Abstract
IS911 is a bacterial insertion sequence composed of two consecutive overlapping open reading frames (ORFs [orfA and orfB]) encoding the transposase (OrfAB) as well as a regulatory protein (OrfA). These ORFs are bordered by terminal left and right inverted repeats (IRL and IRR, respectively) with several differences in nucleotide sequence. IS911 transposition is asymmetric: each end is cleaved on one strand to generate a free 3′-OH, which is then used as the nucleophile in attacking the opposite insertion sequence (IS) end to generate a free IS circle. This will be inserted into a new target site. We show here that the ends exhibit functional differences which, in vivo, may favor the use of one compared to the other during transposition. Electromobility shift assays showed that a truncated form of the transposase [OrfAB(1-149)] exhibits higher affinity for IRR than for IRL. While there was no detectable difference in IR activities during the early steps of transposition, IRR was more efficient during the final insertion steps. We show here that the differential activities between the two IRs correlate with the different affinities of OrfAB(1-149) for the IRs during assembly of the nucleoprotein complexes leading to transposition. We conclude that the two inverted repeats are not equivalent during IS911 transposition and that this asymmetry may intervene to determine the ordered assembly of the different protein-DNA complexes involved in the reaction.
The majority of known bacterial insertion sequences (ISs) are bordered by short imperfect inverted repeat (IR) sequences. The integrity of these IRs is essential for efficient transposition since they provide specificity for transposase binding, strand cleavage, and strand transfer reactions required for IS movement. The IRs of many elements are organized in a simple way into two essential domains: a terminal domain where cleavage occurs and a subterminal transposase-binding domain. However, some transposons carry arrays of sequence elements which differ at each end, and these differences result in a functional asymmetry between the extremities (5). Different activities between the left and right IRs (IRL and IRR, respectively) have also been observed in several of the simplest ISs: for IS10 (Tn10), binding of the integration host factor and H-NS host proteins to subterminal sites close to the outside end plays an important role in regulating transposition (3, 4, 27, 35); the two ends of IS50 also differ in sequence and in activity during Tn5 transposition (7, 29), as do those of IS30 (28). In the case of Mos1, the transposase binds preferentially to the right end, which differs in four positions from the left (1, 2, 36). It is clear that these differences between reactive ends participate in transposition regulation since they are involved in transpososome assembly (for review, see reference 10). As in many ISs, the two IS911 IRs differ at several nucleotide positions. The present study investigates the potential impact of these differences in sequence between the two IRs on IS911 transposition.
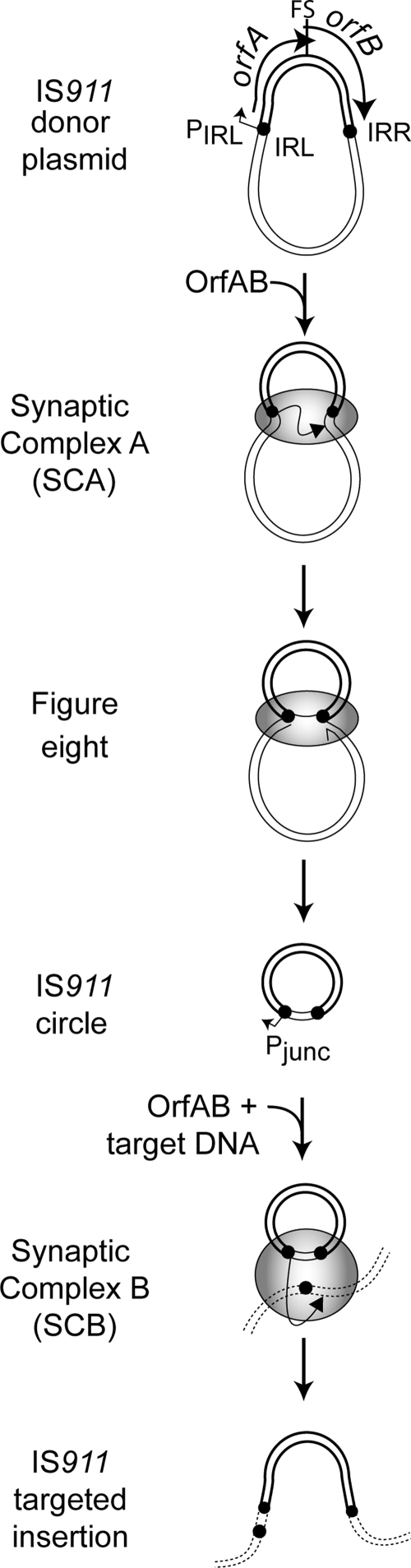
The bacterial IS IS911 is composed of two consecutive open reading frames (ORFs [orfA and orfB]) flanked by two imperfect terminal IRs (Fig. 1). The two ORFs are under the control of a weak promoter (PIRL) and encode two major proteins: OrfA, a regulatory protein, and OrfAB, the transposase. OrfAB is produced by programmed translational frameshifting between orfA and orfB, while OrfA is the product of orfA and shares its first 86 amino acids with OrfAB. OrfAB is composed of 382 residues and can be divided in two parts: residues involved in IR binding (a helix-turn-helix motif [HTH]) (22) and in ensuring protein multimerization (leucine zipper [LZ] and M domains) (12) are located at the N terminus (residues 1 to 149), whereas residues involved in catalysis (DDE) are located at the C terminus (residues 220 to 382) (12). Transposition of IS911 occurs in several steps (Fig. 1) (for review see reference 24). First, the left and right IS911 ends (IRL and IRR) are brought together by OrfAB to generate synaptic complex A (SCA). One strand of one IR (the donor IR) is cleaved to generate a 3′-OH which then attacks 3 nucleotides away from the second IR (the target IR), to generate a figure eight form (20). The figure eight is replicated by host proteins (copy-out transposition) (5) to regenerate the donor plasmid and to generate a closed circular transposon copy: the IS circle (8). The IS circle carries two abutted IRs separated by the 3 bp directly adjacent to the IR targeted during figure eight formation. Coincidently, the newly formed IRR-IRL junction generates a strong promoter, Pjunc, whose integrity is necessary for efficient transposition, and which presumably drives expression of IS911 proteins from the IS circle for the next step in the transposition cycle: insertion into a target DNA (9). This step involves formation of a second synaptic complex, synaptic complex B (SCB), which includes the abutted IRs of the IS circle, the target DNA and OrfAB. The regulatory OrfA protein was shown to stimulate IS911 insertion in vitro and in vivo (30, 31).
FIG. 1.
The IS911 transposition pathway. IS911 is represented as a bold line, donor DNA as a thin line, and target DNA as a dotted line. IRR and IRL are represented by small circles. Promoters Pjunc and PIRL, orfA and orfB, and the programmed translational frameshift site (FS) are indicated. The different steps of IS911 transposition are shown: synapsis of the IRs in the plasmid donor (SCA); cleavage at the terminal 5′-CA-3′ of one of the two IRs (donor IR) to generate a 3′-OH end and transfer of this 3′-OH end at 3 bases from the target IR end to form a figure eight structure; second-strand resolution by the host, which functions to create a covalently closed circle (IS circle); synapsis of the IRL-IRR junction carried by the IS circle with the target DNA (SCB); and single-end cleavage and transfer (SET) of the IRL-IRR junction during IR-targeted insertion.
OrfA behaves as a molecular switch by modulating the targeting activity of OrfAB. In the absence of OrfA, and when presented with a target DNA carrying an isolated IR, 98% of OrfAB-mediated insertion events are directed 3 bp from the IR in a process called IR-targeted insertion. In the presence of OrfA, the insertion events are predominantly non-IR targeted and occur randomly in the target DNA molecule (15, 23). OrfA therefore favors the dispersion of the IS into “nonhomologous” target sites. The mechanisms of IR-targeted and non-IR-targeted insertion events are different. For IR-targeted insertion events, the reaction requires only one catalytically active IR in the IRL-IRR junction (31). We have proposed that the active IR is cleaved and transferred to the target IR, thus generating a branched structure resembling a Holliday junction and called the single-end transfer (SET) intermediate. This intermediate is then processed to give the final insertion product in a reaction which does not require IS911 proteins but involves specialized host factors (14, 33). The pathway for the non-IR-targeted insertion events implies cleavage and transfer of the two ends within the IRL-IRR junction to form a double-end transfer (DET) intermediate leading to nontargeted IS911 insertion (15, 31, 32).
The results presented here demonstrate that IRR and IRL are not equivalent in the insertion step of IS911 transposition. Electrophoretic mobility shift assays (EMSA), demonstrated that a transposase derivative, OrfAB(1-149), specifically truncated for its catalytic domain, binds IRR more efficiently than IRL. This prompted us to investigate whether the two IRs behaved differently during IS911 transposition in vitro. In the first step, leading to IS circle formation, no bias was observed between the two IRs when OrfAB alone was supplied. In contrast, IRR was a more efficient target than IRL in the insertion step in vitro. It seemed possible that this may reflect a higher affinity of the transposase for IRR than for IRL. Since the full-length transposase, OrfAB, binds poorly to the IRs in vitro (12), we used a truncated derivative, OrfAB(1-149), which exhibits significantly higher binding activity (18). EMSA experiments demonstrated that OrfAB(1-149) assembled an SCB-like complex more efficiently when the target DNA carried IRR rather than IRL. Moreover, in in vitro integration assays using full-length OrfAB transposase and a circular IS donor molecule carrying only one active IR (either IRR or IRL) in the junction, the derivative carrying an active IRR was found to be a more efficient donor in generating IR-targeted events than that carrying an active IRL. The same bias was also observed when the regulatory protein OrfA was added to the reaction. The bias observed during IS911 transposition could thus be a consequence of a differential affinity of OrfAB for each IR that controls the assembly of the IS911 nucleoprotein complexes (transposome).
MATERIALS AND METHODS
Bacterial strains and media.
The Escherichia coli strains used were JS238 and DH5α, as described previously (21). Cultures were grown in Terrific broth supplemented, when necessary, with ampicillin (100 μg/ml), tetracycline (12.5 μg/ml), or chloramphenicol (30 μg/ml). Selection was on L plates supplemented with the appropriate antibiotics. Standard MacConkey indicator plates were supplemented with 1% lactose and appropriate antibiotics.
Plasmids.
Plasmid pAPT166 was used as a substrate to study figure eight formation and was described previously (34). Plasmids pBST1, pAPT182, pCL11, pCL12, pCL13, and pCL14 were used as targets in transposition assays. All except pCL13 have been described previously (15). For pCL13, the resident bla gene was removed from pBR322 by an EcoRI-PstI double digestion and replaced by two complementary oligonucleotides (see also “DNA procedures”), with EcoRI and PstI termini and which constitute the mutated IRL, (IRL*). This plasmid is therefore Tcr. Plasmids pAPT99, pAPT177, and pAPT178 were used for production of IS circles carrying, respectively, IRR-IRL, IRR*-IRL, and IRR-IRL* junctions as previously described (14). Plasmids pAPT158, pAPT156, and pLH114, respectively, were used to prepare OrfAB, OrfA and OrfAB(1-149) as described previously (12, 32).
DNA procedures.
Standard techniques were used for DNA manipulation and cloning (25). Restriction and DNA-modifying enzymes were purchased from New England Biolabs. DNA was isolated from agarose gels using the QIAquick gel extraction kit, PCR products were purified using the QIAquick PCR purification kit, and plasmid DNA was extracted using Miniprep or Maxiprep kits (all from Qiagen). Oligonucleotide OCN4 was radiolabeled for use in sequencing as described previously (32).
Oligonucleotides PEL 5′ (5′-GGAAAGTGGCACACTGAATTTGGCCACCTGAACAGA GGTGATATGCTCACCG-3′) and PEL 3′ (5′-ACGTCCTTTCACCGTGTGACTTAAACCGGTGGACTTGTCTCCACTATACGAGTGGCTTAA-3′) were used for pCL13 construction. Oligonucleotides IRLA (5′-TGAAGTGGCACACTGAATTTGGCCACCTGAACAGAG-3′), IRLB (5′-CTCTGTTCAGGTGGCCAAATTCAGTGTGCCACTTCA-3′), IRRA (5′-TGAAGTGGTCAACAAAAACTGGCCACCGAGTTAGAG-5′), and IRRB (5′-CTCTAACTCGGTGGCCAGTTTTTGTTGACCACTTCA-3′) were used for creation of the IRL and IRR fragments.
Cell-free insertion system.
Transposon IS circles were produced in vivo from pAPT99, pAPT177, and pAPT178, gel purified, and used in a standard reaction with purified IS911 proteins as previously reported (31).
Figure eight formation assay.
The standard reaction was performed at 30°C for 45 min in a final volume of 40 μl containing 500 ng of substrate DNA (pAPT1662) and 0.42 μM of OrfAB in 20 mM HEPES (pH 7.5), 5 mM dithiothreitol, 300 mM KCl, 10% glycerol, and 10 mM MnCl2. Reactions were terminated and deproteinized by adding 30 μl of 25 mM EDTA, 0.6 mg/ml proteinase K, and 2% sodium dodecyl sulfate; incubated for 1 h at 37°C; and treated using a QIAquick PCR purification kit (Qiagen).
EMSA.
DNA fragments containing IRL, IRR, or the IRL-IRR junction were generated by PCR and radiolabeled with 32P. In a standard gel retardation assay (12), 7 nM of the DNA fragments was incubated with OrfAB(1-149) in a final volume of 8 μl. Complexes were separated in a 5% polyacrylamide gel in TGE buffer (12 V·cm−1 at 4°C) for 3 h.
Purification of OrfAB, OrfAB(1-149), and OrfA proteins.
The proteins were prepared as described previously (31).
RESULTS
OrfAB(1-149) binds IRR with more affinity than IRL.
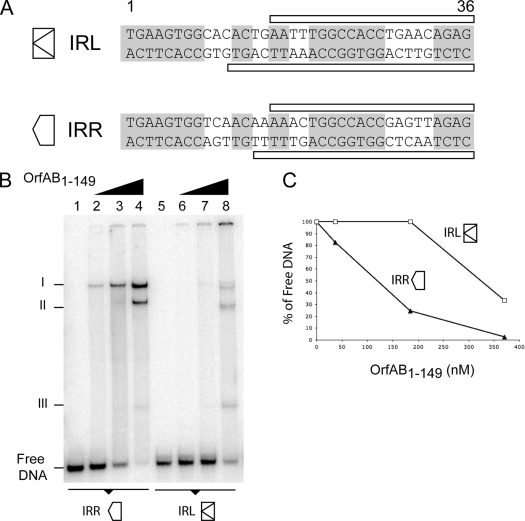
IRL and IRR differ by 12 nucleotides (Fig. 2A). Several of these differences are located in the region of the IR recognized by the transposase (18). This raises the possibility that transposase binds differentially to each end. The relative affinities of transposase for IRR or IRL were compared using an EMSA and a transposase derivative, OrfAB(1-149), with a truncated C-terminal region. OrfAB(1-149) was used because, once translated, the full-length OrfAB binds poorly to IS911 ends, presumably as a result of a folding process which masks the DNA binding domain. On the other hand, OrfAB(1-149) assembles specific DNA-protein complexes with IRR and IRL, which we believe reflect bona fide synaptic complexes or transpososomes (11, 18). Two radiolabeled fragments containing entire IRL or IRR ends together with some flanking DNA (Materials and Methods) were incubated with increasing concentrations of purified OrfAB(1-149). The results of EMSA are presented in Fig. 2B. OrfAB(1-149) generated three complexes (I, II, and III) with either IRR- or IRL-containing DNA as previously described. Complex I is composed of two DNA molecules paired by a yet unknown number of OrfAB(1-149) molecules and is thus thought to resemble the SCA (11). It is clear that specific DNA-protein complexes are formed at lower OrfAB(1-149) concentrations when radiolabeled DNA contains IRR rather than IRL (Fig. 2C). This indicates that OrfAB(1-149) has a higher affinity for IRR than for IRL. If the full-length OrfAB exhibits a similar difference in affinity, this may create a bias in the activity of the two ends during transposition.
FIG. 2.
Comparison of OrfAB(1-149) binding to IRR and OrfAB(1-149) binding to IRL. (A) Nucleotide sequence comparison of the terminal IRs. IRL and IRR are represented as two different arrows. Conserved nucleotides are shown on a gray background. The DNase footprint of OrfAB(1-149) is indicated schematically above and below the sequences. (B) EMSA analysis of the binding of OrfAB(1-149) to IRL and IRR. Equal quantities of radiolabeled IRR- or IRL-containing DNA fragments (100 bp) were incubated with increasing amounts (0.04, 0.19, and 0.37 μM) of OrfAB(1-149). Reaction mixtures were separated on 5% native polyacrylamide gels (12 V·cm−1) to visualize the previously described complexes I, II, and III. Complex I is believed to be the SCA (Fig. 1) (18). (C) Densitometry analysis of the EMSA. Binding of OrfAB(1-149) to DNA was measured by monitoring the decrease of free DNA in the EMSA.
Recruitment of the two IRs in IS circle formation.
Formation of figure eight and the IS circle results from a single-end transfer of the donor IR to a position 3 bp from the target IR end (Fig. 3). The three bases found in the circle junction are therefore those adjacent to the IR that was used as a target in figure eight formation. We investigated the activity of each IR in these initial steps of IS911 transposition: figure eight and IS circle formation.
FIG. 3.
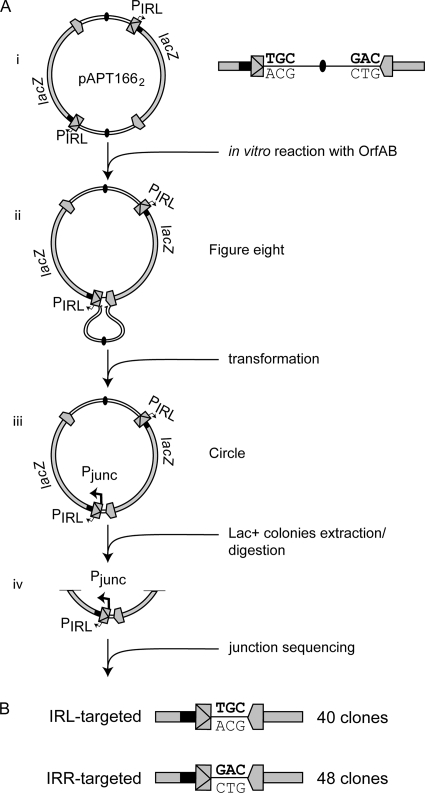
Bias between IRs during IS911 circle formation. (A) Summary of the intramolecular transposition assay used to analyze bias between IRs during early steps of IS911 transposition. IRL and IRR are represented as two different arrows as in Fig. 2. The donor pAPT166 dimer plasmid, pAPT1662, is illustrated. It carries two copies of IS911, each containing IRL and IRR with the endogenous promoter PIRL partially located in IRL and an orfA-lacZ gene fusion. It also contains two copies of the pBR322 origin of replication (filled ovals) and two ampicillin resistance genes (not indicated). In vitro reaction with OrfAB (0.42 μM) generates the figure eight (inter-IS figure eight) which is processed after transformation into MC1061 recA, into an IS circle (IS tandem dimer). The IRL-IRR junction creates the Pjunc promoter, which drives expression of the orfA-lacZ fusion. These colonies are red on MacConkey lactose indicator plates. Plasmid DNA isolated from individual lac+ clones was digested, and the fragment containing the junction was purified and sequenced. (B) Sequences of the IRL-IRR junctions. As expected, two types of junction sequences were obtained: the IRL-TGC-IRR junction corresponds to events in which IRL is used as the target, and the IRL-GAC-IRR junction is representative of events in which IRR is targeted by IRL.
For these purposes, we used a dimer of plasmid pAPT166, pAPT1662 (Fig. 3A), which contains two IS copies in direct orientation with two ampicillin resistance genes and pBR322 origins of replication. The two IS copies contain IRL and IRR in their natural relative orientation and an orfA-lacZ translational fusion (Fig. 3Ai). The purified plasmid dimer was used as a substrate in an in vitro figure eight formation assay in the presence of full-length OrfAB as described previously (20, 21). Products of this reaction containing figure eight forms (Fig. 3Aii) were used to transform competent E. coli JS238 lac mutant strains, and the resulting colonies were selected on MacConkey lactose indicator plates containing ampicillin. After transformation, the figure eight is converted to an IS circle (Fig. 3Aiii) (33) in which one of the plasmid backbones is deleted to generate a new IRL-IRR junction. This reconstitutes a Pjunc promoter correctly positioned to drive expression of the β-galactosidase-encoding gene. These plasmids are equivalent to IS circles but include a single plasmid replication origin and two directly oriented copies of the transposon. Colonies containing transposon “circles” therefore appear red on MacConkey indicator plates. Plasmid DNA from several isolated lac+ colonies was extracted and digested with an appropriate restriction enzyme, and the fragment containing the IRL-IRR junction (which includes the newly formed Pjunc; Fig. 3iv) was gel purified and sequenced to determine which IS end was used in the initial attack leading to figure eight and subsequent circle formation. Reactions were repeated twice and, since the results were comparable, the mixtures were pooled. Of the circle junctions obtained from 88 lac+ colonies, 48 contained the 3 bp originally adjacent to IRR (Fig. 3B) and therefore are events that used IRR as the target end. The other 40 circle junctions carried the 3 bp adjacent to IRL (40 IRL-targeted events). These results clearly show that there is no bias between IS911 IRs during the process of figure eight and circle formation.
Bias between target IRs during insertion.
The results of previous in vitro insertion assays with a nonreplicative IS circle substrate containing a wild-type junction and a target plasmid pAPT182 carrying the two IRs suggested a bias toward insertions near IRR (15). To investigate this in more detail, we used an in vitro insertion competition assay. This included a purified IS circle substrate containing a chloramphenicol resistance gene (Cmr) and an IRR-IRL junction, as well as two target plasmids carrying either the ampicillin resistance gene (bla) and IRL* (pCL12) or the tetracycline resistance gene (Tcr) and IRR* (pCL13). These IRs and resistance genes were cloned into a pBR322 plasmid derivative at the same position but in inverted orientation with respect to each other (Fig. 4). IRR* and IRL* are IS911 ends in which the terminal 2 bp are mutated to prevent their use as donors in cleavage and strand transfer (20). This does not affect their capacity to act as recipient ends in recombination. They were used to avoid creation of a new active junction (an efficient transposition substrate) during IR-targeted insertion (31).
FIG. 4.
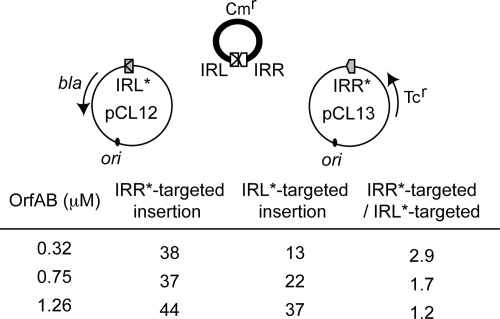
Bias between targeted IRs. In vitro competition insertion experiments were performed using two target plasmids. The symbols are the same that those presented in Fig. 3. Ori (from pBR plasmids) and the ampicillin resistance gene (bla) are indicated. Mutated IRs for the 5′-CA are represented with *. The substrate carries an IRL-IRR junction and a Cmr gene represented in the bold circle. Three OrfAB concentrations (0.32, 0.75, and 1.26 μM) were used for the reactions. The numbers of IRR*-targeted and IRL*-targeted events are indicated as the ratio between the two events.
In vitro insertion reactions, including an equimolar amount of both target plasmids and non-replicative IS Cmr circles, were performed with three different concentrations of purified full-length OrfAB as previously described (31). The reaction mixtures were then used to transform E. coli DH5α. Stable Cmr colonies result from integration of the nonreplicative IS Cmr circles into the replicative target plasmid prior to transformation. Colonies were initially selected on chloramphenicol-containing plates and replicated onto ampicillin- and tetracycline-containing plates. Since, in the presence of OrfAB alone, 98% of insertion events are targeted to IRs (15), the number of Cmr Tcr colonies is representative of IRL*-targeted insertion (pCL12) events, while the number of Cmr Apr colonies is representative of IRR*-targeted insertions (pCL13). Our results have shown that when 0.32 μM of full-length OrfAB was used, the insertion reaction was more efficient if IRR* was used as a target rather than IRL* (Fig. 4): we obtained 38 IRR*-targeted insertion events and only 13 IRL*-targeted insertion events. These results confirm that IRR* is preferred over IRL for IS circle insertion. At higher OrfAB concentrations, a decrease in the bias was observed. This could reflect saturation of the DNA substrates in the reactions. The observed bias was not due to differences in the genetic context of the IRs* in the respective target plasmids since similar results were obtained when IRL* and IRR* were exchanged (generating plasmids pCL14 and pCL11 [Materials and Methods; data not shown]). Therefore, these results show that, with OrfAB alone, IRR* is a more efficient target IR than IRL*. The bias observed here and its dependence on the concentration of full-length OrfAB are in agreement with the observation that the truncated OrfAB(1-149) transposase has a higher affinity for IRR than for IRL.
Synaptic complex formation with the IRR-IRL junction.
To determine whether this insertion bias was due to affinity differences in target selection itself, we developed an EMSA using the truncated form of the transposase to visualize a DNA-protein complex which resembles the SCB (Fig. 1). We incubated a 100-bp radiolabeled DNA fragment containing an IRL-IRR junction (Fig. 5, lane 1) with purified OrfAB(1-149). EMSA analysis revealed that OrfAB(1-149) bound the junction to generate a major complex and several minor bands (Fig. 5, lane 2, complexes a and *). Increasing the protein concentration in the reaction resulted in the loss of all the complexes except for complex a (data not shown). DNase I footprinting performed at high protein concentrations, under conditions in which only complex a was formed, showed that IRL and IRR were protected within the junction (data not shown). This result does not necessarily demonstrate that both IRR and IRL are protected at the same time on the same molecule. It implies that in the population of complex a, at least one of the two IRs is bound by OrfAB(1-149). Furthermore, protection was comparable to that obtained with individual IRs (indicated schematically in Fig. 2A) (18).
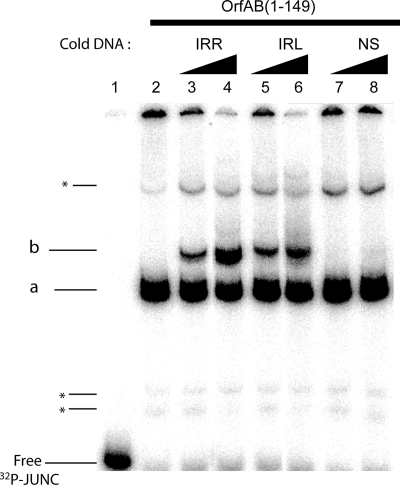
FIG. 5.
SCB formation. EMSA analysis of OrfAB(1-149) binding to the IRL-IRR junction. A radiolabeled DNA fragment (32P-JUNC) containing the IRR-IRL junction (100 bp; lane 1) was incubated with a constant amount of OrfAB(1-149) (0.37 μM; lane 2). Increasing amounts of nonradiolabeled DNA fragment (150 bp) containing either IRR (lanes 3 and 4) or IRL (lanes 5 and 6) or no IRs (lanes 7 and 8) were added to the reaction. Reaction products were separated on 5% native polyacrylamide gels (12 V·cm−1) to visualize the DNA-protein complexes represented as complex *, a, or b. Complex a is thought to be composed of at least two DNA fragments, and complex b is thought to be representative of the SCB (Fig. 1).
Addition of increasing concentrations of unlabeled 150-bp DNA containing either IRR or IRL produced a new DNA-protein complex, b, while complex a disappeared (Fig. 5, lanes 3 to 6). This additional band was not observed if a DNA fragment devoid of IRR or IRL was used (lanes 7 and 8). This is consistent with the idea that the unlabeled 150-bp DNA fragment is incorporated into complex a to form complex b. This suggests that complex b is composed of at least two DNA molecules paired by OrfAB(1-149): complex b would represent the SCB. Appearance of complex b upon addition of cold 150-bp DNA is sequence dependent. Indeed, its formation is more efficient with unlabeled IRR than with unlabeled IRL (Fig. 5, compare lanes 3 and 4 to 5 and 6).
While these results were obtained with OrfAB(1-149), we believe that they will also reflect the behavior of the full-length transposase. They strongly suggest that the target IR is engaged in the SCB and that this is based on the affinity of the transposase for DNA: target IRR would be preferred to target IRL, which in turn, would be preferred to DNA devoid of IR sequences.
Bias between IRs as donors in the insertion step.
To study the activity of the two IRs as donors in the IS circle junction, we used an in vitro insertion assay with three types of purified transposon circle substrates (Fig. 6) (14). All contain a Cmr gene, but each has a different junction: either wild-type IRL-IRR or mutant IRL-IRR* or IRL*-IRR. The target plasmid used was pBST1, which carries IRL*. After an in vitro reaction with purified OrfAB and transformation of E. coli with the reaction products, the resulting colonies were selected on chloramphenicol-containing plates. The results show that the efficiency of insertion was threefold higher with circles carrying the IRL*-IRR junction than with those carrying the IRL-IRR* junction (Fig. 6, line 1). Using an alternative target plasmid, pCL14, which also carries an IRL* but with adjacent sequences different from those in pBST1, a comparable bias was observed (data not shown). Therefore, IRR is a more efficient donor than IRL for targeted insertion.
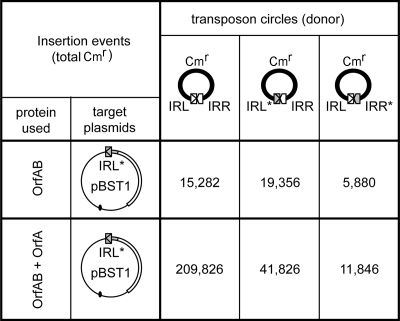
FIG. 6.
Bias between donor IRs. The symbols are the same that those presented in previous figures. The three transposon circles used as substrates are shown. They contain either the IRL-IRR wild-type junction, the IRL*-IRR mutated junction, or the IRL-IRR* mutated junction. The target plasmid is pBST1, and it carries a mutated IRL end (IRL*), an orfA-lacZ gene fusion, a pBR322 origin of replication (black oval), and the ampicillin resistance gene (bla). The OrfAB and OrfA concentrations were, respectively, 0.85 μM and 3.2 μM. The number of total insertion events measured is indicated as the total.
Finally, we also studied the effect of the regulatory protein OrfA on IRR donor bias using similar experimental conditions with the target plasmid pBST1 (Fig. 6, line 2). OrfA is known to stimulate insertion and especially nontargeted insertion (15, 23, 31). As expected, addition of OrfA resulted in a large increase in the number of colonies when the wild-type IRL-IRR junction was used. This is because OrfA stimulates random insertions generated by double-end cleavage and transfer. IS circles including the IRL*-IRR junction generated 41,826 colonies compared to the 11,846 obtained with the IRL-IRR* junction. The bias favoring IRR as the donor IR was therefore clearly conserved (around threefold) in the presence of OrfA.
DISCUSSION
Bias during targeted insertion may reflect differential transposase affinity for IS911 IRs.
The results presented here indicate that bias in end usage occurs during the targeted integration step of IS911 transposition rather than during the formation of the circular transposon intermediate. IRR and IRL appear to behave identically during IS circle formation in vivo from replicative plasmids (Fig. 3), in agreement with results obtained previously from a study of the sequences of a pool of nonreplicative transposon circles obtained in vivo (20). The absence of bias in IR activity during circle formation but differential activity during insertion might lie in differences in the assembly of SCA and SCB (Fig. 1). During the assembly of SCA, the initial step of IS911 transposition, the transposase has no choice: both IRL and IRR must be assembled into the complex. Once formed, the efficiencies of cleavage of IRR or IRL within SCA by the transposase may be equivalent. Bias observed during insertion would thus be a consequence of SCB assembly. During targeted insertion, SCB formation implies transposase binding to at least one of the IRs in the IRL-IRR junction in addition to that carried by the target molecule. Note that our footprinting results with OrfAB(1-149) (not shown) do not reveal whether all three IRs (both junction IRs and the target IR) are bound by the protein at the same time in each SCB-like complex. Since there is a choice during SCB assembly, the transposase would prefer IRR to IRL in the IRL-IRR junction. This would lead to a bias toward IRR being used as the attacking end of the junction.
As underlined above, the affinity differences were observed using a truncated form of the transposase, OrfAB(1-149). Since this lacks the C-terminal DDE catalytic domain, we cannot rule out the possibility that this domain also contributes to the observed bias between IRs. It is clear that the observed differential affinity of OrfAB(1-149) for the two IRs correlates with sequence differences present on the internal part of the IRs which have been shown to be specifically bound by this truncated form of the transposase (see diagram in Fig. 2A). Since the reactive terminal dinucleotide 5′-CA-3′ of the IRs must be recognized by the catalytic domain to permit cleavage, this domain [absent in OrfAB(1-149)] might bind the external part of the IRs. This part of the IR also contains sequence differences, which may be recognized differentially by the DDE domain. The catalytic domain could thus also contribute to the end bias in insertion either by binding efficiency or, less likely since no bias is seen during circle formation, by a bias in cleavage or strand transfer activity. It will therefore be important to characterize more precisely, at the nucleotide level, which part of the IRs contributes to the bias in future experiments.
Stimulation of targeted insertion by OrfA does not change the insertion bias.
OrfA regulates IS911 transposition by stimulating insertion of the IS circle intermediate. Its major effect is to facilitate random insertion (15). It is thought that it accomplishes this by modification of OrfAB activity to stimulate double-end cleavage of the IRL-IRR circle junction and DET (14, 33). This stimulation depends on the OrfA HTH and LZ motifs. The LZ is required for homomultimerization of OrfA and for heteromultimerization with OrfAB, while the HTH motif confers nonspecific DNA binding activity. It is thought that OrfA assists OrfAB in choosing a random target DNA for insertion (23). Consequently, OrfA would not be expected to modify the bias observed to result from SET into a target IR (14, 33). This was indeed found to be the case (Fig. 6). When these insertion reactions were performed with a target plasmid devoid of IR sequences, the donor bias between IRR and IRL was still observable in both the presence and absence of OrfA (data not shown). This is due to some residual SET intermediates formed by OrfAB alone leading to a few random insertions of IS911. While the majority (98%) of integration events catalyzed by OrfAB alone are targeted, 2% of the insertions are nontargeted. These are almost certainly due to low-level formation of SET intermediates between the IRR-IRL junction and a random target DNA (14, 15, 33). For DET intermediates, it is tempting to think that the presence of OrfA in the nucleoprotein complex devoted to random insertion confers the ability to bind a target DNA nonspecifically. OrfA would thus change the activity of the insertion complex and affinity for the target (23).
Bias between reactive ends is observed for other transposons.
A growing number of ISs have been observed to insert next to a sequence resembling their IRs (16). These include IS30, which exhibits dual insertion specificity, inserting both next to sequences resembling its ends but also into a well-defined “hot spot” sequence. This dual target specificity is thought to be due to the presence of two HTH motifs within the transposase: both are involved in insertion next to IRs, while only one appears to be involved in hot spot insertion (13, 17). Here again, targeted insertion could be dependent upon affinity of the transposase for a specific DNA sequence.
In the case of the more elaborate Tn7 transposon, one of the two transposition pathways shows strict target sequence specificity (next to glmS in the E. coli genome). This depends on the sequence-specific DNA binding protein, TnsD (6).
The correlation between higher transposase affinity of a DNA sequence and better reactivity of the sequence has been clearly established for Mos1 and for the bacteriophage Mu. In the case of Mos1, the transposase exhibits a higher affinity for the right than for the left transposon end. Furthermore, an artificial transposon composed of two right ends is more active than the wild-type copy (1, 2). This property seems to be shared by all members of the Tc-Mariner superfamily which have been examined and maybe common for all transposons using the “cut-and-paste” transposition pathway. In the case of Mu, it appears that assembling an active transpososome is more efficient with two right ends rather than with a right and a left end (26). During assembly of a complete transpososome, it is clear that the right end is involved earlier than the left (19). This difference seems to be important in the regulation of transpososome assembly and thus for the initiation of the transposition process (for review, see reference 10).
Together, these data strongly support the idea that the bias observed during IS911 targeted insertion is due to a preferential affinity of the transposase for IRR under conditions in which there is competition between the two ends. The overall transposition process requires an ordered assembly of proteins and DNA substrates to generate a defined transpososome architecture which directs the precise cleavage and strand transfer reactions. In the case of IS911, this first involves formation of the transposon circle intermediate and subsequently the formation of a synaptic complex between the circle and target DNA. The difference in affinity of the transposase for IS911 ends is presumably important for this ordered assembly and therefore in the regulation of these steps.
Footnotes
Published ahead of print on 27 June 2008.
REFERENCES
- 1.Auge-Gouillou, C., M. H. Hamelin, M. V. Demattei, G. Periquet, and Y. Bigot. 2001. The ITR binding domain of the Mariner Mos-1 transposase. Mol. Genet. Genomics 26558-65. [DOI] [PubMed] [Google Scholar]
- 2.Auge-Gouillou, C., M. H. Hamelin, M. V. Demattei, M. Periquet, and Y. Bigot. 2001. The wild-type conformation of the Mos-1 inverted terminal repeats is suboptimal for transposition in bacteria. Mol. Genet. Genomics 26551-57. [DOI] [PubMed] [Google Scholar]
- 3.Chalmers, R., A. Guhathakurta, H. Benjamin, and N. Kleckner. 1998. IHF modulation of Tn10 transposition: sensory transduction of supercoiling status via a proposed protein/DNA molecular spring. Cell 93897-908. [DOI] [PubMed] [Google Scholar]
- 4.Crellin, P., S. Sewitz, and R. Chalmers. 2004. DNA looping and catalysis; the IHF-folded arm of Tn10 promotes conformational changes and hairpin resolution. Mol. Cell 13537-547. [DOI] [PubMed] [Google Scholar]
- 5.Curcio, M. J., and K. M. Derbyshire. 2003. The outs and ins of transposition: from mu to kangaroo. Nat. Rev. Mol. Cell Biol. 4865-877. [DOI] [PubMed] [Google Scholar]
- 6.DeBoy, R. T., and N. L. Craig. 2000. Target site selection by Tn7: attTn7 transcription and target activity. J. Bacteriol. 1823310-3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dodson, K. W., and D. E. Berg. 1989. Factors affecting transposition activity of IS50 and Tn5 ends. Gene 76207-213. [DOI] [PubMed] [Google Scholar]
- 8.Duval-Valentin, G., B. Marty-Cointin, and M. Chandler. 2004. Requirement of IS911 replication before integration defines a new bacterial transposition pathway. EMBO J. 233897-3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duval-Valentin, G., C. Normand, V. Khemici, B. Marty, and M. Chandler. 2001. Transient promoter formation: a new feedback mechanism for regulation of IS911 transposition. EMBO J. 205802-5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gueguen, E., P. Rousseau, G. Duval-Valentin, and M. Chandler. 2005. The transpososome: control of transposition at the level of catalysis. Trends Microbiol. 13543-549. [DOI] [PubMed] [Google Scholar]
- 11.Haren, L., C. Normand, P. Polard, R. Alazard, and M. Chandler. 2000. IS911 transposition is regulated by protein-protein interactions via a leucine zipper motif. J. Mol. Biol. 296757-768. [DOI] [PubMed] [Google Scholar]
- 12.Haren, L., P. Polard, B. Ton-Hoang, and M. Chandler. 1998. Multiple oligomerisation domains in the IS911 transposase: a leucine zipper motif is essential for activity. J. Mol. Biol. 28329-41. [DOI] [PubMed] [Google Scholar]
- 13.Kiss, J., Z. Nagy, G. Toth, G. B. Kiss, J. Jakab, M. Chandler, and F. Olasz. 2007. Transposition and target specificity of the typical IS30 family element IS1655 from Neisseria meningitidis. Mol. Microbiol. 631731-1747. [DOI] [PubMed] [Google Scholar]
- 14.Loot, C., C. Turlan, and M. Chandler. 2004. Host processing of branched DNA intermediates is involved in targeted transposition of IS911. Mol. Microbiol. 51385-393. [DOI] [PubMed] [Google Scholar]
- 15.Loot, C., C. Turlan, P. Rousseau, B. Ton-Hoang, and M. Chandler. 2002. A target specificity switch in IS911 transposition: the role of the OrfA protein. EMBO J. 214172-4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nagy, Z., and M. Chandler. 2004. Regulation of transposition in bacteria. Res. Microbiol. 155387-398. [DOI] [PubMed] [Google Scholar]
- 17.Nagy, Z., M. Szabo, M. Chandler, and F. Olasz. 2004. Analysis of the N-terminal DNA binding domain of the IS30 transposase. Mol. Microbiol. 54478-488. [DOI] [PubMed] [Google Scholar]
- 18.Normand, C., G. Duval-Valentin, L. Haren, and M. Chandler. 2001. The terminal inverted repeats of IS911: requirements for synaptic complex assembly and activity. J. Mol. Biol. 308853-871. [DOI] [PubMed] [Google Scholar]
- 19.Pathania, S., M. Jayaram, and R. M. Harshey. 2003. A unique right end-enhancer complex precedes synapsis of Mu ends: the enhancer is sequestered within the transpososome throughout transposition. EMBO J. 223725-3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Polard, P., and M. Chandler. 1995. An in vivo transposase-catalyzed single-stranded DNA circularization reaction. Genes Dev. 92846-2858. [DOI] [PubMed] [Google Scholar]
- 21.Polard, P., B. Ton-Hoang, L. Haren, M. Betermier, R. Walczak, and M. Chandler. 1996. IS911-mediated transpositional recombination in vitro. J. Mol. Biol. 26468-81. [DOI] [PubMed] [Google Scholar]
- 22.Rousseau, P., E. Gueguen, G. Duval-Valentin, and M. Chandler. 2004. The helix-turn-helix motif of bacterial insertion sequence IS911 transposase is required for DNA binding. Nucleic Acids Res. 321335-1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rousseau, P., C. Loot, C. Guynet, Y. Ah-Seng, B. Ton-Hoang, and M. Chandler. 2007. Control of IS911 target selection: how OrfA may ensure IS dispersion. Mol. Microbiol. 631701-1709. [DOI] [PubMed] [Google Scholar]
- 24.Rousseau, P., C. Normand, C. Loot, C. Turlan, R. Alazard, G. Duval-Valentin, and M. Chandler. 2002. Transposition of IS911, p. 367-383. In N. L. Craig, R. Craigie, M. Gellert, and A. M. Labowitz (ed.), Mobile DNA II. ASM Press, Washington, DC.
- 25.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 26.Savilahti, H., P. Rice, and K. Mizuuchi. 1995. The phage Mu transpososome core: DNA requirements for assembly and function. EMBO J. 144893-4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singh, R. K., J. Liburd, S. J. Wardle, and D. B. Haniford. 2007. The nucleoid binding protein H-NS acts as an anti-channeling factor to favor intermolecular Tn10 transposition and dissemination. J. Mol. Biol. 374950-962. [DOI] [PubMed] [Google Scholar]
- 28.Szabo, M., J. Kiss, Z. Nagy, M. Chandler, and F. Olasz. 2008. Sub-terminal sequences modulating IS30 transposition in vivo and in vitro. J. Mol. Biol. 375337-352. [DOI] [PubMed] [Google Scholar]
- 29.Tomcsanyi, T., and D. E. Berg. 1989. Transposition effect of adenine (Dam) methylation on activity of O end mutants of IS50. J. Mol. Biol. 209191-193. [DOI] [PubMed] [Google Scholar]
- 30.Ton-Hoang, B., M. Betermier, P. Polard, and M. Chandler. 1997. Assembly of a strong promoter following IS911 circularization and the role of circles in transposition. EMBO J. 163357-3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ton-Hoang, B., P. Polard, and M. Chandler. 1998. Efficient transposition of IS911 circles in vitro. EMBO J. 171169-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ton-Hoang, B., P. Polard, L. Haren, C. Turlan, and M. Chandler. 1999. IS911 transposon circles give rise to linear forms that can undergo integration in vitro. Mol. Microbiol. 32617-627. [DOI] [PubMed] [Google Scholar]
- 33.Turlan, C., C. Loot, and M. Chandler. 2004. IS911 partial transposition products and their processing by the Escherichia coli RecG helicase. Mol. Microbiol. 531021-1033. [DOI] [PubMed] [Google Scholar]
- 34.Turlan, C., B. Ton-Hoang, and M. Chandler. 2000. The role of tandem IS dimers in IS911 transposition. Mol. Microbiol. 351312-1325. [DOI] [PubMed] [Google Scholar]
- 35.Ward, C. M., S. J. Wardle, R. K. Singh, and D. B. Haniford. 2007. The global regulator H-NS binds to two distinct classes of sites within the Tn10 transpososome to promote transposition. Mol. Microbiol. 641000-1013. [DOI] [PubMed] [Google Scholar]
- 36.Zhang, L., A. Dawson, and D. J. Finnegan. 2001. DNA-binding activity and subunit interaction of the mariner transposase. Nucleic Acids Res. 293566-3575. [DOI] [PMC free article] [PubMed] [Google Scholar]