Abstract
Of 49 subjects, 21 were colonized with more than one strain of Escherichia coli and 12 subjects had at least one strain present in fewer than 20% of colonies. The ability to accurately characterize E. coli strain diversity is directly related to the number of colonies sampled and the underlying prevalence of the strain.
Efforts to elucidate the epidemiology of antimicrobial resistance have increasingly focused on the critical role of gastrointestinal (GI) tract colonization with resistant bacterial pathogens (3). The use of surveillance cultures (usually employing a perirectal swab approach) is common as a component of both epidemiologic studies and infection control initiatives (5). Although it is recognized that a given individual may be colonized with more than one distinct strain of Escherichia coli at any given time (7), past studies have arbitrarily chosen to select one or more colonies from a plate to identify E. coli strains in fecal specimens and determine strain diversity (2).
It is unknown how many colonies of E. coli bacteria must be characterized in a fecal sample to optimize the likelihood that at least one representative of each colonizing strain is identified. The impact of different sampling strategies on the characterization of E. coli colonization is unclear. Most studies that have investigated this issue have focused on only specific types of E. coli (e.g., diarrheagenic E. coli or E. coli O157) (4, 8, 11), studied nonhuman fecal samples (1, 2, 8, 11), and/or included a small sample size of subjects (<20) (1, 2, 9, 11). Indeed, past studies of E. coli have often selected a relatively small number (i.e., ≤10) of colonies on which to base their investigation of strain diversity (6, 8).
The current study analyzes the genetic diversity of E. coli colonization in fecal samples from long-term care facility (LTCF) residents. The goal of this study was to identify the likelihood of identifying both common and uncommon strains of E. coli using different strategies for colony sampling from fecal specimens.
This study was performed at the Philadelphia Veteran's Administration Medical Center (PVAMC) LTCF. This study was approved by the Institutional Review Boards of the University of Pennsylvania and the PVAMC. This study is based on data originally collected as part of a study investigating the epidemiology of GI tract colonization with fluoroquinolone-resistant and fluoroquinolone-susceptible E. coli strains in LTCF residents (7). As described previously, LTCF residents were recruited for this study between March and July 2002. All residents were considered eligible for inclusion and were enrolled if informed consent could be obtained. For each enrolled subject, one rectal swab was obtained at the time of enrollment.
Swabs were streaked onto MacConkey agar without antimicrobial additives and then triple streaked using a sterile loop to isolate single colonies. Individual colonies were then replica plated to MacConkey agar by using sterile toothpicks. From each MacConkey plate, up to 25 lactose-fermenting colonies (as available) were randomly selected. If fewer than 25 colonies were present, all were sampled. If more than 25 colonies were present, 25 colonies were selected at random. Antimicrobial susceptibilities and species identification were confirmed by automated testing (Vitek; BioMerieux, Hazelwood, MO) (7).
Chromosomal DNA was digested with XbaI and resolved by pulsed-field gel electrophoresis using a CHEF DR II system (Bio-Rad, Hercules, CA) to determine genetic relatedness (7). Strain identity was interpreted according to established criteria (10).
For each sample, pulsed-field gel electrophoresis was performed for all chosen colonies to determine the number of distinct strains present and to calculate the proportion of colonies accounted for by each strain within each subject's swab sample. For each subject colonized with more than one strain, we characterized as “dominant” that strain which was present in the highest proportion of colonies from the swab sample. These data were then used to inform the likelihood of identifying a given strain using different sampling strategies.
The selection probabilities for the different strain types were assumed to follow a multivariate hypergeometric distribution assuming sampling without replacement. However, in the case of an infinite (or very large) population from which to sample (as is the case when sampling bacteria from the GI tract), the probabilities of selection are negligibly altered by the prior selections. Thus, a sampling distribution that considers sampling with replacement and does not alter the sampling probabilities after each selection is an excellent approximation to the hypergeometric distribution, as in this case. Indeed, when the universe size is infinite, the multivariate hypergeometric distribution is well approximated by the multinomial distribution. The probability of selecting at least one colony of a specific type from n sampled colonies, with a proportion p of organisms of this type, is easily computed using the multinomial distribution 1 − (1 − p)n. All statistical calculations were performed using STATA version 9.0 (StataCorp, College Station, TX).
Included in the study were E. coli isolates from 49 LTCF residents (7). The median age of the study subjects was 69 years (range, 38 to 98 years) and two (4.1%) subjects were female. Thirty (61.2%) subjects were white, 18 (36.7%) were African-American, and one (2.0%) was Hispanic.
There were 28 subjects colonized with a single strain of E. coli. Of the 21 subjects colonized with multiple E. coli strains, 11 were colonized with two distinct strains, 8 were colonized with three strains, and 1 subject each was colonized with four strains and five strains. The median number of colonies sampled per subject was 23 (interquartile range [IQR], 17 to 25). The median number of colonies sampled for the 28 subjects with only one strain identified was 22 (IQR, 17 to 25), while the median number of colonies sampled for the 21 subjects with more than one strain identified was 23 (IQR, 16 to 25).
Of the 49 total subjects, 4 (8.2%) subjects had at least one strain present in fewer than 5% of colonies from their swab sample, 7 (14.2%) subjects had at least one strain present in fewer than 10% of colonies, and 12 (24.5%) subjects had at least one strain present in fewer than 20% of colonies.
Among the 21 subjects with more than one E. coli strain identified, 17 (81%) subjects exhibited a dominant strain that comprised ≥50% of all colonies assessed in the swab sample. For the remaining four subjects, the dominant strains comprised between 40 and 46% of all colonies sampled.
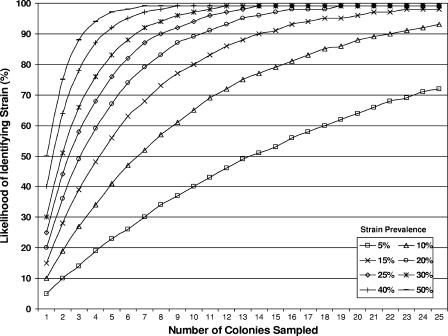
Figure 1 shows the likelihood of identifying an E. coli strain given the number of colonies sampled in a fecal specimen and the underlying strain prevalence. For example, for a strain representing 50% of E. coli bacteria in a given sample, the likelihood of identifying this strain when five sample colonies are picked is measured by the equation 1 − (1 − 0.5)5 = 0.96875, or 97%. For a strain comprising 20% of strains in a given sample, sampling five colonies would identify this strain 67% of the time, as measured by the equation 1 − (1 − 0.2)5 = 0.67232.
FIG. 1.
Likelihood of strain identification.
In this study of 49 LTCF residents, we found that approximately 40% of subjects were colonized with multiple strains of E. coli. Of 83 total strains identified from all subjects, 17 (20.5%) accounted for fewer than 20% of colonies in the fecal samples tested. Of 49 total subjects, 12 (24.5%) had at least one strain present in fewer than 20% of colonies.
The approach to colony sampling from fecal specimens depends on the goals of the study and the genetic diversity in the sample. For epidemiologic studies or infection control initiatives in which identifying the breadth of colonizing strains is important (e.g., to characterize potential person-to-person spread of strains), optimizing the ability to characterize breadth must be balanced against the need for efficiency.
Our results demonstrate that the ability to accurately characterize E. coli strain diversity in human subjects is directly related to the number of colonies sampled and the underlying prevalence of the strain. For example, if one desires at least a 90% likelihood of identifying a strain present in ≥20% of colonies, 11 colonies must be sampled (Fig. 1). If one desires at least a 90% likelihood of identifying a strain present in ≥10% of colonies, 22 colonies must be sampled. Our results provide a characterization of the distribution of the prevalence of different E. coli strains in human subjects and demonstrate that a substantial proportion of strains (i.e., 20.5%) accounted for fewer than one in five colonies in a given fecal specimen. Thus, for efforts designed to provide an estimate of E. coli strain diversity, the ability to detect strains with a prevalence at least this great would seem desirable.
Another goal of such studies might be to ensure identification of the dominant strain of E. coli (1, 6). We found that in over 90% of subjects, the dominant strain comprised ≥50% of all colonies sampled. For the remaining four subjects, the dominant strains comprised between 40 and 46% of all colonies sampled. Thus, in this study, sampling five colonies would provide a 90% likelihood of identifying the dominant strain.
Our study had several potential limitations. Although we sampled up to 25 colonies per subject, it is likely that sampling an even larger number would provide some incremental increase in the ability to determine the breadth of diversity. Our small sample size of subjects may have limited the ability to more precisely calculate the number of colonies required to be sampled. Whether the results of our study, conducted in an LTCF, can be generalized to human subjects in other settings is not known.
In summary, this study presents two important primary findings. First, we provide an approach for determining the likelihood of identifying a particular E. coli strain given two parameters: (i) the number of colonies sampled and (ii) the underlying prevalence of the strain. Second, we used our data set to inform the second of the two assumptions noted above, namely, the prevalence of specific E. coli strains. Taken together, these findings provide important data for informing future research and infection control efforts.
Acknowledgments
This work was supported by Public Health Service grant R01-AG023792 of the National Institute on Aging (NIH) (to E.L.) and a pilot grant from the Mental Illness Research, Education, and Clinical Center (MIRECC) (to J.N.M.). This study was also supported in part by an Agency for Healthcare Research and Quality (AHRQ) Centers for Education and Research on Therapeutics cooperative agreement (U18-HS10399).
E.L. has received research support from Merck Pharmaceuticals and Ortho-McNeil Pharmaceuticals. J.N.M. has received research support from Merck Pharmaceuticals. All other authors report no conflicts of interest.
Footnotes
Published ahead of print on 23 July 2008.
REFERENCES
- 1.Achá, S. J., I. Kühn, G. Mbazima, P. Colque-Navarro, and R. Möllby. 2005. Changes of viability and composition of the Escherichia coli flora in faecal samples during long time storage. J. Microbiol. Methods 63229-238. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, M. A., J. E. Whitlock, and V. J. Harwood. 2006. Diversity and distribution of Escherichia coli genotypes and antibiotic resistance phenotypes in feces of humans, cattle, and horses. Appl. Environ. Microbiol. 726914-6922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Donskey, C. J. 2004. The role of the intestinal tract as a reservoir and source for transmission of nosocomial pathogens. Clin. Infect. Dis. 39219-226. [DOI] [PubMed] [Google Scholar]
- 4.Iijima, Y., S. Tanaka, K. Miki, S. Kanamori, M. Toyokawa, and S. Asari. 2007. Evaluation of colony-based examinations of diarrheagenic Escherichia coli in stool specimens: low probability of detection because of low concentrations, particularly during the early stage of gastroenteritis. Diagn. Microbiol. Infect. Dis. 58303-308. [DOI] [PubMed] [Google Scholar]
- 5.Lautenbach, E., A. D. Harris, E. N. Perencevich, I. Nachamkin, P. Tolomeo, and J. P. Metlay. 2005. Test characteristics of perirectal and rectal swab compared to stool sample for detection of fluoroquinolone-resistant Escherichia coli in the gastrointestinal tract. Antimicrob. Agents Chemother. 49798-800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lidin-Janson, G., B. Kaijser, K. Lincoln, S. Olling, and H. Wedel. 1978. The homogeneity of the faecal coliform flora of normal school-girls, characterized by serological and biochemical properties. Med. Microbiol. Immunol. 164247-253. [DOI] [PubMed] [Google Scholar]
- 7.Maslow, J. N., E. Lautenbach, W. B. Bilker, and J. R. Johnson. 2004. Colonization with extraintestinal pathogenic Escherichia coli among nursing home residents and its relationship to fluoroquinolone resistance. Antimicrob. Agents Chemother. 483618-3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Renter, D. G., J. M. Sargeant, R. D. Oberst, and M. Samadpour. 2003. Diversity, frequency, and persistence of Escherichia coli O157 strains from range cattle environments. Appl. Environ. Microbiol. 69542-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schlager, T. A., J. O. Hendley, A. L. Bell, and T. S. Whittam. 2002. Clonal diversity of Escherichia coli colonizing stools and urinary tracts of young girls. Infect. Immun. 701225-1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 332233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vali, L., A. Hamouda, M. C. Pearce, H. I. Knight, J. Evans, and S. G. Amyes. 2007. Detection of genetic diversity by pulsed-field gel electrophoresis among Escherichia coli O157 isolated from bovine faecal samples by immunomagnetic separation technique. Lett. Appl. Microbiol. 4419-23. [DOI] [PubMed] [Google Scholar]