Abstract
Streptococcus mutans is one of several members of the oral indigenous biota linked with severe early childhood caries (S-ECC). Because most humans harbor S. mutans, but not all manifest disease, it has been proposed that the strains of S. mutans associated with S-ECC are genetically distinct from those found in caries-free (CF) children. The objective of this study was to identify common DNA fragments from S. mutans present in S-ECC but not in CF children. Using suppressive subtractive hybridization, we found a number of DNA fragments (biomarkers) present in 88 to 95% of the S-ECC S. mutans strains but not in CF S. mutans strains. We then applied machine learning techniques including support vector machines and neural networks to identify the biomarkers with the most predictive power for disease status, achieving a 92% accurate classification of the strains as either S-ECC or CF associated. The presence of these gene fragments in 90 to 100% of the 26 S-ECC isolates tested suggested their possible functional role in the pathogenesis of S. mutans associated with dental caries.
The mutans streptococci (MS) are strongly associated with dental caries by virtue of their metabolic, ecological, and epidemiological attributes (23, 46). Among the MS, Streptococcus mutans appears to be a predominant bacterial species in the microbiota of preschool children with severe early childhood caries (S-ECC) (4-6, 49). Although the association between S. mutans and S-ECC seems convincing, most children colonized by S. mutans do not manifest the disease (8), suggesting that among other possibilities, S. mutans vary in their ability to initiate caries.
In our previous study, we demonstrated that strains of S. mutans strains associated with S-ECC differ in their genomic composition compared to caries-free (CF) controls (42). Using the power of suppressive subtractive DNA hybridization (SSH), several unique gene segments were identified from a strain of S. mutans (AF199) that was isolated from a child with S-ECC. The presence of unique genetic loci among S. mutans strains is consistent with the recent work by Waterhouse and Russell (51), as they described the presence of “dispensable genes” distributed among strains of S. mutans. These segments include mobile genetic elements that are widely distributed in S. mutans (2) and have been shown to modulate sucrose (31) and melibiose metabolism (40). S. mutans strains also vary in content in terms of the presence of plasmids (10, 32), mutacin I, II, III, and IV operons (3, 19, 35-37), serotypic antigens (43); competence (34), the comBCD genes (28), and gtfBC (14, 48, 52), among other genetic loci. Based on the wide diversity of genotypes and genetic loci within S. mutans, different strains of S. mutans apparently comprise both common and unique genetic loci, and it seems that these differences are unequally distributed among strains (42, 51). Identifying the unique DNA fragments that are common to most of the strains isolated from S-ECC but not CF children will be important even if their function is unknown because the nucleotide sequences can serve as diagnostic biomarkers for DNA-based detection arrays (38, 41).
Here we report the identification of a hierarchical series of gene biomarkers derived via SSH from strains of S. mutans associated with S-ECC. These biomarkers were then evaluated by machine learning techniques for their ability to classify clinical isolates of S. mutans into one of two categories, CF or S-ECC. Our findings suggest that as few as three SSH biomarkers were sufficient to accomplish this goal.
MATERIALS AND METHODS
Subjects.
Thirty-nine children of Hispanic origin (twenty-three boys and sixteen girls; age range, 2.4 to 8.6 years) were included in the present study. Twenty-two of the subjects were CF children; the remainder were S-ECC children who had a score for decayed, missing, and filled teeth of 9.6 ± 3.6 (mean ± the standard deviation) and a score for decayed, missing, and filled tooth surfaces of 17.9 ± 11.8. The S-ECC children were selected from a list of children who were scheduled for extensive caries restorative treatment under general anesthesia in the operating room at the Bellevue Hospital, New York, NY, from April 2003 to April 2004 (26). CF children of ages comparable to those of the S-ECC cohort were selected from the of the Bellevue Hospital's pediatric dental clinic after having been diagnosed as being free of detectable caries. The study protocol was approved by the Institutional Review Board of New York University School of Medicine and the Bellevue Hospital for human subjects.
Bacterial sample procedures and processing.
Bacterial samples from saliva and pooled plaque of the S-ECC and CF children were collected before any dental treatment was initiated. The supragingival plaque samples were collected and processed as previously described (10, 26). The genomic DNA of pure cultures of isolates of S. mutans were obtained by using a genomic DNA purification kit (Qiagen, Hilden, Germany), as previously described (25, 27). All of the DNA samples from S. mutans were first subjected to chromosomal DNA fingerprinting (9, 24) to identify the genotypes of the isolates of S. mutans and then for subtractive hybridization.
SSH.
SSH was used to isolate DNA fragments present in the strains of S. mutans isolated from plaque samples from S-ECC but not present in the S. mutans of the CF plaque sample. The protocol has been described elsewhere (42). Briefly, the DNA of S. mutans strains isolated from S-ECC subjects, selected as tester strains, was subtracted against the pooled genomic DNA of strains of S. mutans from CF subjects. DNA samples from CF children were mixed (2 μg of each), and 2 μg from this DNA mix was used as driver against 2 μg of each tester DNA. Six subtraction reactions were performed, using the PCR-Select bacterial genome subtraction kit (BD Biosciences, San Jose, CA) with minor modifications (42). Tester and driver samples were digested separately with RsaI. Amplification of tester-specific fragments was performed by using PCR and primers directed at tester-ligated adaptor sequences and the protocol provided by the manufacturer (BD Biosciences). Secondary PCR products (4 μl) were cloned into the pCR4-TOPO (Invitrogen) vector and transformed into Escherichia coli TOP10 cells (Invitrogen). A total of 1,300 transformants were picked at random and grown in 96-deep well plates at 37°C in 1.5 ml of Luria-Bertani medium with kanamycin for 12 h. The plasmid DNA was extracted by using 96 Turbo BioRobot Kit and BioRobot 3000 (Qiagen). False-positive results (SSH fragments present in both S-ECC and CF strains) were identified by dot blot hybridization. Purified plasmid DNA containing cloned SSH fragments were sequenced using the M13 universal primer. Sequencing reactions were performed in both directions on an ABI model 377 DNA sequencer. Nucleic acid and predicted protein compositions were compared to those archived in GenBank using BLAST (National Center for Biotechnology Information [NCBI]). Sequences were also analyzed for protein coding regions via the open reading frame finder (NCBI) and PFAM (http://www.ncbi.nlm.nih.gov/gorf/gorf.html) (47).
Dot blot hybridization.
Dot blots were prepared by using standard procedures (42). PCR products from each of the 1,300 selected transformants were purified by using a PCR purification kit (Qiagen). Portions (10 μl) of the PCR products were diluted with 40 μl of 1 M NaOH, 5 μl of 200 mM EDTA, and 45 μl of sterile water. Diluted PCR products were denatured and spotted directly onto Hybond-N+ nylon membranes (Ambion), using a 96-well manifold (Gibco-BRL). Membranes were UV cross-linked by using Stratalinker (Stratagene) and stored dry before hybridization. Membranes were prehybridized for 15 min in a hybridization oven with 7 ml of warm (68°C) UltraHyb buffer (Ambion). Biotin-labeled probes were made with 100 ng of purified, RsaI-digested driver or tester genomic DNA. Hybridization was carried out for 16 h at 68°C in a rotating hybridization oven. Standard protocols for membrane washing (42) were followed, washing twice under moderate to high-stringency conditions (50 ml of 0.2× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate] plus 0.1% sodium dodecyl sulfate, 15 min, 42 to 65°C). Hybrids were detected with a BrightStar detection kit (Ambion) after exposure to BioMax X-ray films (Kodak).
PCR amplification.
To determine the distribution of putative unique S-ECC-associated sequences among S. mutans, PCR primers and conditions were designed for each of the selected SSH biomarkers for screening other S-ECC and CF genotypes. In addition, each SSH fragment was analyzed for G+C content and then used to query BLAST and protein databases. Standard PCRs were carried out with either S-ECC or CF S. mutans genomic DNA. Typically, a 50-μl PCR included 2.5 μl of 10× PCR buffer (100 mM Tris-HCl [pH 9.0], 15 mM MgCl2, 500 mM KCl), 0.25 μl of 20 mM deoxynucleoside triphosphates, 1 μl of each of the forward and reverse primers (stock concentration, 50 nM), 0.5 μl (5 U) of Taq DNA polymerase (Invitrogen), and 2 μl of template DNA. PCR conditions were as follows: 94°C for 3 min; 94°C for 45 s, 54 to 59°C for 45 s, and 72°C for 60 s for 30 cycles; and 72°C for 7 min. Corresponding tester and driver strains were used as positive and negative controls, respectively. Amplicons were analyzed by electrophoresis on 1.5% agarose gels.
AI.
Two independent forms of artificial intelligence (AI), support vector machine (SVM) and neural network analyses, were used to compare and calculate the sensitivity, specificity, and overall accuracy of each selected SSH biomarkers.
SVM.
The presence or absence of 19 PCR-amplified SSH biomarkers (derived from the original 1,300 clones minus false-positives) from S-ECC strains by SSH versus pooled strains from CF children) was assessed from each of a total of 49 clinically isolated S. mutans strains S-ECC (n = 26) and CF (n = 23). Amplification of each SSH biomarker in each strain was scored as present (“1”) or absent (“0”). SVM was used as a supervised learning method to identify an optimal combination of the S-ECC biomarkers that could correctly classify clinical isolates of S. mutans into one of two categories: S-ECC associated or CF associated. Forty-nine S. mutans strains were randomly assigned to a training set (60%) or a tester set (40%). The SVM classifier program (WEKA, Sequential Minimal Optimization [http://www.cs.waikato.ac.nz/ml/weka/]) was then run on the training set, resulting in an algorithm that defined the S-ECC strains. Due to the limited size of each data set, cross-validation within the original data set was utilized to provide a nearly unbiased estimation of classification. For each classification, the true-positive, true-negative, false-positive, and false-negative values were obtained from which accuracy, sensitivity, and specificity were calculated by using Health Decision Strategies EpiMax software (15).
The SVM analysis was extended by reducing the number of features (SSH biomarkers) used to build the classifier (attribute and/or feature selection). Using the WEKA software, the markers were ranked by information gain; first, the top 10 and then the top 5 markers were chosen to train and classify, cross-validate, and classify the test set.
Neural network and recursive partitioning tree.
A feed-forward, back-propagation, neural network with five input nodes, 20 hidden nodes, and two output nodes was also used to assess the discriminatory capability of the SSH-biomarkers. The five input nodes were for SSH biomarkers 0018, H7, 0102, 0006, and 0004 and were identified from analysis by SVM and the classification tree (see below). A total of 50% of the S-ECC- and CF-associated strains of S. mutans were randomly selected as a training set, and the remaining cases formed the test set. The process of selecting the training set, training the net, and testing was repeated 1,000 times, and the mean sensitivity and specificity and their empirical 95% confidence intervals were calculated.
Finally, a recursive partitioning tree was constructed from the pool of all SSH fragments and pruned to three levels. At each level, a binary decision was applied based on the presence or absence of the (automatically) selected fragment. Neural net analysis and recursive partitioning tree determinations were performed in R version 2.6. (www.R-project.org).
Nucleotide sequence accession numbers.
The nucleotide sequences unique to cariogenic strains of S. mutans were deposited in GenBank under accession numbers EU918292 to EU918301.
RESULTS
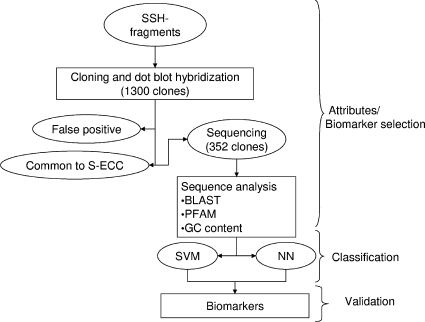
The aim of the present study was to test the utility of SSH to produce strain-specific DNA biomarkers and then examine their distribution among S. mutans strains from children of different disease statuses. Among the 39 children examined, 49 S. mutans genotypes were included in the study. The majority of S-ECC children harbored a single genotype of S. mutans, as determined by screening 10 randomly chosen isolates from mitis-salivarius-bacitracin medium and chromosomal DNA fingerprinting analysis. Using two independent forms of AI, SVM and neural network analysis, we identified the most informative SSH biomarkers that can classify S. mutans strains isolated from either S-ECC or CF children (Fig. 1).
FIG. 1.
Overall study design for subtractive DNA hybridization, attributes/biomarker selection, and classification analysis.
SSH fragments.
To obtain unique gene fragments present in S-ECC S. mutans strain six independent SSH reactions were conducted using genomic DNA and subtracted against the pooled genomic DNA of four S. mutans strains from CF subjects. Approximately 1,300 clones containing SSH fragments were picked (lengths, 300 to 750 bp). To eliminate the false-positives, we used dot blot hybridization with PCR-amplified SSH fragments and biotin-labeled genomic DNA from S. mutans strains used in SSH. The dot blot results indicated that 856 (66%) of DNA fragments were present in most of the six S-ECC strains used in SSH, and 442 (34%) were either absent or present in only a few S-ECC S. mutans strains. A total of 247 clones (19%) were false positives (SSH fragments also present in CF S. mutans strains). Randomly picked 352 clones out of the 856 clones that were present in most of S-ECC strains used in SSH experiment were sequenced. The average size of the SSH fragments was >600 bp: 220 clones (62%) contained useable nonvector sequences with a minimum Phred score of 62. After the duplicate and overlapping fragments were eliminated, a total of 54 unique SSH fragments were obtained containing 42 kb of informative sequence. A BLAST search of the nucleotide or translated sequences of these SSH fragments indicated that 18 did not have significant protein matches to the existing sequences from S. mutans in GenBank. The %G+C content of these SSH fragments ranged from 26 to 50.8%, with an average difference of 4.6% from the genome of S. mutans UA159. Table 1 shows the results from the BLAST query of S. mutans specific SSH fragments from S-ECC obtained from the subtractive library and used in AI.
TABLE 1.
Characterization of S-ECC S. mutans specific DNA biomarkers obtained from SSH librariesa
| Biomarker | Length (bp) | %G+C | Best protein matchb | BLAST e-value or referencec |
|---|---|---|---|---|
| 0001 | 1,193 | 38.57 | Multidrug resistance ABC transporter ATP-binding and permease protein, Streptococcus pyogenes | 3e-82 |
| 0004 | 1,346 | 38.52 | Hypothetical protein SH0211, Staphylococcus haemolyticus | 1e-15 |
| 0006 | 876 | 35.37 | Mobile genetic element, Lactobacillus helveticus | 2e-13 |
| 0007 | 651 | 40.44 | Threonyl-tRNA synthetase, Streptococcus gordonii | 5e-100 |
| 0018 | 506 | 35.81 | Hypothetical protein, Staphylococcus haemolyticus | 8e-32 |
| 0022 | 982 | 36.90 | Putative phosphopantetheinyl transferase, Bacillus subtilis | 4e-31 |
| 0023 | 1,277 | 37.00 | ABC transporter ATP-binding and permease protein, Streptococcus pyogenes | 1e-46 |
| 0024 | 848 | 33.69 | None | |
| 0036 | 736 | 29.77 | Hypothetical protein CLOL250, Clostridium spp. | 4e-25 |
| 0037 | 552 | 29.67 | Transposase orfB, Streptococcus pneumoniae | 1e-32 |
| 0102 | 855 | 30.06 | Hypothetical adenine-specific methylase, Mycoplasma pneumoniae | 9e-06 |
| 0105 | 682 | 36.29 | Hypothetical protein, Finegoldia magna | 2e-87 |
| 0106 | 1,023 | 26.93 | Transcriptional regulator, TetR family, Streptococcus pyogenes | 2e-06 |
| 0145 | 731 | 33.13 | Conserved hypothetical protein, Bacillus anthracis | 2e-40 |
| 0191 | 602 | 27.94 | Predicted protein, Francisella tularensis | 3e-09 |
| 0216 | 706 | 22.81 | Valyl-tRNA synthetase, Streptococcus pneumoniae | 2e-120 |
| G3 | 460 | 56 | TraQ protein (transposon, Bacteroides fragilis YCH46; Porphyromonas gingivalis W83) | Previous study (42) |
| D7 | 493 | 43.6 | None | Previous study (42) |
| H7 | 653 | 49.1 | None | Previous study (42) |
DNA biomarkers used in artificial intelligence analysis are reported in the table.
No significant similarities to archived GenBank sequences were detected.
The e-value represents the number of times this match or a better one would be expected to occur purely by chance in a search of the entire database.
SVM.
The presence or absence of SSH fragments in either S-ECC or CF strains of S. mutans (detected by either dot blot or fragment-specific primers) proved useful in comparing their distribution among S. mutans isolates. In the specific case of SSH fragments derived from S-ECC strains, we wanted to know whether a particular constellation of SSH fragments (biomarkers) could be used to classify additional strains derived from either S-ECC or CF children. To do this, we empirically selected 19 SSH-fragments as biomarkers based upon the their G+C content differing from that of S. mutans, a BLAST match to a possible gene involved with pathogenesis or horizontal transfer, or a mobile genetic element (Table 1). We then surveyed 49 strains of S. mutans from either S-ECC (n = 26) or CF (n = 23) to test the utility of these 19 SSH fragments as biomarkers for S-ECC.
The SVM classifier algorithm generated a model capable of differentiating between S-ECC and CF strains. For each category (S-ECC or CF), the number of true-positive, true-negative, false-positive, and false-negative values were calculated and then used to estimate the overall accuracy, sensitivity, and specificity of the various models (Tables 2 and 3). The resulting classifier correctly partitioned strains into either S-ECC or CF-associated with accuracy of 90% and a sensitivity and specificity of 89 and 90%, respectively (Table 3). The classifier was run again using only the most informative five biomarkers (0018, H7, 0006, 0007, and 0004). With just five biomarkers, the accuracy of classifying strains improved to 92% (Table 2). Biomarker 0018, which was similar (e value of 8e-32) to a hypothetical protein from Staphylococcus haemolyticus, was the most informative of the five and was present in most of the S-ECC S. mutans strains. These five biomarkers were present in 90 to 100% of the 26 S-ECC isolates tested, suggesting that these genes play a functional role in the pathogenic potential of S. mutans.
TABLE 2.
Summary of stratified cross-validation analysis of biomarkersa
| Parameter | No. of biomarkers
|
|
|---|---|---|
| 10 | 5 | |
| Correctly classified instances | 45 | 46 |
| Incorrectly classified instances | 5 | 4 |
| Kappa statistic | 0.7981 | 0.839 |
| Mean absolute error | 0.10 | 0.08 |
| Relative absolute error (%) | 20.05 | 16.04 |
| Total no. of instances | 50 | 50 |
| Sensitivity (%) | 89.3 | 92.6 |
| Specificity (%) | 90.9 | 91.3 |
Cross-validation analysis was done by using the WEKA SVM classifier program and Health Decision Strategies EpiMax software (15).
TABLE 3.
Accuracy of biomarkers by either class S-ECC or CFa
| No. of biomarkers | TP (%) | FP (%) | Precision (%) | Recall (%) | F measure (%) | Class |
|---|---|---|---|---|---|---|
| 10 | 92.6 | 13.0 | 89.3 | 92.6 | 90.9 | S-ECC |
| 87.0 | 7.0 | 90.9 | 87.0 | 88.9 | CF | |
| 5 | 92.6 | 8.7 | 92.6 | 92.6 | 92.6 | S-ECC |
| 91.3 | 7.4 | 91.7 | 91.3 | 91.3 | CF |
The true-positive (TP) or false-positive (FP) status was determined by using the WEKA SVM classifier and Health Decision Strategies EpiMax software (15).
Neural network and recursive partitioning classification.
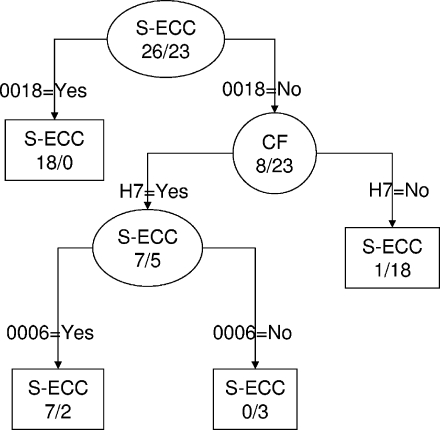
The presence of all possible combinations of the 19 identified biomarkers was explored by classification tree analysis, and the combinations with the highest power to discriminate between S-ECC and CF strains were observed. The tree was pruned to three levels to avoid case-specific decision rules. The optimal tree is shown in Fig. 2 and is based on three biomarkers: 0018, H7, and 0006. This combination of fragments resulted in a sensitivity of 96% and a specificity of 91% for the classification of S-ECC.
FIG. 2.
Recursive partitioning classification of fragment to optimally discriminate caries status. Terminal nodes are shown as squares, and nonterminal nodes are shown as circles. Each node is labeled as either S-ECC or CF depending on the simple majority of cases within the node. Decision rules are shown on the lines connecting the nodes. This analysis shows that fragments 0018, H7, and 0006 used in a decision tree result in a sensitivity of 96% for S-ECC status and 91% for CF status.
Since both the training and test sets were small, there was significant variability in the accuracy of classification from iteration to iteration. The 1,000 iterations of the neural net resulted in a sensitivity of 88.5 and a specificity of 94.2.
DISCUSSION
Our results support the contention that S. mutans strains differ in their genomic compositions and that these differences can be used to classify strains into one of two disease status groups using the power of AI learning. While not the intent of this investigation to characterize the individual SSH fragments, the compositions of many of the SSH fragments suggest that they might have arisen from horizontal gene transfer because they contain either remnants of mobile genetic elements or vary in their G+C content.
That individual strains of S. mutans differ in their genetic composition has been demonstrated in a number of studies (10, 28, 35, 37, 43, 53), and the variation may be as much as 20%, comprising the “dispensable” genome (51). For example, variation in the com genes that mediate quorum sensing and genetic competence shows variation in distribution and genetic composition (4, 50). It may not be a coincidence that all of the loci described above have been linked directly or indirectly with S. mutans' virulence, and all exhibit variation. Some strains of S. mutans harbor 5.6-kb cryptic plasmids, and there are sufficient polymorphisms at the nucleotide level to allow phylogenetic ordering of plasmid-containing strains of S. mutans, giving insight into the evolutionary history of its human host (10).
Genetic variation among strains within a given species is not uncommon (for a review, see reference 1). Escherichia coli, for example, varies in intraspecies genetic composition among natural isolates as much as 20%; this is not surprising given its wide host range (44). Strains of Helicobacter pylori vary (29), with in silico comparisons between genomes of ca. 7%. Comparison of genomes of Staphylococcus aureus showed that strains are “peppered” with mobile elements and contains large blocks of genes in pathogenicity islands, with 6% of the genome being strain specific (13, 16, 20). Comparisons between strains of S. pyogenes (12) and between strains of S. pneumoniae (7) showed intraspecies differences ca. 10%. In both of these close relatives of S. mutans, differences are manifest to a large extent in the presence or absence of large blocks of genes. In many medically important bacteria, strain-specific genes reside in large chromosomal regions called genomic or pathogenicity islands (21-23). S. mutans UA159 contains at least 11 genomic islands (Los Alamos Oralgene site [http://www.oralgen.lanl.gov/]), but the distribution of these and other putative genomic islands among different strains of S. mutans remains unknown. Some of these genomic islands may be directly associated with the expression of virulence.
A novel aspect of the present study was the use of AI learning algorithms to use the presence or absence of SSH fragments to classify each S. mutans isolate by caries state (S-ECC or CF). Recent literature strongly suggests that AI approaches to classification outperform “classical” statistical method (50). This method provides a scaleable solution that can expand to incorporate multiple data types and large numbers of samples. AI, such as SVM, is very commonly used in disease prediction and pattern recognition in microarray data analysis, especially for cancer prediction. SVM algorithms have been successfully used in bacterial proteins (17, 18, 30, 39), metabolites (11), and pattern recognition and yielded >90% accuracy. In the present study the results from two independent forms of AI, SVM and neural network analyses, were compared to the true status of each sample to calculate sensitivity, specificity, and overall accuracy of the output. Exact binomial tests of independent proportions were used to identify fragments that exhibited maximal differentiation of S-ECC and CF. To control for the many multiple comparisons, we used adjusted P values to control for the false discovery rate (45). The number of SSH fragments needed to accurately classify strains can be reduced by considering a two-stage hierarchical classification procedure. As seen in Table 2, this can be achieved with high accuracy (92.0%), with only five biomarkers using a linear SVM. Table 3 shows the average prediction accuracy achieved on other pairwise discriminations, indicating that the CF versus S-ECC distinction can be made with high accuracy using just five SSH fragments. Our studies indicated that virulent clones possess the most important biomarkers, and most of the biomarkers identified are present in various strains. This finding is consistent with others that genetic variation among strains within a given species is common, and these genetic changes are associated with disease. Recently, McMillan et al. (33) reported that reemergence of severe, invasive group A streptococcal diseases could be caused by altered genetic endowment in these organisms. Using similar approach of neural net they identified three genes with a marginal overrepresentation in invasive disease isolates. Significantly, two of these genes, ssa and mf4, encoded superantigens but were only present in a restricted set of group A streptococcal M types. The third gene, spa, was found in variable distributions in all M types in the study (33). Using a similar approach, we identified a small set of SSH fragments that can be used to computationally “predict” S-ECC and CF S. mutans with high accuracy. In addition to SVM we also applied artificial neural networks algorithms to determine the robustness of fragments in classifying strains into S-ECC or CF. The recursive partitioning tree confirms that just three of these fragments (0018, H7, and 0006) can produce a classification accuracy of 94%. Thus, our methodology identifies an optimum combination of genes that may have the highest effect on the characteristic of S. mutans.
These data demonstrated successfully that DNA biomarkers obtained from SSH can be used to classify strains of S. mutans into S-ECC and CF groups. Further independent validation studies with larger sample size are warranted to evaluate the true potentials of these biomarkers. Even though these types of analyses do not tell us what the function or role of particular genetic loci plays in heath or disease, it does provide a panel of biomarkers that may be applicable to risk assessment. In addition, mapping of these fragments or biomarkers onto the chromosome will lead to the possible discovery of genomic islands or other horizontally acquired genetic loci that might be important in contributing to the overall virulence of S. mutans, including those which have yet to be identified. Since S. mutans is largely transferred from mother to child, a chairside test might be devised from these biomarkers capable of indicating potential risk to a child based on their own or their mother's strains of S. mutans. If successful, such a test would have tremendous public health implications for identifying children at risk before they experience this devastating disease.
Acknowledgments
These studies were supported by grant DE013937 from the NIDCR.
We thank Michele Savel and Hareeti R. Gill for assisting in collecting clinical samples and Liying Yang for technical support.
Footnotes
Published ahead of print on 2 July 2008.
REFERENCES
- 1.Achtman, M., and M. Wagner. 2008. Microbial diversity and the genetic nature of microbial species. Nat. Rev. Microbiol. 6431-440. [DOI] [PubMed] [Google Scholar]
- 2.Ajdic, D., W. M. McShan, R. E. McLaughlin, G. Savic, J. Chang, M. B. Carson, C. Primeaux, R. Tian, S. Kenton, H. Jia, S. Lin, Y. Qian, S. Li, H. Zhu, F. Najar, H. Lai, J. White, B. A. Roe, and J. J. Ferretti. 2002. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc. Natl. Acad. Sci. USA 9914434-14439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balakrishnan, M., R. S. Simmonds, M. Kilian, and J. R. Tagg. 2002. Different bacteriocin activities of Streptococcus mutans reflect distinct phylogenetic lineages. J. Med. Microbiol. 51941-948. [DOI] [PubMed] [Google Scholar]
- 4.Becker, M. R., B. J. Paster, E. J. Leys, M. L. Moeschberger, S. G. Kenyon, J. L. Galvin, S. K. Boches, F. E. Dewhirst, and A. L. Griffen. 2002. Molecular analysis of bacterial species associated with childhood caries. J. Clin. Microbiol. 401001-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berkowitz, R. J. 2003. Causes, treatment and prevention of early childhood caries: a microbiologic perspective. J. Can. Dent. Assoc. 69304-307. [PubMed] [Google Scholar]
- 6.Berkowitz, R. J., J. Turner, and C. Hughes. 1984. Microbial characteristics of the human dental caries associated with prolonged bottle-feeding. Arch. Oral Biol. 29949-951. [DOI] [PubMed] [Google Scholar]
- 7.Bruckner, R., M. Nuhn, P. Reichmann, B. Weber, and R. Hakenbeck. 2004. Mosaic genes and mosaic chromosomes—genomic variation in Streptococcus pneumoniae. Int. J. Med. Microbiol. 294157-168. [DOI] [PubMed] [Google Scholar]
- 8.Burt, B. A., W. J. Loesche, and S. A. Eklund. 1985. Stability of selected plaque species and their relationship to caries in a child population over 2 years. Caries Res. 19193-200. [DOI] [PubMed] [Google Scholar]
- 9.Caufield, P. W., K. Ratanapridakul, D. N. Allen, and G. R. Cutter. 1988. Plasmid-containing strains of Streptococcus mutans cluster within family and racial cohorts: implications for natural transmission. Infect. Immun. 563216-3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caufield, P. W., D. Saxena, D. Fitch, and Y. Li. 2007. Population structure of plasmid-containing strains of Streptococcus mutans, a member of the human indigenous biota. J. Bacteriol. 1891238-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Charaniya, S., S. Mehra, W. Lian, K. P. Jayapal, G. Karypis, and W.-S. Hu. 2007. Transcriptome dynamics-based operon prediction and verification in Streptomyces coelicolor. Nucleic Acids Res. 357222-7236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferretti, J., D. Ajdic, and W. McShan. 2004. Comparative genomics of streptococcal species. Indian J. Med. Res. 1191-6. [PubMed] [Google Scholar]
- 13.Goering, R. V., L. K. McDougal, G. E. Fosheim, K. K. Bonnstetter, D. J. Wolter, and F. C. Tenover. 2007. Epidemiologic distribution of the arginine catabolic mobile element among selected methicillin-resistant and methicillin-susceptible Staphylococcus aureus isolates. J. Clin. Microbiol. 451981-1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hazlett, K. R., J. E. Mazurkiewicz, and J. A. Banas. 1999. Inactivation of the gbpA gene of Streptococcus mutans alters structural and functional aspects of plaque biofilm which are compensated by recombination of the gtfB and gtfC genes. Infect. Immun. 673909-3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reference deleted.
- 16.Holden, M. T. G., E. J. Feil, J. A. Lindsay, S. J. Peacock, N. P. J. Day, M. C. Enright, T. J. Foster, C. E. Moore, L. Hurst, R. Atkin, A. Barron, N. Bason, S. D. Bentley, C. Chillingworth, T. Chillingworth, C. Churcher, L. Clark, C. Corton, A. Cronin, J. Doggett, L. Dowd, T. Feltwell, Z. Hance, B. Harris, H. Hauser, S. Holroyd, K. Jagels, K. D. James, N. Lennard, A. Line, R. Mayes, S. Moule, K. Mungall, D. Ormond, M. A. Quail, E. Rabbinowitsch, K. Rutherford, M. Sanders, S. Sharp, M. Simmonds, K. Stevens, S. Whitehead, B. G. Barrell, B. G. Spratt, and J. Parkhill. 2004. Complete genomes of two clinical Staphylococcus aureus strains: evidence for the rapid evolution of virulence and drug resistance. Proc. Natl. Acad. Sci. 1019786-9791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Idicula-Thomas, S., A. J. Kulkarni, B. D. Kulkarni, V. K. Jayaraman, and P. V. Balaji. 2006. A support vector machine-based method for predicting the propensity of a protein to be soluble or to form inclusion body on overexpression in Escherichia coli. Bioinformatics 22278-284. [DOI] [PubMed] [Google Scholar]
- 18.Jian Guo, Y. L. X. L. 2006. GNBSL: a new integrative system to predict the subcellular location for gram-negative bacteria proteins. PROTEOMICS 65099-5105. [DOI] [PubMed] [Google Scholar]
- 19.Kamiya, R. U., M. H. Napimoga, R. T. Rosa, J. F. Hofling, and R. B. Goncalves. 2005. Mutacin production in Streptococcus mutans genotypes isolated from caries-affected and caries-free individuals. Oral Microbiol. Immunol. 2020-24. [DOI] [PubMed] [Google Scholar]
- 20.Kelly, B. G., A. Vespermann, and D. J. Bolton. 14 February 2008, posting date. The role of horizontal gene transfer in the evolution of selected food-borne bacterial pathogens. Food Chem. Toxicol. doi: 10.1016/j.fct.2008.02.006. [DOI] [PubMed]
- 21.Kuramitsu, H. K. 2003. Molecular genetic analysis of the virulence of oral bacterial pathogens: an historical perspective. Crit. Rev. Oral Biol. Med. 14331-344. [DOI] [PubMed] [Google Scholar]
- 22.Kuramitsu, H. K. 1993. Virulence factors of mutans streptococci: role of molecular genetics. Crit. Rev. Oral Biol. Med. 4159-176. [DOI] [PubMed] [Google Scholar]
- 23.Kuramitsu, H. K. 2006. The virulence properties of Streptococcus mutans, 2nd ed. American Society for Microbiology, Washington, DC.
- 24.Li, Y., and P. W. Caufield. 1995. The fidelity of initial acquisition of mutans streptococci by infants from their mothers. J. Dent. Res. 74681-685. [DOI] [PubMed] [Google Scholar]
- 25.Li, Y., Y. Ge, V. M. Barnes, H. M. Trivedi, D. Saxena, P. W. Caufield, and T. Xu. 2005. Novel approach for evaluation of oral microbial reduction using PCR-DGGE, abstr. 3454. Abstr. IADR/AADR Ann. Meet., March 9 to 13. IADR/AADR, Baltimore, MD.
- 26.Li, Y., Y. Ge, D. Saxena, and P. W. Caufield. 2007. Genetic profiling of the oral microbiota associated with severe early-childhood caries. J. Clin. Microbiol. 4581-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li, Y., C. Y. Ku, J. Xu, D. Saxena, and P. W. Caufield. 2005. Survey of oral microbial diversity using PCR-based denaturing gradient gel electrophoresis. J. Dent. Res. 84559-564. [DOI] [PubMed] [Google Scholar]
- 28.Li, Y. H., P. C. Lau, J. H. Lee, R. P. Ellen, and D. G. Cvitkovitch. 2001. Natural genetic transformation of Streptococcus mutans growing in biofilms. J. Bacteriol. 183897-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Linz, B., F. Balloux, Y. Moodley, A. Manica, H. Liu, P. Roumagnac, D. Falush, C. Stamer, F. Prugnolle, S. W. van der Merwe, Y. Yamaoka, D. Y. Graham, E. Perez-Trallero, T. Wadstrom, S. Suerbaum, and M. Achtman. 2007. An African origin for the intimate association between humans and Helicobacter pylori. Nature 445915-918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lorena, A. C., and A. C. P. L. F. de Carvalho. 2007. Protein cellular localization prediction with support vector machines and decision trees. Comput. Biol. Med. 37115-125. [DOI] [PubMed] [Google Scholar]
- 31.Macrina, F. L., K. R. Jones, C. A. Alpert, B. M. Chassy, and S. M. Michalek. 1991. Repeated DNA sequence involved in mutations affecting transport of sucrose into Streptococcus mutans V403 via the phosphoenolpyruvate phosphotransferase system. Infect. Immun. 591535-1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Macrina, F. L., J. L. Reider, S. S. Virgili, and D. J. Kopecko. 1977. Survey of the extrachromosomal gene pool of Streptococcus mutans. Infect. Immun. 17215-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McMillan, D., R. G. Beiko, R. Geffers, J. Buer, L. M. Schouls, B. J. M. Vlaminckx, W. J. B. Wannet, K. S. Sriprakash, and G. S. Chhatwal. 2006. Genes for the majority of group A streptococcal virulence factors and extracellular surface proteins do not confer an increased propensity to cause invasive disease. Clin. Infect. Dis. 43884-891. [DOI] [PubMed] [Google Scholar]
- 34.Murchison, H. H., J. Barrett, G. A. Cardineau, and I. R. Curtiss. 1986. Transformation of Streptococcus mutans with chromosomal and shuttle plasmid (pYA629) DNAs. Infect. Immun. 54273-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qi, F., P. Chen, and P. W. Caufield. 2001. The group I strain of Streptococcus mutans, UA140, produces both the lantibiotic mutacin I and a nonlantibiotic bacteriocin, mutacin IV. Appl. Environ. Microbiol. 6715-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qi, F., P. Chen, and P. W. Caufield. 2000. Purification and biochemical characterization of mutacin I from the group I strain of streptococcus mutans, CH43, and genetic analysis of mutacin I biosynthesis genes. Appl. Environ. Microbiol. 663221-3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qi, F., P. Chen, and P. W. Caufield. 1999. Purification of mutacin III from group III Streptococcus mutans UA787 and genetic analyses of mutacin III biosynthesis genes. Appl. Environ. Microbiol. 653880-3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Radnedge, L., S. Gamez-Chin, P. M. McCready, P. L. Worsham, and G. L. Andersen. 2001. Identification of nucleotide sequences for the specific and rapid detection of Yersinia pestis. Appl. Environ. Microbiol. 673759-3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rashid, M., S. Saha, and G. Raghava. 2007. Support Vector Machine-based method for predicting subcellular localization of mycobacterial proteins using evolutionary information and motifs. BMC Bioinformatics 8337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robinson, W. G., L. Old, Shah, and R. Russell. 2003. Chromosomal insertions and deletions in Streptococcus mutans. Caries Res. 37148-156. [DOI] [PubMed]
- 41.Sawada, K., S. Kokeguchi, H. Hongyo, S. Sawada, M. Miyamoto, H. Maeda, F. Nishimura, S. Takashiba, and Y. Murayama. 1999. Identification by subtractive hybridization of a novel insertion sequence specific for virulent strains of Porphyromonas gingivalis. Infect. Immun. 675621-5625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saxena, D., Y. Li, and P. W. Caufield. 2005. Identification of unique bacterial gene segments from Streptococcus mutans with potential relevance to dental caries by subtraction DNA hybridization. J. Clin. Microbiol. 433508-3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shibata, Y., K. Ozaki, M. Seki, T. Kawato, H. Tanaka, Y. Nakano, and Y. Yamashita. 2003. Analysis of loci required for determination of serotype antigenicity in Streptococcus mutans and its clinical utilization. J. Clin. Microbiol. 414107-4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Steele, M., K. Ziebell, Y. Zhang, A. Benson, P. Konczy, R. Johnson, and V. Gannon. 2007. Identification of Escherichia coli O157:H7 genomic regions conserved in strains with a genotype associated with human infection. Appl. Environ. Microbiol. 7322-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Storey, J. 2003. The positive false discovery rate: a Bayesian interpretation and the q-value. Ann. Stat. 312013-2035. [Google Scholar]
- 46.Tanzer, J. M., J. Livingston, and A. M. Thompson. 2001. The microbiology of primary dental caries in humans. J. Dent. Educ. 651028-1037. [PubMed] [Google Scholar]
- 47.Tatusov, R. L., E. V. Koonin, and D. J. Lipman. 1997. A genomic perspective on protein families. Science 278631-637. [DOI] [PubMed] [Google Scholar]
- 48.Ueda, S., and H. K. Kuramitsu. 1988. Molecular basis for the spontaneous generation of colonization-defective mutants of Streptococcus mutans. Mol. Microbiol. 2135-140. [DOI] [PubMed] [Google Scholar]
- 49.van Houte, J., G. Gibbs, and C. Butera. 1982. Oral flora of children with “nursing bottle caries”. J. Dent. Res. 61382-385. [DOI] [PubMed] [Google Scholar]
- 50.Veltri, R. W., M. Chaudhari, M. C. Miller, E. C. Poole, G. J. O'Dowd, and A. W. Partin. 2002. Comparison of logistic regression and neural net modeling for prediction of prostate cancer pathologic stage. Clin. Chem. 481828-1834. [PubMed] [Google Scholar]
- 51.Waterhouse, J. C., and R. R. B. Russell. Dispensable genes and foreign DNA in Streptococcus mutans. Microbiology 1521777-1788, 2006. [DOI] [PubMed]
- 52.Yamashita, Y., W. H. Bowen, and H. K. Kuramitsu. 1992. Molecular analysis of a Streptococcus mutans strain exhibiting polymorphism in the tandem gtfB and gtfC genes. Infect. Immun. 601618-1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou, X., P. W. Caufield, Y. Li, and F. Qi. 2001. Complete nucleotide sequence and characterization of pUA140, a cryptic plasmid from Streptococcus mutans. Plasmid 4677-85. [DOI] [PubMed] [Google Scholar]