Abstract
The meningococcal Opa proteins play an important role in pathogenesis by mediating invasion of human cells. The aim of this investigation was to determine whether carried and disease-associated meningococci possess different Opa repertoires and whether the diversity of these proteins is associated with clinical severity of disease. Opa repertoires in 227 disease-associated meningococci, isolated in the United Kingdom over a period of 6 years, were compared to the repertoires in 190 asymptomatically carried meningococci isolated in the United Kingdom from a contemporary, nonepidemic period. Multidimensional scaling (MDS) was employed to investigate the association between Opa repertoires and multilocus sequence typing (MLST) genotypes. Associations with clinical severity were also analyzed statistically. High levels of diversity were observed in opa alleles, variable regions, and repertoires, and MDS revealed that MLST genotypes were strongly associated with particular Opa repertoires. Individual Opa proteins or repertoires were not associated with clinical severity, though there was a trend toward an association with the opaD locus. Meningococcal Opa repertoire is strongly linked to MLST genotype irrespective of epidemiological sampling and therefore correlates with invasiveness. It is not, however, strongly associated with severity of meningococcal disease.
Invasive meningococcal disease caused by the gram-negative bacterium Neisseria meningitidis (the meningococcus) has an incidence of one to six cases per 100,000 persons in Europe (40) and a mortality rate of approximately 8% (42). The disease is associated with different clinical presentations, most commonly meningitis or septicemia or a combination of the two. Its clinical severity is likely to be dependent on host and bacterial factors that influence the inflammatory and immunological responses of the host (9, 12). A variety of meningococcal surface and secreted proteins play roles in contact with host cells (30), and some of these regulate host immunological responses (1, 35). Few studies have investigated the distribution of the proteins involved in these interactions in the meningococcal population or their effects on clinical severity of invasive disease.
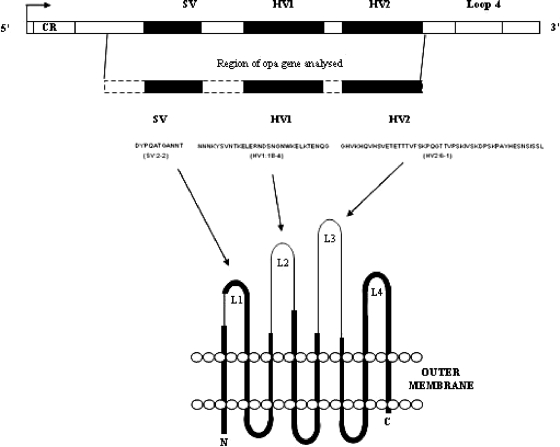
The opacity-associated adhesin (Opa) proteins (Fig. 1), located on the meningococcal surface, promote intimate interaction with the host (44) and modulate host immunological responses. Topologically, Opa proteins display an eight-stranded transmembrane β-barrel structure with four extramembranous putative loop regions (7). A semivariable (SV) and two hypervariable (HV1 and HV2) regions are present in three of the loop regions, and a large number of Opa variants have been observed (5, 22). Sequence variation, particularly in the two HV regions, influences the specificity of Opa variants for different members of the human carcinoembryonic antigen cell adhesion molecule (CEACAM) family of proteins (45, 46) and cell surface saccharides (26). These interactions lead to adhesion between the bacterium and host or adhesion followed by invasion of the host cells dependent on the particular Opa variants and receptors involved (28, 43). The Opa proteins of Neisseria species also modulate the host immune response through interactions with cells of the immune system, including neutrophils and CD4+ T cells (16, 24, 45). Opa proteins also stimulate the secretion of proinflammatory cytokines, the levels of which correlate with severity of meningococcal disease (4, 31), from human macrophages (21).
FIG. 1.
Schematic of an opa gene, including the variable regions sequenced in this study and the relationship of these sequences to the predicted secondary Opa protein structure (reprinted from reference 5). CR, phase variable coding region repeat.
The Opa proteins are encoded by four genetic opa loci in each isolate (32, 39), encoding its Opa repertoire. Population-based studies of Opa diversity have suggested that the meningococcal hyperinvasive clonal complexes, genotypes identified by multilocus sequence typing (MLST) and responsible for the majority of global meningococcal disease (14, 20), are associated with particular Opa repertoires (5, 27). It is unclear whether this association is unique to disease-causing meningococci or whether clonal complexes isolated from asymptomatic carriage that have never or rarely caused disease also exhibit this feature. Furthermore, it is unclear whether these combinations in disease-causing meningococci are associated with differences in clinical severity.
The influence of Opa variation on severity of meningococcal disease, potentially through stimulation of proinflammatory cytokine production or through increases in bacterial invasiveness mediated by different Opa protein variants, is unclear due to the lack of information on the function and frequency distribution of Opa variants among meningococci. The aim of this investigation was to compare the opa gene repertoires of meningococci isolated from invasive disease with those of asymptomatically carried meningococci collected during a contemporary, nonepidemic period and to determine whether opa diversity is associated with clinical severity of meningococcal disease.
MATERIALS AND METHODS
Meningococcal isolate collection.
A total of 417 N. meningitidis isolates, obtained from both asymptomatic carriage and invasive meningococcal disease, were analyzed in this investigation. This was a largely contemporaneous collection, with all carriage isolates collected during 1999 and disease isolates collected during the 6 years between 1996 and 2001. All isolates were from the United Kingdom, but most were from the southeast part of England. The isolates are described more fully below.
A set of 190 isolates were representative of a larger number of isolates collected from individuals carrying the bacterium in Oxfordshire, a county in the southeast part of England, United Kingdom, during 1999. Nasopharyngeal swab samples were taken from 15- to 19-year-old students attending school or college who were recruited as previously described (19), with informed consent obtained in writing. The study was approved by the Trent MultiCentre Research Ethics Committee (study number MREC/99/4/036). Meningococci were identified by positive culture and have been characterized by MLST (18).
A further 227 isolates from invasive meningococcal disease were obtained from two sources of patients. A total of 120 isolates were obtained following admission to the pediatric intensive care unit at St. Mary's Hospital, London, United Kingdom, approximately 50 miles (∼80 km) from Oxford, between 1992 and 2002. A further 107 isolates were collected as part of a national United Kingdom meningococcal disease study overseen by the Royal College of Pediatrics and Child Health (RCPCH) in which all fatal pediatric meningococcal disease cases between 1 December 1997 and 28 February 1999 were investigated. The severity data and disease isolates used in the current study were collected and analyzed under St. Mary's Hospital Local Research Ethics Committee approval number EC3263. This isolate collection was originally assembled for analysis as part of the European Meningococcal Monitoring Network (EUMenNet) (10, 41). Full MLST analysis of disease isolates will be described elsewhere.
For 182 out of the 227 disease isolates, a Glasgow Meningococcal Septicemia Prognostic Score (GMSPS) of clinical severity (37) was assigned retrospectively from case notes. The GMSPS is a widely used, simple, clinical scoring system based on seven clinical parameters, including degree of hypotension, skin/rectal temperature difference, base deficit, extent of hemorrhagic skin lesions, Simpson and Reilly pediatric coma score, presence of meningism, and parents' opinion of any change in the patient's condition in the hour prior to scoring. A score of 8 or above out of a total of 15 indicates 100% sensitivity, 75% specificity, and 29% positive predictive value for death, the risk of which increases as the score rises above 8 (34).
Preparation of meningococcal genomic DNA.
Isolates were revived from storage below −70°C by growth on single plates of Columbia agar in a 5% CO2 atmosphere at 37°C for 18 h or overnight. Meningococcal growth from each individual plate was collected into phosphate-buffered saline solution (1 ml) and heated to 56°C to kill the bacteria before genomic DNA was extracted using a Qiagen DNA mini kit (Qiagen) according to the manufacturer's instructions. Purified genomic DNA was solubilized in distilled/deionized water and stored below −20°C.
Determination of opa gene nucleotide sequences.
The meningococcal Opa proteins are encoded by three or four phase-variable, unlinked genomic opa loci (opaA, opaB, opaD, and opaJ) (32, 39). As previously described (5), the nucleotide sequence of each opa locus was determined. Briefly, each opa locus was amplified by PCR using locus-specific primer sets and nucleotide sequences were determined directly from PCR products. Variable-region amino acid sequence variants and sequence variants within families were identified by comparison with previously published data, and new sequence variants/variant families were assigned arbitrarily in the order of discovery, all according to previously published nomenclature (5).
Alignments of amino acid sequences and the original nucleotide sequences were created using the CLUSTALW algorithm in the program DAMBE (47). Nucleotide and amino acid p distances were calculated using the MEGA version 3.0 software package (15), with sequence alignment gaps compensated for by using a pairwise deletion method. The Opa repertoires were described by the combination of HV1 and HV2 amino acid sequence families at each locus in the same isolate, with each unique HV1-HV2 combination and each unique repertoire of these combinations assigned an arbitrary identification number. This approach reduced the high diversity observed at the allelic level, allowing identification of relationships among repertoires comprising different alleles encoding identical protein sequences. Furthermore, this approach allowed the analyses to focus on the Opa HV region sequences and their combinations, which contain functionally important receptor binding sequences.
Analysis of Opa repertoire clustering.
Classical multidimensional scaling (MDS) analysis (17, 33) was employed to investigate relationships among Opa repertoires as defined by HV1 and HV2 sequence variants (i.e., the functionally and antigenically important regions of Opa) at all four loci. First, the differences in the Opa repertoires among isolates were determined by comparing each isolate to every other and counting the number of different HV region variants between each pair of isolates (with eight being the maximum number different from a total of four HV1 and four HV2 variants). Identical HV regions occurring at multiple opa loci in a pair of isolates were treated as equal, and loci where an opa allele was not detected were treated as different. The resulting “distance matrix” was then used as input for the MDS algorithm. The algorithm determined the relationships among isolates by displaying the distance matrix as a set of data points, with one data point for each isolate that best represented its position in relation to other isolates in Euclidean geometric space. This was represented as a multidimensional scatter plot. The number of spatial dimensions in the MDS analysis was allowed to vary, but the lowest number of dimensions that described the largest proportion of the data was defined as the most efficient at depicting the relationships among isolates on the basis of their Opa repertoires.
Statistical analysis of the association between Opa repertoires and clinical severity of meningococcal disease.
The overall association between Opa and severity was assessed by correlation between distance matrices derived from the Opa repertoires and a distance matrix based on GMSPS scores. These were compared using a Mantel test, and the correlation was tested using a 1,000-iteration permutation test as implemented in the program ARLEQUIN (36). The association between GMSPS and opa alleles or Opa variable-region amino acid sequence families was also assessed using linear regression, with GMSPS as the outcome variable and opa alleles or Opa variable-region amino acid families as a categorical independent variable. Statistical significance was assessed by F tests.
Nucleotide sequence accession numbers.
New opa alleles have been posted in GenBank under accession numbers EU759983 to EU760345.
RESULTS
opa diversity in meningococci isolated from cases of invasive disease.
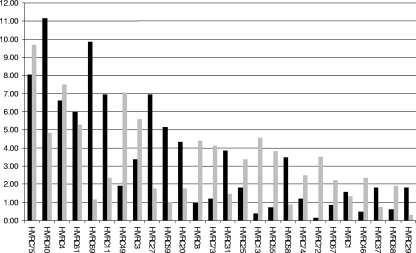
Among the 227 isolates from meningococcal disease analyzed in this investigation, opa gene sequences were detected in 833 of the total 908 loci examined (92%), carrying 205 different opa alleles (Table 1). Meningococci have 3 or 4 opa loci, and in isolates from the invasive disease collection, alleles were not detected at 1 opaA locus, 1 opaB locus, 29 opaD loci (one of which had a frameshift mutation), and 44 opaJ loci (all of which were disrupted by insertion sequence-like elements). PCR and sequencing were repeated with new genomic DNA preparations, alternative primers, and cycling conditions for loci at which an opa allele could not be detected. The 205 alleles encoded 55 HV1-HV2 sequence family combinations (Fig. 2), of which 12 were present at frequencies above 2%, accounting for 705 (85%) of the 833 opa loci. The single most common HV1-HV2 family combination was composed of HV1 family 11 and HV2 family 1 (combination 40), present at 93 loci, of which 82 were present at the opaD locus of sequence type 11 (ST-11) and ST-8 complex isolates. Other occurrences included the opaA loci of eight ST-11 isolates, with these variable regions also present at their opaD loci.
TABLE 1.
Diversity of Opa proteins in isolates from invasive diseasea
| Locus | No. of alleles | No. of SV variants | SV family | No. of HV1 variants | No. of HV1 families | No. of HV2 variants | No. of HV2 families | No. of HVRCs |
|---|---|---|---|---|---|---|---|---|
| opaA | 73 (13.0) | 13 (25.8) | 5 | 39 (44.0) | 17 | 45 (36.0) | 19 | 34 |
| opaB | 92 (11.6) | 15 (27.0) | 5 | 45 (41.1) | 15 | 52 (33.6) | 18 | 37 |
| opaD | 55 (13.9) | 8 (22.7) | 3 | 34 (45.8) | 16 | 37 (40.4) | 17 | 30 |
| opaJ | 50 (15.2) | 12 (23.4) | 5 | 30 (48.6) | 17 | 30 (43.1) | 15 | 24 |
Opa diversity is determined by alleles, variable-region sequences (the numbers of SV, HV1, and HV2 sequence variants observed are given), sequence families, and HV1-HV2 sequence family combinations (HVRCs). Percent distances are given in parentheses for nucleotide alleles and amino acid sequence data.
FIG. 2.
Comparison of HV1-HV2 sequence family combination (HVRC) frequencies in meningococci from invasive disease (black bars) versus those from asymptomatic carriage (gray bars). Only HV1-HV2 sequence family combinations with frequencies above 2% are shown (see Table S1 in the supplemental material for full data set).
opa diversity in meningococci isolated from nasopharyngeal carriage.
Among the 190 meningococcal carriage isolates, 246 different opa allele sequences were detected in 684 (90%) of the total 760 loci analyzed (Table 2). A full-length opa allele was not detected at the opaA loci of 14 isolates (2 of which were disrupted by frameshift mutations), at the opaB loci of 7 isolates, at the opaD loci of 25 isolates, and at the opaJ loci of 30 isolates (2 of which were disrupted by frameshift mutations). A further three isolates had alleles at their opaJ loci that were highly similar by alignment to opa genes from Neisseria lactamica (data not shown). These alleles were identical in isolates 99-30740 and 99-30928, both members of the ST-198 clonal complex. As in the collection of disease isolates, PCR and sequencing were repeated with new genomic DNA preparations, alternative primers, and cycling conditions for isolates at which an opa allele was not detected. There were 65 combinations of HV1-HV2 families (Fig. 2), with 15 combinations present at frequencies above 2%, accounting for 555 (81%) of loci. The single most common HV1-HV2 family combination was combination 75, composed of HV1 family 19 and HV2 family 14, present at 66 loci, equating to 10% of the total loci, appearing in isolates belonging to multiple clonal complexes.
TABLE 2.
Diversity of Opa proteins in isolates from asymptomatic carriagea
| Locus | No. of alleles | No. of SV variants | SV family | No. of HV1 variants | No. of HV1 families | No. of HV2 variants | No. of HV2 families | No. of HVRCs |
|---|---|---|---|---|---|---|---|---|
| opaA | 93 (12.4) | 8 (28.1) | 4 | 47 (48.3) | 13 | 61 (34.7) | 21 | 38 |
| opaB | 88 (12.0) | 11 (26.9) | 4 | 58 (42.4) | 18 | 58 (35.6) | 21 | 43 |
| opaD | 75 (15.0) | 15 (25.7) | 4 | 44 (42.8) | 14 | 51 (43.6) | 19 | 32 |
| opaJ | 61 (15.0) | 15 (30.4) | 5 | 36 (46.7) | 16 | 36 (41.5) | 18 | 30 |
Opa diversity is determined by alleles, variable-region sequences (the numbers of SV, HV1, and HV2 sequence variants observed are given), sequence families, and HV1-HV2 sequence family combinations (HVRCs). Percent distances are given in parentheses for nucleotide alleles and amino acid sequence data.
Comparison of Opa repertoires in carried meningococci versus disease-related isolates.
There were a total of 269 Opa repertoires in the 417 meningococci analyzed, based on the combination of HV1 and HV2 families at each of the four loci per isolate. In isolates from meningococcal disease, there were 133 Opa repertoires (based on the combination of HV1 and HV2 sequence families at each locus), 16 of which were present at frequencies above 1% (appearing in more than 3 isolates). The most frequent repertoire was composed of HV1-HV2 combinations 27 (OpaA), 69 (OpaB), and 40 (OpaD), with the opaJ locus inactivated due to the presence of an insertion sequence element. This repertoire was observed in 39 (17%) of isolates from invasive disease, all of which were members of the ST-11 complex, and was identical to that in carried and disease-associated ST-11 meningococci responsible for an epidemic in the Czech Republic in 1993 (4a). In carried meningococci, there were 142 repertoires, of which 25 were observed at frequencies above 1% (appearing in more than two isolates). The most common of these was characterized by HV1-HV2 combinations 67 (OpaA), 72 (OpaB), and 40 (OpaJ). An opa gene was not detected at the opaD locus in these isolates. This repertoire was present in 10 meningococci, 9 of which were members of the ST-53 complex. A total of five repertoires were present in both collections at low frequencies, appearing in no more than nine disease isolates and two carriage isolates.
Structuring of the Opa repertoires of individual clonal complexes.
Analysis of the Opa repertoires of individual clonal complexes in the combined collection of carriage and disease isolates indicated that the Opa repertoires of some complexes were more conserved, composed of fewer Opa proteins, than others. The three most common clonal complexes were the ST-11, ST41/44, and ST-269 complexes, whose repertoires were composed of six or seven Opa proteins in each complex, with three or four variants above 10% accounting for 71% of the total number of loci analyzed in all isolates from the ST-11 complex (HV1-HV2 combinations 27, 40, and 69), 62% of the total loci in all isolates of the ST-41/44 complex (HV1-HV2 combinations 4, 11, 20, and 31), and 59% of the total loci in all isolates of the ST-269 complex (HV1-HV2 combinations 59, 61, and 69). No differences in the Opa repertoires of individual clonal complexes between asymptomatic carriage and invasive disease were apparent.
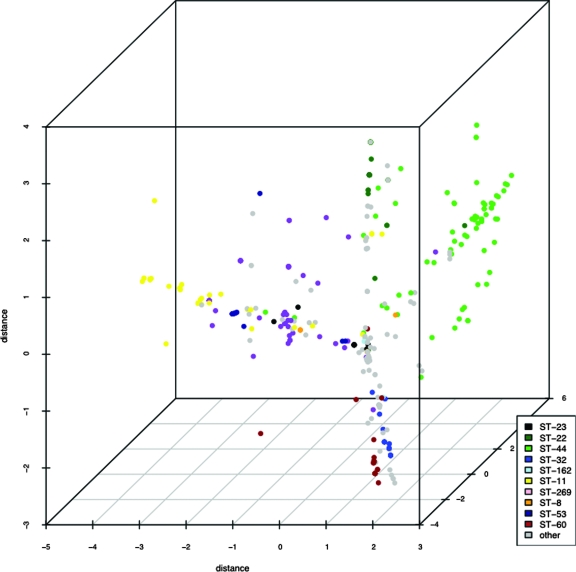
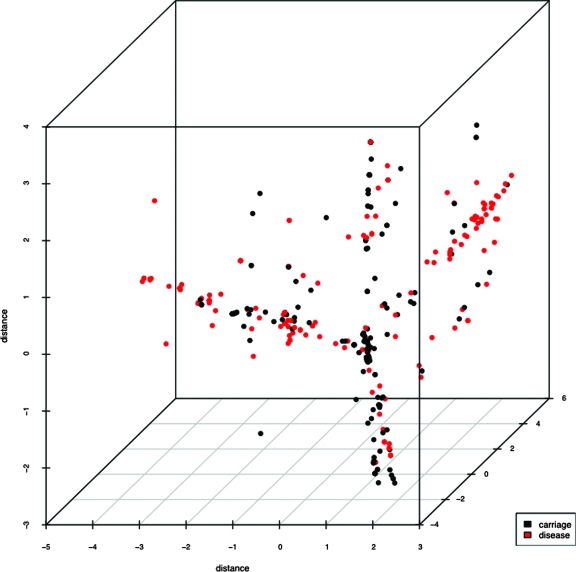
MDS analysis revealed that Opa repertoires of genetically related isolates clustered together (Fig. 3) irrespective of whether they were isolated from disease or carriage. Four major clusters were observed, each containing disease-related and carried isolates (Fig. 4). Two of these clusters were composed mostly of disease-related isolates, one containing the majority of isolates belonging to the ST-41/44 complex, whereas the other included members of the ST-11 and ST-269 complexes. The variance of the scattering of repertoires was larger in isolates from disease, with data points distributed further apart (the mean variance in the three dimensions shown was 3.051), whereas the data points for repertoires from carried isolates clustered tighter (the mean variance in the three dimensions shown was 2.079). The spread of data points along the three axes also differed between disease-related and carried isolate repertoires. Values for carried isolates were 1.777 (x), 1.6499 (y), and 2.809 (z). In disease isolates, the values were 5.889 (x), 2.165 (y), and 1.098 (z). These data indicated that the repertoires of disease isolates mostly clustered along the x axis, whereas those of carried isolates tended toward the y axis.
FIG. 3.
MDS scatter plot showing distances between Opa repertoires of carriage and disease isolates in three dimensions of sequence space. Isolates have been colored according to clonal complex to indicate associations between antigenic and genotypic repertoires.
FIG. 4.
MDS scatter plot showing distances between Opa repertoires of carriage and disease isolates in three dimensions of sequence space. Isolates have been colored according to carriage or disease status.
Association of Opa diversity and clinical severity.
GMSPS data were available for 182 patients of the 227 for whom meningococcal isolates were collected (80.2%). Of these, 96 out of 182 (52.75%) had scores of 0 to 7, whereas 86 of 182 (47.25%) had scores of 8 or higher. The null hypothesis, that the opa gene or Opa protein repertoire of meningococci is not associated with clinical severity of meningococcal disease, was investigated. No evidence for a correlation between the distance matrices constructed from the Opa repertoire (HV1-HV2 family combinations) and the severity scores was found (correlation coefficient of −0.001 and P value of 0.51). There was no significant relationship observed between any component of opa alleles or Opa proteins and severity as measured by the GMSPS. A trend was observed, however, for an association across the data set between the three variable regions encoded by alleles at the opaD locus and severity, with p values of approximately 0.1 (data not shown).
DISCUSSION
The data presented here show that meningococcal Opa repertoires and MLST genotypes are closely linked, irrespective of epidemiological sampling. Isolates from carried genotypes, as well as those from hyperinvasive genotypes, had highly structured, similar Opa repertoires. An association between Opa diversity and clinical severity of meningococcal disease was not found, although there was a trend toward an association at the opaD locus.
MDS analysis of the diversity of the Opa repertoires in this investigation indicated an association with meningococcal MLST sequence types and clonal complexes. Previous studies have identified combinations of Opa proteins consistently observed in meningococcal hyperinvasive clonal complexes over decades of epidemic spread (5, 27). Hyperinvasive clonal complexes are not representative of the genetic diversity observed in asymptomatically carried meningococcal populations, however, which are composed of numerous, diverse genotypes (13, 14). Consequently, the association between Opa repertoire and MLST genotype cannot be extended to carriage populations by way of inferences made using data solely from hyperinvasive complexes. In this investigation, the asymptomatically carried meningococci were all genetically diverse serogroup B and C isolates collected from asymptomatic carriers during a contemporary, nonepidemic period in the United Kingdom. Furthermore, the majority of these isolates belonged to clonal complexes that are never or have rarely been isolated from disease. These data now provide additional evidence that meningococcal isolates belonging to the same sequence type or clonal complex are consistently associated with particular combinations of Opa adhesins. This association appears to be epidemiologically stable, maintained whether isolates are collected from carriage or disease. Furthermore, MDS analysis supported a trend toward differential clustering of Opa repertoires in meningococci isolated from carriage and disease, also indicated by the low number of repertoires appearing in both groups of isolates.
Different Opa proteins exhibit binding specificities for a range of human receptors, including members of the CEACAM family (28, 43). The data presented here are consistent with a view in which particular combinations of Opa proteins confer different meningococcal genotypes with particular sets of binding specificities and these specificities are stably associated with each genotype. In the case of disease-causing meningococci, these combinations and specificities may contribute to invasiveness, for example, particular Opa proteins may enhance colonization and invasion of the host. Different variants may confer different effects on host immunity, since Opa proteins are known to elicit the release of proinflammatory cytokines from monocytes (21) and downregulate the activation and proliferation of CD4+ T cells via CEACAM1 binding (2, 16). Currently, little is known about the function of more than a few Opa variants and it is difficult to infer the binding specificities of the variants identified here from the few that have been studied. By determining that different Opa variants are associated with carried clonal complexes compared to those causing invasive disease, the population data in this investigation may inform future studies aimed at understanding the functional roles of different Opa variants in meningococcal pathogenesis.
An alternative explanation for the association between Opa repertoire and MLST genotype involves antigenic Opa diversity rather than function. The evolution and diversity of the meningococcal Opa repertoire are likely to be influenced by diversifying selection pressure from host antibody responses. Indeed, Opa-specific antibodies have been detected after meningococcal carriage and after invasive disease (23, 38). Host population antibody responses against the meningococcal PorA protein, a porin located in the outer membrane, have been suggested to shape the diversity of the meningococcal population (11). We propose that host population antibody responses also affect the structuring of the Opa repertoire in a similar way, by selecting against meningococci sharing parts of their Opa repertoires with other, unrelated genotypes that may have been previously recognized by host immune defenses. An immunological reason for the structuring of the diversity would not be mutually exclusive with a functional explanation, since the parts of Opa proteins containing epitopes for antibodies may be different from those conferring functional specificity. Furthermore, the structuring and evolution of the meningococcal Opa repertoire is highly likely to be influenced by the strength of both functional and immunological selective forces during carriage.
The association of particular Opa repertoires with hyperinvasive clonal complexes may explain also why these meningococci cause most invasive disease (6). Different Opa repertoires have different receptor binding specificities (8, 28, 43), which are likely to influence both meningococcal invasiveness and the ability of the bacteria to regulate human immunity during carriage and disease. Consequently, it may be possible to target hyperinvasive meningococci by using Opa protein-based vaccines, especially since Opa proteins elicit antibodies after immunization with outer membrane vesicle vaccines and after meningococcal infection (25, 38). There are still important immunological questions surrounding this strategy, however, such as those concerning the effects of Opa proteins on arrest of proliferation of CD4+ T lymphocytes (16) and whether these effects are clinically relevant during vaccination.
A trend was observed for an association between disease severity and opaD locus across the set of disease isolates but was not statistically supported. The reason for this potential link, which requires further investigation with a larger data set, is unclear due to the number and diversity of Opa variants at this locus. No other associations between opa alleles or Opa repertoires and severity were observed. The factors affecting meningococcal clinical severity are incompletely understood, and previous investigations have explored the effects of genomic DNA load and lipooligosaccharide (LOS) levels and their influence on host cytokine profiles (4, 12, 31). Cytokine levels are critically important in influencing meningococcal disease severity. In contrast to the association between LOS levels in the blood and clinical severity of meningococcal disease, cytokine levels have been found to be identical in LOS-expressing and LOS-negative bacteria, suggesting a LOS-independent pathway (4). Given the biological activities mediated by Opa proteins, including promotion of invasion of host cells and stimulation of cytokine release from monocytes (21), the lack of an Opa effect on disease severity is interesting. One caveat regarding this is that although we did not detect an association between clinical severity and particular Opa sequence variants, differences in Opa expression levels among different clonal complexes, as opposed to sequence diversity, have yet to be fully explored and may be important in pathogenesis and severity. For example, previous studies have found that expression of Opa proteins upregulates the expression of CEACAM receptors on host cells (29), which leads to increased adhesion of meningococci (3). Furthermore, while this study has focused on the diversity of Opa proteins, little is also known about genetic variation in CEACAM, the major Opa receptor, and whether particular genotypes or haplotypes influence severity or susceptibility to invasive meningococcal disease, perhaps by allowing increased uptake of meningococci via Opa interaction.
In conclusion, the meningococcal Opa repertoire was found to be strongly linked to MLST genotypes, irrespective of epidemiological sampling. Since MLST genotype also identifies hyperinvasive meningococci, particular Opa repertoires correlate with disease. The Opa repertoire was not, however, strongly associated with severity of meningococcal disease, suggesting that other host and meningococcal factors may be more important than variation in Opa in determining differences in clinical severity of disease.
Supplementary Material
Acknowledgments
We acknowledge the aid of staff at St. Mary's Hospital, London, United Kingdom, for collecting human blood samples, purifying genomic DNA, and assembling clinical severity scores. The assistance of Roisin Ure for DNA extraction and the aid of the departmental DNA sequencing service at the University of Oxford, Department of Zoology, for separation of nucleotide sequence reactions are also acknowledged.
This study was funded by the Meningitis Research Foundation (MRF), Bristol, United Kingdom, as part of project 02/02.
A.J.P. and M.C.J.M. are named as inventors and M.J.C. is named as a contributor on patent applications in the area of serogroup B meningococcal vaccine development. No other authors report conflicts of interest.
Footnotes
Published ahead of print on 28 May 2008.
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Al-Bader, T., M. Christodoulides, J. E. Heckels, J. Holloway, A. E. Semper, and P. S. Friedmann. 2003. Activation of human dendritic cells is modulated by components of the outer membranes of Neisseria meningitidis. Infect. Immun. 715590-5597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boulton, I. C., and S. D. Gray-Owen. 2002. Neisserial binding to CEACAM1 arrests the activation and proliferation of CD4+ T lymphocytes. Nat. Immunol. 3229-236. [DOI] [PubMed] [Google Scholar]
- 3.Bradley, C. J., N. J. Griffiths, H. A. Rowe, R. S. Heyderman, and M. Virji. 2005. Critical determinants of the interactions of capsule-expressing Neisseria meningitidis with host cells: the role of receptor density in increased cellular targeting via the outer membrane Opa proteins. Cell. Microbiol. 71490-1503. [DOI] [PubMed] [Google Scholar]
- 4.Brandtzaeg, P., P. Kierulf, P. Gaustad, A. Skulberg, J. N. Bruun, S. Halvorsen, and E. Sorensen. 1989. Plasma endotoxin as a predictor of multiple organ failure and death in systemic meningococcal disease. J. Infect. Dis. 159195-204. [DOI] [PubMed] [Google Scholar]
- 4a.Callaghan, M. J., C. O. Buckee, K. A. Jolley, P. Kriz, M. C. Maiden, and S. Gupta. 14 March 2008. The effect of immune selection on the structure of the meningococcal Opa protein repertoire. PLoS Pathog. 4e1000020. [Epub ahead of print.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Callaghan, M. J., K. A. Jolley, and M. C. J. Maiden. 2006. The opacity-associated adhesin repertoire in hyperinvasive Neisseria meningitidis. Infect. Immun. 745085-5094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caugant, D. A. 1998. Population genetics and molecular epidemiology of Neisseria meningitidis. APMIS 106505-525. [PubMed] [Google Scholar]
- 7.de Jonge, M. I., M. P. Bos, H. J. Hamstra, W. Jiskoot, P. van Ulsen, J. Tommassen, L. van Alphen, and P. van der Ley. 2002. Conformational analysis of opacity proteins from Neisseria meningitidis. Eur. J. Biochem. 2695215-5223. [DOI] [PubMed] [Google Scholar]
- 8.de Jonge, M. I., H. J. Hamstra, L. van Alphen, J. Dankert, and P. van der Ley. 2003. Mapping the binding domains on meningococcal Opa proteins for CEACAM1 and CEA receptors. Mol. Microbiol. 501005-1015. [DOI] [PubMed] [Google Scholar]
- 9.Emonts, M., J. A. Hazelzet, R. de Groot, and P. W. Hermans. 2003. Host genetic determinants of Neisseria meningitidis infections. Lancet Infect. Dis. 3565-577. [DOI] [PubMed] [Google Scholar]
- 10.Frosch, M., and M. Maiden. 2007. The European networking for combating meningococcal disease. FEMS Microbiol. Rev. 311-2. [DOI] [PubMed] [Google Scholar]
- 11.Gupta, S., M. C. Maiden, I. M. Feavers, S. Nee, R. M. May, and R. M. Anderson. 1996. The maintenance of strain structure in populations of recombining infectious agents. Nat. Med. 2437-442. [DOI] [PubMed] [Google Scholar]
- 12.Hackett, S. J., M. Guiver, J. Marsh, J. A. Sills, A. P. Thomson, E. B. Kaczmarski, and C. A. Hart. 2002. Meningococcal bacterial DNA load at presentation correlates with disease severity. Arch. Dis. Child. 8644-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jolley, K. A., J. Kalmusova, E. J. Feil, S. Gupta, M. Musilek, P. Kriz, and M. C. Maiden. 2000. Carried meningococci in the Czech Republic: a diverse recombining population. J. Clin. Microbiol. 384492-4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jolley, K. A., D. J. Wilson, P. Kriz, G. McVean, and M. C. Maiden. 2005. The influence of mutation, recombination, population history, and selection on patterns of genetic diversity in Neisseria meningitidis. Mol. Biol. Evol. 22562-569. [DOI] [PubMed] [Google Scholar]
- 15.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief. Bioinform. 5150-163. [DOI] [PubMed] [Google Scholar]
- 16.Lee, H. S., I. C. Boulton, K. Reddin, H. Wong, D. Halliwell, O. Mandelboim, A. R. Gorringe, and S. D. Gray-Owen. 2007. Neisserial outer membrane vesicles bind the coinhibitory receptor carcinoembryonic antigen-related cellular adhesion molecule 1 and suppress CD4+ T lymphocyte function. Infect. Immun. 754449-4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ligges, U., and M. Maechler. 2003. Scatterplot3d—an R package for visualizing multivariate data. J. Stat. Softw. 81-20. [Google Scholar]
- 18.Maiden, M. C., A. B. Ibarz-Pavon, R. Urwin, S. J. Gray, N. J. Andrews, S. C. Clarke, A. M. Walker, M. R. Evans, J. S. Kroll, K. R. Neal, D. A. Ala'aldeen, D. W. Crook, K. Cann, S. Harrison, R. Cunningham, D. Baxter, E. Kaczmarski, J. Maclennan, J. C. Cameron, and J. M. Stuart. 2008. Impact of meningococcal serogroup C conjugate vaccines on carriage and herd immunity. J. Infect. Dis. 197737-743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maiden, M. C., and J. M. Stuart. 2002. Carriage of serogroup C meningococci 1 year after meningococcal C conjugate polysaccharide vaccination. Lancet 3591829-1831. [DOI] [PubMed] [Google Scholar]
- 20.Maiden, M. C. J., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 953140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Makepeace, B. L., P. J. Watt, J. E. Heckels, and M. Christodoulides. 2001. Interactions of Neisseria gonorrhoeae with mature human macrophage opacity proteins influence production of proinflammatory cytokines. Infect. Immun. 691909-1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malorny, B., G. Morelli, B. Kusecek, J. Kolberg, and M. Achtman. 1998. Sequence diversity, predicted two-dimensional protein structure, and epitope mapping of neisserial Opa proteins. J. Bacteriol. 1801323-1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mandrell, R. E., and W. D. Zollinger. 1989. Human immune response to meningococcal outer membrane epitopes after natural infection or vaccination. Infect. Immun. 571590-1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McNeil, G., and M. Virji. 1997. Phenotypic variants of meningococci and their potential in phagocytic interactions: the influence of opacity proteins, pili, PilC and surface sialic acids. Microb. Pathog. 22295-304. [DOI] [PubMed] [Google Scholar]
- 25.Milagres, L. G., M. C. Gorla, C. T. Sacchi, and M. M. Rodrigues. 1998. Specificity of bactericidal antibody response to serogroup B meningococcal strains in Brazilian children after immunization with an outer membrane vaccine. Infect. Immun. 664755-4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moore, J., S. E. Bailey, Z. Benmechernene, C. Tzitzilonis, N. J. Griffiths, M. Virji, and J. P. Derrick. 2005. Recognition of saccharides by the OpcA, OpaD, and OpaB outer membrane proteins from Neisseria meningitidis. J. Biol. Chem. 28031489-31497. [DOI] [PubMed] [Google Scholar]
- 27.Morelli, G., B. Malorny, K. Muller, A. Seiler, J. F. Wang, J. del Valle, and M. Achtman. 1997. Clonal descent and microevolution of Neisseria meningitidis during 30 years of epidemic spread. Mol. Microbiol. 251047-1064. [DOI] [PubMed] [Google Scholar]
- 28.Muenzner, P., C. Dehio, T. Fujiwara, M. Achtman, T. F. Meyer, and S. D. Gray-Owen. 2000. Carcinoembryonic antigen family receptor specificity of Neisseria meningitidis Opa variants influences adherence to and invasion of proinflammatory cytokine-activated endothelial cells. Infect. Immun. 683601-3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muenzner, P., M. Naumann, T. F. Meyer, and S. D. Gray-Owen. 2001. Pathogenic Neisseria trigger expression of their carcinoembryonic antigen-related cellular adhesion molecule 1 (CEACAM1; previously CD66a) receptor on primary endothelial cells by activating the immediate early response transcription factor, nuclear factor-kappaB. J. Biol. Chem. 27624331-24340. [DOI] [PubMed] [Google Scholar]
- 30.Nassif, X. 1999. Interaction mechanisms of encapsulated meningococci with eucaryotic cells: what does this tell us about the crossing of the blood-brain barrier by Neisseria meningitidis? Curr. Opin. Microbiol. 271-77. [DOI] [PubMed] [Google Scholar]
- 31.Øvstebo, R., P. Brandtzaeg, B. Brusletto, K. B. Haug, K. Lande, E. A. Høiby, and P. Kierulf. 2004. Use of robotized DNA isolation and real-time PCR to quantify and identify close correlation between levels of Neisseria meningitidis DNA and lipopolysaccharides in plasma and cerebrospinal fluid from patients with systemic meningococcal disease. J. Clin. Microbiol. 422980-2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parkhill, J., M. Achtman, K. D. James, S. D. Bentley, C. Churcher, S. R. Klee, G. Morelli, D. Basham, D. Brown, T. Chillingworth, R. M. Davies, P. Davis, K. Devlin, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, S. Leather, S. Moule, K. Mungall, M. A. Quail, M. A. Rajandream, K. M. Rutherford, M. Simmonds, J. Skelton, S. Whitehead, B. G. Spratt, and B. G. Barrell. 2000. Complete DNA sequence of a serogroup A strain of Neisseria meningitidis Z2491. Nature 404502-506. [DOI] [PubMed] [Google Scholar]
- 33.R Development Core Team. 2006. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria.
- 34.Riordan, F. A., O. Marzouk, A. P. Thomson, J. A. Sills, and C. A. Hart. 2002. Prospective validation of the Glasgow Meningococcal Septicaemia Prognostic Score. Comparison with other scoring methods. Eur. J. Pediatr. 161531-537. [DOI] [PubMed] [Google Scholar]
- 35.Robinson, K., M. Taraktsoglou, K. S. Rowe, K. G. Wooldridge, and D. A. Ala'Aldeen. 2004. Secreted proteins from Neisseria meningitidis mediate differential human gene expression and immune activation. Cell. Microbiol. 6927-938. [DOI] [PubMed] [Google Scholar]
- 36.Schneider, S., D. Roessli, and L. Excoffier. 2000. ARLEQUIN: a software for population genetics data analysis. Version 2.000. Genetics and Biometry Laboratory, Dept. of Anthropology, University of Geneva, Geneva, Switzerland.
- 37.Sinclair, J. F., C. H. Skeoch, and D. Hallworth. 1987. Prognosis of meningococcal septicaemia. Lancet ii38. [DOI] [PubMed] [Google Scholar]
- 38.Sjursen, H., E. Wedege, E. Rosenqvist, A. Naess, A. Halstensen, R. Matre, and C. O. Solberg. 1990. IgG subclass antibodies to serogroup B meningococcal outer membrane antigens following infection and vaccination. APMIS 981061-1069. [DOI] [PubMed] [Google Scholar]
- 39.Tettelin, H., N. J. Saunders, J. Heidelberg, A. C. Jeffries, K. E. Nelson, J. A. Eisen, K. A. Ketchum, D. W. Hood, J. F. Peden, R. J. Dodson, W. C. Nelson, M. L. Gwinn, R. DeBoy, J. D. Peterson, E. K. Hickey, D. H. Haft, S. L. Salzberg, O. White, R. D. Fleischmann, B. A. Dougherty, T. Mason, A. Ciecko, D. S. Parksey, E. Blair, H. Cittone, E. B. Clark, M. D. Cotton, T. R. Utterback, H. Khouri, H. Qin, J. Vamathevan, J. Gill, V. Scarlato, V. Masignani, M. Pizza, G. Grandi, L. Sun, H. O. Smith, C. M. Fraser, E. R. Moxon, R. Rappuoli, and J. C. Venter. 2000. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science 2871809-1815. [DOI] [PubMed] [Google Scholar]
- 40.Trotter, C., S. Samuelsson, A. Perrocheau, S. de Greeff, H. de Melker, S. Heuberger, and M. Ramsay. 2005. Ascertainment of meningococcal disease in Europe. Euro Surveill. 10247-250. [PubMed] [Google Scholar]
- 41.Trotter, C. L., M. Chandra, R. Cano, A. Larrauri, M. E. Ramsay, C. Brehony, K. A. Jolley, M. C. Maiden, S. Heuberger, and M. Frosch. 2007. A surveillance network for meningococcal disease in Europe. FEMS Microbiol. Rev. 3127-36. [DOI] [PubMed] [Google Scholar]
- 42.van Deuren, M., P. Brandtzaeg, and J. W. van der Meer. 2000. Update on meningococcal disease with emphasis on pathogenesis and clinical management. Clin. Microbiol. Rev. 13144-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Virji, M., D. Evans, A. Hadfield, F. Grunert, A. M. Teixeira, and S. M. Watt. 1999. Critical determinants of host receptor targeting by Neisseria meningitidis and Neisseria gonorrhoeae: identification of Opa adhesiotopes on the N-domain of CD66 molecules. Mol. Microbiol. 34538-551. [DOI] [PubMed] [Google Scholar]
- 44.Virji, M., K. Makepeace, D. J. Ferguson, M. Achtman, and E. R. Moxon. 1993. Meningococcal Opa and Opc proteins: their role in colonization and invasion of humanepithelial and endothelial cells. Mol. Microbiol. 10499-510. [DOI] [PubMed] [Google Scholar]
- 45.Virji, M., K. Makepeace, D. J. Ferguson, and S. M. Watt. 1996. Carcinoembryonic antigens (CD66) on epithelial cells and neutrophils are receptors for Opa proteins of pathogenic neisseriae. Mol. Microbiol. 22941-950. [DOI] [PubMed] [Google Scholar]
- 46.Virji, M., S. M. Watt, S. Barker, K. Makepeace, and R. Doyonnas. 1996. The N-domain of the human CD66a adhesion molecule is a target for Opa proteins of Neisseria meningitidis and Neisseria gonorrhoeae. Mol. Microbiol. 22929-939. [DOI] [PubMed] [Google Scholar]
- 47.Xia, X., and Z. Xie. 2001. DAMBE: software package for data analysis in molecular biology and evolution. J. Hered. 92371-373. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.