Abstract
The present investigation was undertaken to assess the proportion of methicillin-resistant Staphylococcus aureus (MRSA) strains among hospital-acquired isolates and to determine the clones of MRSA currently circulating in Poland by using a number of molecular techniques. Between January and May 2005, methicillin resistance was investigated among a total of 915 S. aureus isolates collected from 39 hospitals. A total of 208 (22.7%) isolates were positive for the mecA gene by PCR. The molecular characterization of MRSA isolates was carried out by the multiple-locus variable-number tandem repeat fingerprinting, pulsed-field gel electrophoresis, multilocus sequence typing, and staphylococcal chromosomal cassette mec (SCCmec) typing methods. The Hungarian (PFGE B; ST239, SCCmec type III [ST239-III]), Iberian (ST247-I), and Berlin (ST45-IV) clones were predominant, representing approximately 52.9, 11.5, and 10.0% of the MRSA isolates, respectively. A decline in the proportion of earlier MRSA clones, such as ST5-IV (a Pediatric clone), ST80-IV) (a Mediterranean clone), ST239-III (a Polish and Brazilian clone), and ST30-IV (a southwest Pacific clone) was observed. Additionally, the emergence of an MRSA clone with SCCmec type V, possibly representing a community-acquired strain, was observed in two hospitals during this study.
Staphylococcus aureus is one of the leading human pathogens that can cause various diseases, ranging from the trivial to the rapidly fatal. A great array of virulence factors and often multidrug resistance can make the treatment of this pathogen a major challenge. Since the introduction of methicillin in 1959, methicillin-resistant S. aureus (MRSA) strains were first isolated in the United Kingdom in the early 1960s and subsequently in all parts of the world. MRSA strains emerged from methicillin-susceptible S. aureus (MSSA) by the acquisition of an exogenous mobile genetic element, designated the staphylococcal chromosomal cassette mec (SCCmec). Currently, five main types (I to V) of SCCmec have been described (23, 25, 32, 38). All SCCmec types carry the mecA gene, which encodes the low-affinity penicillin-binding protein 2a and confers resistance to methicillin (and other beta-lactams). SCCmec types I, II, and III are the most common among hospital-acquired MRSA strains. These cassettes confer multiresistance, as they contain both plasmid sequences and transposons that comprise drug resistance genes (23, 25).
Epidemiological studies have revealed clonal dissemination to be the major mechanism of MRSA spread (42). Multiple MRSA clones have been identified in different countries and continents, and a number of these have been described as international or pandemic clones (39, 40). These pandemic MRSA clones (Iberian, Brazilian, Hungarian, New York/Japan, Pediatric, EMRSA-15, and EMRSA-16), belonging to five major lineages, have been designated the major causes of hospital-acquired MRSA infections worldwide (51). However, global studies have shown that the clonal structure of MRSA can be dynamic and variable over time (4, 10, 41).
In the late 1990s, community-acquired MRSA (CA-MRSA) infections were increasingly reported in the literature (17, 18, 33, 37, 46). Several studies already have reported the increasing incidence of CA-MRSA infections in hospitals (7, 45). Such a situation highlights the importance of monitoring the clonal structure and the modes of transmission of MRSA within hospitals. Epidemiological typing provides data to aid the development and application of effective measures to control the presence and dissemination of important clones of S. aureus, including MRSA, that are responsible for hospital- and community-acquired infections (8, 14, 15, 40, 43, 44). Combinations of different typing methods allow the assignment of the isolates to known MRSA clones or the determination of new ones (15).
Despite a number of original works, there has been no systematic molecular survey on the countrywide collection of Polish MRSA isolates (26, 28, 35). In order to monitor the dynamic process of changes in the resistance profile of clinical isolates of S. aureus, the National Reference Centre for Antibiotics (NRCA) in the National Medicines Institute has initiated a surveillance program providing data on the level of MRSA and its clonal structure in Polish hospitals.
MATERIALS AND METHODS
Study design.
Thirty-nine hospitals in Poland, representing secondary and tertiary referral centers only, participated in the study. All medical centers were asked to send 25 consecutive S. aureus isolates (the first isolate per patient) between January and May 2005. Hospitals were asked to collect isolates from all wards that were located in a particular hospital. Clinical isolates were recovered from well-defined infections (20) in adults (>16 years old) 48 h after hospitalization.
Identification of isolates.
Isolates were reidentified in the NRCA to the species level using standard microbiological methods (5).
MRSA screening.
All staphylococcal isolates were examined for methicillin resistance by using a 30-μg cefoxitin disk. Resistance was confirmed by PCR to detect the mecA gene (36). Further phenotypic and molecular investigations were performed on MRSA isolates only.
Susceptibility testing.
The MICs of gentamicin, linezolid, quinupristin-dalfopristin, rifampin, tetracycline, trimethoprim-sulfamethoxazole, vancomycin, and teicoplanin were determined by the broth microdilution method according to CLSI guidelines (9), with the exception of daptomycin, for which MICs were determined using Etests (AB-Biodisk, Solna, Sweden). S. aureus ATCC 29213 was used as the reference strain. MICs of fusidic acid were determined according to French guidelines (47). Erythromycin and clindamycin also were tested by the disc diffusion method to differentiate the inducible or constitutive macrolide-lincosamide-streptogramin B resistance phenotype (MLSB) according to CLSI guidelines (9).
Glycopeptide susceptibility testing.
All isolates for which the vancomycin MICs were ≥2 mg/liter were examined for heterogeneous glycopeptide resistance using a strategy described previously (26). Population analysis profiles were performed according to the method of Trakulsomboon et al. (49) to confirm the heteroglycopeptide-intermediate S. aureus (hGISA) phenotype. S. aureus ATCC 29213, S. aureus ATCC 700698 (Mu3), and S. aureus ATCC 700699 (Mu50) were used as reference strains.
Preparation of total DNA for PCR.
Total DNA of the isolates was purified using the Genomic Mini DNA kit (A&A Biotechnology, Gdynia, Poland) as previously described (41).
MLVF.
Preliminary typing was performed by multiple-locus variable-number tandem repeat fingerprinting (MLVF), a PCR-based method that is routinely used in the NRCA to determine the clonality of S. aureus isolates (43, 44). An analysis was performed according to criteria previously described (34).
PFGE.
Total DNA preparation, digestion with the SmaI restriction enzyme (MBI Fermentas, Vilnius, Lithuania), and pulsed-field gel electrophoresis (PFGE) were performed as described by Chung et al. (8). A CHEF-DR III system (Bio-Rad, Hercules, CA) was used for the electrophoresis. Restriction patterns were compared using the Molecular Analyst software Fingerprinting, version 1.12 (Bio-Rad), and were classified into PFGE types and subtypes according to the criteria of Tenover et al. (48).
MLST.
Multilocus sequence typing (MLST) analysis was performed as described by Enright et al. (14). The allele sequences of the seven loci were compared to those submitted to the S. aureus MLST database (http://www.mlst.net).
SCCmec analysis.
The SCCmec types were determined by multiplex PCR as described by Oliveira and de Lencastre (38). The following reference strains were used in the analysis: the Iberian clone PER 184 and PER 88 isolates (38) for SCCmec I type and its type IA variant, respectively; the New York/Japan strain Mu 50 (4) for SCCmec type II; the Hungarian strain J405 (http://www.mlst.net) and the Brazilian strain HU25 (40) for SCCmec III type and its IIIA variant, respectively; and the Pediatric strain POL3 (40) for SCCmec type IV.
PCR for ccrC.
The detection of ccrC, which is characteristic of the V type of SCCmec, was performed as described by Zhang et al. (52). The control strain for type V SCCmec was WIS (WBG 8318-JCSC3624) (25).
Accessory gene regulator (agr) typing.
The agr types were determined by the multiplex PCR strategy as described by Lina et al. (29).
Detection of lukS-PV and lukF-PV.
The presence of the lukS-PV and lukF-PV genes, encoding Panton-Valentine leukocidin, was determined by PCR as previously described (30).
Statistical analysis.
The statistical software STATISTICA 5.0 (Statsoft Inc.) was used in Windows 98. Statistical analysis was performed by the χ2 test, and the Fisher exact test was used to assess the variation in MRSA prevalence among observed wards and point resistance to selected antibiotics. The Spearman correlation was used to analyze associations between SCCmec types and the total patterns of resistance to different antibiotics. P < 0.05 was considered statistically significant.
RESULTS
Clinical isolates.
Between January and May 2005, 915 consecutive clinical isolates of S. aureus (the first isolate per patient) were sent to the NRCA. Each laboratory sent from between 7 and 47 isolates of S. aureus (an average of 24 isolates per laboratory). Clinical isolates were recovered from well-defined infections (20) in adults hospitalized in intensive care units (ICUs); surgical, haematological, and orthopedic wards; dialysis centers; and other specialized units. Data from the following classes of specimens were further analyzed: blood, lower respiratory tract specimens, surgical site infections, bone and joint specimens, and cerebrospinal fluid.
MRSA isolates.
The proportion of MRSA isolates was 22.7% (n = 208), ranging from 3.7 to 63.1% in individual hospitals. In 32 (82%) centers, the proportion of MRSA exceeded 10%. The proportion of MRSA was higher in tertiary-care institutions (average, 33%) than in secondary care (average, 19%). As shown in Table 1, variation in the proportion of MRSA also was noticeable in different hospital wards, with the highest proportions in ICUs (36%; P < 0.01; n = 49). MRSA proportions were highest among respiratory tract specimens (29%; n = 40) (Table 2).
TABLE 1.
Hospital departments from which Staphylococcus aureus was isolated
| Ward | No. of isolates (n = 915) | No. of strainsa
|
Proportion (%) of all strains | |
|---|---|---|---|---|
| MSSA (n = 707) | MRSA (n = 208) | |||
| ICU | 135 | 86 | 49 | 36.2 |
| Surgery | 630 | 490 | 140 | 22.2 |
| Hematology | 45 | 37 | 8 | 17.8 |
| Internal medicine | 42 | 38 | 4 | 9.5 |
| Dialysis centers | 46 | 42 | 4 | 8. |
| Other | 17 | 14 | 3 | 17.6 |
MSSA isolates accounted for 77.3% of all isolates; MRSA isolates accounted for the other 22.7%.
TABLE 2.
Source of Staphylococcus aureus isolates from hospitalized patients
| Specimen source | No. of isolates (n = 915) | No. of strainsa
|
Proportion (%) of all strains | |
|---|---|---|---|---|
| MSSA (n = 707) | MRSA (n = 208) | |||
| Respiratory tract | 137 | 97 | 40 | 29.2 |
| Blood | 156 | 118 | 38 | 24.3 |
| Surgical site | 575 | 450 | 125 | 21.7 |
| Bone and joint | 33 | 31 | 2 | 6.1 |
| Other | 14 | 11 | 3 | 21.4 |
MSSA isolates accounted for 77.3% of all isolates; MRSA isolates accounted for the other 22.7%.
Antimicrobial susceptibility.
All cefoxitin-resistant isolates were mecA positive (n = 208). MRSA isolates were fully susceptible to quinupristin-dalfopristin, fusidic acid, linezolid, glycopeptides, and daptomycin. Susceptibility was more variable for the following agents: tetracycline (24.5% susceptible), gentamicin (34.1% susceptible), trimethoprim-sulfamethoxazole (54.8% susceptible), and rifampin (87.5% susceptible). Among MRSA isolates, 75.5% were erythromycin and clindamycin resistant (encompassing both constitutive and inducible phenotypes). No isolates with the M phenotype (resistant to erythromycin but susceptible to clindamycin) were detected in this study. Among all MRSA, the hGISA phenotype was confirmed in 4 isolates (1.9%) from those primary selected 17 isolates for which the vancomycin MICs were ≥2 mg/liter.
MLVF typing.
All isolates were grouped into 20 MLVF types, with the most common being B (n = 113 isolates; 17 subtypes), G (n = 24; 6 subtypes), and A (n = 21; 4 subtypes) (Table 3).
TABLE 3.
Molecular characteristics and resistance of MRSA isolates
| MLVF type(s) (no. of isolates studied, no. of subtypes) | PFGE type (no. of isolates studied, no. of subtypes) | ST(s) (no. of isolates studied) | SCCmec type(s)/subtype(s) (no. of isolates studied) | CCa | agr group (no. of isolates studied) | Previous clone name(s) (current ST and SCCmec nomenclature) | Antimicrobial resistanceb,c | Sourced (no. of isolates) | Hospital warde (no. of isolates) | No. of centers/no. of cities from which the strains were isolated |
|---|---|---|---|---|---|---|---|---|---|---|
| B (110, 15) | B (23, 15) | 239 and 585f (17) | III (20); IIIA (58); IIIC (19); IIID (13) | CC8 | I (110) | Hungarian (ST239/585; III/IIIA/IIIC/IIID) | ERY, CLI, GEN, SXT, TZP, TET | w (69), rts (21), b (28) un (2) | surf (75), ICU (25), int. med (4), neu (2), dc (2), ha, un | 28/21 |
| B (3, 2) | K (2, 2) | 239 (2) | IIIA; IIIC (2) | CC8 | I (3) | Polish (ST239; IIIA/IIIC) | ERY, CLI, GEN, TET | w (1), rts (1), b (1) | sur, ICU (2) | 2/2 |
| C (2, 2) | W (2, 1) | 239 (1) | I (2) | CC8 | I (2) | (ST239; I)g,h | ERY, CLI, GEN, RIF, SXT, TET | w (2) | sur (2) | 2/2 |
| X (2, 1) | X (1) | 239 (1) | II (2) | CC8 | I (2) | (ST239; II)g,h | ERY, CLI, TET | rts (1), un (1) | sur, un | 2/2 |
| F (1); G (23, 6) | D (7, 6) | 247 (6) | IA (4); IB (20) | CC8 | I (24) | Iberian (ST247; IA/IB) | ERY, CLI GEN, RIF, SXT, TET | w (14), rts (7), b (3) | sur (17), ICU (7), | 12/8 |
| F (1); L (6,3) | D (4, 2) | 8 (2) | IB (7) | CC8 | I (7) | (ST8; IB)g,h | ERY, CLI | w (3), rts (3), b (1) | sur (3), ICU (2), ha (2) | 6/4 |
| G (1) | F (1) | 8 (1) | IV (1) | CC8 | I (1) | EMRSA-2, EMRSA-6, Irish-2 (ST8; IV)h | ERY, CLI, GEN, TET | rts (1) | ICU | 1/1 |
| H (5, 2) | H (2, 1) | 8 (1) | II (5) | CC8 | II (5) | Irish-1 (ST8; II)h | ERY, CLI, TET | w (2), rts (3) | sur (3), ICU (2) | 4/4 |
| I (5, 1) | I (1) | 225f (1) | II (5) | CC5 | II (5) | Rhine Hesse subclone (ST225; II)g,h | ERY, CLI | w (3), b (2) | sur (3), ICU, ha | 4/4 |
| E (8, 1); S (1) | F (2, 2) | 225f (2) | IV (9) | CC5 | II (9) | (ST225; IV)g,h | ERY, CLI | w (4), rts (1), b (4) | sur (6), ICU, ha (2) | 5/2 |
| U (5, 1) | U (1) | 496f (1) | II (5) | CC5 | II (5) | (ST496; II)g,h | ERY, CLI, SXT, | w (1), b (4) | sur (3), ICU (2), | 1/1 |
| O (2, 1) | O (1) | 496f (1) | IIIC (2) | CC5 | II (2) | (ST496; IIIC)g,h | ERY, CLI | b (1), cf (1) | sur, ICU | 1/1 |
| D (3, 2) | Z (2, 2) | 228f (2) | I (3) | CC5 | II (3) | Southern Germany (ST228; I)h | ERY, CLI, GEN, RIF, SXT, TET | w (2), rts (1) | sur,(2) | 2/2 |
| D (3, 1) | P (1) | 5 (1) | III (3) | CC5 | II (3) | (ST5; III)g,h | ERY, CLI, GEN | b (2), rts (1) | sur (2), ICU | 2/2 |
| A (21, 4) | A (4, 1) | 45 (1) | IV (21) | CC45 | I (21) | Berlin (ST45; IV) | w (16), b (2), rts (2), bjs (1) | sur (14), ICU (3), ha (3), dc | 12/8 | |
| Y (1) | Y (1) | 45 (1) | II (1) | CC45 | I (1) | (ST45; II)g,h | ERY, CLI | w (1) | sur | 1/1 |
| J (5, 3) | J (3, 2) | 338f (5) | Vi (5) | CC59 | I (5) | CA-MRSA (ST338; V)g,h,i | ERY, CLI, GEN, SXT | w (4), b (1) | sur (3), gyn, int. med | 2/2 |
CCs are based on previous works.
Antimicrobial abbreviations: ERY, erythromycin; CLI, clindamycin; GEN, gentamicin; RIF, rifampin; SXT, trimethoprim-sulfamethoxazole; TZP, quinupristin-dalfopristin; and TET, tetracycline.
Antimicrobial resistance patterns are shown only if they were observed in more than 50% of the isolates of the clone tested.
Abbreviations: w, wound; rts, respiratory tract specimen; b, blood; cf, cerebrospinal fluid; bjs, bone and joint specimen; and un, unknown.
Abbreviations: int. med, internal medicine ward; sur, surgery; ha, hematology; dc, dialysis center; gyn, gynecology; neu, neurology; and un, unknown.
New ST in Poland.
No previous clone name.
New clone in Poland.
New SCCmec type in Poland.
PFGE typing.
For isolates with identical MLVF subtypes, agr types, and SCCmec types/subtypes, only the first isolate was analyzed by PFGE. PFGE was carried out on 58 isolates (Table 3). Altogether, 15 PFGE types were distinguished for the most common types: B (n = 110 isolates; 15 subtypes), D (n = 31 isolates; 8 subtypes), and A (n = 21 isolates; 1 subtype) (Table 3).
MLST.
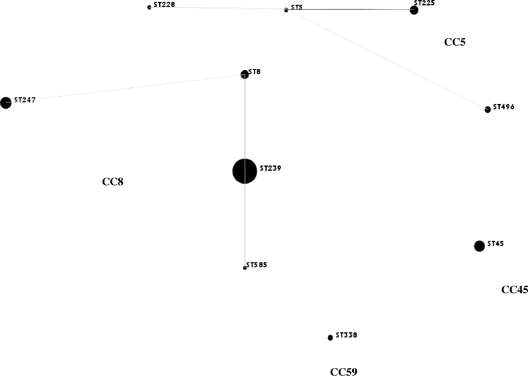
For isolates with identical PFGE subtypes, agr types, and SCCmec types/subtypes, only the first isolate was analyzed by MLST. MLST was carried out on 46 isolates, representing all of the types and subtypes defined by PFGE. Additionally, MLST was determined for all isolates assigned to MLVF and PFGE type J (Table 3). Ten different STs were distinguished, ST5, ST8, ST45, ST225, ST228, ST239, ST247, ST338, ST496, and ST585, which were grouped into four clonal complexes (CCs) (Table 3, Fig. 1). The most numerous (n = 154; 74%) was CC8 (Table 4). CC5 (n = 27; 13%) and CC45 (n = 22; 10%) comprised similar numbers of isolates. The least abundant was CC59 (n = 5; 2.4%) (Table 3).
FIG. 1.
An eBURST snapshot of MRSA isolates from Poland. The size of a spot is proportional to the number of isolates.
TABLE 4.
Point differences in resistance to selected antimicrobials
| Antimicrobial | % Sensitivity of isolates for SCCmec typea:
|
|||
|---|---|---|---|---|
| I | II | III | IV | |
| Gentamicin | 25* | 72 | 23** | 58 |
| Trimethoprim-sulfamethoxazole | 28* | 61 | 15** | 71 |
| Rifampin | 50*** | 94 | 96 | 100 |
| Tetracycline | 22* | 50 | 6** | 77 |
| MLSB | 14* | 44 | 6** | 71 |
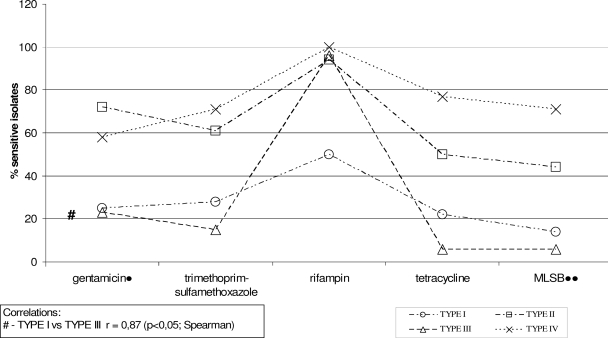
SCCmec type V was excluded from the analysis because of the small number of isolates (n = 5). Quinupristin-dalfopristin, fusidic acid, linezolid, glycopeptides, and daptomycin were excluded from the analysis, because all isolates were susceptible to those antimicrobials. *, type I versus types II and IV; **, type III versus types II and IV; ***, type I versus types II, III, and IV (in each case, P < 0.01 as determined by χ2 and Fisher exact tests).
SCCmec types.
Four main types of SCCmec were found (I to IV) among 98% (n = 203) of the isolates. SCCmec types III (n = 118) and I (n = 36) were predominant, accounting for 57 and 17%, respectively. These two types of SCCmec also were the most variable, with I, IA, and IB and III, IIIA, IIIC, and IIID variants, respectively. Types II (n = 18) and IV (n = 31) were less common, representing 9 and 15% of the isolates in the collection, respectively. As the remaining isolates (n = 5), characterized by ST338, were nontypeable using this method, the SCCmec type of these was determined by the detection of the ccrC gene, which is characteristic for SCCmec type V (23) (Table 3).
agr types.
Only two agr groups were found among all of the isolates tested (Table 3). agr type I was found among all but five isolates characterized by CC8, as well as all isolates characterized by CC45 and CC59. All representatives of CC5 belonged to agr type II. Five isolates characterized as ST8 and SCCmec type II (CC8) also belonged to agr type II.
lukS-PV/lukF-PV PCR.
The luk-PV genes were found exclusively in five isolates belonging to ST338 and SCCmec type V (n = 5).
Statistical analysis.
The statistical analysis results are shown in Fig. 2 and Table 4. The variation in MRSA prevalence among specific wards was significant only for ICUs (P < 0.001). There were significant correlations between SCCmec types and resistance to selected antimicrobials (gentamicin, trimethoprim-sulfamethoxazole, rifampin, tetracycline, tomacrolides, lincosamides, and streptogramin B).
FIG. 2.
Associations between SCCmec types and resistance patterns to selected antimicrobial agents. Quinupristin-dalfopristin, fusidic acid, linezolid, glycopeptides, and daptomycin were excluded from the analysis, because all isolates were susceptible to those antimicrobials.
DISCUSSION
In contrast to previous Polish studies on S. aureus, this was the broadest molecular investigation and the first molecular survey of a countrywide collection of MRSA isolates in Poland. In an antimicrobial survey carried out from 1999 to 2000, no molecular data were included (35). Earlier molecular studies on Polish MRSA focused exclusively on nonconsecutive isolates associated with hospital outbreaks (26, 28). Although the main purpose of the present study was to evaluate the dissemination and structure of MRSA clones, important additional information was obtained regarding the proportion of MRSA throughout the country as well as the proportion of MRSA with regard to the site of infection and the type of hospital ward. Overall, methicillin resistance among S. aureus isolates was 22.7% and was higher than that found in a previous antimicrobial surveillance study, in which it was 15.3% (33). However, the proportions of MRSA from the same sources of isolation examined in the present study and in that previous study were 21.1 and 21.6%, respectively. The proportion of MRSA from patients with S. aureus bacteremia reached 24.3% in this study (Table 1). This places Poland in the midrange of countries participating in the European Antimicrobial Surveillance Study in 2005 (16). Similarly, as in other European countries, the MRSA proportion was highest and statistically significant among S. aureus isolates from ICUs (36.2%; P < 0.01) (Table 2).
Methicillin resistance in S. aureus usually is accompanied by resistance to other groups of antimicrobial agents, so therapeutic options are limited. Our study revealed a relatively high proportion of isolates resistant to trimethoprim-sulfamethoxazole (45.2%) among Polish MRSA isolates. The proportion of strains with resistance to rifampin was 12.5%, which was lower than the 38.3% reported previously by Matynia et al. (35). It is noteworthy that in this survey no strains resistant to quinupristin-dalfopristin (compared with 9.1% previously), linezolid, or daptomycin were detected. The proportion of other antimicrobials tested did not change significantly compared to that of the previous survey. One major change in the resistance profile of the MRSA population compared to that of the study carried out from 1999 to 2000 was the presence of four (1.9%) hGISA isolates. These have emerged from three different clones belonging to ST247-IB, ST239-IIIA/C, and ST8-II in contrast to earlier Polish isolates that belonged mainly to ST247-IA (27, 31). Three of the isolates were characterized by agr type I, whereas one isolate of ST8-II was characterized by agr type II. These results were similar to those described previously by Howe et al. (21), who demonstrated that hGISA/GISA strains emerged from lineages with agr types I and II (27, 31).
There was correlation among isolates harboring SCCmec type I and type III in total resistance patterns to gentamicin, trimethoprim-sulfamethoxazole, and tetracycline, but not to rifampin; there was also correlation in resistance patterns in terms of the MLSB phenotype (r = 0.87 [Spearman correlation]; P < 0.01 [Spearman correlation]). There also were several similarities between isolates harboring SCCmec types II and IV concerning five antimicrobials, but there was no total pattern of statistical significance (Fig. 2, Table 4). The overall resistance pattern showed that isolates harboring SCCmec type I were in the largest proportion of multiresistant strains, whereas isolates with the smallest proportion of resistance harbored SCCmec type IV. However, it was hard to determine if resistance associations to selected antibiotics in our collection could be explained by the SCCmec type. We did find several STs with different SCCmec types; however, we had to exclude several clones from the analysis because of the low numbers of isolates. The multiresistance correlation of SCCmec types I and III probably was associated with the predomination of Iberian and Hungarian clones among isolates harboring appropriate cassettes (Table 3). It has been shown that SCCmec subtypes IA and IB harbor aminoglycoside resistance genes only (40), so the multiresistance of the Iberian clone (ST247-IA/IB) could be explained by the presence of several resistance genes (or mutations, as in case of rifampin resistance [3]) integrated in a different location on the chromosome than the mec cassette (24). In the case of the Hungarian clone (ST239-III and its subtypes), the resistance pattern could be explained by the presence of several resistance genes in the cassette. However, both clones have been shown previously to be multiresistant (26).
Due to the lack of molecular studies to investigate the clonal structure associated with the resistance profiles of MRSA in Matynia et al.'s work (35), it is not possible to comment on whether changes in the resistance profile of MRSA in 2005 were associated with the appearance and dissemination of clones not seen previously in Polish hospitals. Such a situation highlights the importance of the continued monitoring of the clonal structure of MRSA in the future.
Controlling the dissemination of MRSA in Polish hospitals is of great concern. Since Poland joined the European Union (EU) in 2004, the increased migration of people both to and from other EU countries has been observed. Because of free access to healthcare facilities within the EU in general, there is a risk that clones previously existing in other countries will appear in Poland, and vice versa. It has been shown that easy cross-border patient mobility may be a major reason for the appearance of new clones in the hospital setting (12).
Among all isolates examined, 18 MRSA clones were determined on the basis of SCCmec and MLST typing, with the exception of Hungarian and Polish clones, which were characterized by the same ST239 and SCCmec type III and type III subtypes. In this case, PFGE patterns were essential for the determination of the clone. All isolates belonged to four CCs: CC5, CC8, CC45, and CC59 (Table 3, Fig. 1), of which CC5, CC8, and CC45 comprise three of the five major hospital MRSA lineages (15); CC59 comprises both a hospital and community MRSA lineage (14).
The most numerous CC8, comprising 74% of MRSA isolates, was found in multiple medical centers. This CC grouped four STs, namely ST8, ST239, ST247, and ST585 (Fig. 1), with different SCCmec types and subtypes that had been detected among isolates belonging to the same ST (Table 3). This resulted in the identification of five clones and singletons not previously described in Poland (Table 3). Most of these clones (Irish-2 [ST8-I, ST8-II] and Irish-1 [ST8-IV]) were found in Europe, some of which were epidemic (19, 51). It should be emphasized that all of these clones were represented by single isolates only and therefore were not defined as epidemic. It is possible that their emergence in Poland was due to either the acquisition of different SCCmec types by particular MSSA clones already present or the introduction of these clones from other countries into Polish hospitals.
CC8 comprised the two most numerous clones, ST239-III (and its SCCmec III variants) and ST247-IA/B, which were found in multiple medical centers and were clearly epidemic (Table 3, Fig. 1). The relatedness of these clones to the international Hungarian and Iberian clones, as well as their previous dominance among MRSA in Poland, was confirmed previously (25, 40). ST585-IIIC, a single-locus variant (slv) of ST239, was included in the Hungarian clone group according to Nimmo et al. (37). The widespread and prolonged existence of both clones in Polish hospitals could have facilitated their evolution, which is reflected in the high diversity of PFGE and SCCmec subtypes (Table 3). This high degree of variability has been reported previously for the Iberian and Hungarian clones (1, 26, 40).
CC5 and CC45 grouped similar numbers of isolates; however, their clonal composition was completely different. Five STs, namely ST5, ST225, ST228, and ST496, and six newly observed clones in Poland belonged to CC5 (Table 3, Fig. 1). In contrast to CC8, CC5 constituent clones were represented by a small number of isolates. Two of the clones (Rhine-Hesse [ST225-II] and South German [ST228-I]) recently have been observed in neighboring countries as epidemic clones (12). For one of the novel clones in Poland, ST225, 9 of 14 isolates harbored SCCmec type IV. ST225 is an slv of ST5 (Fig. 1); thus, ST225-IV could be a derivative of ST5-IV (Pediatric clone), which was previously detected in Poland (26). Two other newly observed genotypes in Poland, ST496-II and ST496-IIIC, were identified in five and two isolates, respectively (Table 3). ST496 is a double-locus variant of ST5 (Fig. 1) and, in the case of ST496-II, most likely arose from ST5 through point mutations. ST5-II (New York/Japan clone) was reported previously as the dominant clonal type among Hungarian S. aureus isolates and as widely disseminated in French hospitals (10, 13). However, the presence of ST496-IIIC and ST5-III isolates may have occurred due to the acquisition of different SCCmec subtypes by MSSA belonging to CC5.
In contrast to the two CCs described above, CC45, with only one ST, ST45, was much more genetically consistent. All but one isolate harbored SCCmec type IV. ST45-IV previously was identified as the Berlin clone (15, 26, 42). This clearly epidemic clone showed high homogeneity, especially in PFGE subtypes (Table 3), which seems to be its characteristic feature (42). The mechanism by which the non-multidrug-resistant (MDR) Berlin clone emerged as the third most predominant in Poland remains unknown. It has been shown elsewhere that non-MDR clones have successfully emerged and replaced MDR clones (2, 10, 41).
A single isolate of ST45 that harbored SCCmec type II was reported in Poland for the first time; however, such isolates already have been reported in the United States, Hong Kong, and Finland (22, 42).
CC59 grouped five isolates of ST338, which is an slv of ST59. All of the isolates possessed SCCmec type V and were Panton-Valentine leukocidin positive. ST59-IV, ST59-V, and ST59-Vt were reported as common CA-MRSA clones on the west coast of the United States and in Taiwan, Hong Kong, and Australia (6, 11, 14). Such isolates were associated with skin and soft-tissue infections, severe sepsis, necrotizing pneumonia, and necrotizing fasciitis (14). In 2005, the presence of ST59-IV in The Netherlands was described (50). According to molecular characteristics, the isolates detected in this study were probably CA-MRSA. However, we did not have all of the epidemiological data necessary to unambiguously state the incidence of CA-MRSA in two Polish hospitals. Nonetheless, this is the first report of MRSA from CC59 carrying SCCmec type V in Poland.
Although ST239-III (Hungarian) and ST247-IA/IB (Iberian) were dominant clones, a decline in the proportion of earlier-described MRSA clones in Poland, such as ST239-III (PFGE type K; Polish clone), was observed (23) (Table 3). Additionally, no isolates representing the previously present ST239-III (Brazilian clone), ST5-IV (Pediatric clone), ST30-IV (southwest Pacific clone), and ST80-IV (Mediterranean clone) were found in the present survey (Table 3) (24).
In summary, MRSA strains belonging to CC8 and CC5 were the most disseminated in Poland, and ST239-III and ST247-IA/IB were the predominant clones. ST45-IV has emerged as the third most predominant clone. It also seems that some previously widely disseminated clones have been replaced by novel ones not previously encountered in Poland. From the data presented here, we can hypothesize that a number of the new clones identified in Polish hospitals have been introduced into Polish hospitals due the acquisition of different SCCmec types by MSSA isolates or from abroad as a result of increased international travel in recent years.
Acknowledgments
We are grateful to Keichii Hiramatsu for kindly providing strains Mu50, Mu3, and WIS (WBG 8318) and also to Herminia de Lencastre and Alexander Tomasz for kindly providing representatives of the MRSA international clones. We acknowledge the use of the S. aureus MLST database, which is located at Imperial College, London, United Kingdom, and is funded by the Wellcome Trust. We thank Stephen Murchan for the critical reading of the manuscript and for English language editing.
These studies were partially supported by grant 2 P05A 052 30 (to W.H.) from the Committee of Scientific Research (KBN, Poland).
Footnotes
Published ahead of print on 9 July 2008.
REFERENCES
- 1.Aires de Sousa, M., C. Bartzavali, I. Spiliopoulou, I. Santos Sanches, M. I. Crisostomo, and H. De Lancastre. 2003. Two international methicillin-resistant Staphylococcus aureus clones endemic in a University Hospital in Patras, Greece. J. Clin. Microbiol. 412027-2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amorim, M. L., N. A. Faria, D. C. Oliveira, C. Vasconcelos, J. C. Cabeda, A. C. Mendes, E. Calado, A. P. Castro, M. H. Ramos, J. M. Amorim, and H. de Lencastre. 2007. Changes in the clonal nature and antibiotic resistance profiles in methicillin-resistant Staphylococcus aureus isolates associated with the spread of EMRSA-15 clone in a tertiary-care Portuguese hospital. J. Clin. Microbiol. 452881-2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aubry-Damon, H., C. J. Soussy, and P. Courvalin. 1998. Characterization of mutations in the rpoB gene that confer rifampin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 422590-2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baba, T., F. Takeuchi, M. Kuroda, H. Yuzawa, K. Aoki, A. Oguchi, Y. Nagai, N. Iwama, K. Asano, T. Naimi, H. Kuroda, L. Cui, K. Yamamoto, and K. Hiramatsu. 2002. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 3591819-1827. [DOI] [PubMed] [Google Scholar]
- 5.Bannerman, T. L. 2003. Staphylococcus, Micrococcus, and other catalase-positive cocci that grow aerobically, p. 264-282. In P. R. Murray, E. J. Baron, J. H. Jorgensen, M. A. Pfaller, and R. H. Yolken (ed.), Manual of clinical microbiology, 8th ed., vol. 1. ASM Press, Washington, DC. [Google Scholar]
- 6.Boyle-Vavra, S., B. Ereshefsky, C. Wang, and R. S. Daum. 2005. Successful multiresistant community-associated methicillin-resistant Staphylococcus aureus lineage from Taipei, Taiwan, that carries either the novel staphylococcal chromosome cassette mec (SCCmec) type Vt or SCCmec type IV. J. Clin. Microbiol. 434719-4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bratu, S., A. Eramo, R. Kopec, E. Coughlin, M. Ghitan, R. Yost, E. K. Chapnick, D. Landman, and J. Quale. 2005. Community-associated methicillin-resistant Staphylococcus aureus in hospital nursery and maternity units. Emerg. Infect. Dis. 11808-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chung, M., H. de Lencastre, P. Matthews, A. Tomasz, I. Adamsson, M. Aires de Sousa, T. Camou, C. Cocuzza, A. Corso, I. Couto, A. Dominguez, M. Gniadkowski, R. Goering, A. Gomes, K. Kikuchi, A. Marchese, R. Mato, O. Melter, D. Oliveira, R. Palacio, R. Sa-Leao, I. Santos Sanches, J. H. Song, P. T. Tassios, and P. Villari. 2000. Molecular typing of methicillin-resistant Staphylococcus aureus by pulsed-field gel electrophoresis: comparison of results obtained in a multilaboratory effort using identical protocols and MRSA strains. Microb. Drug Resist. 6189-198. [DOI] [PubMed] [Google Scholar]
- 9.Clinical and Laboratory Standards Institute. 2006. Performance standards for antimicrobial susceptibility testing, 16th informational supplement, M7-A7. Clinical and Laboratory Standards Institute, Wayne, PA.
- 10.Conceição, T., M. Aires-de-Sousa, M. Füzi, Á. Tóth, J. Pászti, E. Ungvári, W. B. van Leeuwen, A. van Belkum, H. Grundmann, and H. de Lencastre. 2007. Replacement of methicillin-resistant Staphylococcus aureus clones in Hungary over time: a 10-year surveillance study. Clin. Microbiol. Infect. 13971-979. [DOI] [PubMed] [Google Scholar]
- 11.Coombs, G. W., J. C. Pearson, F. G. O'Brien, R. J. Murray, W. B. Grubb, and K. J. Christiansen. 2006. Methicillin-resistant Staphylococcus aureus clones, western Australia. Emerg. Infect. Dis. 12241-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deurenberg, R. H., C. Vink, G. J. Oudhuis, J. E. Mooij, C. Driessen, G. Coppens, J. Craeghs, E. De Brauwer, S. Lemmen, H. Wagenvoort, A. W. Friedrich, J. Scheres, and E. E. Stobberingh. 2005. Different clonal complexes of methicillin-resistant Staphylococcus aureus are disseminated in the Euregio Meuse-Rhine region. Antimicrob. Agents Chemother. 494263-4271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Durand, G., M. Bes, H. Meugnier, M. C. Enright, F. Forey, N. Liassine, A. Wenger, K. Kikuchi, G. Lina, F. Vandenesch, and J. Etienne. 2006. Detection of new methicillin-resistant Staphylococcus aureus clones containing the toxic shock syndrome toxin 1 gene responsible for hospital- and community-acquired infections in France. J. Clin. Microbiol. 44847-853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Enright, M. C., N. P. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 381008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Enright, M. C., D. A. Robinson, G. Randle, E. J. Feil, H. Grundmann, and B. G. Spratt. 2002. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). Proc. Natl. Acad. Sci. USA 997687-7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.European Antimicrobial Resistance Surveillance System. 2005. Annual report. On-going surveillance of S. pneumoniae, S. aureus, E. coli, E. faecium, E. faecalis, K. pneumoniae, P. aeruginosa. EARSS, Bilthoven, The Netherlands.
- 17.Gomes, A. R., H. Westh, and H. de Lencastre. 2006. Origins and evolution of methicillin-resistant Staphylococcus aureus clonal lineages. Antimicrob. Agents Chemother. 503237-3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Groom, A. V., H. D. Wolsey, T. S. Naimi, K. Smith, S. Johnson, D. Boxrud, K. A. Moore, and J. E. Cheek. 2001. Community-acquired methicillin-resistant Staphylococcus aureus in a rural American Indian community. JAMA 2861201-1205. [DOI] [PubMed] [Google Scholar]
- 19.Hanssen, A.-M., A. Fossum, J. Mikalsen, D. S. Halvorsen, G. Bukholm, and J. U. Ericson Sollid. 2005. Dissemination of community-acquired methicillin-resistant Staphylococcus aureus clones in northern Norway: sequence types 8 and 80 predominate. J. Clin. Microbiol. 432118-2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horan, T. C., M. Andrus, and M. A. Dudeck. 2004. CDC/NHSN surveillance definition of health care-associated infection and criteria for specific types of infections in the acute care setting 2004. National Healthcare Safety Network, Division of Healthcare Quality Promotion, Centers for Disease Control and Prevention, Atlanta, GA. http://www.cdc.gov/ncidod/dhqp/pdf/NNIS/NosInfDefinitions.pdf.
- 21.Howe, R. A., A. Monk, M. Wootton, T. R. Walsh, and M. C. Enright. 2004. Vancomycin susceptibility within methicillin-resistant Staphylococcus aureus lineages. Emerg. Infect. Dis. 10855-857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ip, M., R. W. H. Yung, T. K. Ng, W. K. Luk, C. Tse, P. Hung, M. Enright, and D. J. Lyon. 2005. Contemporary methicillin-resistant Staphylococcus aureus clones in Hong Kong. J. Clin. Microbiol. 435069-5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ito, T., Y. Katayama, K. Asada, N. Mori, K. Tsutsumimoto, C. Tiensasitorn, and K. Hiramatsu. 2001. Structural comparison of three types of staphylococcal cassette chromosome mec integrated in the chromosome in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 451323-1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ito, T., K. Okuma, X. X. Ma, H. Yuzawa, and K. Hiramatsu. 2003. Insights on antibiotic resistance of Staphylococcus aureus from its whole genome: genomic island SCC. Drug Resist. Updat. 4141-52. [DOI] [PubMed] [Google Scholar]
- 25.Ito, T., X. X. Ma, F. Takeuchi, K. Okuma, H. Yuzawa, and K. Hiramatsu. 2004. Novel type V staphylococcal cassette chromosome mec driven by a novel cassette chromosome recombinase, ccrC. Antimicrob. Agents Chemother. 482637-2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krzysztoñ-Russjan, J., J. Empel, T. Leski, M. Gniadkowski, and W. Hryniewicz. 2005. Clonal structure of methicillin-resistant Staphylococcus aureus (MRSA) population in Poland: revision and update. Microb. Drug Resist. 11127-136. [DOI] [PubMed] [Google Scholar]
- 27.Krzysztoñ-Russjan, J., M. Gniadkowski, H. Polowiniak-Pracka, E. Hagmajer, and W. Hryniewicz. 2002. The first Staphylococcus aureus isolates with reduced susceptibility to vancomycin in Poland. J. Antimicrob. Chemother. 501065-1069. [DOI] [PubMed] [Google Scholar]
- 28.Leski, T., D. Oliveira, K. Trzcinski, I. Santos Sanches, M. Aires de Sousa, W. Hryniewicz, and H. de Lencastre. 1998. Clonal distribution of methicillin-resistant Staphylococcus aureus in Poland. J. Clin. Microbiol. 263532-3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lina, G., F. Boutite, A. Tristan, M. Bes, J. Etienne, and F. Vandenesch. 2003. Bacterial competition for human nasal cavity colonization: role of staphylococcal agr alleles. Appl. Environ. Microbiol. 6918-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lina, G., Y. Piemont, F. Godail-Gamot, M. Bes, M. Peter, V. Gauduchon, F. Vandenesch, and J. Etienne. 1999. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin. Infect. Dis. 291128-1132. [DOI] [PubMed] [Google Scholar]
- 31.Luczak-Kadlubowska, A., J. Krzyszton-Russjan, and W. Hryniewicz. 2006. Characteristics of Staphylococcus aureus strains isolated in Poland in 1996 to 2004 that were deficient in species-specific proteins. J. Clin. Microbiol. 444018-4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma, X. X., T. Ito, C. Tiensasitorn, M. Jamklang, P. Chongtrakool, S. Boyle-Vavra, R. S. Daum, and K. Hiramatsu. 2002. Novel type of staphylococcal cassette chromosome mec identified in community-acquired methicillin-resistant Staphylococcus aureus strains. Antimicrob. Agents Chemother. 461147-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maguire, G. P., A. D. Arthur, P. J. Boustead, B. Dwyer, and B. J. Currie. 1996. Emerging epidemic of community-acquired methicillin-resistant Staphylococcus aureus in the Northern Territory. Med. J. Aust. 164721-723. [DOI] [PubMed] [Google Scholar]
- 34.Malachowa, N., A. Sabat, M. Gniadkowski, J. Krzyszton-Russjan, J. Empel, J. Miedzobrodzki, K. Kosowska-Shick, P. C. Appelbaum, and W. Hryniewicz. 2005. Comparison of multiple-locus variable-number tandem repeat analysis with pulsed-field gel electrophoresis, spa typing, and multilocus sequence typing for clonal characterization of Staphylococcus aureus isolates. J. Clin. Microbiol. 433095-3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matynia, B., E. Młodzinska, and W. Hryniewicz. 2005. Antimicrobial susceptibility patterns of Staphylococcus aureus in Poland obtained by the National Quality Assurance Programme. Clin. Microbiol. Infect. 11379-385. [DOI] [PubMed] [Google Scholar]
- 36.Murakami, K., W. Minamide, K. Wada, E. Nakamura, H. Teraoka, and S. Watanabe. 1991. Identification of methicillin-resistant strains of staphylococci by polymerase chain reaction. J. Clin. Microbiol. 292240-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nimmo, G. R., G. W. Coombs, J. C. Pearson, F. G. O'Brien, K. J. Christiansen, J. D. Turnidge, I. B. Gosbell, P. Collignon, and M. L. McLaws. 2006. Methicillin-resistant Staphylococcus aureus in the Australian community: an evolving epidemic. Med. J. Aust. 184384-388. [DOI] [PubMed] [Google Scholar]
- 38.Oliveira, D. C., and H. de Lencastre. 2002. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 462155-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oliveira, D. C., I. Santos-Sanches, R. Mato, M. Tamayo, G. Ribeiro, D. Costa, and W. de Lancastre. 1998. Virtually all methicillin-resistant Staphylococcus aureus (MRSA) infections in the largest Portuguese teaching hospital are caused by two internationally spread multiresistant strains: the “Iberian” and the “Brazilian” clones of MRSA. Clin. Microbiol. Infect. 4373-384. [DOI] [PubMed] [Google Scholar]
- 40.Oliveira, D. C., A. Tomasz, and H. De Lancastre. 2001. The evolution of pandemic clones of methicillin-resistant Staphylococcus aureus: identification of two ancestral genetic backgrounds and the associated mec elements. Microb. Drug Resist. 7349-361. [DOI] [PubMed] [Google Scholar]
- 41.Pérez-Roth, E., F. Lorenzo-Díaz, N. Batista, A. Moreno, and S. Méndez-Álvarez. 2004. Tracking methicillin-resistant Staphylococcus aureus clones during a 5-year period (1998 to 2002) in a Spanish hospital. J. Clin. Microbiol. 424649-4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Robinson, A., and M. C. Enright. 2003. Evolutionary models of the emergence of methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 473926-3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sabat, A., J. Krzyszton-Russjan, W. Strzalka, R. Filipek, K. Kosowska, W. Hryniewicz, J. Travis, and J. Potempa. 2003. New method for typing Staphylococcus aureus strains: multiple-locus variable-number tandem repeat analysis of polymorphism and genetic relationships of clinical isolates. J. Clin. Microbiol. 411801-1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sabat, A., N. Malachowa, J. Miedzobrocki, and W. Hryniewicz. 2006. Comparison of PCR-based methods for typing Staphylococcus aureus isolates. J. Clin. Microbiol. 443804-3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saiman, L., M. O'Keefe, P. L. Graham III, F. Wu, B. Saïd-Salim, B. Kreiswirth, A. LaSala, P. M. Schlievert, and P. Della-Latta. 2003. Hospital transmission of community-acquired methicillin-resistant Staphylococcus aureus among postpartum women. Clin. Infect. Disease. 371313-1319. [DOI] [PubMed] [Google Scholar]
- 46.Saravolatz, L. D., N. Markowitz, L. Arking, D. Pohloh, and E. Fisher. 1982. Methicillin-resistant Staphylococcus aureus. Epidemiologic observations during a community-acquired outbreak. Ann. Intern. Med. 9611-16. [DOI] [PubMed] [Google Scholar]
- 47.Soussy, C. J., G. Carret, J. D. Cavallo, H. Chardon, C. Chidiac, P. Choutet, P. Courvalin, H. Dabernat, H. Drugeon, L. Dubreuil, F. Goldstein, V. Jarlier, R. Leclercq, M. H. Nicolas-Chanoine, A. Philippon, C. Quentin, B. Rouveix, and J. Sirot. 2000. Antibiogram Committee of the French Microbiology Society. Report 2000-2001. Pathol. Biol. (Paris) 48832-871. (In French.) [PubMed] [Google Scholar]
- 48.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 332233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Trakulsomboon, S., S. Danchaivijitr, Y. Rongrungruang, C. Dhiraputra, W. Susaemgrat, T. Ito, and K. Hiramatsu. 2001. First report of methicillin-resistant Staphylococcus aureus with reduced susceptibility to vancomycin in Thailand. J. Clin. Microbiol. 39591-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wannet, W. J. B., E. Spalburg, M. E. O. C. Heck, G. N. Pluister, E. Tiemersma, R. J. L. Willems, X. W. Huijsdens, A. J. de Neeling, and J. Etienne. 2005. Emergence of virulent methicillin-resistant Staphylococcus aureus strains carrying Panton-Valentine leucocidin genes in the Netherlands. J. Clin. Microbiol. 433341-3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wisplinghoff, H., B. Ewertz, S. Wisplinghoff, D. Stefanik, G. Plum, F. Perdreau-Remington, and H. Seifert. 2005. Molecular evolution of methicillin-resistant Staphylococcus aureus in the metropolitan area of Cologne, Germany, from 1984 to 1998. J. Clin. Microbiol. 435445-5451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang, K., J. A. McClure, S. Elsayed, T. Louie, and J. M. Conly. 2005. Novel multiplex PCR assay for characterization and concomitant subtyping of staphylococcal cassette chromosome mec types I to V in methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 435026-5033. [DOI] [PMC free article] [PubMed] [Google Scholar]