Abstract
We investigated associations between the genotypic and phenotypic features of Staphylococcus aureus bloodstream isolates and the clinical characteristics of bacteremic patients enrolled in a phase III trial of S. aureus bacteremia and endocarditis. Isolates underwent pulsed-field gel electrophoresis, PCR for 33 putative virulence genes, and screening for heteroresistant glycopeptide intermediate S. aureus (hGISA). A total of 230 isolates (141 methicillin-susceptible S. aureus and 89 methicillin-resistant S. aureus [MRSA]) were analyzed. North American and European S. aureus isolates differed in their genotypic characteristics. Overall, 26% of the MRSA bloodstream isolates were USA 300 strains. Patients with USA 300 MRSA bacteremia were more likely to be injection drug users (61% versus 15%; P < 0.001), to have right-sided endocarditis (39% versus 9%; P = 0.002), and to be cured of right-sided endocarditis (100% versus 33%; P = 0.01) than patients with non-USA 300 MRSA bacteremia. Patients with persistent bacteremia were less likely to be infected with Panton-Valentine leukocidin gene (pvl)-constitutive MRSA (19% versus 56%; P = 0.005). Although 7 of 89 MRSA isolates (8%) exhibited the hGISA phenotype, no association with persistent bacteremia, daptomycin resistance, or bacterial genotype was observed. This study suggests that the virulence gene profiles of S. aureus bloodstream isolates from North America and Europe differ significantly. In this study of bloodstream isolates collected as part of a multinational randomized clinical trial, USA 300 and pvl-constitutive MRSA strains were associated with better clinical outcomes.
Staphylococcus aureus is an emerging cause of nosocomial and community acquired bacteremia throughout the world. A recent population-based study estimated the incidence of invasive methicillin-resistant S. aureus (MRSA) infections in the United States to be 31.8/100,000 persons (12). USA 300 strains, originally associated with infections originating in the community (19), are increasingly recognized as a cause of healthcare-associated infections (12, 16, 26). Panton-Valentine leukocidin gene (pvl)-constitutive MRSA strains (27), including USA 300 strain types (9, 13, 14, 25, 31), have been reported from many regions of the world. Although pvl is generally associated with skin infections and necrotizing pneumonia, less is known about its significance in bacteremia. As a result, there is a growing need to understand the impact of specific bacterial genotypic and phenotypic characteristics on clinical outcome (7, 10, 17).
In the present investigation, we characterized a large number of S. aureus isolates collected as part of a multinational phase III randomized, evaluator-blinded clinical trial comparing daptomycin to standard therapy for the treatment of S. aureus bacteremia and endocarditis. We then evaluated potential associations between the presence of specific bacterial genotypic and phenotypic characteristics and the clinical features and outcomes of bacteremic patients.
MATERIALS AND METHODS
Patient characteristics and settings.
The design and results of the multinational, open-label randomized controlled trial of therapy for S. aureus bacteremia have been previously reported (5). The institutional review board at each site approved the protocol, and all patients or their authorized representatives provided written informed consent. Briefly, patients were randomized to treatment with daptomycin or standard therapy consisting of either vancomycin or antistaphylococcal penicillin. All patients randomly assigned to standard treatment were also scheduled to receive gentamicin for the first 4 days. Clinical data and corresponding S. aureus bloodstream isolates were prospectively collected from enrolled patients. Final evaluations were performed by an adjudication committee that was blinded to treatment group.
Study definitions.
Uncomplicated bacteremia was defined as isolation of S. aureus from enrollment blood cultures in patients without endocarditis and without evidence of hematogenous spread. In patients without endocarditis, complicated bacteremia was defined by the isolation of S. aureus from blood cultures on at least 2 days through study day 5, the presence of spread of infection, or infection involving prostheses not removed within 4 days. Right- and left-sided endocarditis were defined according to the criteria of Li et al. (15) Persistent bacteremia was defined as persisting or relapsing S. aureus bacteremia as determined by the adjudication committee. Patients who failed due to persisting or relapsing S. aureus infection, clinical failure, or death were considered failures in this analysis.
Bacterial isolates.
A total of 230 baseline S. aureus isolates were collected from patients with S. aureus bacteremia enrolled in the phase III trial during 2002 to 2005 (5). Only a single isolate per patient was included in this investigation. The isolates were identified as S. aureus on the basis of positive catalase, coagulase and DNase tests, and antimicrobial susceptibility testing performed according to the Clinical and Laboratory Standards Institute (formerly the National Committee for Clinical Laboratory Standards) guidelines (20). The isolates were stored at −70°C from the time of identification until they were tested in the current investigation.
PFGE.
Pulsed-field gel electrophoresis (PFGE) with SmaI was performed on all isolates, and the gels were analyzed by using the BioNumerics software (Applied Maths, Kortrijk, Belgium) as previously described (18). Dice coefficients (pairwise similarity) were calculated for each pair of isolates, and a dendrogram was constructed by using an optimization value of 0.50% and a position tolerance ranging from 1.25 to 1.35% (end of the fingerprint). A similarity coefficient of 80% was used to define pulsed-field type clusters.
PCR assays for genotyping.
Genomic DNA from S. aureus was extracted by using an Ultraclean microbial DNA kit (MolBio Laboratories, Carlsbad, CA). Thirty-three bacterial determinants including toxins (sea, seb, sec, sed, see, seg, seh, sei, sej, tst, eta, etb, hlg, and pvl), adhesins (bbp, clfA, clfB, cna, ebpS, fnbA, fnbB, map [also known as eap], sdrC, sdrD, sdrE, and spa), and other virulence genes (chp, efb, icaA, V8, arcA, the agr group, and SCCmec type) were examined by using PCR assays. Primers and conditions used to amplify the genes of interest have been previously described (1). PCR amplifications were performed in a Dyad thermal cycler (Bio-Rad, Hercules, CA) with HotStart Taq polymerase (Qiagen, Valencia, CA). Genomic DNA (∼100 ng) was added to a 1× multiplex PCR mix containing 3 mM MgCl2, 10 mM deoxynucleoside triphosphates, and 0.2 μM concentrations of each of the forward and reverse primers and Taq polymerase. PCR products were analyzed by 2% agarose gel electrophoresis. A positive control and a negative control (ATCC 6358) were included in each PCR run (1).
Twenty-nine genes were screened by using multiplex PCR. Genes not detected in the multiplex reaction were subsequently reanalyzed by uniplex PCR for confirmation of presence or absence of the gene. spa, arcA, and the agr groups I to IV were detected by using uniplex PCR alone. This was performed due to the presence of multiple repeats in the spa gene that interfere with the detection of other genes. agr groups I to IV were detected serially using primers described by Peacock et al. under conditions described previously (23). arcA was detected by using methods described by Diep et al. (3); no positive control was available for this gene.
SCCmec typing.
SCCmec typing was performed by using multiplex PCR as described by Oliveira et al. (22). SCCmec type I genes were further validated by using uniplex PCR by the Zhang et al. and Okuma et al. methods, since they could not be differentiated from type IV by multiplex PCR (21, 35).
Identification of hGISA.
The presence of the heterogeneously glycopeptide-intermediate S. aureus (hGISA) phenotype was determined in all MRSA isolates using modified PAP-AUC analysis. The modified PAPs were performed as previously described (29, 33). The AUC for each strain was determined by trapezoidal rule using Sigma Plot 9.0 (Richmond, CA). Each strain was run in conjunction with Mu3 as the control hGISA strain. A ratio was then calculated by dividing the AUC of the test strain by the AUC of Mu3. A PAP-AUC ratio of >0.90 was used to define the hGISA phenotype (32).
Statistical methods.
Continuous variables were compared between groups by using the Wilcoxon rank-sum test. Categorical variables were analyzed with the Fisher exact test. Significance levels were corrected for multiple tests based on a false discovery rate (FDR) of <20%.
RESULTS
Patient characteristics.
A total of 235 patients comprised the modified intention to treat population for the clinical trial. Baseline S. aureus isolates were available for 230 of these patients: 57 patients (25%) with uncomplicated bacteremia, 120 patients (52%) with complicated bacteremia, and 35 patients (15%) with right-sided and 18 patients (8%) with left-sided endocarditis (Table 1). Right-sided endocarditis was associated with younger age (median, 38 years; range, 27 to 91) and injection drug use (80%).
TABLE 1.
Characteristics of 230 patients with uncomplicated bacteremia, complicated bacteremia, and right- and left-sided S. aureus endocarditis
| Characteristica | Bacteremia
|
Endocarditis
|
||
|---|---|---|---|---|
| Uncomplicated (n = 57) | Complicated (n = 120) | Right sided (n = 35) | Left sided (n = 18) | |
| Median age in yr (range) | 57 (26-87) | 57 (21-88) | 38 (27-91) | 63 (33-90) |
| No. female (%) | 21 (37) | 42 (35) | 19 (54) | 9 (50) |
| No. of patients (%)* of race or ethnic group | ||||
| Caucasian | 36 (63) | 87 (73) | 18 (51) | 13 (72) |
| Black | 12 (21) | 23 (19) | 15 (43) | 4 (22) |
| No. of patients (%)* with risk factor | ||||
| Diabetes mellitus | 22 (39) | 53 (44) | 4 (11) | 5 (28) |
| Systemic inflammatory response syndrome | 42 (74) | 90 (75) | 29 (83) | 14 (78) |
| Injection drug use | 7 (12) | 11 (9) | 28 (80) | 3 (17) |
| Surgery within prior 30 days | 18 (32) | 54 (45) | 5 (14) | 6 (33) |
| Human immunodeficiency virus positive | 2 (4) | 1 (1) | 4 (11) | 0 |
| Intravascular devices | 7 (12) | 25 (21) | 4 (11) | 2 (11) |
| Extravascular foreign material | 13 (23) | 34 (28) | 2 (6) | 8 (44) |
| Creatinine clearance < 50 ml/min | 11 (19) | 21 (18) | 4 (11) | 4 (22) |
| Prior history of endocarditis | 1 (2) | 2 (2) | 6 (17) | 3 (17) |
| Presumed skin source | 25 (44) | 35 (29) | 4 (11) | 1 (6) |
| No. of patients (%) with MRSA infection | 21 (37) | 44 (37) | 15 (43) | 9 (50) |
*, percentage of patients within each diagnostic category with the indicated characteristic.
Bacterial characteristics.
Of the 230 baseline S. aureus strains isolated, 89 (39%) were MRSA and 141 (61%) were methicillin-susceptible S. aureus (MSSA). Twenty-six clusters of closely related strains were identified on PFGE in the total sample. One MRSA isolate was nontypeable. The predominant clones were USA 300 (28 isolates; 12%), USA 600 (28 isolates; 12%), and USA 100 (26 isolates; 11%). Among the 88 typeable MRSA isolates, 23 (26%) were USA 300 clones, and 41 (46%) were non-USA 300 (25 [28%] USA 100; 8 [9%] USA 600; 3 [3%] USA 400; 3 [3%] USA 500; 1 [1%] USA 200; and 1 [1%] USA 800). Twenty-four MRSA isolates did not belong to any specific USA type. All USA 300 MRSA isolates were positive for the pvl and arcA genes (Table 2). Among the 141 MSSA isolates, 5 (4%) were USA 300 and 49 (35%) were non-USA 300 isolates (20 [14%] USA 200; 20 [14%] USA 600; 6 [4%] USA 400; 1 [1%] USA 100; and 2[1%] USA 700). Eighty-seven MSSA isolates did not belong to any specific USA type. Twenty-six MSSA isolates were positive for the pvl gene.
TABLE 2.
Characteristics of patients with USA 300 and non-USA 300 MRSA bacteremiaa
| Characteristicsb | Strain type
|
Pc | |
|---|---|---|---|
| USA 300 (n = 23) | Non-USA 300 (n = 65) | ||
| Median age in yr (range) | 43 (25-58) | 62 (22-91) | <0.001†d |
| No. female (%) | 7 (30) | 34 (52) | 0.09 |
| No. of patients (%)* of race or ethnic group | 0.002‡† | ||
| Caucasian | 9 (39) | 51 (78) | |
| Black | 10 (43) | 9 (14) | |
| Hispanic | 3 (13) | 2 (3) | |
| Other | 1 (4) | 3 (5) | |
| No. of patients (%)* in: | 0.018† | ||
| North America | 23 (100) | 52 (80) | |
| Europe | 0 | 13 (20) | |
| No. of patients (%)* with risk factor | |||
| Diabetes mellitus | 6 (26) | 29 (45) | 0.14 |
| Systemic inflammatory response syndrome | 20 (87) | 47 (72) | 0.25 |
| Injection drug use | 14 (61) | 10 (15) | <0.001† |
| Surgery in prior 30 days | 7 (30) | 29 (45) | 0.33 |
| Human immunodeficiency virus positive | 1 (4) | 1 (2) | 0.46 |
| Intravascular devices | 1 (4) | 13 (20) | 0.10 |
| Extravascular foreign material | 4 (17) | 24 (37) | 0.12 |
| Creatinine clearance < 50 ml/min | 2 (9) | 15 (23) | 0.22 |
| Prior history of endocarditis | 3 (13) | 3 (5) | 0.18 |
| Presumed skin source | 7 (30) | 20 (31) | >0.99 |
| No. of patients (%)* undergoing therapy | 0.27‡ | ||
| Daptomycin | 10 (44) | 34 (52) | |
| Vancomycin | 12 (52) | 31 (48) | |
| Semisynthetic penicillin ± vancomycin | 1 (4) | 0 | |
| No. of patients (%)* with persistent bacteremiae | 2 (9) | 18 (28) | 0.08 |
| No. of patients/total no. (%)* with success at test of curef | |||
| Uncomplicated bacteremia | 3/4 (75) | 15/17 (88) | 0.49 |
| Complicated bacteremia | 3/8 (38) | 22/35 (63) | 0.25 |
| Right-sided endocarditis | 9/9 (100) | 2/6 (33) | 0.01† |
| Left-sided endocarditis | 1/2 (50) | 1/7 (14) | 0.42 |
| No. of patients (%)* with bacterial characteristics | |||
| pvl | 23 (100) | 18 (28) | <0.001† |
| arcA (for arginine mobile catabolic element) | 23 (100) | 2 (3) | <0.001† |
One MRSA isolate was nontypeable and was excluded from the analysis.
*, percentage of patients within each diagnostic category with the indicated characteristic.
Calculated using the Fisher exact test unless otherwise noted. †, statistically significant result with an FDR of <20%; ‡, comparison of USA 300 and non-USA 300 distributions across all levels of this characteristic.
This value was calculated using the Wilcoxon rank-sum test.
Persistent bacteremia was defined as persisting or relapsing S. aureus bacteremia as determined by the adjudication committee.
Failures due to persisting or relapsing S. aureus infection, clinical failure, or death were used to calculate success rates.
Geographic variation.
A total of 195 isolates were obtained from patients in North America (76 MRSA and 119 MSSA) and 35 isolates (13 MRSA and 22 MSSA) were obtained from patients in Europe (Table 3). MRSA isolates from North America were more frequently USA 300 (23 isolates; 30%) and USA100 (22 isolates; 29%); USA 600 was the predominant clone for European MRSA isolates (6 isolates; 46%). North American and European isolates had similar distributions of adhesion genes. Based on an FDR of <20%, North American MRSA isolates were significantly more likely to contain arcA (33% versus 0%; P = 0.016), eta (91% versus 31%; P < 0.001), pvl (55% versus 0%; P < 0.001), and seb (83% versus 31%; P < 0.001) and less likely to contain tst (63% versus 100%; P = 0.008) and sec (14% versus 46%; P = 0.015) than European isolates. Among MSSA isolates, eta (84% versus 36%; P < 0.001), pvl (22% versus 0%; P = 0.014), and seb (81% versus 5%; P < 0.001) were significantly more common in North American isolates than in European isolates. No difference in the distribution of the SCCmec type and agr type among MRSA isolates was observed in the two cohorts. Most S. aureus isolates in North America (88 isolates; 45%) and Europe (18 isolates; 51%) were of agr type I.
TABLE 3.
Differences in toxin gene profile of North American and European S. aureus isolates on univariate analysis
| Virulence factor and gene | No. (%) of isolatesa
|
||
|---|---|---|---|
| North American | European | Pb | |
| MRSA | 76 | 13 | |
| eta | 69 (91) | 4 (31) | <0.001 |
| pvl | 42 (55) | 0 | <0.001 |
| seb | 63 (83) | 4 (31) | <0.001 |
| sec | 11 (14) | 6 (46) | 0.015 |
| tst | 48 (63) | 13 (100) | 0.008 |
| arcA | 25 (33) | 0 | 0.016 |
| MSSA | 119 | 22 | |
| eta | 100 (84) | 8 (36) | <0.001 |
| pvl | 26 (22) | 0 | 0.014 |
| seb | 96 (81) | 1 (5) | <0.001 |
No significant differences were observed among North American and European isolates for the following genes: bbp, clfA, clfB, cna, ebpS, fnbA, fnbB, map (eap), sdrC, sdrD, sdrE, spa, etb, hlg, sea, sed, see, seg, seh, sei, sej, agr I to IV, chp, efb, icaA, V8, and SCCmec I to IV (MRSA) and bbp, clfA, clfB, cna, ebpS, fnbA, fnbB, map (eap), sdrC, sdrD, sdrE, spa, etb, hlg, sea, sec, sed, see, seg, seh, sei, sej, tst, agr I to IV, chp, efb, icaA, and V8 (MSSA; SCCmec and arcA analyses were not performed for MSSA isolates). The percentages refer to the percentage of patients within each S. aureus subset with the indicated virulence gene.
As determined by the Fisher exact test; all P values shown in this table (referring to a comparison of values for North American and European isolates) are statistically significant with an FDR of <20%.
Associations between bacterial and clinical characteristics.
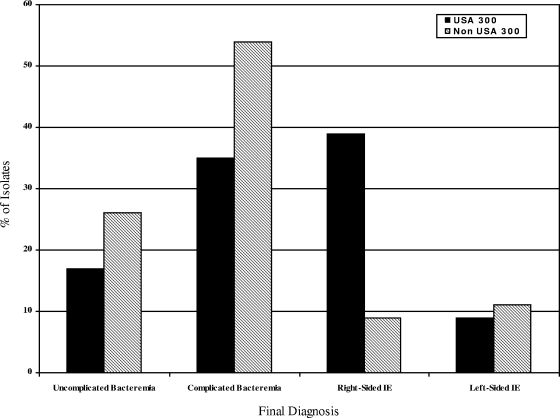
Among MRSA isolates, USA 300 genotype was associated with younger age (median 43 versus 62 years; P < 0.001), male gender (70% versus 48%; P = 0.09), black race (43% versus 14%; P = 0.002 [for the overall race comparison]) and injection drug use (61% versus 15%; P < 0.001) compared to non-USA 300 isolates (Table 2). The presence of USA 300 isolates differed by final diagnosis (P = 0.020). USA 300 isolates were more common in patients with right-sided endocarditis, whereas non-USA 300 isolates were more common in the other diagnoses (39% versus 9%; P = 0.02 [for the overall comparison of final diagnosis for USA 300 and non-USA 300 isolates]) (Fig. 1). USA 300 MRSA isolates were also significantly more likely to have a successful treatment outcome than patients with MRSA right-sided endocarditis due to non-USA 300 MRSA (100% versus 33%; P = 0.01).
FIG. 1.
Distribution of USA 300 and non-USA 300 MRSA bloodstream isolates according to final diagnosis. The P value for overall comparison of the final diagnosis for USA 300 and non-USA 300 isolates is 0.02, which is statistically significant and corresponds to an FDR of <20%. “% of Isolates” refers to the percentage of isolates based on the total number of USA 300 and non-USA 300 isolates.
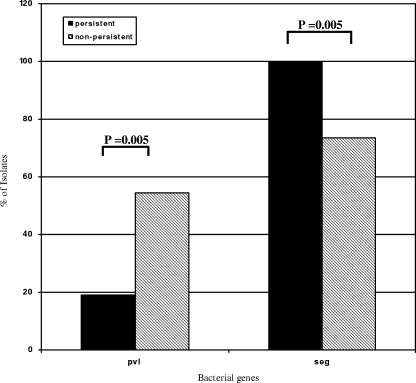
Univariate analysis with the FDR correction revealed several associations between bacterial virulence factors and outcomes. MRSA isolates associated with persistent bacteremia were significantly less likely to contain the gene for pvl (19% persistent versus 56% nonpersistent; P = 0.005) and significantly more likely to contain seg (100% persistent versus 74% nonpersistent bacteremia; P = 0.005) (Fig. 2). No association between virulence genes and failure or success of therapy was observed.
FIG. 2.
Bacterial genes significantly associated (FDR < 20%) with persistent bacteremia among MRSA bloodstream isolates. “% of Isolates” refers to the percentage of isolates based on the total number of MRSA isolates positive for the virulence gene among patients with persistent (n = 21) and nonpersistent (n = 68) bacteremia.
Genotypic and clinical characteristics associated with hGISA isolates.
Of the 89 MRSA isolates, 7 hGISA strains were identified by using the PAP-AUC ratio method. All seven isolates were obtained from patients in North America and were associated with the “hospital acquired” profile: all were positive for SCCmec type II and most belonged to the USA 100 (three isolates), 200 (one isolate), or 600 (one isolate) PFGE profiles. Two of the hGISA strains did not fit a USA PFGE profile. Two isolates were positive for the pvl gene.
Of the seven patients infected with hGISA strains, two had uncomplicated bacteremia, four had complicated bacteremia, and one had left-sided endocarditis. Persistent bacteremia was observed in four patients with hGISA infection. Four patients with hGISA isolates received vancomycin, while three received daptomycin. Of these, one patient treated with daptomycin for left-sided endocarditis developed an increased MIC to daptomycin (MIC = 2 μg/ml). Three of the seven patients with hGISA bacteremia (two with complicated bacteremia [one treated with vancomycin and one treated with daptomycin] and one patient with left-sided endocarditis [treated with daptomycin]) failed therapy.
DISCUSSION
S. aureus is a leading cause of bacteremia and endocarditis in much of the industrialized world (4, 6, 30). In the present study, we investigated associations between the genotypic and phenotypic characteristics of bloodstream S. aureus isolates and the outcome of bacteremic patients. The results of this investigation demonstrate that bacterial genotypic properties differed by geographic region and were significantly associated with clinical characteristics, infection type, and outcome.
The predominant strain in this investigation, USA 300, accounted for approximately one-third of all MRSA bloodstream isolates from the United States. This finding is consistent with recent reports (12, 26) and confirms the emergence of the USA 300 strain type as an important cause of bacteremia, as well as skin infection. Although recent reports have documented an increasing incidence of USA 300 MRSA infections in European countries (14, 25, 31), no isolates from this clone were identified in the European cohort. This finding may be due in part to the small sample size of our European cohort.
The distribution of putative virulence genes among S. aureus isolates differed by geographic region. For example, all European isolates lacked the pvl and arcA genes, suggesting that these genes remain relatively uncommon among S. aureus bloodstream isolates from that region (8). In contrast, all MRSA isolates and over three-quarters of MSSA isolates from European countries were positive for the tst gene, an observation that is consistent with recent reports (28). The presence of specific bacterial virulence genes among S. aureus isolates from the same geographical region may reflect a rapid expansion of specific clones or horizontal transfer of mobile genetic elements among strains.
The severity of bacteremia due to USA 300 strains in our study was no greater than that observed in non-USA 300 strains and was significantly associated with better outcomes in certain clinical settings. For example, patients with right-sided MRSA endocarditis caused by USA 300 strains were significantly more likely to be cured of their infection than patients with right-sided MRSA endocarditis due to non-USA 300 strains. In addition, isolates from patients with persistent MRSA bacteremia were significantly less likely to contain the pvl gene than isolates from patients with nonpersistent MRSA bacteremia. These findings are consistent with several recent reports. For example, Popovich et al. (24) reported similar outcomes, including rates of persistent bacteremia, length of hospitalization, and hospital readmission rates, among patients with MRSA bacteremia due to “community genotype” strains and those infected with the “hospital genotype” strains. In another study evaluating the genotypic characteristics of U.S. and South African S. aureus isolates from patients with skin and soft tissue infections, Campbell et al. reported that infections due to pvl-constitutive S. aureus isolates were significantly more likely to be cured than similar infections caused by S. aureus isolates without the pvl gene (1). Taken together, these findings suggest that the presence of pvl, per se, does not confer a worse clinical course in all forms of infection caused by S. aureus.
Although 8% of the MRSA bloodstream isolates, all from U.S. patients, exhibited the hGISA phenotype, no major associations with clinical outcomes, such as persistent bacteremia or treatment failure, were identified. This is in contrast to some prior reports regarding hGISA bacteremia. For example, Charles et al. observed significant differences in the outcomes of 5 patients with heterogeneously vancomycin-intermediate S. aureus (hVISA) bacteremia compared to 47 patients with vancomycin-susceptible MRSA bacteremia (2). hVISA bacteremia was associated with higher bacterial loads, longer duration of bacteremia, and treatment failure. However, this association between persistent bacteremia and hGISA isolates has not been observed in other studies. Khosrovaneh et al. evaluated the vancomycin susceptibilities of MRSA isolates obtained from 22 patients with persistent or recurrent bacteremia (11). Three (13.6%) isolates were found to exhibit heteroresistance to vancomycin. However, most patients had other reasons for persistent infection, including inadequate removal of infected foci. Taken together, these findings suggest that the clinical relevance of vancomycin heteroresistance in defining the course and outcomes of bacteremic patients remains uncertain. Although our findings did not suggest any potential associations between the hGISA phenotype and clinical outcomes, this may in part be due to the small sample of hGISA patients and the high degree of variability in the clinical and genotypic characteristics of our cohort.
The clinical and microbiological data in the present study were highly accurate, having been collected as part of a registrational clinical trial on a well-defined cohort of patients with S. aureus bacteremia and endocarditis from more than 40 centers in four countries. The present study was limited by the relatively small sample size of isolates from Europe. In addition, assessment of virulence determinants was based on the qualitative assessment by PCR. We did not evaluate quantitative expression of these genes or the presence of single nucleotide polymorphisms, which may influence the function of gene products (34). Clinical outcomes may be correlated with in vivo gene expression, but that analysis is beyond available technology. Some genes, such as pvl and arcA, were found to be highly clonal to USA 300 isolates and therefore associations with right-sided endocarditis might reflect a “hitchhiker effect” due to linkage disequilibrium between these genes and other virulence determinants rather than a causal association (23). Finally, we were unable to assess the frequency of USA 300 isolates in patients with community-acquired versus hospital-acquired bacteremia due to the unavailability of relevant clinical data.
The present study documents several important findings, including the increasing incidence of USA 300 isolates among North American patients with S. aureus bloodstream infections and the higher incidence of tst gene-positive isolates causing S. aureus bacteremia in Europe than previously reported. The present study also suggests potential associations between S. aureus genotype and clinical outcomes. MRSA isolates positive for the pvl gene were associated with nonpersistent bacteremia, while USA 300 isolates were associated with right-sided endocarditis. In addition, right-sided endocarditis due to USA 300 strains of MRSA was associated with better treatment success rates compared to non-USA 300 MRSA isolates. Larger international collections of S. aureus isolates are required to fully characterize the geographic distribution of virulence factors and potentially validate the observations made in the present study.
Acknowledgments
The following represent potential conflicts of interest. H.W.B., G.R.C., G.S., V.G.F., and M.C. have served as consultants for Cubist Pharmaceuticals. V.G.F. has served as a consultant for Astellas, Biosynexus, Inhibitex, Merck, Johnson & Johnson, and Theravance. M.E.S. has served as a consultant to Theravance and received honoraria from Astellas. T.L. has received research support from Cubist Pharmaceuticals. M.J.R. has received grant support, has served as a consultant, or has participated as a speaker for Astellas, Cubist, Theravance, Targanta, Cerexa, Forrest, Johnson & Johnson, Pfizer, and Wyeth Pharmaceuticals. V.G.F. has received grant support from Cerexa, Cubist, Inhibitex, Merck, Nabi, the National Institutes of Health, and Theravance; V.H.C. has received grant support from the American Heart Association; M.E.J. has received grant support from Theravance; G.S. has received grant support from Pfizer and Cubist; and G.R.C. has received grant support from Inhibitex, Merck, and Theravance. V.G.F. has received lecture fees from Cubist, Nabi, and Pfizer; H.W.B. has received lecture fees from Novartis, Pfizer, Schering-Plough, and Cubist; and G.S. has received lecture fees from Cubist, Pfizer, and Wyeth. J.A., J.N.S., and S.A.L. are employees of Cubist Pharmaceuticals.
This study was supported by Cubist Pharmaceuticals.
Footnotes
Published ahead of print on 2 July 2008.
REFERENCES
- 1.Campbell, S. J., H. S. Deshmukh, C. L. Nelson, I. G. Bae, M. E. Stryjewski, J. J. Federspiel, G. T. Tonthat, T. H. Rude, S. L. Barriere, R. Corey, and V. G. Fowler, Jr. 2007. Genotypic characteristics of Staphylococcus aureus isolates from a multinational trial of complicated skin and skin structure infections. J. Clin. Microbiol. 46678-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Charles, P. G., P. B. Ward, P. D. Johnson, B. P. Howden, and M. L. Grayson. 2004. Clinical features associated with bacteremia due to heterogeneous vancomycin-intermediate Staphylococcus aureus. Clin. Infect. Dis. 38448-451. [DOI] [PubMed] [Google Scholar]
- 3.Diep, B. A., S. R. Gill, R. F. Chang, T. H. Phan, J. H. Chen, M. G. Davidson, F. Lin, J. Lin, H. A. Carleton, E. F. Mongodin, G. F. Sensabaugh, and F. Perdreau-Remington. 2006. Complete genome sequence of USA 300, an epidemic clone of community-acquired methicillin-resistant Staphylococcus aureus. Lancet 367731-739. [DOI] [PubMed] [Google Scholar]
- 4.Fluit, A. C., M. E. Jones, F. J. Schmitz, J. Acar, R. Gupta, and J. Verhoef. 2000. Antimicrobial susceptibility and frequency of occurrence of clinical blood isolates in Europe from the SENTRY antimicrobial surveillance program, 1997 and 1998. Clin. Infect. Dis. 30454-460. [DOI] [PubMed] [Google Scholar]
- 5.Fowler, V. G., Jr., H. W. Boucher, G. R. Corey, E. Abrutyn, A. W. Karchmer, M. E. Rupp, D. P. Levine, H. F. Chambers, F. P. Tally, G. A. Vigliani, C. H. Cabell, A. S. Link, I. DeMeyer, S. G. Filler, M. Zervos, P. Cook, J. Parsonnet, J. M. Bernstein, C. S. Price, G. N. Forrest, G. Fatkenheuer, M. Gareca, S. J. Rehm, H. R. Brodt, A. Tice, and S. E. Cosgrove. 2006. Daptomycin versus standard therapy for bacteremia and endocarditis caused by Staphylococcus aureus. N. Engl. J. Med. 355653-665. [DOI] [PubMed] [Google Scholar]
- 6.Fowler, V. G., Jr., J. M. Miro, B. Hoen, C. H. Cabell, E. Abrutyn, E. Rubinstein, G. R. Corey, D. Spelman, S. F. Bradley, B. Barsic, P. A. Pappas, K. J. Anstrom, D. Wray, C. Q. Fortes, I. Anguera, E. Athan, P. Jones, J. T. van der Meer, T. S. Elliott, D. P. Levine, and A. S. Bayer. 2005. Staphylococcus aureus endocarditis: a consequence of medical progress. JAMA 2933012-3021. [DOI] [PubMed] [Google Scholar]
- 7.Fowler, V. G., Jr., M. K. Olsen, G. R. Corey, C. W. Woods, C. H. Cabell, L. B. Reller, A. C. Cheng, T. Dudley, and E. Z. Oddone. 2003. Clinical identifiers of complicated Staphylococcus aureus bacteremia. Arch. Intern. Med. 1632066-2072. [DOI] [PubMed] [Google Scholar]
- 8.Holmes, A., M. Ganner, S. McGuane, T. L. Pitt, B. D. Cookson, and A. M. Kearns. 2005. Staphylococcus aureus isolates carrying Panton-Valentine leucocidin genes in England and Wales: frequency, characterization, and association with clinical disease. J. Clin. Microbiol. 432384-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kearns, A. M., G. M. Hill, R. L. R. East, C. McCormick, I. Smith, et al. 2007. Community-associated MRSA ST8-SCCmecIVa (USA-300): experience in England and Wales. 25th ECCMID, Munich, Germany.
- 10.Khatib, R., L. B. Johnson, M. G. Fakih, K. Riederer, A. Khosrovaneh, M. Shamse Tabriz, M. Sharma, and S. Saeed. 2006. Persistence in Staphylococcus aureus bacteremia: incidence, characteristics of patients, and outcome. Scand. J. Infect. Dis. 387-14. [DOI] [PubMed] [Google Scholar]
- 11.Khosrovaneh, A., K. Riederer, S. Saeed, M. S. Tabriz, A. R. Shah, M. M. Hanna, M. Sharma, L. B. Johnson, M. G. Fakih, and R. Khatib. 2004. Frequency of reduced vancomycin susceptibility and heterogeneous subpopulation in persistent or recurrent methicillin-resistant Staphylococcus aureus bacteremia. Clin. Infect. Dis. 381328-1330. [DOI] [PubMed] [Google Scholar]
- 12.Klevens, R. M., M. A. Morrison, J. Nadle, S. Petit, K. Gershman, S. Ray, L. H. Harrison, R. Lynfield, G. Dumyati, J. M. Townes, A. S. Craig, E. R. Zell, G. E. Fosheim, L. K. McDougal, R. B. Carey, and S. K. Fridkin. 2007. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA 2981763-1771. [DOI] [PubMed] [Google Scholar]
- 13.Krziwanek, K., C. Luger, B. Sammer, S. Stumvoll, M. Stammler, S. Metz-Gercek, and H. Mittermayer. 2007. PVL-positive MRSA in Austria. Eur. J. Clin. Microbiol. Infect. Dis. 26931-935. [DOI] [PubMed] [Google Scholar]
- 14.Larsen, A., M. Stegger, R. Goering, M. Sorum, and R. Skov. 2007. Emergence and dissemination of the methicillin resistant Staphylococcus aureus USA300 clone in Denmark (2000-2005). Eur. Surveill. 1222-24. [Google Scholar]
- 15.Li, J. S., D. J. Sexton, N. Mick, R. Nettles, V. G. Fowler, Jr., T. Ryan, T. Bashore, and G. R. Corey. 2000. Proposed modifications to the Duke criteria for the diagnosis of infective endocarditis. Clin. Infect. Dis. 30633-638. [DOI] [PubMed] [Google Scholar]
- 16.Maree, C. L., R. S. Daum, S. Boyle-Vavra, K. Matayoshi, and L. G. Miller. 2007. Community-associated methicillin-resistant Staphylococcus aureus isolates causing healthcare-associated infections. Emerg. Infect. Dis. 13236-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McClelland, R. S., V. G. Fowler, Jr., L. L. Sanders, G. Gottlieb, L. K. Kong, D. J. Sexton, K. Schmader, K. D. Lanclos, and R. Corey. 1999. Staphylococcus aureus bacteremia among elderly versus younger adult patients: comparison of clinical features and mortality. Arch. Intern. Med. 1591244-1247. [DOI] [PubMed] [Google Scholar]
- 18.McDougal, L. K., C. D. Steward, G. E. Killgore, J. M. Chaitram, S. K. McAllister, and F. C. Tenover. 2003. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J. Clin. Microbiol. 415113-5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Naimi, T. S., K. H. LeDell, K. Como-Sabetti, S. M. Borchardt, D. J. Boxrud, J. Etienne, S. K. Johnson, F. Vandenesch, S. Fridkin, C. O'Boyle, R. N. Danila, and R. Lynfield. 2003. Comparison of community- and health care-associated methicillin-resistant Staphylococcus aureus infection. JAMA 2902976-2984. [DOI] [PubMed] [Google Scholar]
- 20.National Committee for Clinical Laboratory Standards. 2000. Methods for dilution antimicrobial tests for bacteria that grow aerobically, 5th ed., approved standard M7-A5, vol. 20, no. 2. NCCLS, Wayne, PA.
- 21.Okuma, K., K. Iwakawa, J. D. Turnidge, W. B. Grubb, J. M. Bell, F. G. O'Brien, G. W. Coombs, J. W. Pearman, F. C. Tenover, M. Kapi, C. Tiensasitorn, T. Ito, and K. Hiramatsu. 2002. Dissemination of new methicillin-resistant Staphylococcus aureus clones in the community. J. Clin. Microbiol. 404289-4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oliveira, D. C., and H. de Lencastre. 2002. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 462155-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peacock, S. J., C. E. Moore, A. Justice, M. Kantzanou, L. Story, K. Mackie, G. O'Neill, and N. P. Day. 2002. Virulent combinations of adhesin and toxin genes in natural populations of Staphylococcus aureus. Infect. Immun. 704987-4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Popovich, K. J., R. A. Weinstein, and B. Hota. 2008. Are community-associated methicillin-resistant Staphylococcus aureus (MRSA) strains replacing traditional nosocomial MRSA strains? Clin. Infect. Dis. 46787-794. [DOI] [PubMed] [Google Scholar]
- 25.Ruppitsch, W., A. Stoger, D. Schmid, R. Fretz, A. Indra, F. Allerberger, and W. Witte. 2007. Occurrence of the USA300 community-acquired Staphylococcus aureus clone in Austria. Eur. Surveill. 12E0710251. [DOI] [PubMed] [Google Scholar]
- 26.Seybold, U., E. V. Kourbatova, J. G. Johnson, S. J. Halvosa, Y. F. Wang, M. D. King, S. M. Ray, and H. M. Blumberg. 2006. Emergence of community-associated methicillin-resistant Staphylococcus aureus USA300 genotype as a major cause of health care-associated blood stream infections. Clin. Infect. Dis. 42647-656. [DOI] [PubMed] [Google Scholar]
- 27.Tristan, A., M. Bes, H. Meugnier, G. Lina, B. Bozdogan, P. Courvalin, M. E. Reverdy, M. C. Enright, F. Vandenesch, and J. Etienne. 2007. Global distribution of Panton-Valentine leukocidin-positive methicillin-resistant Staphylococcus aureus, 2006. Emerg. Infect. Dis. 13594-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van der Mee-Marquet, N., C. Epinette, J. Loyau, L. Arnault, A. S. Domelier, B. Losfelt, N. Girard, and R. Quentin. 2007. Staphylococcus aureus strains isolated from bloodstream infections changed significantly in 2006. J. Clin. Microbiol. 45851-857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walsh, T. R., A. Bolmstrom, A. Qwarnstrom, P. Ho, M. Wootton, R. A. Howe, A. P. MacGowan, and D. Diekema. 2001. Evaluation of current methods for detection of staphylococci with reduced susceptibility to glycopeptides. J. Clin. Microbiol. 392439-2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wisplinghoff, H., T. Bischoff, S. M. Tallent, H. Seifert, R. P. Wenzel, and M. B. Edmond. 2004. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin. Infect. Dis. 39309-317. [DOI] [PubMed] [Google Scholar]
- 31.Witte, W., C. C. Strommenger, and U. Nübel. Community MRSA ST8 (“USA300”) has arrived in Central Europe. 25th ECCMID, Munich, Germany.
- 32.Wootton, M., R. A. Howe, R. Hillman, T. R. Walsh, P. M. Bennett, and A. P. MacGowan. 2001. A modified population analysis profile (PAP) method to detect hetero-resistance to vancomycin in Staphylococcus aureus in a UK hospital. J. Antimicrob. Chemother. 47399-403. [DOI] [PubMed] [Google Scholar]
- 33.Wootton, M., A. P. MacGowan, T. R. Walsh, and R. A. Howe. 2007. A multicenter study evaluating the current strategies for isolating Staphylococcus aureus strains with reduced susceptibility to glycopeptides. J. Clin. Microbiol. 45329-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu, Y., J. M. Rivas, E. L. Brown, X. Liang, and M. Hook. 2004. Virulence potential of the staphylococcal adhesin CNA in experimental arthritis is determined by its affinity for collagen. J. Infect. Dis. 1892323-2333. [DOI] [PubMed] [Google Scholar]
- 35.Zhang, K., J. A. McClure, S. Elsayed, T. Louie, and J. M. Conly. 2005. Novel multiplex PCR assay for characterization and concomitant subtyping of staphylococcal cassette chromosome mec types I to V in methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 435026-5033. [DOI] [PMC free article] [PubMed] [Google Scholar]