 35.Strub, S., J. R. van der Ploeg, K. Nuss, C. Wyss, A. Luginbühl, and A. Steiner. 2007. Quantitation of Guggenheimella bovis and treponemes in bovine tissues related to digital dermatitis. FEMS Microbiol. Lett. 269:48-53. [DOI] [PubMed] [Google Scholar]
- 36.Trott, D. J., M. R. Moeller, R. L. Zuerner, J. P. Goff, W. R. Waters, D. P. Alt, R. L. Walker, and M. J. Wannemuehler. 2003. Characterization of Treponema phagedenis-like spirochetes isolated from papillomatous digital dermatitis lesions in dairy cattle. J. Clin. Microbiol. 41:2522-2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walker, R. L., D. H. Read, K. J. Loretz, and R. W. Nordhausen. 1995. Spirochetes isolated from dairy cattle with papillomatous digital dermatitis and interdigital dermatitis. Vet. Microbiol. 47:343-355. [DOI] [PubMed] [Google Scholar]
- 38.Weisburg, W. G., S. M. Barns, D. A. Pelletier, and D. J. Lane. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wyss, C., F. E. Dewhirst, B. J. Paster, T. Thurnheer, and A. Luginbühl. 2005. Guggenheimella bovis gen. nov., sp. nov., isolated from lesions of bovine dermatitis digitalis. Int. J. Syst. Evol. Microbiol. 55:667-671. [DOI] [PubMed] [Google Scholar]
- 40.Zuerner, R. L., M. Heidari, M. K. Elliott, D. P. Alt, and J. D. Neill. 2007. Papillomatous digital dermatitis spirochetes suppress the bovine macrophage innate immune response. Vet. Microbiol. 125:256-264. [DOI] [PubMed] [Google Scholar]