Abstract
Bacterial spores are resistant to a wide range of chemical and physical insults that are normally lethal for the vegetative form of the bacterium. While the integrity of the protein coat of the spore is crucial for spore survival in vitro, far less is known about how the coat provides protection in vivo against predation by ecologically relevant hosts. In particular, assays had characterized the in vitro resistance of spores to peptidoglycan-hydrolyzing enzymes like lysozyme that are also important effectors of innate immunity in a wide variety of hosts. Here, we use the bacteriovorous nematode Caenorhabditis elegans, a likely predator of Bacillus spores in the wild, to characterize the role of the spore coat in an ecologically relevant spore-host interaction. We found that ingested wild-type Bacillus subtilis spores were resistant to worm digestion, whereas vegetative forms of the bacterium were efficiently digested by the nematode. Using B. subtilis strains carrying mutations in spore coat genes, we observed a correlation between the degree of alteration of the spore coat assembly and the susceptibility to the worm degradation. Surprisingly, we found that the spores that were resistant to lysozyme in vitro can be sensitive to C. elegans digestion depending on the extent of the spore coat structure modifications.
Bacilli are soil bacteria that exist in their habitat either as vegetative cells or, more abundantly, as spores (23, 30). The spore is a dormant state that confers in vitro resistance to a broad range of insults that are bactericidal for the vegetative form of bacteria, including extreme temperatures, mechanical stresses, or exposure to chemicals (5). The ability of spores to resist these treatments is at least partially attributed to the spore coat, a proteinaceous layer composed of at least 70 protein species in Bacillus subtilis (11). The coat is organized in two distinct layers, an outer coat and an inner coat. The proper assembly of the spore coat requires the expression of at least five morphogenetic proteins (11), including CotE that controls the recruitment and the binding of a large subset of proteins to the coat (36). A ΔcotE mutation results in spores that lack the outer coat and frequently display an aberrant inner coat that appears to be disconnected from the outer surface of the forespore (6, 36). Further, the precise timing of CotE expression is critical for the proper recruitment of proteins to the coat during assembly, and delaying CotE expression during sporulation results in spores lacking the outer spore structures (3).
The modifications in the coat structure observed in the ΔcotE mutant spores are assumed to be responsible for their sensitivity to peptidoglycan-degrading enzymes, such as hen egg white lysozyme (36). Lysozymes are widely distributed among organisms and are considered effectors of innate immunity in metazoa or as digestive enzymes in unicellular and multicellular eukaryotes (27). In soil, Bacillus species encounter different types of predator organisms, including protozoa and bacteriophagous nematodes. Recently, the relevance of in vivo studies in understanding the role of the spore coat structures (15) was demonstrated using the protozoan Tetrahymena thermophila. The bacteriovorous nematode Caenorhabditis elegans appears to interact with soil bacteria like the endospore-forming Bacillus cereus in the wild (9). C. elegans expresses and presumably secretes into its intestinal lumen bacteriolytic proteins, including lysozyme-like proteins (19, 20) encoded by numerous genes (>10) in its genome that show homology to either c-type, i-type, or Ch-type lysozymes (M.-H. Laaberki and J. Dworkin, unpublished data). In the present study, we investigated the fate of Bacillus subtilis cells during C. elegans predation. We tested the ability of C. elegans to ingest and digest vegetative Bacillus subtilis cells or spores. In addition, we characterized the role of coat structures in the resistance to C. elegans ingestion.
MATERIALS AND METHODS
Care of bacterial strains and C. elegans strains.
Bacillus subtilis strains used in this study (Table 1) were all derivatives of B. subtilis PY79 (35). Genetic manipulations of B. subtilis were performed by standard methods (4). The gerD-cwlD (JDB1486) mutant was obtained by transforming B. subtilis PY79 with genomic DNA from strain TB1 (kindly provided by Simon Cutting, University of London, United Kingdom) and by selecting for resistance to neomycin (2.5 μg/ml). The germination deficiency in comparison to the wild-type strain on rich medium of the gerD-cwlD mutant was as low (<0.0001%) as described previously (34). The strains ectopically expressing cotE at amyE under the control of different promoters in a ΔcotE::cat background were obtained after transformation of the laboratory ΔcotE (JDB1323) mutant with the genomic DNA from strains AH2914, AH2915, AH2921, AH2920, and AH2942 to give the strains JDB2122, JDB2124, JDB2127, JDB2125, and JDB2129, respectively. AH strains were kindly provided by A. Henriques (Universidade Nova de Lisboa, Portugal) and are in a different B. subtilis background (3). The sensitivity of the AH strains in C. elegans was the same as for the PY79 derivative strains (data not shown). The bacteria were maintained in Luria broth liquid media or agar media. B. subtilis strains were sporulated in liquid Difco sporulation medium, and spores were purified as described previously (10). Spores were purified using a 33% diatrizoate meglumine-8% diatrizoate sodium solution and washed with cold water until 100% of phase-bright spores were obtained. The spores were stored at 4°C in double-distilled H2O and used within a week following the initial growth in Difco sporulation medium. The C. elegans N2 strain (a kind gift of Iva Greenwald, Columbia University, New York) and the JM90 strain (kindly provided by Jim McGhee, The University of Calgary, Alberta, Canada) were maintained on nematode growth medium agar (NGM) with Escherichia coli OP50 (JDE304) as the food source (33).
TABLE 1.
Bacterial strains used in this study
| Strain | Relevant genotype | Reference(s) or source |
|---|---|---|
| JDB3 | Wild-type PY79 strain, prototrophic | 35 |
| JDE304 | Escherichia coli OP50 | I. Greenwald |
| JDB1931 | Listeria innocua | D. Portnoy |
| JDB1323 | cotE::cat | 14 |
| JDB1380 | safA::tet | 14 |
| JDB1371 | spoVID::kan | 14 |
| JDB1343 | cotA::cat | 14 |
| JDB1345 | cotG::erm | 14 |
| JDB1346 | cotH::cat | 14 |
| JDB1348 | cotO::tet | 14 |
| JDB1381 | yutH::tet | 14 |
| JDB1382 | ysxE::erm | 14 |
| JDB1582 | amyE::PspoIIQ-rfp cat | This study |
| JDB1584 | cotE::cat::tet | This study |
| JDB1586 | cotE::cat::tet amyE::PspoIIQ-rfp cat | This study |
| JDB1641 | spoVID::kan amyE::PspoIIQ-rfp cat | This study |
| JDB1334 | amyE::Pspanc-gfp cat | 16 |
| JDB1486 | gerD-cwlD::neo | This study and reference 34 |
| JDB1494 | cotE::cat gerD-cwlD::neo | This study |
| JDB2122 | cotE::cat amyE::PgerE-cotE neo | This study and reference 3 |
| JDB2124 | cotE::cat amyE::PcotG-cotE neo | This study and reference 3 |
| JDB2127 | cotE::cat amyE::PcotEP1-cotE neo | This study and reference 3 |
| JDB2125 | cotE::cat amyE::PcotEP2-cotE neo | This study and reference 3 |
| JDB2129 | cotE::cat amyE::PcotEP1P2-cotE neo | This study and reference 3 |
Culturing bacteria from worms.
C. elegans eggs were isolated using a 10% commercial bleach and 1 N NaOH solution followed by three washes with 1× M9 (33). The eggs were allowed to hatch overnight at 20°C in 1× M9 while rocking. The number of larval stage 1 (L1) larvae was scored, and L1 larvae were added to 100-mm NGM plates containing a lawn of E. coli OP50 with about 2,000 L1 larvae per plate (33). Development then proceeded until the L4/young adult stage (36 h at 20°C). The larvae were washed four times in a large volume of 1× M9 with 0.01% Tween 20 (M9-T) and incubated for 8 h in M9-T at room temperature while rocking to allow excretion of E. coli from the worm intestine. After two washes with M9-T, 400 to 600 worms were added to a 60-mm plate containing nematode minimal medium agar (NMM) (3 g/liter NaCl, 17 g/liter agar, 5 mg/liter cholesterol, 1 mM CaCl2, 1 mM MgSO4, 25 mM KH2PO4 [pH 6]) on which 100 μl of spore preparation at 109 spores/ml had been spread. The worms were incubated at 20°C for 14 h, and the survival of vegetative cells of B. subtilis, E. coli, and Listeria innocua, was tested with 1-day-old adult worms placed on NGM plates seeded with 100 μl of a culture grown for 6 hours that had 1 CFU in 3 ml of LB at 37°C. The adult worms were incubated for 24 h on the bacteria, harvested, and washed with M9-T, and incubated in M9-T with 30 mM sodium azide to induce paralysis. The surface of the worms was sterilized using 1× M9 with 3% commercial bleach (20 min) and washed twice with M9-T. Since the worms are impermeant to any liquid following paralysis of the pharynx muscles by sodium azide, only the outside of the worms is subjected to sterilization. The worms resumed movement following washings, indicating that sodium hypochlorite had not penetrated their gut. The worms were resuspended in a final volume of 600 μl, and the number of worms in two separate 50-μl aliquots was determined. The outside contamination was estimated by plating 100 μl of this solution. The worms were disrupted by vortexing with high-density 1-mm zirconia beads (BioSpec Products) until completion as determined by microscopy. Dilutions of the worm lysates were made in T-base (24) containing 1 mM MgSO4 and were plated on LB agar to determine the bacterial yield per worm (CFU/worm). The heat resistance of the bacteria in the disrupted worms was determined by exposing the samples to 80°C for 20 min. All experiments were performed a minimum of three times.
Microscopy.
To visualize paralyzed worms, agar pads containing melted 2% agar in double-distilled H2O were prepared on microscope slides. The worms were pipetted from the assay plates with M9-T and 30 mM NaN3, dropped on the agar pad, and covered with a glass coverslip. Nomarski differential interference contrast (DIC) and fluorescence images were obtained using a Nikon Eclipse 90i microscope with a 100× objective. Pictures were captured with a Hamamatsu ORCA-ER digital camera using Nikon Elements BR software. Green fluorescent protein fluorescence and mCherry fluorescence were detected with EX460-500 BA510-560 and EX530-560 BA590-650 filter sets, respectively (Chroma). The exposure was 500 ms for all fluorescence pictures and 2 ms for DIC pictures.
Spore pulse-chase experiment.
A large population of C. elegans N2 (L4-young adults) was synchronized, washed, and transferred to a 100-mm NMM agar plate containing a lawn of spores of strain JDB1334 (a PY79 derivative with a spectinomycin resistance gene) as described previously (16). After 2 h of incubation at 20°C, the worms were washed five times with M9-T. A total of 500 to 600 worms were then added to 60-mm NMM plates containing no bacteria (“no food”), ΔcotE spores, wild-type spores, or an overnight LB culture of E. coli OP50. The number of spectinomycin-resistant CFU per worm was determined as described above.
Measurement of growth rate.
About 30 eggs of the C. elegans N2 strain were placed on a 60-mm plate containing NMM agar and incubated overnight at 20°C to allow L1 larvae to hatch. An equivalent amount of each bacterial strain (based on optical density) was added to the L1 larvae. Following subsequent incubation at 20°C, the developmental stage was checked every 12 h by microscope observations (4× magnification) until approximately 50% of the animals reached the adult stage. Adults were recognized by the appearance of a mature vulva and by the production, after several hours, of eggs (2). The growth rates were calculated and corresponded to the number of days required for 50% of the population to reach the adulthood at 20°C. t tests were performed to compare the three sets of growth rate measurements.
RESULTS
Sensitivity of B. subtilis to digestion by C. elegans.
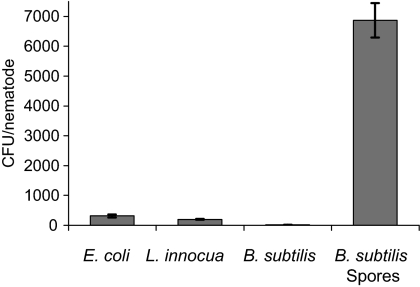
We tested the resistance of vegetative B. subtilis cells to C. elegans digestion by feeding a population of young adult C. elegans N2 worms (about 600 for each sample) with vegetative cells of wild-type B. subtilis (JDB3). We determined the number of surviving bacteria per worm and recovered only 16 ± 1 CFU per worm (mean ± standard deviation), which indicates that few viable bacteria were recovered from the nematode intestines (Fig. 1). We compared this recovery with that obtained following incubation of the worms with cultures of Listeria innocua (JDB1931), another nonpathogenic gram-positive bacterium, or a gram-negative bacterium, Escherichia coli OP50 (JDE304). In both cases, the number of recovered bacteria was at least 10-fold greater than for B. subtilis vegetative cells with 311 ± 48 CFU/worm obtained for E. coli and 199 ± 15 CFU obtained for L. innocua (Fig. 1). Vegetative cells of B. subtilis were detected at less than one CFU per worm in worms at earlier stages (larval stages 1 to 3) of development (data not shown). Although we cannot rule out the possibility that vegetative cells of B. subtilis are less taken up by the nematode, this result suggests that vegetative B. subtilis cells were highly sensitive to digestion by the nematode.
FIG. 1.
Survival of B. subtilis spores in the C. elegans intestine. Adult C. elegans worms were fed with either B. subtilis PY79 vegetative cells or spores or Listeria innocua or Escherichia coli OP50 cells. After mechanical disruption of the nematodes (∼600 worms per sample), the surviving bacteria were determined after growth on LB agar. The values represent the mean CFU/worm ± standard deviation (error bars) for three experiments.
In contrast with vegetative B. subtilis cells, large numbers of bacteria were easily recovered from the worms when C. elegans were fed wild-type B. subtilis spores (6,869 ± 575 CFU/worm [Fig. 1]). We confirmed that the bacteria recovered were spores and not vegetative cells by demonstrating their ability to survive high heat treatment (80°C, 20 min). Although there are numerous differences between spores and vegetative cells that could account for the increased survival of spores following ingestion by C. elegans, we focused on the possible protective role of the spore coat. We examined three strains carrying mutations in coat morphogenetic proteins (cotE, safA, and spoVID) necessary for lysozyme resistance (15). In addition, we examined the correlation between the in vitro chemical sensitivity of spores and their survival following ingestion by C. elegans by testing strains carrying mutations in the cotG, cotH, cotO, yutH, and ysxE genes that exhibit increased sensitivity to various reagents. We also tested the spores of the cotA mutant that do not exhibit any sensitivity in vitro using standard assays (28). CotH, CotO, and CotG are required for the recruitment of a small subset of proteins to the coat and are considered minor morphogenetic proteins (21, 29, 37). While cotO spores, unlike cotG mutant spores, have been reported to be sensitive in vitro to lysozyme (21), we found using the same lysozyme assay (24) that cotO mutant spores were as resistant as wild-type spores were (data not shown). The cotH, yutH, and ysxE mutant spores are slightly sensitive to hypochlorite, and the yutH and ysxE spores were also partially digested by T. thermophila (15).
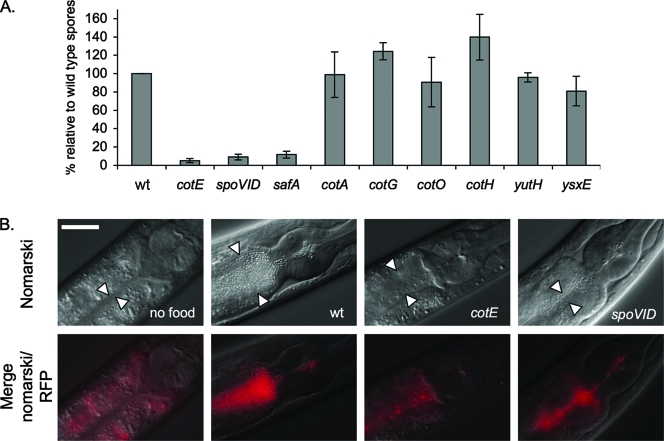
We compared the survival of these mutant spores with wild-type spores by determining the number of recovered bacteria per worm after 16 h of incubation at 20°C with L4-young adult C. elegans. In this assay, there was no significant difference between the values obtained for the wild-type spores and for the spores of the cotA (JDB1343), cotG (JDB1345), cotO (JDB1348), cotH (JDB1346), yutH (JDB1381), and ysxE (JDB1382) strains (98% ± 25%, 125% ± 9%, 91% ± 27%, 139% ± 25%, 96% ± 5%, and 81% ± 16% as a percentage of the wild-type spores, respectively [Fig. 2A]). In contrast, between 10- and 20-fold-less cotE (JDB1323), spoVID (JDB1371), and safA (JDB1380) spores were recovered from the worm intestine (5% ± 2%, 9% ± 3%, and 12% ± 4% as a percentage of wild-type spores, respectively [Fig. 2A]). There was no significant difference observed between the recovery of cotE spores and either spoVID or safA spores.
FIG. 2.
Role of the coat morphogenetic proteins in the resistance of spores to C. elegans digestion. (A) Measurement of the survival of B. subtilis wild-type and coat mutant spores in C. elegans. L4-young adults were fed with wild-type spores (wt) or cotE, safA, spoVID, cotA, cotG, cotO, yutH, and ysxE mutant spores. Survival of mutant spores is represented relative to the survival of wild-type spores. The experiment was performed three times in duplicate, and error bars indicate the standard deviations. (B) Survival of B. subtilis spores within the C. elegans intestine. Merged Nomarski and red fluorescence and Nomarski photomicrographs of the upper intestine of C. elegans L3 larvae are shown. The worms were maintained without food (“no food”) or were fed with spores carrying the PspoIIQ-mCherry reporter in a wild-type background, ΔcotE PspoIIQ-mCherry spores. and ΔspoVID PspoIIQ-mCherry spores. Note the autofluorescence of the intestinal granules on the merged Nomarski and red fluorescence images. The white arrowheads indicate the borders of the intestinal lumen. RFP, red fluorescent protein. Bar = 0.025 mm.
In the course of the experiment, we observed that the densities of the spores on the agar plates for the cotE, spoVID, and safA strains were lower in comparison to the plates containing the wild-type spores, consistent with the digestion of the cotE, spoVID, and safA spores by the nematode (data not shown). We confirmed that the number of spores of cotE, safA, and spoVID mutants were indeed reduced in the worm intestine by monitoring the fate of spores containing red fluorescent protein from the wild-type, cotE, spoVID, and safA strains in the gut (Fig. 2B, cotE and spoVID) (safA strain not shown). These observations indicated that the overall structure of wild-type spores is maintained in the worm intestine and that wild-type spores accumulated in the worm intestine (Fig. 2B). In contrast, fluorescence associated with the cotE, safA, and spoVID spores was considerably reduced in the worm intestine (Fig. 2B and data not shown). Differential interference contrast microscopy of nematodes fed with spores of the cotA, cotO, cotG, yutH, and ysxE mutants confirmed that the spores of these strains accumulate in the intestine of worms as in the case of wild-type spores (data not shown). Thus, only the major alterations in coat formation that are seen in mutants of the major morphogenetic proteins like CotE, SafA, and SpoVID render spores sensitive to digestion by the worm.
Analysis of the ingestion and digestion of cotE mutant spores.
We examined the possibility that the lower recovery of ΔcotE spores was due to decreased ingestion by the nematodes by measuring the kinetics of spore ingestion and excretion. We fed nematodes with spectinomycin-resistant (Specr) spores of B. subtilis strain JDB1334 that has a spectinomycin resistance cassette inserted in a locus (amyE) that does not affect sporulation. We transferred these worms to agar plates containing either no bacteria or a lawn with either E. coli OP50, wild-type B. subtilis, or ΔcotE mutant spores. We assumed that ingestion of new bacteria would chase the Specr spores out of the intestine, and therefore, we monitored the kinetics of excretion of the Specr spores from the nematode intestine. After 20 min, less than 20% of the Specr spores were recovered from the worms shifted to plates containing either E. coli, ΔcotE spores, or wild-type spores (Table 2). After 40 min, less than 10% of the Specr spores remained in the intestine (Table 2). In contrast, the number of wild-type spores decreased at a slower rate in the absence of food, with almost 50% of the Specr spores detected in the intestine of the nematodes 3 h after the transfer (Table 2). After 3 h on E. coli, wild-type or ΔcotE spores, few (2%) of the Specr spores were detected in the worms (Table 2). Given that the ΔcotE spores were as efficient as the wild-type spores at chasing the initial spores out of the worm intestine, we concluded that the ΔcotE spores seemed to be as efficiently ingested as wild-type spores or E. coli and so the low CFU/worm observed (Fig. 2A) was likely due to the sensitivity of the spores to the nematode digestive process.
TABLE 2.
Ingestion of cotE spores by C. elegansa
| Time after transfer to plate | No. of spectinomycin-resistant CFU/worm (% of CFU/worm before transfer)
|
|||
|---|---|---|---|---|
| OP50 | cotE | wt | Without food | |
| 20 min | 12 | 17 | 17 | NDb |
| 40 min | 7 | 5 | 10 | 79 |
| 3 h | 0 | 0 | 2 | 49 |
L4 larvae were fed spectinomycin-resistant B. subtilis spores and transferred to agar plates containing either a lawn of E. coli OP50, a lawn of ΔcotE spores, a lawn of wild-type spores (wt), or without any source of food (without food). The number of spectinomycin-resistant CFU per worm was determined before transfer and at 20 min, 40 min, and 3 h following the transfer to each plate. The results are represented as a percentage of the number of CFU per worm before the transfer. The data shown are from one experiment but are representative of two independent experiments.
ND, not done.
Given that ΔcotE spores were more sensitive to worm digestion, we hypothesized that they would be more easily used as a food source by C. elegans than wild-type spores. We examined this possibility by comparing the development of L1 larvae at 20°C on either metabolically active bacteria (E. coli OP50 and vegetative B. subtilis cells) or on dormant wild-type and ΔcotE spores. The worms reached adulthood on either E. coli strain OP50 or B. subtilis vegetative cells in less than 3 days (Table 3), which was similar to the results reported previously (2) and confirming both the sensitivity of vegetative B. subtilis to worm ingestion and that these bacteria are indeed taken up by the nematode. Consistent with our hypothesis, worms developed faster on ΔcotE spores than on wild-type spores (5.3 and 4.3 days, respectively [Table 3]; P < 0.05). Surprisingly, however, the worms developed on wild-type spores that are presumably resistant to C. elegans digestion, albeit more slowly than on vegetative cells (5.3 and 2.6 days, respectively [Table 3]). In fact, nematodes grown on wild-type spores manifested the hallmarks of development under caloric restriction described for C. elegans mutants that have an eating defect (1). Specifically, we observed that C. elegans fed on spores were smaller and thinner than the nematode fed on vegetative cells of B. subtilis or E. coli OP50 and that adult hermaphrodites generally contained fewer eggs (data not shown). This observation indicated that growth on spores compared to metabolically active bacteria led to limited nutrient availability and suggested that spores become sensitive to the worm's digestion.
TABLE 3.
Growth rate of C. elegans on B. subtilis sporesa
| Strain | Mean no. of days to adulthood (±SD)b |
|---|---|
| E. coli OP50 | 2.6 (±0.11) |
| B. subtilis | |
| Vegetative cells | 2.6 (±0.17) |
| Spores | 5.3 A (±0.29) |
| ΔcotE mutant spores | 4.3 AC (±0.43) |
| gerD-cwlD mutant spores | 8.5 B (±0.71) |
| ΔcotE gerD-cwlD mutant spores | 6.3 BC (±0.32) |
C. elegans L1 larvae were fed with either E. coli OP50 strain, B. subtilis PY79 wild-type strain vegetative cells or spores, cotE mutant spores, gerD-cwlD mutant spores, or cotE gerD-cwlD spores.
The number of days for C. elegans to reach adulthood at 20°C is indicated. The values are the means from three independent experiments. Values with the same letter (A, B, or C) are significantly different (P < 0.05).
The ability of worms to develop on spores, albeit more slowly than on vegetative cells, may indicate that ingested spores had germinated and were therefore sensitive to digestion. We therefore investigated the role of spore germination on nematode growth rate by incubating L1 nematodes with spores of B. subtilis that are defective for germination due to an insertion in the gerD-cwlD genes (strain JDB1486) (34). The gerD-cwlD spores are indeed highly defective in germination, with less than 0.0002% of the spores germinating on rich medium in comparison to wild-type spores (not shown) (34). The growth of C. elegans was slower in the presence of gerD-cwlD spores (8.5 days [Table 3]) than on either wild-type or ΔcotE spores (5.3 and 4.3 days, respectively [Table 3]), suggesting that the germination of B. subtilis spores and their subsequent sensitivity to digestion permitted the growth of C. elegans. We excluded the possibility that the nematode could develop on NMM agar by demonstrating that L1 larvae incubated on a plate without any source of food (spores or metabolically active bacteria) did not grow in the course of the experiment (data not shown). Finally, ΔcotE gerD-cwlD spores (JDB1494) supported a growth rate (6.3 days [Table 3]) intermediate to those of the ΔcotE and gerD-cwlD spores (4.3 and 8.5 days, respectively [Table 3]). This result indicates that part of the growth rate on ΔcotE spores is due to germination of the spores during the incubation, similarly to the wild-type spores, but also that the ΔcotE gerD-cwlD spores are a better source of nutrient than the gerD-cwlD spores, indicating that both ΔcotE mutant spores are digested due to their increased sensitivity to digestion.
Role of the spore coat structures in sensitivity to C. elegans digestion.
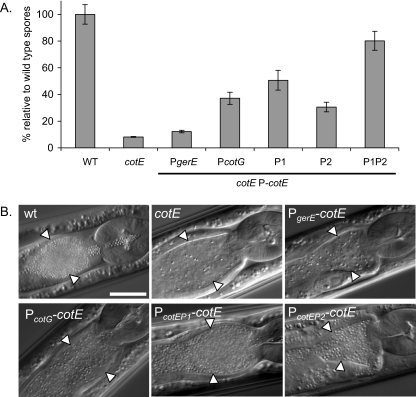
The sensitivity of ΔcotE spores to C. elegans digestion prompted us to study how coat protein composition afforded protection during the digestive process. This composition can be affected by modulating the timing of CotE protein expressed ectopically in a ΔcotE mutant background (3). Early expression of CotE from the amyE locus is observed when it is under the control of its own promoters (PcotEP1, PcotEP2, or PcotEP1P2), whereas late expression is observed when it is under the control of the gerE and cotG promoters (PgerE and PcotG) (3). Expression of cotE under PgerE or PcotG and, to a lesser extent under the PcotEP2 promoter, led to a similar, but not identical, defect in overall protein composition and the structure of the coat to that observed in a ΔcotE null mutant strain (3). However, coat composition and structure were more similar to those in the wild-type spores in strains expressing cotE under PcotEP1 or PcotEP1P2 (3).
While these experiments indicated that early expression of cotE from either PcotEP1, PcotEP2, or PcotEP1P2 is required for the proper assembly of the outer coat, spores derived from these strains were as resistant to lysozyme as the wild-type spores were (3). Thus, these strains uncoupled, for the first time, lysozyme resistance observed in vitro and outer coat assembly (3) and allowed us to test the distinct role of each of these phenotypes in sensitivity to digestion by C. elegans. Surprisingly, the spores of all the strains tested except for the ΔcotE PcotEP1P2-cotE strain (JDB2129) were susceptible to ingestion by worms (Fig. 3). Using the measurements of spores per worm and further supported by data obtained in growth rate experiments and microscopy observations, we can distinguish three classes of mutants. The first class comprises ΔcotE PgerE-cotE spores (JDB2122) recovered at a level comparable to that obtained for the ΔcotE spores (12% ± 1%,and 8% ± 0.3% as a percentage of wild-type spores, respectively [Fig. 3A]). These spores supported the growth of worms as well as ΔcotE spores did (data not shown) and are not distinguishable from ΔcotE spores in the intestine (Fig. 3B). The second class comprises the ΔcotE PcotG-cotE (JDB2124) and ΔcotE PcotEP2-cotE (JDB2125) spores which were recovered at fourfold- and threefold-higher levels, respectively, than the level of ΔcotE spores (37% ± 4% and 31% ± 4%, respectively [Fig. 3A]). These spores supported nematode growth at a rate similar to the ΔcotE spores (data not shown) and are clearly seen in the worm intestine (Fig. 3B). The third class of mutants, including the ΔcotE PcotEP1-cotE (JDB2127) and ΔcotE PcotEP1P2-cotE (JDB2129) spores, was recovered at 6- and 10-fold-higher levels than the level of ΔcotE spores (51% ± 7%, 80% ± 7%, and 8% ± 0.3%, respectively [Fig. 3A]) and supported the growth of the worms as well as wild-type spores did (data not shown) and accumulated in the intestine (Fig. 3B). Thus, the viability of these three classes of mutant spores following ingestion by the worm correlates with their respective defect in coat protein composition and structural modification of the coat.
FIG. 3.
Role of outer coat formation in the resistance of spores to C. elegans. The sensitivity of B. subtilis wild-type spores and various cotE mutant spores in C. elegans was measured. (A) L4 larvae were fed with wild-type spores (WT), ΔcotE mutant spores, or spores of a ΔcotE mutant expressing cotE from five promoters at the amyE locus: PgerE, PcotG, PcotEP1 (P1), PcotEP2 (P2), PcotEP1P2 (P1P2). Data are represented in comparison to the number of CFU/worm obtained for the wild-type spores. The experiment was performed four times in duplicate, and a representative experiment is shown (standard deviations are indicated by error bars). (B) Nomarski pictures of the upper intestine of N2 L4 larvae fed with wild-type (wt), ΔcotE, ΔcotE PgerE-cotE, ΔcotE PcotG-cotE, ΔcotE PcotEP1-cotE, and ΔcotE PcotEP2-cotE spores. Worms fed with the ΔcotE PcotEP1P2-cotE spores appeared similar to worms fed with the wild-type spores or ΔcotE PcotEP1-cotE spores. The white arrowheads indicate the borders of the intestinal lumen. Bar = 0.025 mm.
DISCUSSION
Bacillus species are abundant soil bacteria (∼106 organisms/gram [18]) and likely serve as a source of nutrients for the bacteriovorous nematode C. elegans (9). Indeed, B. subtilis vegetative cells are efficiently digested by the worm (Fig. 1) (7), consistent with the longer life span observed for C. elegans grown on vegetative Bacillus subtilis compared to the life span for C. elegans grown on E. coli OP50 (8). In contrast, we observed that wild-type B. subtilis spores were highly resistant to worm digestion (Fig. 1) as would be predicted from their resistance to phagocytosis by the protozoan T. thermophila and by their survival in the mammalian gastrointestinal tract (11, 15, 34). In a previous study, we found that other Bacillus species (Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis) are also highly sensitive to the worm ingestion and that B. anthracis spores are fully resistant to the worm digestive process (16). In B. subtilis, the proper recruitment and assembly of the spore coat are crucial for the spore resistance properties, and in particular, mutations in coat morphogenetic proteins (CotE, SpoVID, and SafA) result in lysozyme sensitivity (25, 26, 36). Our finding that spores carrying mutations in the genes encoding these proteins were sensitive to digestion (Fig. 2A) is consistent with the role of the overall structure and composition of the coat in providing resistance to nematode predation. In contrast, spores carrying mutations in minor morphogenetic proteins, (CotG, CotO, and CotH) exhibited little or no effect on coat structure (21, 29) and were as resistant as the wild-type spores were (Fig. 2A).
The precise expression of cotE and the subsequent proper assembly of the cotE-dependent proteins in the coat appear to be crucial steps in achieving resistance to digestion (Fig. 3). Increased defects in coat structure, particularly the outer coat, led to increased digestion by the worm (Fig. 3). However, even ΔcotE PcotEP2-cotE spores that have a coat structure and composition similar to those wild-type spores and in which only a few protein species were found missing by sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis (3) are still somewhat sensitive to digestion. Thus, either the defect in this small subset of proteins is sufficient to enable the digestion of the spore, or the structure or composition of the coat is more affected in this strain than previously explored. Whatever the ultimate explanation, this result clearly highlights the usefulness of the C. elegans model for uncovering subtle coat protein mutant phenotypes.
In contrast to previous experiments using T. thermophila (15), we did not observe a correlation between in vitro lysozyme sensitivity and C. elegans digestibility. Indeed, the yutH and ysxE spores that are sensitive both to lysozyme and to digestion by T. thermophila (15) were as resistant to C. elegans digestion as the wild-type spores were. One explanation could be that these two studies used nonisogenic B. subtilis 168 strains. This discrepancy could also be explained by a longer time of residency of the spores in the protozoan digestive vacuole compared to the intestine of a L4 worm (∼1 h) (Table 2). Longer persistence in the T. thermophila vacuoles would result in longer exposure to digestive enzymes and thus increased degradation of the yutH and ysxE spores.
ΔcotE mutant spores are degraded during incubation with a high concentration of purified lysozyme (36) and are also readily digested by C. elegans (Fig. 2A and B). However, spores produced by cells in which cotE expression was delayed were resistant to lysozyme (3) but sensitive to digestion (Fig. 3). Thus, the timing of expression of cotE appeared critical for the survival of the spore in the worm gut and may indicate more subtle defects in spore coat assembly in these cotE expression mutants.
The C. elegans digestive process has been suggested to involve both mechanical disruption mediated by a pharyngeal grinder and enzymatic digestion of the ingested microorganisms in the intestine (19). However, since ΔcotE spores do not exhibit increased survival in mutant worms with a defective grinder (M.-H. Laaberki and J. Dworkin, unpublished data), they, and presumably other defective spores, are likely to be degraded by enzymes secreted into the intestinal lumen. This result is consistent with a previous study showing that wild-type and cotE spores exhibit essentially identical resistance to mechanical disruption (12). C. elegans expresses and potentially secretes into its digestive system several enzymes that could degrade sensitive spores, including 10 lysozyme genes (lys-1 to lys-10) and 5 insect-lysozyme genes (ilys-1 to ilys-5) (19, 31), and 5 of the lys genes and 2 of the ilys genes are expressed in intestinal cells (13, 17, 20, 32). In addition, numerous peptidases and lipases are expressed and also presumably secreted in the intestine of the nematode (20). A synergistic activity of these enzymes could be responsible not only for the degradation of ΔcotE spores but also for the degradation of the ectopically expressed cotE spores that are lysozyme resistant in vitro (3). An understanding of the enzymatic basis for digestion of spore coat mutants as well as vegetative Bacillus cells in C. elegans will be greatly facilitated by the genetic tools available in this organism. For example, RNA interference could be used to reduce the expression of potential candidate bacteriolytic enzymes (17, 22), and then the effect of these manipulations on bacterial survival could be determined using the assays we have described. Finally, this knowledge will help us refine our understanding of the role of the spore coat structures in resistance to an ecologically relevant predator.
Acknowledgments
We thank Simon Cutting, Patrick Eichenberger, and Adriano Henriques for providing B. subtilis strains. We thank Allison Fay for constructing the JDB1582 strain.
This work was supported by start-up funds from the Department of Microbiology of Columbia University.
Footnotes
Published ahead of print on 27 June 2008.
REFERENCES
- 1.Avery, L. 1993. The genetics of feeding in Caenorhabditis elegans. Genetics 133897-917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Avery, L., and B. B. Shtonda. 2003. Food transport in the C. elegans pharynx. J. Exp. Biol. 2062441-2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Costa, T., M. Serrano, L. Steil, U. Volker, C. P. Moran, Jr., and A. O. Henriques. 2007. The timing of cotE expression affects Bacillus subtilis spore coat morphology but not lysozyme resistance. J. Bacteriol. 1892401-2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cutting, S. M., and P. B. V. Horn. 1990. Genetic analysis, p. 27-74. In C. R. Harwood and S. M. Cutting (ed.), Molecular biological methods for Bacillus. John Wiley and Sons, Chichester, England.
- 5.Driks, A. 1999. Bacillus subtilis spore coat. Microbiol. Mol. Biol. Rev. 631-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Driks, A., S. Roels, B. Beall, C. P. Moran, Jr., and R. Losick. 1994. Subcellular localization of proteins involved in the assembly of the spore coat of Bacillus subtilis. Genes Dev. 8234-244. [DOI] [PubMed] [Google Scholar]
- 7.Garsin, D. A., C. D. Sifri, E. Mylonakis, X. Qin, K. V. Singh, B. E. Murray, S. B. Calderwood, and F. M. Ausubel. 2001. A simple model host for identifying Gram-positive virulence factors. Proc. Natl. Acad. Sci. USA 9810892-10897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garsin, D. A., J. M. Villanueva, J. Begun, D. H. Kim, C. D. Sifri, S. B. Calderwood, G. Ruvkun, and F. M. Ausubel. 2003. Long-lived C. elegans daf-2 mutants are resistant to bacterial pathogens. Science 3001921. [DOI] [PubMed] [Google Scholar]
- 9.Grewal, P. S. 1991. Influence of bacteria and temperature on the reproduction of Caenorhabditis elegans (Nematoda: Rhabditiae) infesting mushrooms (Agaricus bisporus). Nematologica 3772-82. [Google Scholar]
- 10.Henriques, A. O., B. W. Beall, K. Roland, and C. P. Moran, Jr. 1995. Characterization of cotJ, a σE-controlled operon affecting the polypeptide composition of the coat of Bacillus subtilis spores. J. Bacteriol. 1773394-3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Henriques, A. O., and C. P. Moran, Jr. 2007. Structure, assembly, and function of the spore surface layers. Annu. Rev. Microbiol. 61555-588. [DOI] [PubMed] [Google Scholar]
- 12.Jones, C. A., N. L. Padula, and P. Setlow. 2005. Effect of mechanical abrasion on the viability, disruption and germination of spores of Bacillus subtilis. J. Appl. Microbiol. 991484-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kerry, S., M. TeKippe, N. C. Gaddis, and A. Aballay. 2006. GATA transcription factor required for immunity to bacterial and fungal pathogens. PLoS One 1e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim, H., M. Hahn, P. Grabowski, D. C. McPherson, M. M. Otte, R. Wang, C. C. Ferguson, P. Eichenberger, and A. Driks. 2006. The Bacillus subtilis spore coat protein interaction network. Mol. Microbiol. 59487-502. [DOI] [PubMed] [Google Scholar]
- 15.Klobutcher, L. A., K. Ragkousi, and P. Setlow. 2006. The Bacillus subtilis spore coat provides “eat resistance” during phagocytic predation by the protozoan Tetrahymena thermophila. Proc. Natl. Acad. Sci. USA 103165-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laaberki, M. H., and J. E. Dworkin. 2008. Death and survival of spore-forming bacteria in the Caenorhabditis elegans intestine. Symbiosis 4695-100. [Google Scholar]
- 17.Mallo, G. V., C. L. Kurz, C. Couillault, N. Pujol, S. Granjeaud, Y. Kohara, and J. J. Ewbank. 2002. Inducible antibacterial defense system in C. elegans. Curr. Biol. 121209-1214. [DOI] [PubMed] [Google Scholar]
- 18.Martin, P. A., and R. S. Travers. 1989. Worldwide abundance and distribution of Bacillus thuringiensis isolates. Appl. Environ. Microbiol. 552437-2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McGhee, J. D. 2007. The C. elegans intestine. WormBook 20071-36. http://www.wormbook.org/chapters/www_intestine/intestine.html. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McGhee, J. D., M. C. Sleumer, M. Bilenky, K. Wong, S. J. McKay, B. Goszczynski, H. Tian, N. D. Krich, J. Khattra, R. A. Holt, D. L. Baillie, Y. Kohara, M. A. Marra, S. J. Jones, D. G. Moerman, and A. G. Robertson. 2007. The ELT-2 GATA-factor and the global regulation of transcription in the C. elegans intestine. Dev. Biol. 302627-645. [DOI] [PubMed] [Google Scholar]
- 21.McPherson, D. C., H. Kim, M. Hahn, R. Wang, P. Grabowski, P. Eichenberger, and A. Driks. 2005. Characterization of the Bacillus subtilis spore morphogenetic coat protein CotO. J. Bacteriol. 1878278-8290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murphy, C. T., S. A. McCarroll, C. I. Bargmann, A. Fraser, R. S. Kamath, J. Ahringer, H. Li, and C. Kenyon. 2003. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature 424277-283. [DOI] [PubMed] [Google Scholar]
- 23.Nicholson, W. L. 2002. Roles of Bacillus endospores in the environment. Cell. Mol. Life Sci. 59410-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nicholson, W. L., and P. Setlow. 1990. Sporulation, germination and outgrowth., p. 391-450. In C. R. Harwood and S. M. Cutting (ed.), Molecular biological methods for Bacillus. John Wiley and Sons, Chichester, England.
- 25.Ozin, A. J., A. O. Henriques, H. Yi, and C. P. Moran, Jr. 2000. Morphogenetic proteins SpoVID and SafA form a complex during assembly of the Bacillus subtilis spore coat. J. Bacteriol. 1821828-1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ozin, A. J., C. S. Samford, A. O. Henriques, and C. P. Moran, Jr. 2001. SpoVID guides SafA to the spore coat in Bacillus subtilis. J. Bacteriol. 1833041-3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prager, E. M., and P. Jolles. 1996. Lysozyme: a model enzyme in protein chemistry, p. 9-87. In P. Jolles (ed.), Lysozymes: model enzymes in biochemistry and biology. Birkhauser-Verlag, Basel, Switzerland.
- 28.Rogolsky, M. 1968. Genetic mapping of a locus which regulates the production of pigment associated with spores of Bacillus subtilis. J. Bacteriol. 952426-2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sacco, M., E. Ricca, R. Losick, and S. Cutting. 1995. An additional GerE-controlled gene encoding an abundant spore coat protein from Bacillus subtilis. J. Bacteriol. 177372-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saile, E., and T. M. Koehler. 2006. Bacillus anthracis multiplication, persistence, and genetic exchange in the rhizosphere of grass plants. Appl. Environ. Microbiol. 723168-3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schulenburg, H., and C. Boehnisch. 2008. Diversification and adaptive sequence evolution of Caenorhabditis lysozymes (Nematoda: Rhabditidae). BMC Evol. Biol. 8114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shapira, M., B. J. Hamlin, J. Rong, K. Chen, M. Ronen, and M. W. Tan. 2006. A conserved role for a GATA transcription factor in regulating epithelial innate immune responses. Proc. Natl. Acad. Sci. USA 10314086-14091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sulston, J. E., and J. Hodgkin. 1988. Methods, p. 587-606. In W. B. Wood (ed.), The nematode Caenorhabditis elegans. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 34.Tam, N. K., N. Q. Uyen, H. A. Hong, H. le Duc, T. T. Hoa, C. R. Serra, A. O. Henriques, and S. M. Cutting. 2006. The intestinal life cycle of Bacillus subtilis and close relatives. J. Bacteriol. 1882692-2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Youngman, P., J. B. Perkins, and R. Losick. 1984. A novel method for the rapid cloning in Escherichia coli of Bacillus subtilis chromosomal DNA adjacent to Tn917 insertions. Mol. Gen. Genet. 195424-433. [DOI] [PubMed] [Google Scholar]
- 36.Zheng, L. B., W. P. Donovan, P. C. Fitz-James, and R. Losick. 1988. Gene encoding a morphogenic protein required in the assembly of the outer coat of the Bacillus subtilis endospore. Genes Dev. 21047-1054. [DOI] [PubMed] [Google Scholar]
- 37.Zilhão, R., G. Naclerio, A. O. Henriques, L. Baccigalupi, C. P. Moran, Jr., and E. Ricca. 1999. Assembly requirements and role of CotH during spore coat formation in Bacillus subtilis. J. Bacteriol. 1812631-2633. [DOI] [PMC free article] [PubMed] [Google Scholar]