Abstract
Branched-chain amino acids are the most abundant amino acids in proteins. The Bacillus subtilis ilv-leu operon is involved in the biosynthesis of branched-chain amino acids. This operon exhibits a RelA-dependent positive stringent response to amino acid starvation. We investigated this positive stringent response upon lysine starvation as well as decoyinine treatment. Deletion analysis involving various lacZ fusions revealed two molecular mechanisms underlying the positive stringent response of ilv-leu, i.e., CodY-dependent and -independent mechanisms. The former is most likely triggered by the decrease in the in vivo concentration of GTP upon lysine starvation, GTP being a corepressor of the CodY protein. So, the GTP decrease derepressed ilv-leu expression through detachment of the CodY protein from its cis elements upstream of the ilv-leu promoter. By means of base substitution and in vitro transcription analyses, the latter (CodY-independent) mechanism was found to comprise the modulation of the transcription initiation frequency, which likely depends on fluctuation of the in vivo RNA polymerase substrate concentrations after stringent treatment, and to involve at least the base species of adenine at the 5′ end of the ilv-leu transcript. As discussed, this mechanism is presumably distinct from that for B. subtilis rrn operons, which involves changes in the in vivo concentration of the initiating GTP.
Branched-chain amino acids are the most abundant amino acids in proteins and form the hydrophobic cores of the proteins. Moreover, these amino acids are precursors for the biosynthesis of iso- and anteiso-branched fatty acids, which represent the major fatty acid species of the membrane lipids in Bacillus species (5). The initial step of isoleucine or valine synthesis is the condensation of 2-oxobutanoate derived from threonine and pyruvate or two pyruvates, leading to the formation of branched-chain keto acids (8). Leucine is synthesized from one of the branched-chain keto acids, i.e., α-ketoisovalerate. The Bacillus subtilis ilv-leu operon comprises seven genes (ilvBHC and leuABCD) necessary for the biosynthesis of branched-chain amino acids (12). The expression of the ilv-leu operon is under positive regulation involving the CcpA protein (36, 41), which is involved in carbon catabolite control of not only hundreds of the catabolic operons and genes but also many cellular processes (6, 11). This CcpA-dependent positive regulation of ilv-leu links carbon metabolism to amino acid anabolism. Recent global gene expression studies of amino acid availability (23) and CodY regulation (25), as well as studies of metabolic linking of ilv-leu expression to nitrogen metabolism (40), revealed that the ilv-leu operon is under direct negative transcriptional control through two major global regulators of nitrogen metabolism (TnrA and CodY). TnrA is known to both activate and repress nitrogen-regulated genes during nitrogen-limited growth (43). The CodY protein is a GTP-binding repressor of several operons, including ilv-leu, that are normally quiescent when cells are growing in a nutrient-rich medium (32). A high concentration of GTP activates the CodY repressor, which serves as a gauge of the general energetic capacity of cells. CodY is also activated through direct interaction with branched-chain amino acids to bind to the promoter regions of its target genes for their repression (35). Furthermore, proteome and transcriptome analyses of the stringent response revealed that the ilv-leu operon exhibited positive stringent control in response to amino acid starvation provoked by dl-norvaline addition (7).
The stringent response is one of the most important adaptations by which bacteria survive under harsh conditions. Of the various occasions of the stringent response resulting from the synthesis of guanosine-5′-diphosphate-3′-diphosphate (ppGpp) from GTP, which is catalyzed by the RelA protein associated with ribosomes, the most prominent is the repression of stable RNA synthesis (4). This response includes direct and indirect activation of the expression of certain genes, including those involved in amino acid biosynthesis. B. subtilis relA mutants, like those of other microorganisms, are unable to synthesize ppGpp (39). Even under nutrient excess conditions, the accumulation of ppGpp caused by amino acid depletion results in a reduction in intracellular GTP, which eventually leads to the induction of sporulation and the transcription of stationary-phase genes in the stringent (relA+) but not in the relaxed (relA) strain (14, 21, 30, 32). The deleterious effects of the relA mutation can be suppressed by the addition of decoyinine, a GMP synthase inhibitor, or by the inactivation of CodY through lowering of the level of intracellular GTP (32). Thus, a low level of intracellular GTP can stimulate the transcription of stationary-phase genes, and ppGpp plays a role by accentuating the reduction in the level of GTP probably through ppGpp inhibition of IMP dehydrogenase, the first enzyme in the GMP synthesis pathway (17, 21, 29, 31).
Krásný and Gourse (18) recently reported that B. subtilis rRNA promoters, for which the initiating nucleoside triphosphate (NTP) for transcription is GTP, appear to be regulated through changes in the GTP pool size, without the mediation of the CodY protein. In contrast to the situation for Escherichia coli, where ppGpp decreases rRNA promoter activity directly (3), it appears that ppGpp may not inhibit B. subtilis RNA polymerase (RNAP) directly. Rather, an increase in the ppGpp concentration might reduce the GTP concentration, thereby modulating rRNA promoter activity indirectly.
In the present work, we investigated the positive stringent response of the expression of ilv-leu upon lysine starvation, which is dependent on RelA. This amino acid starvation led to RelA-dependent positive stringent control, as observed upon dl-norvaline treatment (7). We found two molecular mechanisms underlying this positive stringent control of ilv-leu, i.e., CodY-dependent and -independent mechanisms. The former was triggered by a decrease in the in vivo concentration of GTP, a corepressor of the CodY protein, causing derepressed ilv-leu expression. The latter (CodY-independent) mechanism comprised the modulation of the transcription initiation frequency, likely depending on fluctuation of the in vivo RNAP substrate concentrations upon stringent treatment, and involved at least the base species of adenine at the 5′ end of the ilv-leu transcript.
MATERIALS AND METHODS
Bacterial strains and their construction.
The B. subtilis strains used in this work are listed in Table 1. To construct strains FU737 and FU739, FU771 and FU772, FU769 and FU770, and FU844 and FU845, plasmid pCRE-test2 derivatives carrying the respective ilv-leu regions comprising nucleotides −248, −187, −100, and −55 to +26, the preparation of which was described previously (41), were each linearized with PstI and then used for double-crossover transformation of strains 1A765 and 1A766 to chloramphenicol resistance (5 μg/ml) on tryptose blood agar base (Difco) plates supplemented with 10 mM glucose (TBABG). Strains FU737 and FU739 were further transformed with PCR products containing the tnrA62::Tn917 and ccpA::neo regions, which had been amplified using the respective primer pairs TNRA-F/TNRA-R and CCPA-F/CCPA-R (Table 2) and DNAs of strains FU659 and FU402 as templates to erythromycin (0.3 μg/ml) and neomycin (15 μg/ml) resistance on TBABG to produce strains FU775 and FU776 and strains FU773 and FU774, respectively. To obtain strains FU745, FU747, FU810, and FU809, strains FU737, FU739, FU844, and FU845, respectively, were transformed with chromosomal DNA of strain PS37 at a low concentration (10 ng/ml) to spectinomycin resistance (60 μg/ml). The presence of ΔcodY in the resulting transformants was confirmed by the appearance in ΔcodY strains of a PCR product shorter by 250 bp than that of codY+ strains, as described previously (25). The disruption of the gid gene, present in the ΔcodY strains, does not affect the ilv-leu repression involving CodY (41). The relA1 mutation was reported to be an amino acid substitution at position 240 (Gly to Glu) (14). This substitution was confirmed to be present not only in the chromosomal DNA of strain 1A766 but also in those of strains FU747 and FU809 by means of DNA sequencing of the PCR products of the corresponding regions.
TABLE 1.
B. subtilis strains used in this work
| Strain | Genotypeb | Reference or source |
|---|---|---|
| 168 | trpC2 | 1 |
| 1A765 (BR16)a | trpC2 lys | 39 |
| 1A766 (BR17)a | trpC2 lys relA1 | 39 |
| PS37 | trpC2 gid::spt ΔcodY (Sptr) | 34 |
| FU402 | trpC2 ccpA::neo (Neor) | 41 |
| FU659 | trpC2 tnrA62::Tn917 | 41 |
| NIG2001 | trpC2 pheA1 rpoCHis6 (Neor) | 9 |
| FU737 | trpC2 lys amyE::[cat Pilv-leu(−248/+26)-lacZ] | This work |
| FU739 | trpC2 lys relA1 amyE::[cat Pilv-leu(−248/+26)-lacZ] | This work |
| FU745 | trpC2 gid::spt ΔcodY lys amyE::[cat Pilv-leu(−248/+26)-lacZ] | This work |
| FU747 | trpC2 gid::sptΔcodY lys relA1 amyE::[cat Pilv-leu(−248/+26)-lacZ] | This work |
| FU769 | trpC2 lys amyE::[cat Pilv-leu(−100/+26)-lacZ] | This work |
| FU770 | trpC2 lys relA1 amyE::[cat Pilv-leu(−100/+26)-lacZ] | This work |
| FU771 | trpC2 lys amyE::[cat Pilv-leu(−187/+26)-lacZ] | This work |
| FU772 | trpC2 lys relA1 amyE::[cat Pilv-leu(−187/+26)-lacZ] | This work |
| FU773 | trpC2 lys ccpA::neo amyE::[cat Pilv-leu(−248/+26)-lacZ] | This work |
| FU774 | trpC2 lys relA1 ccpA::neo amyE::[cat Pilv-leu(−248/+26)-lacZ] | This work |
| FU775 | trpC2 lys tnrA62::Tn917 amyE::[cat Pilv-leu(−248/+26)-lacZ] | This work |
| FU776 | trpC2 lys relA1 tnrA62::Tn917 amyE::[cat Pilv-leu(−248/+26)-lacZ] | This work |
| FU809 | trpC2 lys relA1 gid::spt ΔcodY amyE::[cat Pilv-leu(−55/+26)-lacZ] | This work |
| FU810 | trpC2 lys gid::spt ΔcodY amyE::[cat Pilv-leu(−55/+26)-lacZ] | This work |
| FU844 | trpC2 lys amyE::[cat Pilv-leu(−55/+26)-lacZ] | This work |
| FU845 | trpC2 lys relA1 amyE::[cat Pilv-leu(−55/+26)-lacZ] | This work |
| FU895 | trpC2 lys amyE::{cat Pilv-leu[−55/+26(C+1G)]-lacZ} | This work |
| FU904 | trpC2 lys amyE::{cat Pilv-leu[−55/+26(C+1G)(A+2G)]-lacZ} | This work |
| FU905 | trpC2 lys amyE::{cat Pilv-leu[−55/+26(A+2G)]-lacZ} | This work |
The strain was obtained from the Bacillus Genetic Stock Center (Columbus, OH).
Sptr, spectinomycin resistance; Neor, neomycin resistance.
TABLE 2.
Oligonucleotide primers used in this work
| Purpose of oligonucleotide primer | Name of oligonucleotide primer | Sequence of oligonucleotide primera |
|---|---|---|
| tnrA::Tn917 and ccpA::neo transfer | TNRA-F | ATAGAGTTTTTCAGAATAATGGCGTCG |
| TNRA-R | GCATTATCAGCTATTTTGAAGACGCGC | |
| CCPA-F | AGAAACGCATTTGCCAGTCTTTGTTG | |
| CCPA-R | TCGGTGCCGTTCCTCCATTGCTGCGA | |
| Random base substitution | D-248F3 | GCGCTCTAGATGATCTGTCAGACTCAATCCAT |
| D+26RR | GATGATTTGGATCCGTGAAGCTTGCATTTATCTTTTGTNNNNCTCATA | |
| Construction of strains FU895, FU904, and FU905 | D-55F | GCGCGCGCTCTAGAAATAATTTTAAAAAATGCTG |
| D+26R (GA) | GCGCGGATCCGTGAAGCTTGCATTTATCTTTTGTTCAA | |
| D+26R (GG) | GCGCGGATCCGTGAAGCTTGCATTTATCTTTTGTCCAA | |
| D+26R (CG) | GCGCGGATCCGTGAAGCTTGCATTTATCTTTTGTCGAA | |
| Primer extension | PEpF | CCAGTTAAAGGATTTGAGCGTAGCGAAb |
| PEpR | TCCACAGTAGTTCACCACCTTTTCCCTATAb | |
| In vitro transcription (D-55F/ITlacZ-R) | ITlacZ-R | CAGGAAACAGCTATGACCTGCGGGCCTCTTCGCTATTA |
Underlining indicates restriction enzyme sites.
Sequence from plasmid pCRE-test2.
To construct strains FU895, FU904, and FU905 carrying the respective base substitutions at +1 and +2 (C+1G, C+1G and A+2G, and A+2G; the wild-type ilv-leu transcription initiation base of C, reported by Grandoni et al. [12], is arbitrarily assigned as +1), the promoter regions (nucleotides −55 to +26) were amplified using primer pairs D-55F/D+26R (GA), D-55F/D+26 (GG), and D-55F/D+26R (CG) (Table 2), respectively, and chromosomal DNA of strain 168 as a template. The PCR products trimmed with XbaI and BamHI were cloned into plasmid pCRE-test2 (24) in E. coli strain DH5α, as described previously (41). The correct construction of the fusions in the resulting plasmids was confirmed by sequencing. The plasmids carrying the region with the base substitution(s) were each linearized with PstI and then used for double-crossover transformation of strains 1A765 to chloramphenicol resistance (5 μg/ml) on TBABG plates, which produced strains FU895, FU904, and FU905, respectively.
Cell growth and β-Gal assay.
The lacZ fusion strains were grown at 30°C overnight on TBABG containing the following resistance antibiotic(s): erythromycin (0.3 μg/ml), chloramphenicol (5 μg/ml), spectinomycin (60 μg/ml), and/or neomycin (15 μg/ml). The cells were inoculated into 100 ml of minimal medium comprising 0.4% glucose, 0.2% glutamine, and 50 μg/ml tryptophan (MM medium) (45) supplemented with a mixture of 16 amino acids (glutamine, histidine, tyrosine, and asparagine being omitted) (2) at an optical density at 600 nm (OD600) of 0.08 and then incubated at 37°C. When the OD600 reached approximately 0.5, 45-ml aliquots of the culture were harvested, and the cells were spun down at 25°C (2,000 × g for 10 min). The cells were suspended in 45 ml MM medium supplemented with the above-described amino acid mixture with and without lysine, and then the cultures were incubated further. During culture incubation before and after cell resuspension, 1-ml aliquots of the culture were withdrawn at most at 15-min intervals, and then β-galactosidase (β-Gal) activity in crude cell extracts was spectrophotometrically measured as described previously (45).
In the case that the effect of decoyinine on lacZ expression in the fusion strains was examined, a culture was divided into two 45-ml portions when a culture reached an approximate OD600 of 0.5, and decoyinine (500 μg/ml) was added to one of the portions. The β-Gal activity was monitored as described above.
Determination of in vivo concentrations of nucleotides.
The intracellular concentrations of nucleotides, including ppGpp, were determined by high-performance liquid chromatography (HPLC) after extraction with 1 N formic acid, essentially as previously described (28, 30). The cells were grown in MM medium with 16 amino acids to an approximate OD600 of 0.5 and then spun down as described above. The cells were resuspended at the same cell density in MM medium supplemented with the amino acid mixture with and without lysine as that of the original culture. In the case of decoyinine treatment, a portion of the cells was exposed to 500 μg/ml decoyinine that had been added directly to the culture, and the other portion of the cells was incubated without decoyinine addition. After incubation for 30 min, 10 ml of each culture was filtered through a Millipore polyvinylidene difluoride membrane filter (0.45-μm pore size, 47 mm in diameter) for 10 s and then rapidly washed with a small amount of distilled water through vacuum aspiration. Each membrane on which the cells had been collected was soaked in 2 ml of 1 M formic acid in a plastic petri dish on ice, and then the cells were detached from the membrane and kept on ice for 60 min. The cell debris was removed by ultrafiltration with a Millipore Ultrafree-MC instrument (5,000 nominal molecular weight limit [NMWL]), and the filtrates were freeze-dried. After being resolved in distilled water, the samples were subjected to HPLC. The nucleotides were eluted at a flow rate of 1.5 ml/min with a gradient made up of a low-ionic-strength buffer (7 mM KH2PO4, pH 4.0, with H3PO4) and a high-ionic-strength buffer (0.5 M KH2PO4 plus 0.5 M Na2SO4, pH 5.4, with KOH). By comparison with the peak areas of standards, the amount of each nucleotide was determined.
The in vivo concentrations of metabolites including nucleotides were determined by means of capillary electrophoresis mass spectrometry (CE/MS) after extraction with methanol, essentially as described previously (37). The cells were subjected to lysine starvation or decoyinine treatment as described above. The cells were collected on a Millipore Isopore membrane filter HTTP (0.4-μm pore size, 47 mm in diameter). Each of the cell-bearing membranes was put into a heat-sealable plastic bag containing 2 ml of ice-chilled methanol, and then the cells were well detached from the membrane and the bag was completely sealed. The sealed bags were incubated at 70°C for 1 h. After the cell suspensions had been centrifuged to spin down cell debris, the supernatants were treated with chloroform to remove lipids. After the upper layer had been subjected to ultrafiltration (Millipore Ultrafree-MC [5,000 NMWL]), the filtrates were freeze-dried. After the pellets had been dissolved in distilled water, samples were subjected to CE/MS to identify the metabolites including NTP and ppGpp in the samples and to measure their concentrations, as described previously (37); this metabolome analysis was carried out by Human Metabolome Technologies, Inc., Japan. We determined the in vivo concentrations of not only ATP but also ADP and AMP in this metabolome analysis. So, energy charges [(ATP + 1/2 ADP)/(ATP + ADP + AMP)] were calculated to be roughly 0.7, which is comparable to those obtained by the extraction method without filtering (13).
The in vivo molar concentrations of metabolites were calculated from the relationship of the aqueous volume of 1 OD600 unit (OD600 × ml) corresponding to 0.83 μl (10).
Base substitution of nucleotides −2 to +2 in close vicinity to the ilv-leu transcription initiation base as to the CodY-independent positive stringent response.
The ilv-leu promoter region (nucleotides −248 to +26) was amplified by PCR using primer set D-248F3/D+26RR (Table 2); D+26RR contains any bases at the positions corresponding to nucleotides −2, −1, +1, and +2. The PCR product was trimmed with XbaI and BamHI and then cloned into plasmid pCRE-test2, and the resulting plasmid was linearized and used for the transformation of strain 1A765 to chloramphenicol resistance, as described above.
The chloramphenicol-resistant transformants possessing the ilv-leu promoter (Pilv-leu)-lacZ fusions carrying randomly substituted bases at nucleotides −2 to +2 in the amyE locus were screened to find those which exhibit low inducibility of lacZ upon decoyinine addition, as follows. Each of the transformants was inoculated into MM medium with lysine (50 μg/ml) in the wells of a microplate (96 wells) to an OD600 of 0.05; CodY does not function in cells growing in this medium (41). The microplate with cultures in each well was incubated at 25°C on a microplate shaker (Bioshaker; Taitec), and the OD600 was monitored with a microplate reader (Nalge Nunc International) until the OD600 reached 0.2, when decoyinine was added to 500 μg/ml to some wells. After further incubation with and without decoyinine for 1 h, 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (50 μg/ml) was added to the cultures, followed by further incubation for 30 min. After reading the OD630 of the supernatant of each culture, tens of transformants that exhibited the lowest inducibilities of lacZ with decoyinine were selected, and their ilv-leu promoter sequences including nucleotides −2 to +2 were determined.
Primer extension analysis.
Primer extension analysis was performed as described previously (44). RNA samples were prepared as described previously (46) from cells of strains FU844, FU895, FU904, and FU905 that had been grown in MM medium containing the 16-amino-acid mixture. Reverse transcription using the above-described four RNA samples was initiated from the PEpR primer (Table 2), which had been labeled at its 5′ end by use of a Megalabel kit (Takara Bio) and [γ-32P]ATP (GE Healthcare). A template for the dideoxy sequencing reactions for a ladder preparation starting from the same end-labeled primer was prepared by PCR using the primer pair PEpF/PEpR (Table 2) and DNA from strain FU844 as a template.
In vitro transcription analysis.
To prepare His-tagged RNAP, cells of strain NIG2001 (9) were grown in LB medium (33) containing neomycin (5 μg/ml) (total, 1 liter) to an OD600 of 1. The harvested cells were washed with 145 mM NaCl and then suspended in buffer I (10 mM Tris-Cl [pH 8.0], 10% [vol/vol] glycerol, and 5 mM imidazole). The cells were broken by sonication, and the extract was centrifuged for 30 min at 28,000 × g to obtain 30 ml of supernatant in total. The supernatant was applied to a column of 30 ml Ni2+-nitrilotriacetic acid resin (Qiagen), washed with buffer I, and then eluted with buffer II (10 mM Tris-Cl [pH 8.0], 10% glycerol, and 100 mM imidazole). The peak fractions exhibiting absorbance at 280 nm were pooled and concentrated by ultrafiltration to 1 mg/ml protein; RNAP in this preparation was judged to be roughly 50% pure by means of sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The same volume of glycerol as that of the RNAP preparation was added to give 0.5 mg/ml protein, followed by storing at −80°C.
The DNA templates for in vitro transcription were prepared by PCR using primer pair D-55F/ITlacZ-R (Table 2) and chromosomal DNAs of strains FU844, FU895, FU904, and FU905 as templates. Multiple-round transcription was performed essentially as described previously (18). The standard reaction mixtures (10 μl) comprised approximately 0.2 μM RNAP, 1 nM DNA template, 40 mM Tris-Cl (pH 8.0), 10 mM MgCl2, 1 mM dithiothreitol, 150 mM KCl, 100 μg/ml bovine serum albumin, 200 μM each of ATP, CTP, and GTP, 10 μM UTP, and 2 μM [α-32P]UTP (110 TBq/mmol) (GE Healthcare). Reactions were initiated with RNAP, allowed to proceed at 30°C for 15 min, and terminated by the addition of an equal volume of stop solution (loading buffer consisted of 80% [vol/vol] formamide, 0.1% [wt/vol] sodium dodecyl sulfate, 8% [vol/vol] glycerol, 8 mM EDTA, 0.05% [wt/vol] bromophenol blue, and 0.05% [wt/vol] xylene cyanol). Runoff transcripts were electrophoresed on a 7 M urea-5% polyacrylamide gel, and then the radioactivities were quantified with an image analyzer (Typhoon 9400; GE Healthcare).
RESULTS
Involvement of CodY and RelA in the positive stringent response of the ilv-leu operon upon starvation of a required amino acid, lysine.
The B. subtilis ilv-leu operon involved in the biosynthesis of branched-chain amino acids is under CcpA-mediated positive regulation and is also under negative regulation mediated by TnrA and CodY, which recognize and bind to their respective cis elements located upstream of the ilv-leu promoter (Fig. 1) (36, 41). The proteome and transcriptome analyses suggested that the expression of the ilv-leu operon might be under RelA-dependent positive regulation on the stringent response evoked by dl-norvaline addition (7). This regulation might have resulted from a decrease in the GTP concentration, which caused the relief from CodY repression, as described previously (14, 15, 21, 31). This hypothesis was tested by means of lacZ fusion experiments to see whether the stringent response enhances the expression of ilv-leu in either the relA+ or the relA1 strain in the genetic backgrounds of a codY deletion (ΔcodY) as well as a ccpA deletion and a tnrA disruption (Fig. 1 and 2). The relA1 mutation, isolated by Swanton and Edlin (39), is an amino acid substitution at position 240 of the RelA protein (Gly to Glu) (14), which largely abolished ppGpp synthesis, as described below. However, a relA deletion mutant was auxotrophic for valine, leucine, and isoleucine, in contrast to the prototrophic phenotype of a relA1 mutant, which is unexplainable (42). We used strain 1A766 (relA1 lys) in this study because its isogenic strain 1A765 (lys) exhibits a stringent response upon lysine starvation (39). We constructed relA+ relA1 sets of Lys− strains FU737/FU739, FU775/FU776, FU773/FU774, and FU745/FU747, carrying a lacZ fusion of an ilv-leu promoter region (nucleotides −246 to +26) at the amyE locus, in the wild-type, tnrA::Tn917, ccpA::neo, and ΔcodY backgrounds, respectively. When cells of the relA+ Lys− strains (FU737 [tnrA+] and FU775 [tnrA::Tn917]) were subjected to lysine starvation, β-Gal synthesis was largely induced in the two strains as their growth began to stop (Fig. 2A). However, this induction upon lysine starvation was not observed in cells of the relA1 Lys− strains (FU739 [tnrA+] and FU776 [tnrA::Tn917]) (Fig. 2B). When cells of the relA+ Lys− strains (FU737 [ccpA+] and FU773 [ccpA::neo]) were subjected to lysine starvation, β-Gal induction was observed in both strains. However, β-Gal synthesis in the ccpA::neo cells was considerably reduced and the growth of these cells was also relatively slow, because of the lack of CcpA-mediated positive regulation (Fig. 2C). Also, no β-Gal induction upon lysine starvation was observed in cells of the relA1 Lys− strains (FU739 [ccpA+] and FU774 [ccpA::neo]) (Fig. 2D). These results indicated that no induction of β-Gal synthesis under the control of the ilv-leu promoter upon lysine starvation occurs without RelA, although this induction occurs without TnrA and CcpA.
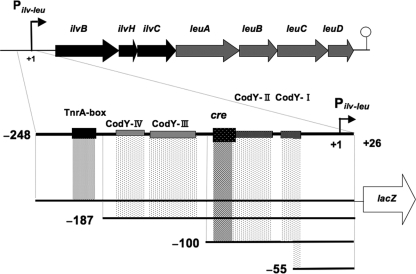
FIG. 1.
B. subtilis ilv-leu operon and deletion analysis for the positive stringent response. The ilv-leu operon, consisting of seven genes (ilvBHC and leuABCD) (12, 20), was transcribed from the ilv-leu promoter (Pilv-leu) to a terminator downstream of leuD (41). The locations of the TnrA box (40), a catabolite-responsive element (cre) for the binding of the complex of the CcpA and P-Ser-HPr proteins (36, 41), and the CodY binding sites (CodY-I, -II, -III, and -IV) (35) are indicated. To perform deletion analysis of the ilv-leu promoter region for the positive stringent response, the respective promoter regions comprising bases −248 to +26, −187 to +26, −100 to +26, and −55 to +26 were fused with lacZ and then integrated into the amyE locus, as described in the text.
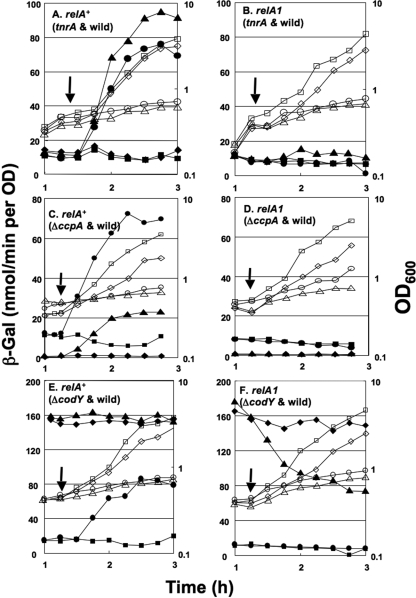
FIG. 2.
Involvement of CodY and RelA in the positive stringent response of ilv-leu transcription upon lysine starvation. To monitor the ilv-leu promoter activity, all of the Lys− strains used in this analysis carried the lacZ fusion with the promoter region comprising nucleotides −248 to +26 in the amyE locus. Cells of strains FU737 (relA+ tnrA+) and FU775 (relA+ tnrA::Tn917) (A), FU739 (relA1 tnrA+) and FU776 (relA1 tnrA::Tn917) (B), FU737 and FU773 (relA+ ccpA::neo) (C), FU739 and FU774 (relA1 ccpA::neo) (D), FU737 and FU745 (relA+ ΔcodY) (E), and FU739 and FU747 (relA1 ΔcodY) (F) were grown in MM medium containing a mixture of 16 amino acids (15 amino acids plus lysine) to the logarithmic growth phase (OD600 of 0.5) and then spun down, as described in the text. A portion of the cells was suspended and further incubated in MM medium containing 15 amino acids plus lysine, and the other part of the cells was suspended and incubated in MM medium containing 15 amino acids to subject the cells to lysine starvation, as described in the text; arrows indicate the start times of incubation with and without lysine. Cell growth (OD600, open symbols) and lacZ expression (β-Gal activity, filled symbols) were monitored during growth and lysine starvation; results for wild-type strains (tnrA+ ccpA+ codY+) with starvation (circles) and without starvation (squares) and mutant strains (tnrA::Tn917, ccpA::neo, or ΔcodY) without starvation (diamonds) and with starvation (triangles) are shown.
When cells of the relA+ Lys− strains (FU737 [codY+] and FU745 [ΔcodY]) were subjected to lysine starvation, very high constitutive β-Gal synthesis was observed in the ΔcodY cells, with some significant induction upon lysine starvation, although the synthesis was well induced in the codY+ cells upon lysine starvation (Fig. 2E). Furthermore, no further induction of β-Gal synthesis over the constitutive level was observed upon lysine starvation in cells of the relA1 Lys− strains (FU739 [codY+] and FU747 [ΔcodY]) (Fig. 2F). Interestingly, β-Gal synthesis in cells of strain FU747 was greatly decreased after lysine starvation; this drastic decrease due to a lack of CodY and RelA cannot properly be explained at present. The overall results suggested that the induction of β-Gal synthesis was most likely triggered by CodY inactivation, because only the codY deletion rendered it constitutive.
Occurrence of another positive stringent response of the ilv-leu promoter upon lysine starvation which is CodY independent but RelA dependent.
We performed deletion analysis of the ilv-leu promoter region for the positive stringent response of the ilv-leu operon to confirm the involvement of CodY in this response, which unexpectedly led us to find another positive ilv-leu stringent response involving only RelA, as follows. For deletion analysis, we used relA+ relA1 sets of strains FU737/FU739, FU771/FU772, FU769/FU770, and FU844/FU845 carrying lacZ fusions with promoter regions comprising nucleotides −248 to +26 with all cis elements, −187 to +26 with cre and CodY-I to -IV, −100 to +26 with cre and CodY-I and -II, and −55 to +26 with only part of CodY-I, respectively, as illustrated in Fig. 1. As shown in Fig. 3A, panels a, b, c, and d, the relA+ strains, FU737, FU771, FU769, and FU844 carrying lacZ fusions with various promoter regions, exhibited the positive stringent response of β-Gal synthesis upon lysine starvation, whereas the corresponding relA1 strains, FU739, FU772, FU770, and FU845, exhibited no positive stringent response. As described previously (41), CcpA-mediated positive and CodY-mediated negative regulation did not occur in strains carrying the lacZ fusions with the ilv-leu promoter regions comprising nucleotides −100 to +26 and −55 to +26 and nucleotides −55 to +26, respectively, so the basal levels of β-Gal synthesis were the lowest for strains FU769 and FU770 (Fig. 3A, panel c) and the highest for strains FU844 and FU845 (Fig. 3A, panel d). Thus, the induction of β-Gal synthesis upon lysine starvation was only 2.5-fold for FU844 (relA+) (Fig. 3A, panel d). This positive stringent response observed with the ilv-leu promoter region comprising nucleotides −55 to +26 was RelA dependent but most likely CodY independent because there are no CodY-binding sites in this promoter region (Fig. 1).
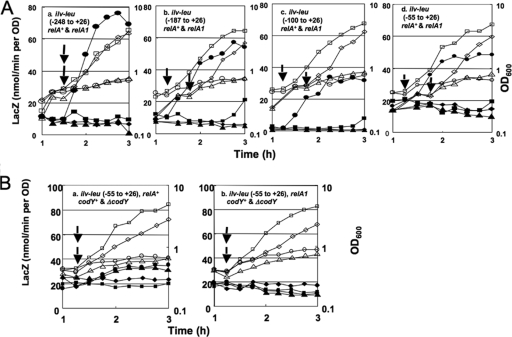
FIG. 3.
Deletion analysis of the ilv-leu promoter region as to its positive stringent response. (A) Lys− strains (FU737 [relA+] and FU739 [relA1], FU771 [relA+] and FU772 [relA1], FU769 [relA+] and FU770 [relA1], and FU844 [relA+] and FU845 [relA1]), carrying the lacZ fusions of the ilv-leu promoter regions comprising nucleotides −248 to +26 (a), −187 to +26 (b), −100 to +26 (c), and −55 to +26 (d), were used for the analysis. (B) Lys− relA+ strains (FU844 [codY+] and FU810 [ΔcodY]) (a) and Lys− relA1 strains (FU845 [codY+] and FU809 [ΔcodY]) (b), carrying the ilv-leu promoter region (nucleotides −55 to +26), were used. Cells of each strain were grown and subjected to lysine starvation, as described in the legend to Fig. 2. Each arrow indicates the start time of further incubation with and without lysine after cell suspension. Cell growth (OD600, open symbols) and lacZ expression (β-Gal activity, filled symbols) were monitored during growth and lysine starvation. (A) relA+ strains with starvation (circles) and without starvation (squares) and relA1 strains with starvation (triangles) and without starvation (diamonds). (B) codY+ strains with starvation (circles) and without starvation (squares) and ΔcodY strains with starvation (triangles) and without starvation (diamonds).
To confirm the occurrence of this CodY-independent positive stringent response requiring only the promoter region of nucleotides comprising −55 to +26, we constructed ΔcodY strains without and with relA1 carrying the lacZ fusion of this promoter region. We performed lysine starvation experiments using the codY+ and ΔcodY fusion strains FU844 (relA+ codY+) and FU810 (relA+ ΔcodY) and FU845 (relA1 codY+) and FU809 (relA1 ΔcodY) and confirmed that this positive stringent response, which was lower than that involving CodY, occurred in the relA+ strains with and without CodY (Fig. 3B, panel a) but did not occur in the relA1 strains with and without CodY (Fig. 3B, panel b). However, it is notable that this positive response was somewhat diminished in the ΔcodY strain (Fig. 3B, panel a). This was likely due to the indirect effect of the codY deletion on the ilv-leu promoter activity, which was decreased without mediation by the CodY-binding sites of ilv-leu. The overall results clearly indicate that there are two molecular mechanisms underlying the RelA-dependent positive stringent response of ilv-leu; one is CodY dependent and the other is CodY independent, and the latter requires only the limited promoter region very close to the ilv-leu transcription initiation site (nucleotides −55 to +26).
Positive response of ilv-leu expression to decoyinine addition.
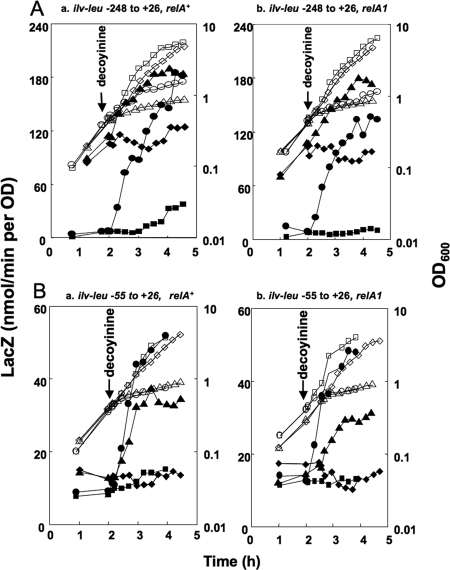
Decoyinine is a specific inhibitor of GMP synthase (38), so decoyinine addition can decrease guanine nucleotides without the production of (p)ppGpp (14, 15, 21, 22, 31). Thus, we examined the effect of decoyinine addition on ilv-leu expression in the relA+ strains (FU737 [codY+] and FU745 [ΔcodY]) carrying the ilv-leu promoter region (nucleotides −248 to +26) (Fig. 4A, panel a). Decoyinine addition remarkably induced ilv-leu expression in strain FU737 (codY+). Also, it induced the expression even in strain FU745 (ΔcodY), constitutively expressing ilv-leu due to the absence of CodY. This induction pattern upon decoyinine addition was observed in the relA1 strains (FU739 [codY+] and FU747 [ΔcodY]) (Fig. 4A, panel b), indicating that the β-Gal induction is independent of RelA. We also examined the effect of decoyinine addition on ilv-leu expression by using relA+ strains FU844 (codY+) and FU810 (ΔcodY) (Fig. 4B, panel a) and relA1 strains FU845 (codY+) and FU809 (ΔcodY) (Fig. 4B, panel b), carrying the lacZ fusion of the ilv-leu promoter region (nucleotides −55 to +26). Also, we observed similar CodY-independent induction patterns in both the relA+ and the relA1 strains, although a lower lacZ induction was observed for the ΔcodY strain (Fig. 4B, panels a and b), as in the case of lysine starvation (Fig. 3B, panel a). This CodY- and RelA-independent positive response upon decoyinine addition, likely occurring very close to the transcription initiation site, was considered to be essentially the same as the CodY-independent but RelA-dependent positive response which occurred upon lysine starvation.
FIG. 4.
Effect of decoyinine addition on ilv-leu expression. (A) relA+ strains (FU737 [codY+] and FU745 [ΔcodY]) (a) and relA1 strains (FU739 [codY+] and FU747 [ΔcodY]) (b), carrying the ilv-leu promoter region (nucleotides −248 to +26), and (B) relA+ strains (FU844 [codY+] and FU810 [ΔcodY]) (a) and relA1 strains (FU845 [codY+] and FU809 [ΔcodY]) (b), carrying the ilv-leu promoter region (nucleotides −55 to +26), were used for the analysis. Each of the codY+ ΔcodY set strains was grown as two cultures, and decoyinine was added to only one culture. Cell growth (OD600, open symbols) and lacZ expression (β-Gal activity, filled symbols) were monitored during incubation before and after decoyinine addition (codY+ strains with decoyinine [circles] and without decoyinine [squares] and ΔcodY strains with decoyinine [triangles] and without decoyinine [diamonds]), as described in the text.
Fluctuation of in vivo concentrations of NTP and ppGpp upon lysine starvation and decoyinine addition.
The fluctuation of the in vivo concentrations of NTP and (p)ppGpp upon amino acid (isoleucine, methionine, or aspartate) starvation (30) or decoyinine addition (14, 22) was investigated. It was reported that the in vivo GTP and ATP concentrations decreased or increased approximately two to three times upon starvation of the above-mentioned amino acids (30) or upon decoyinine addition (14, 22), respectively, whereas (p)ppGpp temporally increased in a relA+ strain only upon amino acid starvation.
We measured the concentrations of NTP and ppGpp in cells of strains 1A765 (relA+) and 1A766 (relA1) which had been subjected to lysine starvation and decoyinine treatment for 30 min as described in Materials and Methods (Table 3). We adopted two methods to determine the intracellular concentrations of nucleotides, including ppGpp; one involves extraction of the nucleotides from cells with formic acid and then determination of their concentrations by means of HPLC (28, 30), and the other involves extraction with methanol and determination by CE/MS (37). Table 3 shows the in vivo millimolar concentrations of four NTPs and ppGpp, which were obtained by the former and latter methods, respectively. The two methods gave similar fluctuations of the in vivo concentrations of these nucleotides, as follows. The GTP concentrations in the relA+ cells (approximately 1.3 mM with the former method and approximately 0.4 mM with the latter method) decreased two to six times upon decoyinine addition and lysine starvation, whereas those in the relA1 cells decreased two to four times only upon decoyinine addition. The ATP (approximately 2.7 mM) and CTP (approximately 0.5 mM) concentrations increased two to five times in both the relA+ and the relA1 cells upon either decoyinine addition or lysine starvation. The UTP concentrations in the relA+ and relA1 cells (roughly 1 mM) remained rather constant even under these stress conditions. ppGpp was detected only in the relA+ cells suffering from lysine starvation, its concentration being approximately 50 μM. These fluctuations in the in vivo concentrations of NTP and ppGpp were essentially the same as those in the cells subjected to amino acid starvation and decoyinine treatment under the distinct growth conditions (14, 22, 30).
TABLE 3.
In vivo concentrations of NTP and ppGppa
| Stress treatment | Strain genotype (condition)b | Concn (mM) of:
|
||||
|---|---|---|---|---|---|---|
| GTP | ATP | CTP | UTP | ppGpp | ||
| HPLC | relA+ (+K) | 1.37 ± 0.30 | 2.33 ± 0.63 | 0.60 ± 0.12 | 1.25 ± 0.25 | NDc |
| relA+ (−K) | 0.23 ± 0.12 | 7.13 ± 1.1 | 1.53 ± 0.56 | 1.55 ± 0.21 | 0.067 ± 0.020 | |
| relA1 (+K) | 1.26 ± 0.31 | 2.14 ± 0.48 | 0.47 ± 0.09 | 1.02 ± 0.15 | ND | |
| relA1 (−K) | 1.09 ± 0.39 | 8.51 ± 2.12 | 1.81 ± 0.22 | 2.50 ± 0.12 | ND | |
| relA+ (−Dc) | 1.24 ± 0.41 | 3.40 ± 1.2 | 0.78 ± 0.15 | 1.57 ± 0.51 | ND | |
| relA+ (+Dc) | 0.36 ± 0.22 | 5.30 ± 1.9 | 1.37 ± 0.30 | 0.83 ± 0.15 | ND | |
| relA1 (−Dc) | 0.71 ± 0.24 | 3.04 ± 1.02 | 0.49 ± 0.08 | 0.80 ± 0.12 | ND | |
| relA1 (+Dc) | 0.25 ± 0.18 | 5.50 ± 0.45 | 1.20 ± 1.20 | 1.70 ± 0.35 | ND | |
| CE/MS | relA+ (+K) | 0.35 ± 0.15 | 2.15 ± 1.08 | 0.32 ± 0.06 | 0.81 ± 0.15 | ND |
| relA+ (−K) | 0.089 ± 0.042 | 11.94 ± 2.50 | 1.07 ± 0.07 | 1.04 ± 0.35 | 0.027 ± 0.015 | |
| relA+ (−Dc) | 0.44 ± 0.16 | 3.13 ± 1.24 | 0.38 ± 0.05 | 1.19 ± 0.08 | ND | |
| relA+ (+Dc) | 0.19 ± 0.03 | 7.88 ± 2.02 | 0.75 ± 0.11 | 1.29 ± 0.05 | ND | |
The in vivo concentrations of NTP and ppGpp in cells of strains 1A765 (relA+) and 1A766 (relA1) 30 min after stress treatment were determined as described in the text.
+K and −K denote without and with lysine starvation, respectively. −Dc and +Dc denote without and with decoyinine treatment, respectively.
ND, not detected.
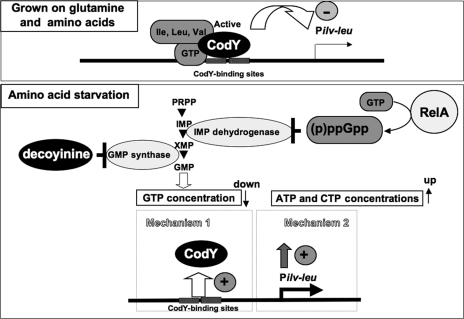
The overall results obtained so far clearly indicate that the positive stringent response to lysine starvation and decoyinine treatment involves two molecular mechanisms; one comprises relief from CodY-dependent repression of the ilv-leu promoter (mechanism 1) (Fig. 5), and the other comprises a CodY-independent enhancement of the ilv-leu promoter activity, which occurs in close vicinity to the ilv-leu transcription initiation site (mechanism 2) (Fig. 5). The two forms of positive regulation are most likely evoked by the lowering of the in vivo GTP concentration; mechanism 1 involves the detachment of CodY from the CodY-binding sites through a decrease in GTP, a CodY corepressor, whereas mechanism 2, which is independent of the CodY-binding sites, probably involves positive modulation of the ilv-leu promoter activity, possibly through the fluctuation of the in vivo concentrations of the RNAP substrates, especially GTP, under the stress conditions, as described below.
FIG. 5.
Positive stringent response of the ilv-leu operon to amino acid (lysine) starvation involves two molecular mechanisms. As shown at the top, when B. subtilis cells grow in MM medium containing glucose and glutamine and supplemented with 16 amino acids, the CodY protein interacting with GTP and branched-chain amino acids, corepressors of CodY, represses the transcription from the ilv-leu promoter through interference of RNAP entry to it by the complex. As shown at the bottom, when cells are subjected to lysine starvation, the RelA protein synthesizes (p)ppGpp from GTP. The promoter (p)ppGpp inhibits IMP dehydrogenase, forming XMP in the de novo synthesis pathway starting from 5-phosphoribosyl-1-pyrophosphate (PRPP), resulting in lowering of the in vivo GMP and subsequent GTP concentrations. On the other hand, addition of decoyinine to the same medium causes the inhibition of GMP synthase without the production of (p)ppGpp, resulting in lowering of the in vivo GMP and subsequent GTP concentrations. The lowering of the GTP concentration most likely causes relief from CodY repression of the ilv-leu promoter (mechanism 1), whereas lowering of the GTP concentration as well as raising of the ATP and CTP concentrations likely causes the activation of the transcription from the ilv-leu promoter catalyzed by RNAP (mechanism 2).
Identification of bases involved in the positive stringent response of ilv-leu which requires no CodY-binding site.
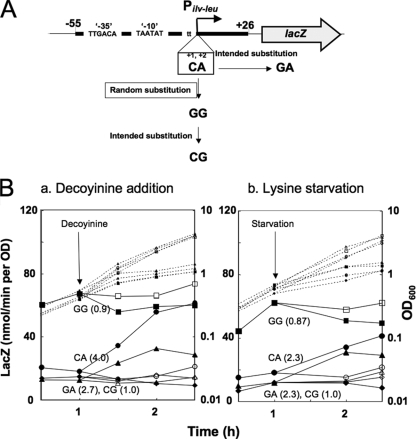
Krásný and Gourse (18) reported that the initiating NTP for transcription from B. subtilis rRNA promoters is most likely GTP and that changes in the promoter activity always correlate with changes in the intracellular GTP concentration. This communication led us to identify the bases involved in the CodY-independent positive stringent response of ilv-leu, which might be the transcription initiation base or one in close vicinity to it. The cytosine at nucleotide +1 has been reported to be the transcription initiation base of ilv-leu (12), which was confirmed by us as described below, so we replaced this C with G, A, and T; the replacement of C with T greatly enhanced the promoter strength (data not shown). However, the replacement of C with A and T did not affect the positive stringent response of ilv-leu (data not shown), although the replacement of C with G significantly affected it (Fig. 6, compare CA and GA).
FIG. 6.
Nucleotide sequence dependency of the CodY-independent positive stringent response in the vicinity of the transcription initiation site. (A) The wild-type ilv-leu TTCA sequence (CA) (nucleotides −2 to +2) in the vicinity of the transcription initiation site was randomly substituted, as described in the text. As a result, the promoter possessing the TTGG sequence did not exhibit the CodY-dependent positive stringent response at all. Based on this fact, we constructed a series of strains carrying the promoter region (nucleotides −55 to +26) possessing the TTGG (GG), TTGA (GA), or TTCG (CG) sequence, which was fused with lacZ in addition to the wild-type strain possessing the CA sequence. (B) Cells of strains FU844 (CA) (circles), FU859 (GA) (triangles), FU905 (CG) (diamonds), and FU904 (GG) (squares) were grown in MM medium supplemented with 16 amino acids as two cultures. When the OD600 of the cultures reached 0.5, decoyinine was added to one culture, and then the cultures with and without decoyinine were incubated further. On the other hand, the four strains were grown in the same medium until the OD600 was 0.5 as two cultures, and then one culture was subjected to lysine starvation for more than 1 h, as described in the text. Cell growth (OD600, dotted lines) and lacZ expression (β-Gal activity, solid lines) were monitored without stringent treatment (open symbols) and with treatment (filled symbols). Arrows indicate the start times of stringent treatment. The numbers in parentheses are ratios, which were obtained by dividing the β-Gal activities of the cells subjected to stringent treatment for 1 h by those of the cells with no treatment after 1 h.
We therefore decided to introduce random base substitutions at nucleotides −2 to +2 to examine whether they affect this positive stringent response. At first, the ilv-leu promoter region (nucleotides −248 to +26) was amplified by PCR using a primer set, the reverse of which carried the random base substitutions at nucleotides −2 to +2, fused with lacZ, and then used for the transformation of strain 1A765 (lys relA+) for the fusion to be inserted into the amyE locus. The transformants were screened to find those which exhibited low inducibility of lacZ upon decoyinine addition under the growth medium conditions where CodY does not function, as described in Materials and Methods. Out of tens of transformants carrying the lacZ fusions with the base substitutions, which showed low inducibility of lacZ upon decoyinine addition, the transformant that exhibited the lowest inducibility of lacZ was found to possess a lacZ fusion with a TTGG sequence at nucleotides −2 to +2 instead of the wild-type TTCA (Fig. 6A) (data not shown).
The TTGG sequence of ilv-leu carries two base substitutions at nucleotides +1 and +2 compared to the wild-type sequence TTCA. We further constructed two strains, FU904 and FU905, which carry the GG and CG sequences at nucleotides +1 and +2 in the ilv-leu promoter region (nucleotides −55 to +26) fused with lacZ, respectively (Fig. 6A), in addition to strains FU844 and FU895, carrying the CA and GA sequences in the same promoter region. As shown in Fig. 6B (refer to the β-Gal activities on the vertical axes), the strength of these base-substituted promoters was highly dependent on their sequences, with the strength decreasing in the order of GG, CA, and GA and CG; the promoters carrying the latter two sequences exhibited the lowest activities.
β-Gal synthesis in cells of strains FU844 (CA), FU895 (GA), FU905 (CG), and FU904 (GG) was monitored during their growth in the presence and absence of decoyinine (Fig. 6B, panel a). Decoyinine addition induced β-Gal synthesis in strain FU844 (CA) by 4.0-fold compared with the β-Gal activities during the first 1 h of incubation with and without decoyinine, whereas this addition induced β-Gal synthesis in strain FU895 (GA) by 2.7-fold. However, decoyinine addition did not induce β-Gal synthesis in FU905 (CG), and it slightly reduced that in FU904 (GG) by 0.9-fold. As shown in Fig. 6B, panel b, β-Gal synthesis in cells of Lys− strains FU844 (CA), FU895 (GA), FU905 (CG), and FU904 (GG) was also monitored in the presence and absence of lysine. Lysine starvation induced β-Gal synthesis in strains FU844 (CA) and FU895 (GA) by 2.3-fold compared with the β-Gal activities during the first 1 h of incubation without and with lysine. However, lysine starvation did not induce β-Gal synthesis in strain FU905 (CG), and it rather reduced β-Gal synthesis in strain FU904 (GG) by 0.87-fold.
These results indicate that the CodY-independent positive stringent response of ilv-leu, which is evoked by amino acid (lysine) starvation or decoyinine treatment, involves the nucleotide sequence at nucleotides +1 and +2, especially the nucleotide species at nucleotide +2, adenine (Fig. 6B). If the wild-type A at nucleotide +2 was replaced with G (CG and GG sequences at nucleotides +1 and +2), the positive stringent response completely disappeared.
Identification of the transcription initiation nucleotides of base-substituted ilv-leu promoters.
The cytosine at nucleotide +1 has been reported to be the transcription initiation base of ilv-leu (12), whereas either or both adenines at +2 and +3 are also reported to be the transcription initiation bases (16). However, we have no experimental results indicating the transcription initiation nucleotide of base-substituted ilv-leu promoters. Hence, we performed promoter extension analysis to map the 5′ end of the lacZ transcript whose synthesis is under the control of each of the base-substituted ilv-leu promoters, using total RNAs from strains FU844 (CA), FU895 (GA), FU905 (CG), and FU904 (GG) (Fig. 7). As shown in Fig. 7, the transcription initiation base of the wild-type ilv-leu promoter was supposed to be C at nucleotide +1, as reported previously (12). However, the initiation bases of the base-substituted ilv-leu promoters were varied, except that the initiation base of the CG promoter was the same as that of the wild-type CA promoter (nucleotide +1). The initiation base of the GG promoter could be G at nucleotide +2, whereas those of the GA promoter could be U, G, and A at nucleotides −1, +1, and +2, respectively, with a predominance of U. These results imply that C at nucleotide +1 might tend to be an initiation nucleotide but that G at nucleotide +1 can hardly be. It is notable that the transcription initiation bases of the CG and GG promoters were C at nucleotide +1 and G at nucleotide +2, respectively; nevertheless, the CG and GG promoters completely lost the positive stringent response of ilv-leu.
FIG. 7.
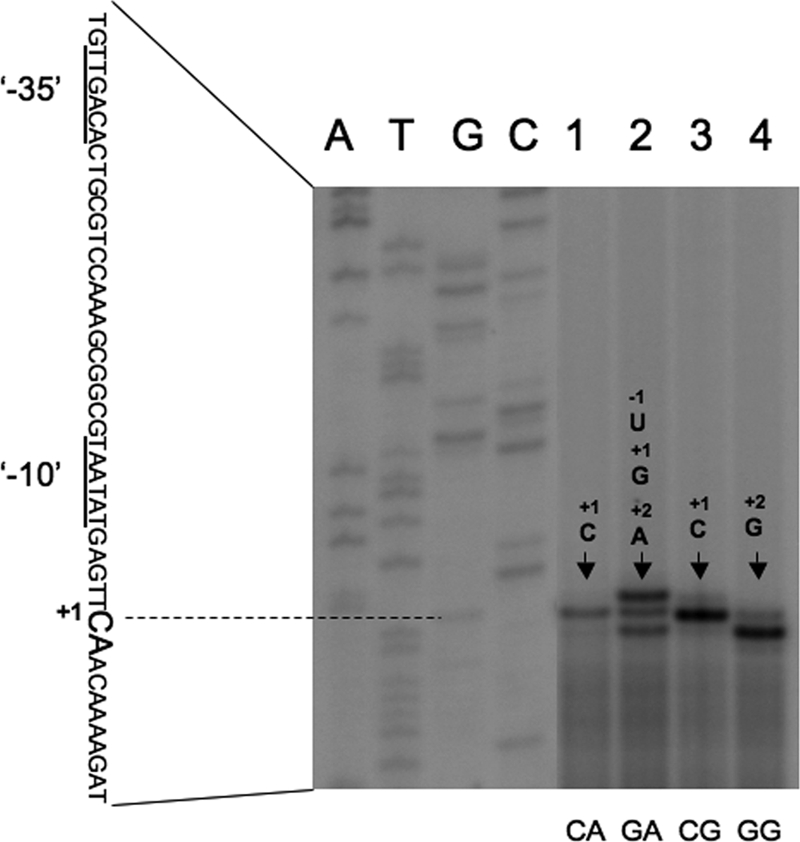
Mapping of the 5′ end of the ilv-leu transcripts derived from the base-substituted promoters by means of primer extension analysis. Total RNAs from strains FU844 (CA), FU895 (GA), FU905 (CG), and FU904 (GG) grown in MM medium supplemented with 16 amino acids were annealed with the PEpR primer (Table 2), and then primer extension was performed as described in the text. Lanes A, T, G, and C contained the products of the respective dideoxy sequencing reactions, with the PCR product as the template, as described in the text. The part of the wild-type nucleotide sequence of the coding strand corresponding to the ladder is shown with the transcription initiation base (+1) (enlarged CA sequence [nucleotides +1 to +2]), and the corresponding −10 and −35 regions for the ilv-leu promoter are underlined.
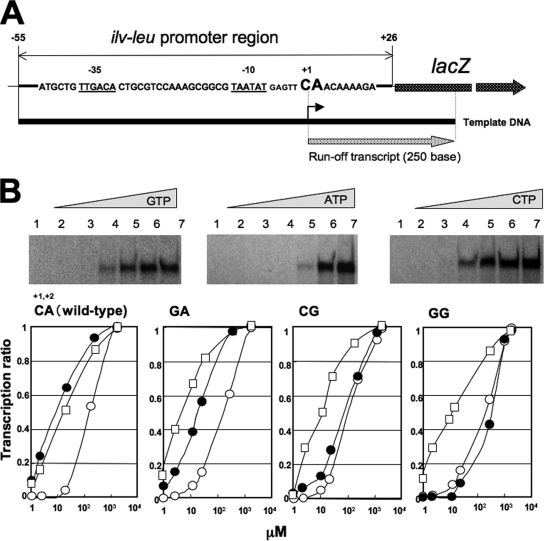
Dependency of in vitro ilv-leu transcription from base-substituted promoters on the concentrations of NTP.
Krásný and Gourse (18) reported that the rRNA promoters in B. subtilis appear to be regulated through changes in the pool sizes of the initiating NTP, GTP. Thus, we examined whether in vitro transcription from the ilv-leu promoter depends on the NTP concentrations. As shown in Fig. 8A, we constructed an in vitro transcription system involving His-tagged B. subtilis RNAP and a DNA template (CA [nucleotides +1 and +2]) covering the wild-type ilv-leu promoter region (nucleotides −55 to +26) and the 5′ part of the lacZ gene, which produced a 250-base runoff transcript. When this in vitro transcription was carried out with various concentrations of each of GTP, ATP, and CTP and a fixed concentration of the other three NTPs, it was more tolerant to lowering of the NTP concentrations, in the order of GTP, CTP, and ATP (Fig. 8B, top). When the GA, CG, and GG templates possessing GA, CG, and GG besides CA at nucleotides +1 and +2 were utilized, the in vitro transcription was more tolerant at lower concentrations of GTP in the order of the CA, GA, CG, and GG templates; the transcription on the DNA templates possessing A at nucleotide +2 was more tolerant than that on those possessing G at the same position (Fig. 8B, bottom). However, the in vitro transcription with various ATP and CTP concentrations was essentially independent of the DNA template no matter which was utilized. This tolerance to a decrease in the in vivo GTP concentration is most likely one explanation for the positive stringent response of the CodY-independent positive stringent control, because the in vivo GTP concentration decreased from roughly the 1,000 μM range to the 100 μM range upon the stringent response (Table 3), which caused in vitro transcription on the CG and GG templates carrying G at nucleotide +2 to drastically decrease.
FIG. 8.
In vitro transcription from base-substituted ilv-leu promoters. (A) In vitro transcription from the wild-type ilv-leu promoter. His-tagged B. subtilis RNAP was prepared, and the DNA template used for in vitro transcription was a PCR product covering the ilv-leu promoter region (nucleotides −55 to +26) and a 5′ portion of lacZ, as described in the text. Multiple rounds of transcription producing a 250-base runoff transcript were performed with the substrate concentrations of 200 μM ATP, CTP, and GTP and 12 μM of UTP, as described in the text. The runoff transcripts were electrophoresed on a urea-polyacrylamide gel and then quantified with an image analyzer, as described in the text. (B) (Top) In vitro transcription using a CA template (wild type) possessing C and A at nucleotides +1 and +2 was carried out with various concentrations—0 μM (lane 1), 0.02 μM (lane 2), 0.2 μM (lane 3), 2 μM (lane 4), 20 μM (lane 5), 200 μM (lane 6), and 2,000 μM (lane 7)—of GTP, ATP, and CTP, and fixed concentrations of the other three NTPs. (Bottom) Concentration dependencies for GTP (filled circles), ATP (open circles), and CTP (open squares) of in vitro transcription on the CA, GA, CG, and GG templates, where a transcription ratio of 1 indicates the maximal synthesis of the runoff transcript.
The factors which might explain the enhancement of in vivo ilv-leu transcription upon the stringent response might be the increases in the in vivo concentrations of ATP and CTP (Table 3), although these concentration changes did not occur within the sensitive range in our in vitro transcription system (Fig. 8B). Moreover, we found that high concentrations of ATP (more than 7.5 mM) severely inhibited the in vitro transcription (data not shown), so we were unable to increase the ATP concentration to its in vivo concentration of 10 mM after the stringent treatment (Table 3). Thus, we failed to reproduce the in vivo positive stringent response of ilv-leu with the in vitro transcription system, even if any alteration in the system was tried (data not shown).
DISCUSSION
We have investigated the mechanisms underlying the positive stringent response of the expression of ilv-leu upon lysine starvation and decoyinine addition. This amino acid starvation triggered RelA-dependent positive stringent control of ilv-leu (Fig. 2 and 3), whereas decoyinine addition triggered RelA-independent positive stringent control (Fig. 4). Lysine starvation triggers RelA-catalyzed ribosome-mediated synthesis of ppGpp, which probably inhibits IMP dehydrogenase (17, 21, 29, 31), leading to a decrease in the in vivo concentration of GTP (Table 3), as described previously for treatment with dl-α-oxo-β-methyl-n-valerate (30) and serine hydroxamate (18) and for aspartate or methionine starvation (30). On the other hand, decoyinine treatment directly inhibits GMP synthase, which causes the decrease of the in vivo GTP concentration (Table 3) (14, 22, 30). Thus, lysine starvation and decoyinine addition triggered a decrease in the concentration of GTP, a corepressor of the CodY protein, which caused derepressed ilv-leu expression through detachment of the CodY protein from its cis elements upstream of the ilv-leu promoter (Fig. 5), just as suggested for the above-described amino acid starvation. However, the results of the deletion and CodY requirement analyses on this stringent response of ilv-leu expression (Fig. 1 to 4) suggested another molecular mechanism underlying this positive stringent response of ilv-leu, which is CodY independent (Fig. 5). This CodY-independent mechanism most likely comprises the modulation of the transcription initiation frequency, which requires at least the base species of adenine at the 5′ end of the ilv-leu transcript (nucleotide +2) (Fig. 6).
The ilv-leu promoter region (nucleotides −55 to +26) does not contain CodY-binding sites CodY-IV, -III, and -II and contains only part of the CodY-I site (Fig. 1), so CodY does not repress the expression of the lacZ gene fused with this promoter region (41) (also refer to the basal levels for β-Gal synthesis of the wild-type and ΔcodY strains in Fig. 3B and 4B). Moreover, the CodY protein did not repress the transcription of the ilv-leu promoter if only CodY-binding site CodY-II was deleted (Shigeo Tojo and Yasutaro Fujita, unpublished observation). However, we observed a greater induction of β-Gal synthesis in strain FU844 (relA+ codY+) than in strain FU810 (relA+ ΔcodY) upon lysine starvation (Fig. 3B, panel a) and upon decoyinine addition (Fig. 4B, panel a) and also more in strain FU845 (relA1 codY+) than in strain FU809 (relA1 ΔcodY) upon decoyinine addition (Fig. 4B, panel b). This indirect enhancing effect of the CodY protein on β-Gal induction might be attributed to some modulation of the NTP concentrations which presumably affects the ilv-leu promoter activity.
We determined the transcription initiation base of the ilv-leu by means of primer extension, suggesting the cytosine at nucleotide +1 to be a transcription initiation base, as described by Grandoni et al. (12). However, this transcription initiation site is contradictory to the fact that adenines at +2 and +3 are likely the initiation bases (16). Thus, the transcription initiation site of ilv-leu remained to be confirmed by means of other methods more reliable than the primer extension. Nevertheless, it is unlikely that the position of the transcription initiation base is absolutely fixed for this stringent response because the 5′ ends of the transcripts derived from the base-substituted ilv-leu promoters were found to be altered depending on the base-substituted sequence (Fig. 7).
Krásný and Gourse (18) suggested that B. subtilis and E. coli use different strategies to control rRNA synthesis. The P1 and P2 promoters of the B. subtilis rrnB and rrnO operons start with GTP (18). All of the P1 and P2 promoters of the other rrn operons also start with GTP (Yousuke Natori and Fujio Kawamura, personal communication). The initiation NTP for transcription from B. subtilis rRNA promoters is GTP, which is in clear contrast to the case of E. coli. Changes in promoter activity always correlate with changes in the intracellular GTP concentration, and they are usually dependent on RelA (18). In contrast to the situation for E. coli, where ppGpp decreases rRNA promoter activity by directly inhibiting RNAP (3), ppGpp might not inhibit B. subtilis RNAP directly (18). Rather, an increase in the ppGpp concentration might reduce the available GTP pool, thereby modulating rRNA promoter activity indirectly. Recently, this proposal was verified to be applicable to Thermus thermophilus rRNA transcription as well (17).
The molecular mechanism underlying the CodY-independent positive stringent response resembles that underlying the negative stringent response of rRNA synthesis in that both mechanisms involve the transcription initiation frequency, which likely depends on the RNAP substrate concentrations and the base sequence of the 5′ end of the transcript. However, the positive stringent response of ilv-leu is substantially different from the negative stringent response of rRNA synthesis in the following two ways. (i) When both C (nucleotide +1) and A (nucleotide +2) of the ilv-leu promoter were replaced with G, the positive stringent response of ilv-leu became a negative stringent response upon both lysine starvation and decoyinine treatment (Fig. 6B), although the substitution of G for C at nucleotide +1 affected this positive stringent response only slightly, while the substitution of G for A at nucleotide +2 greatly affected it (Fig. 6B). When the 5′ ends of the transcripts derived from the base-substituted ilv-leu promoters were determined, their transcription initiation bases were found to be altered depending on the base-substituted sequence (Fig. 7). Thus, unlike the stringent response of rRNA synthesis, for which the base species of the transcription initiation nucleotide is a determinant (18), the base species A or G at nucleotide +2, which is not always the transcription initiation base, is more critical as to whether there is a positive or negative stringent response, respectively. (ii) Changes in the concentrations of NTP and ppGpp account for much of the regulation of E. coli rRNA synthesis (26, 27). E. coli rRNA promoters require higher concentrations of NTP for transcription than other promoters in vitro, and ppGpp moderately but specifically inhibits transcription from rRNA promoters in vitro. Like E. coli rRNA promoters, B. subtilis rRNA promoters exhibit NTP dependence in vitro, which is characteristic of the regulation of promoter activity through changes in NTP concentrations in vivo (18). The rrnB and rrnO P1 promoters required high levels of GTP but not ATP, whereas these P1 promoters in which G at nucleotide +1 is replaced with A required high levels of ATP but not GTP. In contrast to the rrn promoters, the ilv-leu promoters (wild-type CA and base-substituted GA [nucleotides +1 and +2]) did not require high levels of GTP. The base-substituted CG and GG promoters required high levels of GTP; the latter promoter appeared to require a higher level of GTP (Fig. 8B). On the other hand, these four kinds of ilv-leu promoter required almost the same levels of ATP and CTP.
We attempted to reproduce the CodY-independent positive stringent response of ilv-leu in vitro transcription by using the in vivo concentrations of the RNAP substrates before and after stringent treatment. However, we could not reproduce the enhancement of the in vitro transcription through such modulation of these substrate concentrations. We consider that this is due to the current in vitro transcription system being significantly distinct from actual in vivo transcription. Moreover, we do not exclude the possibility that there are other transcription factors, possibly associated with RNAP, for the CodY-independent positive stringent response of ilv-leu.
An interesting question arises, i.e., whether this kind of CodY-independent stringent response of ilv-leu prevails for the regulation of various catabolic and anabolic operons; the molecular mechanism underlying this stringent response of ilv-leu is substantially different from that for the rrn operons. We performed DNA microarray analysis using a ΔcodY strain grown with and without decoyinine, which implied that several metabolic operons, which are involved in critical stages of metabolic regulation, might be subject to this kind of stringent response (Kanako Kumamoto and Yasutaro Fujita, unpublished observation). The molecular mechanism underlying this specific and unique stringent response, including the identification of additional sequence conservation besides adenine at nucleotide +2 in close vicinity to the transcription initiation base, as well as the extent of the contribution of this response to the framework of the global metabolic network in B. subtilis, is currently under detailed investigation.
After the completion of the current work, a communication described by Krásný et al. (19) was posted online, open to the public. Their work, dealing only with the CodY-independent stringent response of the ilv-leu promoter to amino acid starvation in B. subtilis, was performed independently from our current work. There is a clear difference between the two works in the results of the identification of the initiation base of ilv-leu transcription. Although it is unlikely that the position of the transcription initiation is absolutely fixed for this stringent response, as inferred from the fact that the 5′ ends of the transcripts derived from the base-substituted ilv-leu promoters were altered depending on the base-substituted sequence (Fig. 7), the transcription initiation base of the wild-type ilv-leu promoter remains to be determined by other methods more reliable than the primer extension which was adopted in both works. Another clear difference between the two works is that our four kinds of the base-substituted ilv-leu promoters showed almost the same concentration dependencies of ATP in the in vitro transcription assay (Fig. 8), in contrast to their results. This might stem from the unknown difference between our in vitro transcription system and theirs.
Acknowledgments
We are grateful to Y. Sadaie (Saitama University, Japan) for providing the B. subtilis strain and to Y. Ohashi (Human Metabolome Technologies, Inc., Japan) for his efforts determining the in vivo nucleotide concentrations. We also thank K. Morisaki, S. Miyazaki, A. Shimobayashi, S. Yamashita, Y. Hikita, and H. Yamamoto for their help.
This work was supported by Grants-in-Aid for Scientific Research on Priority Areas, Scientific Research (B), and the High-Tech Research Center Project for Private Universities from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Footnotes
Published ahead of print on 18 July 2008.
REFERENCES
- 1.Anagnostopoulos, C., and J. Spizizen. 1961. Requirements for transformation in Bacillus subtilis. J. Bacteriol. 81741-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atkinson, M. R., L. V. Wray, Jr., and S. H. Fisher. 1990. Regulation of histidine and proline degradation enzymes by amino acid availability in Bacillus subtilis. J. Bacteriol. 1724758-4765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barker, M. M., T. Gaal, and R. L. Gourse. 2001. Mechanism of regulation of transcription initiation by ppGpp. II. Models for positive control based on properties of RNAP mutants and competition for RNAP. J. Mol. Biol. 305689-702. [DOI] [PubMed] [Google Scholar]
- 4.Cashel, M., D. R. Gentry, V. J. Hernandez, and D. Vinella. 1996. The stringent response, p. 1458-1496. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, DC.
- 5.de Mendoza, D., G. E. Schujman, and P. S. Aguilar. 2002. Biosynthesis and function of membrane lipids, p. 43-55. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, DC.
- 6.Deutscher, J., A. Galinier, and I. Martin-Verstraete. 2002. Carbohydrate uptake and metabolism, p. 129-150. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, DC.
- 7.Eymann, C., G. Homuth, C. Scharf, and M. Hecker. 2002. Bacillus subtilis functional genomics: global characterization of the stringent response by proteome and transcriptome analysis. J. Bacteriol. 1842500-2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fink, P. S. 1993. Biosynthesis of the branched-chain amino acids, p. 307-317. In A. L. Sonenshein, J. A. Hoch, and R. Losick, (ed.), Bacillus subtilis and other gram-positive bacteria. American Society for Microbiology, Washington, DC.
- 9.Fujita, M., and Y. Sadaie. 1998. Rapid isolation of RNA polymerase from sporulating cells of Bacillus subtilis. Gene 221185-190. [DOI] [PubMed] [Google Scholar]
- 10.Fujita, Y., and E. Freese. 1979. Purification and properties of fructose-1,6-bisphosphatase of Bacillus subtilis. J. Biol. Chem. 2545340-5349. [PubMed] [Google Scholar]
- 11.Fujita, Y., Y. Miwa, S. Tojo, and K. Hirooka. 2007. Carbon catabolite control and metabolic networks mediated by the CcpA protein in Bacillus subtilis, p. 91-110. In Y. Fujita (ed.), Global regulatory networks in Bacillus subtilis. Transworld Research Network, Kerala, India.
- 12.Grandoni, J. A., S. A. Zahler, and J. M. Calvo. 1992. Transcriptional regulation of the ilv-leu operon of Bacillus subtilis. J. Bacteriol. 1743212-3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hutchison, K. W., and R. S. Hanson. 1974. Adenine nucleotide changes associated with the initiation of sporulation in Bacillus subtilis. J. Bacteriol. 11970-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Inaoka, T., and K. Ochi. 2002. RelA protein is involved in induction of genetic competence in certain Bacillus subtilis strains by moderating the level of intracellular GTP. J. Bacteriol. 1843923-3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Inaoka, T., K. Takahashi, M. Ohnishi-Kameyama, M. Yoshida, and K. Ochi. 2003. Guanine nucleotides guanosine 5′-diphosphate 3′-diphosphate and GTP co-operatively regulate the production of an antibiotic bacilysin in Bacillus subtilis. J. Biol. Chem. 2782169-2176. [DOI] [PubMed] [Google Scholar]
- 16.Juang, Y.-L., and J. D. Helmann. 1994. The δ-subunit of Bacillus subtilis RNA polymerase. An allosteric effector of the initiation and core-recycling phases of transcription. J. Mol. Biol. 2391-14. [DOI] [PubMed] [Google Scholar]
- 17.Kasai, K., T. Nishizawa, K. Takahashi, T. Hosaka, H. Aoki, and K. Ochi. 2006. Physiological analysis of the stringent response elicited in an extreme thermophilic bacterium, Thermus thermophilus. J. Bacteriol. 1887111-7122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krásný, L., and R. L. Gourse. 2004. An alternative strategy for bacterial ribosome synthesis: Bacillus subtilis rRNA transcription regulation. EMBO J. 234473-4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krásný, L., H. Tišerová, J. Jonák, D. Rejman, and H. Sanderová. 2008. The identity of the transcription +1 position is crucial for changes in gene expression in response to amino acid starvation in Bacillus subtilis. Mol. Microbiol. 6942-54. [DOI] [PubMed] [Google Scholar]
- 20.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, P. Bessières, A. Bolotin, S. Borchert, R. Borriss, L. Boursier, A. Brans, M. Braun, S. C. Brignell, S. Bron, S. Brouillet, C. V. Bruschi, B. Caldwell, V. Capuano, N. M. Carter, S.-K. Choi, J.-J. Codani, I. F. Connerton, et al. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390249-256. [DOI] [PubMed] [Google Scholar]
- 21.Lopez, J. M., A. Dromerick, and E. Freese. 1981. Response of guanosine 5′-triphosphate concentration to nutritional changes and its significance for Bacillus subtilis sporulation. J. Bacteriol. 146605-613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lopez, J. M., C. L. Marks, and E. Freese. 1979. The decrease of guanine nucleotides initiates sporulation of Bacillus subtilis. Biochim. Biophys. Acta 587238-252. [DOI] [PubMed] [Google Scholar]
- 23.Mäder, U., G. Homuth, C. Scharf, K. Büttner, R. Bode, and M. Hecker. 2002. Transcriptome and proteome analysis of Bacillus subtilis gene expression modulated by amino acid availability. J. Bacteriol. 1844288-4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miwa, Y., and Y. Fujita. 2001. Involvement of two distinct catabolite-responsive elements in catabolite repression of the Bacillus subtilis myo-inositol (iol) operon. J. Bacteriol. 1835877-5884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Molle, V., Y. Nakaura, R. P. Shivers, H. Yamaguchi, R. Losick, Y. Fujita, and A. L. Sonenshein. 2003. Additional targets of the Bacillus subtilis global regulator CodY identified by chromatin immunoprecipitation and genome-wide transcript analysis. J. Bacteriol. 1851911-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murray, H. D., and R. L. Gourse. 2004. Unique roles of the rrn P2 rRNA promoters in Escherichia coli. Mol. Microbiol. 521375-1387. [DOI] [PubMed] [Google Scholar]
- 27.Murray, H. D., D. A. Schneider, and R. L. Gourse. 2003. Control of rRNA expression by small molecules is dynamic and nonredundant. Mol. Cell 12125-134. [DOI] [PubMed] [Google Scholar]
- 28.Ochi, K. 1987. Metabolic initiation of differentiation and secondary metabolism by Streptomyces griseus: significance of the stringent response (ppGpp) and GTP content in relation to A factor. J. Bacteriol. 1693608-3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ochi, K. 1987. Changes in nucleotide pools during sporulation of Streptomyces griseus in submerged culture. J. Gen. Microbiol. 1332787-2795. [Google Scholar]
- 30.Ochi, K., J. Kandala, and E. Freese. 1981. Initiation of Bacillus subtilis sporulation by the stringent response to partial amino acid deprivation. J. Biol. Chem. 2566866-6875. [PubMed] [Google Scholar]
- 31.Ochi, K., J. Kandala, and E. Freese. 1982. Evidence that Bacillus subtilis sporulation induced by the stringent response is caused by the decrease in GTP or GDP. J. Bacteriol. 1511062-1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ratnayake-Lecamwasam, M., P. Serror, K. W. Wong, and A. L. Sonenshein. 2001. Bacillus subtilis CodY represses early-stationary-phase genes by sensing GTP levels. Genes Dev. 151093-1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 34.Serror, P., and A. L. Sonenshein. 1996. CodY is required for nutritional repression of Bacillus subtilis genetic competence. J. Bacteriol. 1785910-5915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shivers, R. P., and A. L. Sonenshein. 2004. Activation of the Bacillus subtilis global regulator CodY by direct interaction with branched-chain amino acids. Mol. Microbiol. 53599-611. [DOI] [PubMed] [Google Scholar]
- 36.Shivers, R. P., and A. L. Sonenshein. 2005. Bacillus subtilis ilvB operon: an intersection of global regulons. Mol. Microbiol. 561549-1559. [DOI] [PubMed] [Google Scholar]
- 37.Soga, T., Y. Ohashi, Y. Ueno, H. Naraoka, M. Tomita, and T. Nishioka. 2003. Quantitative metabolome analysis using capillary electrophoresis mass spectrometry. J. Proteome Res. 2488-494. [DOI] [PubMed] [Google Scholar]
- 38.Suhadolnik, R. J. 1970. Nucleoside antibiotics, p.96-122. John Wiley & Sons, Inc., New York, NY.
- 39.Swanton, M., and G. Edlin. 1972. Isolation and characterization of an RNA relaxed mutant of B. subtilis. Biochem. Biophys. Res. Commun. 46583-588. [DOI] [PubMed] [Google Scholar]
- 40.Tojo, S., T. Satomura, K. Morisaki, K. Yoshida, K. Hirooka, and Y. Fujita. 2004. Negative transcriptional regulation of the ilv-leu operon for biosynthesis of branched-chain amino acids through the Bacillus subtilis global regulator TnrA. J. Bacteriol. 1867971-7979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tojo, S., T. Satomura, K. Morisaki, J. Deutscher, K. Hirooka, and Y. Fujita. 2005. Elaborate transcription regulation of the Bacillus subtilis ilv-leu operon involved in the biosynthesis of branched-chain amino acids through global regulators of CcpA, CodY and TnrA. Mol. Microbiol. 561560-1573. [DOI] [PubMed] [Google Scholar]
- 42.Wendrich, T. M., and M. A. Marahiel. 1997. Cloning and characterization of a relA/spoT homologue from Bacillus subtilis. Mol. Microbiol. 2665-79. [DOI] [PubMed] [Google Scholar]
- 43.Wray, L. V., Jr., J. M. Zalieckas, and S. H. Fisher. 2001. Bacillus subtilis glutamine synthetase controls gene expression through a protein-protein interaction with transcription factor TnrA. Cell 107427-435. [DOI] [PubMed] [Google Scholar]
- 44.Yoshida, K., D. Aoyama, I. Ishio, T. Shibayama, and Y. Fujita. 1997. Organization and transcription of the myo-inositol operon, iol, of Bacillus subtilis. J. Bacteriol. 1794591-4598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yoshida, K., I. Ishio, E. Nagakawa, Y. Yamamoto, M. Yamamoto, and Y. Fujita. 2000. Systematic study of gene expression and transcription organization in the gntZ-ywaA region of the Bacillus subtilis genome. Microbiology 146573-579. [DOI] [PubMed] [Google Scholar]
- 46.Yoshida, K., K. Kobayashi, Y. Miwa, C. Kang, M. Matsunaga, H. Yamaguchi, S. Tojo, M. Yamamoto, R. Nishi, N. Ogasawara, T. Nakayama, and Y. Fujita. 2001. Combined transcriptome and proteome analysis as a powerful approach to study genes under glucose repression in Bacillus subtilis. Nucleic Acids Res. 29683-692. [DOI] [PMC free article] [PubMed] [Google Scholar]