Abstract
Nothing is currently known regarding the global regulatory networks of Treponema denticola and other oral spirochetes. In this report, we assess the properties and potential phosphotransfer capability of a putative two-component regulatory system (TCS) of T. denticola that is formed by the products of open reading frames tde0032 (a sensor kinase) and tde0033 (a response regulator), henceforth designated AtcS and AtcR, respectively. Using PCR and DNA sequence analyses, atcS and atcR were demonstrated to be widely distributed and conserved among T. denticola isolates. Reverse transcription-PCR (RT-PCR) analyses revealed that these genes are cotranscribed and may also be expressed as part of a larger operon that includes several flanking genes. Analyses using 5′ rapid amplification of cDNA ends identified the transcriptional start sites for these operons and provided evidence that some of these genes may be independently transcribed from internal promoters. Real-time RT-PCR and Western blot analysis revealed significant upregulation of atcRS during late-stage growth, indicating growth-phase-dependent expression. Lastly, the phosphorelay capability of the AtcRS system was assessed and demonstrated using recombinant proteins. AtcS was found to undergo autophosphorylation and to transfer phosphate to AtcR. These analyses represent the first description of a functional TCS in an oral spirochetes and provide insight into the transcriptional regulatory mechanisms of these important bacteria.
Periodontal disease is a progressive disease that begins with the formation of a polymicrobial biofilm that ultimately consists of several hundred species of endogenous bacteria. Treponema denticola, a member of the “red microbial complex,” occurs in high numbers in periodontal lesions (42). Recent data suggest that T. denticola may also be associated with low birth weight (28) and esophageal cancers (25). In spite of the established importance of this organism in human health, little is know regarding its global regulatory networks and the dynamics of its transcriptional expression patterns. Two-component systems (TCS), which are ubiquitous in the bacterial world (49), serve as important sensory systems that allow living organisms to respond to changing environmental conditions. The importance of these systems in biofilm formation and in the regulation of the expression of virulence factors has been demonstrated for numerous pathogens (14, 24, 26, 34, 43). Classical TCS consist of a histidine kinase and a response regulator. Upon receiving the appropriate stimulus or upon ligand binding, the kinase autophosphorlyates at a conserved His residue. This is followed by the transfer of the phosphate to a conserved Asp residue present in the receiver domain of the response regulator (33). This results in conformational changes in the output domain of the response regulator that allow it to mediate DNA binding, specific protein-protein interactions, or enzymatic activities that influence regulation of transcription and cellular activity (9).
Annotation of the T. denticola genome has identified eight putative histidine kinases and nine putative response regulators (40). In this study we have initiated the analysis of one of these putative TCS, consisting of the T. denticola open reading frames (ORFs) tde0032 and tde0033, which encode a putative sensor kinase and response regulator, respectively. Here we demonstrate that both tde0032 and tde0033 are universal among T. denticola isolates, highly conserved, cotranscribed, and regulated by growth phase. The tde0032 product was found to autophosphorylate and to transfer phosphate to the tde0033 product. Based on the properties of these proteins and the proximity of the genes that encode them to tde0037, encoding a member of the AbrB protein family (a DNA binding, growth-phase-dependent transcriptional regulator) (10, 35, 39, 41, 44, 45, 47), we henceforth refer to the tde0033 product as AtcR (AbrB-associated two-component response regulator) and to the tde0032 product as AtcS (AbrB-associated two-component sensor kinase). The data presented here suggest that AtcR and AtcS may form a TCS that serves as an important regulator of T. denticola growth-phase-specific metabolism. This study is the first to demonstrate a functional TCS in an oral spirochete and to explore the possible regulatory networks in the periodontal pathogen T. denticola.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
T. denticola strains 33520, N17A1, MS25, N16B1, 35404, 35405, and GM1 were cultivated in NOS medium (ATCC medium 1494) under anaerobic conditions (5% H2, 20% CO2, 75% N2, 37°C). Growth was monitored by dark-field microscopy. All strains were isolated previously from human periodontal pockets.
Ligase-independent cloning, production of r-proteins, and generation of antisera.
Recombinant proteins (r-proteins) were generated using a ligase-independent cloning approach as previously described (18). The entire AtcR protein was cloned, while only the kinase domain of AtcS (amino acids 27 to 248) was used in the study. The genes were amplified using standard PCR conditions with primers harboring tail sequences that complement the single-stranded overhangs of the pET46 Ek/LIC vector (Novagen). The resulting amplicons were treated with T4 DNA polymerase to regenerate single-strand overhangs and then annealed into the vector as instructed by the supplier (Novagen). The resulting plasmids were propagated in Escherichia coli NovaBlue cells (Novagen). Expression of r-protein was conducted in E. coli BL21(DE3) cells by induction with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) (3 h, 37°C). Proteins expressed from the pET46 Ek/LIC vector possess an N-terminal fusion of 1.7 kDa consisting of a hexahistidine tag. The His-tagged r-proteins were purified to homogeneity using an Ni-nitrilotriacetic acid affinity matrix as instructed by the supplier (Qiagen).
To generate antisera, 50 μg of each r-protein in Freund's complete adjuvant (Pierce) was subcutaneously injected into C3H-HeJ mice (4 to 6 weeks old). Boosts were administered at 2 and 4 weeks in Freund's incomplete adjuvant. The mice were sacrificed at week 6, blood was collected, and the specificity of the antiserum was confirmed by immunoblot analyses.
Qualitative and quantitative reverse transcription-PCR (RT-PCR).
RNA was extracted from T. denticola 35405 propagated in NOS medium using the RNeasy RNA extraction kit (Qiagen). cDNA was generated using random hexamer primers and Superscript III reverse transcriptase (Invitrogen). Qualitative PCR was conducted using GoTaq master mix (Promega) and the primer sets shown in Table 1. Amplicons were resolved by gel electrophoresis using 2% Metaphor agarose (Cambrex) and Tris-acetate-EDTA buffer. Real-time PCRs were carried out using Sybr green PCR master mix (Applied Biosciences) in an MJ Research Opticon 2 real-time thermocycler with the following cycling conditions: 95°C for 10 min followed by 40 cycles of 94°C for 15 s; 60°C for 30 s, and 72°C for 30 s. To generate standard curves for quantitation, amplicons of the genes of interest were cloned into the pCR2.1 TOPO vector (Invitrogen) and serial dilutions of the purified plasmids were used as template in real-time PCR. Data were expressed as the total number of calculated transcripts as a percentage of the total number of flaA transcripts.
TABLE 1.
Primers used in this study
| Primera | Sequence |
|---|---|
| tde0029-F | CAGACTCAAATGCAAGGAGAAGCAG |
| tde0029-R | AACTTTAGCAACCTTACCATCAGGG |
| Int 30/29-F | CCATCAAAATATTTCGAATTGATTGCAG |
| Int 30/29-R | CAGCATCGTAGTCATTCGCAGTC |
| tde0030-F | GACTGCGAATGACTACGATGCTG |
| tde0030-R | CCAAATCGTCTCTCGAAGTAAACC |
| Int 31/30-F | GGTTTACTTCGAGAGACGATTTGG |
| Int 31/30-R | CAGTATGGCTGCCGCTAAATGG |
| tde0031-F | CCATTTAGCGGCAGCCATACTG |
| tde0031-R | GGTGATTCCTTCGTTATGTGTTTTCC |
| Int atcS/31-F | CTCTTATTTACTGAATATGACGGAAGCC |
| Int atcS/31-R | GAACTTCATTATCTTCGATGACGGC |
| atcS-F | GCCGTCATCGAAGATAATGAAGTTC |
| atcS-R | CCTGTTATAAAGATGATAACGGCATCG |
| Int atcR/atcS-F | GCGAGTACATAACAAGGTTGAATTTATCGGAGG |
| Int atcR/atcS-R | GAAAGTCATGCCGCCAGCC |
| atcR-F | TTTTTGATAATGCCATTGAGGGTGC |
| atcR-R | GCTTTGGCGGTTTATAAAGGCAG |
| Int 34/atcR-F | ATATTCTGCCTTTATAAACCGCCAAAGCG |
| Int 34/atcR-R | CCCAATACCAAAGATGCGACAGCC |
| tde0034-F | GGCTGTCGCATCTTGGTATTGGG |
| tde0034-R | GCAGCTGCAGTTCCTGCAAGG |
| Int 35/34-F | CTTGCAGGAACTGCAGCTGC |
| Int 35/34-R | CCAAGAGCAATCATTAACCCATAGG |
| tde0035-F | CCTATGGGTTAATGATTGCTCTTGG |
| tde0035-R | GTTTAAGTCGAATATTTTTGACCACTCAATTG |
| Int 36/35-F | CACTGCATATTTTCAAAAAACCGTTTGCG |
| Int 36/35-R | GCCCCAATTCCGTAAAATACCTCG |
| Int 37/36-F | CGAGGTATTTTACGGAATTGGGGC |
| Int 37/36-R | GTTTGTTCTAAAAGCACTGTAACGGC |
| atcS-LIC-F* | TTGTCAATAAGCGTTTTGATGATTTTGAAAACGGCC |
| atcS-LIC-R* | TCTATAAAACATTCGGAAGATTAGGGAGCAAAACAAGC |
| atcR-LIC-F* | ATGCTTAAAATAGCCGTCATCGAAGATAATGAAG |
| atcR-LIC-R* | TTCAGGTTTCTCCTTGGGTAAAAAAGCCG |
| flaA-F | GCTCAGGGTTGATGATCAGG |
| flaA-R | GCAATTGATTTGATAACGCCG |
Int denotes the intergenic region between the two ORF numbers listed. All LIC cloning primers also contain the published overhang sequences.
TSS identification using 5′ RACE.
Transcriptional start sites (TSS) were identified using 5′ rapid amplification of cDNA ends (RACE) (Invitrogen) as previously described (53). Briefly, cDNA was generated using primers specific for the upstream regions of tde0037 (abrB), atcR (tde0033), and tde0031 and purified using Snap columns (Invitrogen). Purified cDNAs were 3′ poly(C) tailed using terminal deoxyribonucleoside transferase and dCTP. The amplicons were then PCR amplified using the abridged anchor primer (Invitrogen), which anneals to the poly(C) tail, and nested gene-specific primers. A second round of PCR was then performed using a primer set consisting of the gene specific primer used in round 1 and the universal anchor primer (Invitrogen). The resulting amplicons were cloned into the pCR2.1 TOPO vector and the inserts sequenced on a fee-for-service basis (MWG Biotech).
Northern blot analysis.
Total RNA was isolated from 100-ml cultures using the RNeasy Midi kit (Qiagen). RNA was fractionated in 1% agarose-formaldehyde gels (15 μg per lane) using 1× MOPS (morpholinepropanesulfonic acid) buffer and transferred to Hybond N+ nylon membranes (Amersham) by vacuum blotting (Pharmacia). Blots were screened with either oligonucleotide or PCR-generated probes. Oligonucleotides (30 pmol) were 5′ end labeled using [γ-32P]ATP (6,000 Ci mmol−1; Perkin-Elmer) and T4 polynucleotide kinase. PCR-generated probes were internally labeled using the NEBlot kit (NEB) and [[α-32P]dATP (3,000 Ci mmol−1; Perkin-Elmer). All other methods, buffers, hybridization conditions, and washes were as previously described (17). Hybridization was detected by autoradiography at −70°C with intensifying screens.
Autophosphorylation and phosphotransfer assays.
To assess autophosphorylation of AtcS, r-AtcS (20 μg ml−1) was incubated with 2 μCi of [γ-32P]ATP (3,000 Ci mmol−1; Perkin-Elmer) in phosphorylation buffer (30 mM HEPES, 50 mM KCl, 10 mM MgCl2, 0.5 mM EDTA, 2 mM DTT, pH 8.0) at 37°C. Aliquots of the reaction mixture were removed at 0, 10, 20, and 30 min. FhbB (a factor H binding protein) (18) and AtcR, proteins that are not expected to undergo autophosphorylation, served as negative control proteins. The reaction components were then solubilized in 2× sodium dodecyl sulfate (SDS) sample buffer and immediately placed on ice. The samples were then subjected to SDS-polyacrylamide gel electrophoresis (SDS-PAGE) using Criterion precast 12% gels (200 V, 1 h) and transferred to polyvinylidene difluoride membranes by electroblotting for subsequent autoradiography (at −70°C with intensifying screens).
The ability of AtcS to transfer phosphate to AtcR was assessed as follows. AtcS (20 μg/ml) was incubated with 2 μCi [γ-32P]ATP for 30 min in phosphorylation buffer, and then AtcR was added at a 3:1 ratio. Aliquots were then removed at 2- to 5-min intervals up to 55 min. The samples were then treated exactly as described in the preceding paragraph.
RESULTS
Distribution, sequence conservation, and functional domains of AtcS and AtcR.
Annotation of the T. denticola genome sequence identified several putative sensor kinase and response regulator pairs (40). The AtcRS pair (encoded by ORFs tde0032 and tde0033) is the focus of this report. The predicted functional domains and residues of both AtcS and AtcR are indicated in Fig. 1.
FIG. 1.
Sequence and functional domains of AtcS and AtcR. The amino acid sequences of AtcS (A) and AtcR (B) are shown. Possible sites of phosphorylation for both AtcS (His) and AtcR (Asp) are marked by black circles above the corresponding residue. The H, N, and G boxes (described in the text) of AtcS are underlined, and the putative N-terminal transmembrane domain is boxed. The receiver and LytTR domains of AtcR are labeled by a solid underline and a dashed underline, respectively. Conserved residues of AtcR that have been demonstrated in LytTR-containing proteins to be required for binding to DNA are marked by diamonds (22, 23). Other residues that are conserved in LytTR domains but have not been characterized functionally are indicated with an X (26).
AtcS is predicted to be a 29-kDa protein sensor kinase. The Tmap and TMpred algorithms indicate that the N-terminal domain of the protein has strong predictive probability of forming a transmembrane domain (Fig. 1A), a feature that would be expected for a sensor kinase. AtcS harbors conserved H (His phosphorylation) and N and G box (nucleotide binding) functional domains that are typically found in histidine kinases (Fig. 1A) (4, 43). Three His residues are found within the predicted H box (Fig. 1A). Initial de novo structural modeling based on the HMMSTR/Rosetta algorithm (5, 6) suggests that of these residues, His-57 and His-60 are the most likely to be potential phosphorylation sites since they are oriented into the nucleotide binding pocket. AtcR is predicted to be a 28-kDa response regulator, and as expected for a protein within this functional category, it lacks a leader peptide and transmembrane domains. AtcR has a CheY-like receiver domain (Fig. 1B) that contains three Asp residues at positions 52, 54, and 59 that have the potential to serve as phosphate acceptors (Fig. 1B) (13, 27, 38, 48). AtcR also possesses a LytTR DNA binding domain (26) that spans the C-terminal 100 residues of the protein (Fig. 1B). The potential significance of specific functional domains of AtcS and AtcR in T. denticola biology are discussed in detail below.
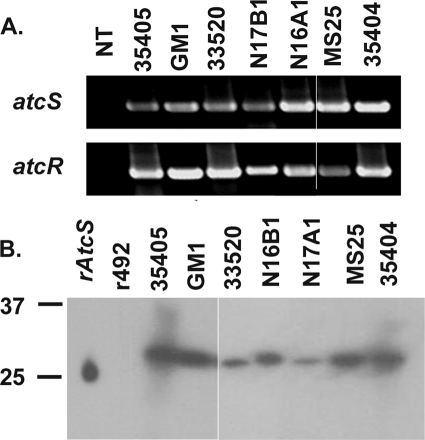
To assess the distribution and potential sequence conservation of atcRS, PCR analyses of seven T. denticola strains were performed using gene-specific primer sets. Amplicons of the predicted size were obtained from all strains tested, suggesting that this TCS is universal (Fig. 2A). Cloning of the amplicons and subsequent DNA sequence analyses revealed that both atcR and atcS are highly conserved. Amino acid similarity and identity values for both proteins among isolates were greater than 98% and 94%, respectively. The three histidine residues predicted to be possible sites of phosphorylation for AtcS are conserved in all strains. While the Asp residues of AtcR at positions 52 and 59 are generally invariant among the seven strains tested, in strain N16B1, the Asp at position 54 is replaced by an Asn. All other residues thought to be essential in the functional activities of histidine kinases and response regulators are conserved.
FIG. 2.
Conservation of AtcS and AtcR among T. denticola strains. PCR amplicons obtained with atcS and atcR gene-specific primer sets from seven different strains are shown in panel A. Lane NT shows PCRs with no template added. Western blotting analysis of AtcS is shown in panel B. Cell lysates of T. denticola strains were screened with anti-AtcS antiserum. Purified r-AtcS served as a positive control, and an unrelated His6-tagged histidine kinase (tde0492 product) cloned from strain 35405 served as a negative control.
To assess the production of AtcS and AtcR by T. denticola, antiserum was generated using r-proteins and used to screen immunoblots of cell lysates from seven different strains. All strains were cultivated for 6 to 8 days and were highly motile at the time of harvesting. The anti-AtcS antiserum reacted with a single protein band of the appropriate size in all strains (Fig. 2B). The anti-AtcR antiserum also reacted with a protein of the predicted size (data not shown). R-AtcS and r-AtcR were used as the positive controls for immunodetection, and an unrelated His-tagged protein (ORF tde0492) served as the negative control.
Transcriptional analysis of atcS and atcR.
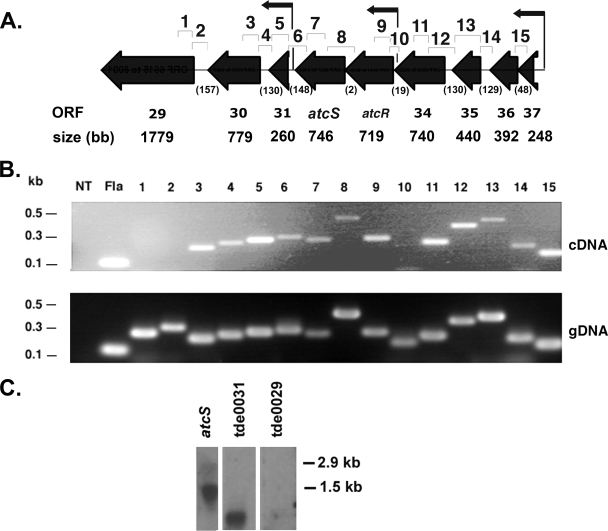
Sensor kinase and response regulator pairs of TCS are typically cotranscribed as a polycistronic mRNA. To determine if atcR and atcS are expressed during in vitro cultivation as an operon, RT-PCR analyses were conducted using internal primer sets specific for each ORF and a primer set designed to amplify across the intergenic spacers (Fig. 3A). These genes were found to be expressed in vitro and to be cotranscribed. To determine if additional genes (Table 2) are cotranscribed with atcR-atcS, further RT-PCR analyses were performed. A separate and distinct operon consisting of the four genes that are located just upstream of the atcRS locus was also detected. This operon consisted of ORFs tde0034 through tde0037. The potential functional roles of the proteins encoded by these ORFs are described in Table 2. The RT-PCR analyses also revealed that the two ORFs downstream of atcS (tde0030 and tde0031) are expressed independently but can also be cotranscribed with the atcRS TCS.
FIG. 3.
RT-PCR and Northern blot analysis of the atcR/atcS locus. Panel A shows a schematic of atcR-atcS and their up- and downstream genes (TIGR annotation). The lengths of the intergenic regions are shown in parentheses. Primer binding sites and the amplicons used for RT-PCR are shown above, with the numbers correlating to the gel lanes below. The TSS are indicated by the small arrows. In panel B the PCR amplicons obtained from RT-PCR analyses or from PCR using genomic DNA as a template (positive control) are shown. NT denotes the negative controls of either no reverse transcriptase (cDNA) or no template (genomic DNA [gDNA]). Panel C shows the results of Northern blot analyses using probes to atcR, tde0031, and tde0029 (as indicated). The migration positions of the 23S and 16S rRNA bands (2.9 kb and 1.5 kb) are shown.
TABLE 2.
Description of ORFs analyzed in this study
| ORF designation | Gene name | Annotated function and description |
|---|---|---|
| tde0029 | hlyB | ABC transporter HlyB family |
| tde0030 | Prolipoprotein diacylglyceryl transferase | |
| tde0031 | Hypothetical; protein harbors two predicted transmembrane domains | |
| tde0032 | atcS | Histidine kinase |
| tde0033 | atcR | Response regulator |
| tde0034 | Hypothetical | |
| tde0035 | Acetyltransferase, GNAT family | |
| tde0036 | Hypothetical | |
| tde0037 | abrB | Transcriptional regulator |
As a means of assessing the relative abundances of the different transcripts derived from the atcRS operon and its surrounding genes and to confirm the operonic structure of these transcriptional units, Northern hybridization analyses were performed. RNA was harvested from stationary-phase spirochetes, separated by formaldehyde-agarose gel electrophoresis, and probed with radiolabeled PCR products targeting tde0029 or tde0031 and an oligonucleotide targeting atcS. A probe to tde0029 was used as a negative control, as this ORF was not found to be expressed by T. denticola under any of the conditions tested as inferred from RT-PCR analyses. As predicted, the tde0029 probe did not hybridize. Probes targeting the atcRS operon hybridized with a 1.5-kb transcript, which corresponds with the predicted size of a transcript consisting of atcR and atcS. The probe to tde0031 bound to a transcript that is consistent with the size of tde0031 and tde0030 together (1 kb). The fact that these probes did not detect larger transcripts suggests that the colinkage of some genes inferred by RT-PCR may indicate that these larger transcripts are minor species expressed at low levels. Focusing on the atcR and atcS genes, the primary transcriptional unit for these genes appears to be a two-gene operon.
To identify the TSS for the genes and operons in the atcRS gene cluster (tde0031 through tde0037) and to further localize the transcriptional control elements, 5′ RACE analyses were performed. The amplicons obtained from these analyses were TA cloned and sequenced. Three TSS were analyzed (indicated in Fig. 3A). One TSS mapped just upstream of atcR and presumably serves as the primary TSS for the atcRS operon. A second TSS mapped upstream of tde0031 and thus appears to be the initiation site for the tde0031-tde0030 operon. The final TSS, which localized upstream of tde0037, serves as the TSS for the tde0037-tde0034 operon. Analysis of the sequence upstream from each of these TSS (conducted using the virtual footprint algorithm at www.prodoric.de) revealed the existence of conserved δ70-like −10 and −35 sites (Fig. 4). Virtual footprint analysis also predicts an AbrB family transcription factor binding site that overlaps the −35 site in the promoter region of tde0037. Since tde0037 encodes an AbrB homolog, the presence of this binding site just upstream of the coding sequence suggests that AbrB may autoregulate, consistent with what is seen for Bacillus (46).
FIG. 4.
Analysis of the upstream promoter regions. The nucleotide sequences of the upstream regions of tde0037, atcR, and tde0031 are shown. The start of transcription is indicated by an arrow, with +1 denoting the first base of the transcript. The −35 and −10 sites, putative AbrB binding site, and ribosome binding site (RBS) are shown. The start codon is indicated by the star.
Analysis of AtcR and AtcS expression levels in log- and stationary-phase spirochetes.
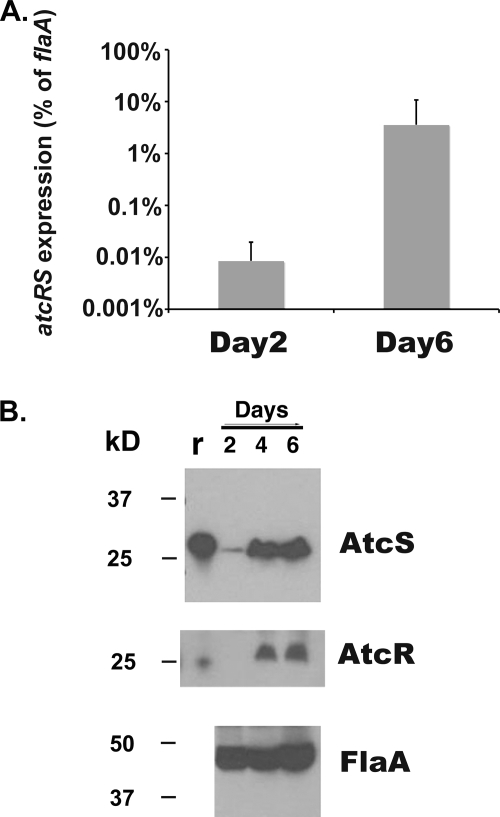
The data above demonstrate that both the AbrB homolog (encoded by tde0037) and AtcR-AtcS are expressed during in vitro cultivation. In other bacteria, AbrB transcription is influenced by growth phase (39, 47). To determine if the expression of atcRS is similarly influenced by growth phase, RNA was extracted from spirochetes after 2 days (middle exponential phase) or 6 days (early stationary phase) of growth and the relative amount of the atcRS transcript was quantified by real-time RT-PCR. The flaA gene served as the control for a constitutively expressed gene. The expression levels of flaA have been demonstrated to be constitutive in all spirochetes analyzed (7, 31, 32, 36). The expression level of atcRS was low at day 2 (relative to that of flaA) (Fig. 5A). However, at day 6 a 500-fold induction in atcRS transcript levels was observed, indicating that expression of this operon is responsive to growth phase (Fig. 5A). To determine if the increase in mRNA levels seen during later stages of growth correlates with increased protein production, AtcS and AtcR protein levels were assessed in cells collected at days 2, 4, and 6. Immunoblots were generated and screened with antisera to AtcR and AtcS. Detection of the constitutively produced FlaA protein served as a loading control. Antisera raised against each protein detected a single band of the appropriate size in cells grown for 4 and 6 days, with little or no protein detected at day 2 (Fig. 5B). The protein levels observed for AtcS and AtcR are consistent with those inferred from the real-time RT-PCR analyses, verifying that the TCS is indeed regulated by growth phase.
FIG. 5.
Growth-phase-dependent expression of atcS and atcR. Expression of atcS and atcR was analyzed by quantitative real-time PCR (A) or Western blotting (B). Transcriptional levels of atcS and atcR during early- and late-stage growth (2 and 6 days, respectively) were determined and are expressed as percentages of flaA transcript levels. The data presented represent the averages of two independent experiments each done in triplicate. Error bars represent standard deviations. Western blotting was done as described in Materials and Methods. Lane r shows purified recombinant protein. Equivalent protein loading for each lane was confirmed by screening a blot with anti-FlaA antiserum.
Demonstration of the phosphotransfer capabilities of the AtcRS TCS.
To determine if AtcS undergoes autophosphorylation and can then transfer phosphate to AtcR, in vitro phosphorylation assays were conducted. Purified recombinant AtcS was incubated with [γ-32P]ATP as described in Materials and Methods (Fig. 6A). A purified r-His-tagged T. denticola protein (FhbB) that binds complement regulatory proteins and AtcR served as the negative controls (18). Specific and time-dependent incorporation of radiolabeled phosphate into the AtcS band was observed. There was no incorporation of label into the negative control proteins. These data demonstrate that AtcS undergoes autophosphorylation.
FIG. 6.
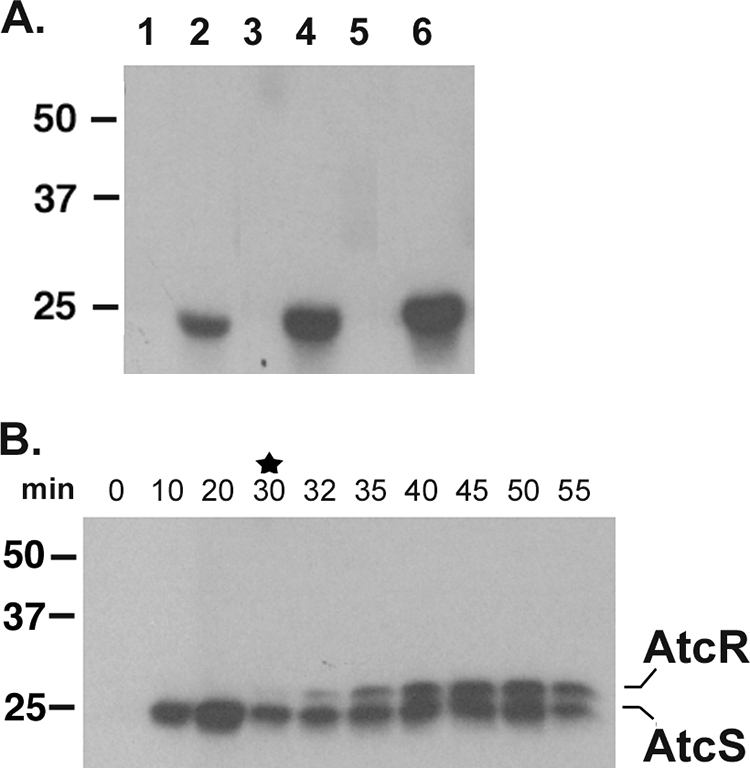
Autophosphorylation and phosphotransfer activities of AtcS. The ability of AtcS to autophosphorylate (A) was assessed as described in the text. AtcS (lanes 2, 4, and 6) and the negative control factor H binding protein FhbB (lanes 1, 3, and 5) were incubated with [γ-32P]ATP. Aliquots of each reaction mixture were removed at 10 (lanes 1 and 2), 20 (lanes 3 and 4), or 30 (lanes 5 and 6) min; fractionated by SDS-PAGE; transferred to membranes; and subjected to autoradiography (A). In panel B the results of phosphotransfer analyses are presented. AtcS was preloaded with phosphate by a 30-min incubation with [γ-32P]ATP, and then AtcR was added (denoted by the star). Aliquots were taken at the time points indicated. Phosphate incorporation was assessed by SDS-PAGE and autoradiography.
Next, the ability of AtcS to transfer phosphate to AtcR was assessed. First, AtcS was allowed to autophosphorylate as described above. Then purified AtcR was added with a 1:3 stoichiometry of AtcS to AtcR. Aliquots of the reaction mixture were removed over time and analyzed by SDS-PAGE and autoradiography. Rapid transfer of phosphate from AtcS to AtcR was observed (Fig. 6B). By 10 min after addition of AtcR, no further accumulation of phosphate on AtcR was detected. Importantly, phosphate labeling of AtcR occurred only in the presence of phosphorylated AtcS indicating the specificity of the labeling and phosphotransfer reaction (data not shown). It is noteworthy that phosphotransfer occurred only if AtcS was preloaded with phosphate. When AtcR and AtcS were combined prior to the addition of [γ-32P]ATP, no labeling of AtcR was observed even after an hour of coincubation (data not shown). These results are discussed in detail below.
DISCUSSION
T. denticola persists in a highly competitive environment in terms of both microbial diversity and nutrient fluctuation. As such, it must be able to rapidly adapt to changing environmental conditions. Little is know about the global regulatory networks of this important human pathogen. Here we present the first analysis of a TCS in any oral spirochete. The focus of this study is the putative TCS encoded by ORFs tde0033 and tde0032, whose products we have designated AtcR (response regulator) and AtcS (sensor kinase), respectively. The genes encoding these proteins were found to be conserved and universal among T. denticola isolates, suggesting an important role in T. denticola biology.
As is typical for kinase-response regulator pairs, atcR and atcS were determined to be cotranscribed. The orientation and intergenic spacer lengths of genes upstream and downstream of atcRS raised the possibility that additional genes may be cotranscribed with atcRS or separately as polycistronic mRNAs. RT-PCR analyses provided suggestive evidence that ORF tde0031 is cotranscribed with atcRS. However, mRNA of a size that would correspond to this larger polycistronic mRNA was not detected by Northern blotting in in vitro-cultivated spirochetes. Hence, during in vitro cultivation the dominant transcriptional unit for atcRS is as a bicistronic mRNA. Surrounding ORFs also form operons. The downstream ORFs tde0031 and tde0030 were cotranscribed, and the four immediately upstream genes (ORFs tde0034 through tde0037) also formed a transcriptional unit. ORF tde0037 (designated abrB) encodes a putative stationary-phase transcription factor of the AbrB family (39, 47). It is part of a three-member gene family in T. denticola 35405. Based on the proximity of the atcRS operon to abrB, we hypothesized that atcRS expression might be responsive to growth state or cell density. Consistent with this, a nearly 500-fold increase in the atcRS transcript level was observed at day 6 of in vitro growth compared with the levels seen during early log phase (day 2). Immunoblot analyses confirmed that the increase in mRNA level correlates with a significant increase in AtcR and AtcS protein levels. The specific signals that initiate the upregulation of this TCS and the downstream effects of its upregulation are not yet known.
To further understand the molecular basis of the transcriptional regulation of the atcRS operon and its flanking genes, TSS analyses were performed. TSS were identified upstream of tde0037 (abrB), upstream of tde0031 (within the intergenic spacer), and upstream of atcR (within the 3′ end of the tde0034 coding sequence). The predicted −10 and −35 regions associated with each TSS are consistent with δ70 binding sites (based on the virtual footprinting algorithm). A consensus AbrB binding site that may allow for autoregulation through AbrB is present within the −35 promoter region of the tde0037-tde0034 operon (46).
The AtcRS TCS has significant homology with the VirRS TCS of Clostridium perfringens (2, 22, 23, 30) and the C4-dicarboxylate TCS of Bacillus (51). AtcR displays ∼60% amino acid similarity with VirR, while AtcS is ∼43 to 65% similar to the CitA domains of the C4-dicarboxylate sensors. The homology between AtcS and CitA domain proteins resides primarily in the H box of the kinase domain, a domain that is involved in the interaction of the sensor kinase with the response regulator (29, 50). Hence, the homology between AtcS and dicarboxylate sensors may be attributed more to selective structural pressures and may not necessarily reflect a role of AtcS (and hence the AtcRS system) in sensing dicarboxylates. It is noteworthy that AtcR contains the unique LytTR-type DNA binding domain. LytTR domains have been identified in VirS and other transcriptional regulators of the AlgR/AgrA/LytR family (26). While LytTR domains are abundant in oral bacteria, including the streptococci and Prevotella (www.tigr.org), T. denticola is the only spirochete that has been shown to possess this domain. Structural predictions of LytTR domains indicate that they are distinct from the helix-turn-helix and winged helix-turn-helix DNA binding domains of most transcriptional regulators. LytTR domains appear to bind to a specific DNA sequence pattern in the upstream regions of target genes. Transcriptional regulators that possess the LytTR domain have been demonstrated to influence the biosynthesis of extracellular polysaccharides (15), fimbriation, expression of exoproteins, quorum sensing (26), and production of virulence factors (8, 30). A search of the T. denticola genome sequence revealed the presence of nearly 50 potential LytTR domain interaction sites (unpublished data). However, considerable variation was observed in these putative sites. This is not surprising in light of the deep evolutionary branching of the spirochetes. Hence, it is premature to assume that these sites define the basis of transcriptional regulation by ActRS. Nonetheless, the presence of these related sequence motifs supports the hypothesis that the AtcRS TCS may be an important global regulator in T. denticola.
To further assess the potential ability of AtcR and AtcS to function as a TCS, r-proteins were generated and tested for their ability to autophosphorylate and or undergo phosphotransfer. Autophosphorylation of AtcS was demonstrated, and the phosphorylated protein was able to transfer phosphate to AtcR. The general paradigm for phosphotransfer is that the functional kinase exists as a dimer and cross-phosphorylation of the individual histidines by the activated kinase domains allows for a conformational change in the H box. The phosphorylated dimer forms a stable interaction surface whereby the response regulator is able to interact and phosphotransfer occurs (21, 29, 43, 50). No well-defined dimerization domain is evident in AtcS. However, we noted that there is an alpha helix with high coiled-coil formation probability located in the N-terminal domain of the protein (16). In spirochetes, coiled-coil domains have been demonstrated to be involved in the protein-protein interactions (11, 12, 19, 20, 37). In other bacteria, coiled coils have been shown to be central to the dimerization of several microbial proteins, including some transcriptional regulators (1, 3, 52). As detailed above, we found that phosphotransfer from AtcS to AtcR requires that AtcS first be preloaded with phosphate. A possible basis for this observation is that the rate of phosphotransfer is low and a threshold of phosphorylated dimers must be reached before phosphotransfer can occur. Upon addition of AtcR to the preloaded AtcS, phosphate appears to be quickly transferred, but then a state is reached where no further phosphotransfer occurs. This could be due to the concentration of phosphorylated dimers of AtcS falling below the essential threshold that is required for phosphotransfer.
In conclusion, AtcRS is the first functional TCS characterized in an oral spirochete. TCS most certainly play a key regulatory role in the ability of T. denticola to survive within a complex polymicrobial community and adapt to ever-changing environmental variables. With the demonstration of a functional TCS in T. denticola, it will now be possible to begin to dissect the global regulatory networks of this important disease-causing agent of humans.
Acknowledgments
This work was supported in part by a grant from NIDCR, NIH. J.R.F. was supported in part by a training grant from NIAID, NIH.
We thank E. Peter Greenberg for providing a number of the T. denticola strains.
Footnotes
Published ahead of print on 11 July 2008.
REFERENCES
- 1.Barbara, K. E., K. A. Willis, T. M. Haley, S. J. Deminoff, and G. M. Santangelo. 2007. Coiled coil structures and transcription: an analysis of the S. cerevisiae coilome. Mol. Genet. Genomics 278135-147. [DOI] [PubMed] [Google Scholar]
- 2.Ba-Thein, W., M. Lyristis, K. Ohtani, I. T. Nisbet, H. Hayashi, J. I. Rood, and T. Shimizu. 1996. The virR/virS locus regulates the transcription of genes encoding extracellular toxin production in Clostridium perfringens. J. Bacteriol. 1782514-2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belogurov, G. A., M. N. Vassylyeva, V. Svetlov, S. Klyuyev, N. V. Grishin, D. G. Vassylyev, and I. Artsimovitch. 2007. Structural basis for converting a general transcription factor into an operon-specific virulence regulator. Mol. Cell 26117-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bilwes, A. M., C. M. Quezada, L. R. Croal, B. R. Crane, and M. I. Simon. 2001. Nucleotide binding by the histidine kinase CheA. Nat. Struct. Biol. 8353-360. [DOI] [PubMed] [Google Scholar]
- 5.Bystroff, C., and Y. Shao. 2002. Fully automated ab initio protein structure prediction using I-SITES, HMMSTR and ROSETTA. Bioinformatics 18(Suppl. 1)S54-S61. [DOI] [PubMed] [Google Scholar]
- 6.Bystroff, C., V. Thorsson, and D. Baker. 2000. HMMSTR: a hidden Markov model for local sequence-structure correlations in proteins. J. Mol. Biol. 301173-190. [DOI] [PubMed] [Google Scholar]
- 7.Caimano, M. J., R. Iyer, C. H. Eggers, C. Gonzalez, E. A. Morton, M. A. Gilbert, I. Schwartz, and J. D. Radolf. 2007. Analysis of the RpoS regulon in Borrelia burgdorferi in response to mammalian host signals provides insight into RpoS function during the enzootic cycle. Mol. Microbiol. 651193-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carter, G. P., D. Lyras, D. L. Allen, K. E. Mackin, P. M. Howarth, J. R. O'Connor, and J. I. Rood. 2007. Binary toxin production in Clostridium difficile is regulated by CdtR, a LytTR family response regulator. J. Bacteriol. 1897290-7301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Galperin, M. Y., A. N. Nikolskaya, and E. V. Koonin. 2001. Novel domains of the prokaryotic two-component signal transduction systems. FEMS Microbiol. Lett. 20311-21. [DOI] [PubMed] [Google Scholar]
- 10.Hamon, M. A., N. R. Stanley, R. A. Britton, A. D. Grossman, and B. A. Lazazzera. 2004. Identification of AbrB-regulated genes involved in biofilm formation by Bacillus subtilis. Mol. Microbiol. 52847-860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hovis, K. M., J. C. Freedman, H. Zhang, J. L. Forbes, and R. T. Marconi. 2008. Identification of an antiparallel coiled-coil/loop domain required for ligand binding by the Borrelia hermsii FhbA protein: additional evidence for the role of FhbA in the host-pathogen interaction. Infect. Immun. 762113-2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hovis, K. M., J. P. Jones, T. Sadlon, G. Raval, D. L. Gordon, and R. T. Marconi. 2006. Molecular analyses of the interaction of Borrelia hermsii FhbA with the complement regulatory proteins factor H and factor H-like protein 1. Infect. Immun. 742007-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee, J., J. T. Owens, I. Hwang, C. Meares, and S. Kustu. 2000. Phosphorylation-induced signal propagation in the response regulator NtrC. J. Bacteriol. 1825188-5195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li, Y. H., P. C. Lau, N. Tang, G. Svensater, R. P. Ellen, and D. G. Cvitkovitch. 2002. Novel two-component regulatory system involved in biofilm formation and acid resistance in Streptococcus mutans. J. Bacteriol. 1846333-6342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lizewski, S. E., J. R. Schurr, D. W. Jackson, A. Frisk, A. J. Carterson, and M. J. Schurr. 2004. Identification of AlgR-regulated genes in Pseudomonas aeruginosa by use of microarray analysis. J. Bacteriol. 1865672-5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lupas, A., M. Van Dyke, and J. Stock. 1991. Predicting coiled coils from protein sequences. Science 2521162-1164. [DOI] [PubMed] [Google Scholar]
- 17.Marconi, R. T., D. S. Samuels, and C. F. Garon. 1993. Transcriptional analyses and mapping of the ospC gene in Lyme disease spirochetes. J. Bacteriol. 175926-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McDowell, J. V., J. Frederick, L. Stamm, and R. T. Marconi. 2007. Identification of the gene encoding the FhbB protein of Treponema denticola, a highly unique factor H-like protein 1 binding protein. Infect. Immun. 751050-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McDowell, J. V., M. E. Harlin, E. A. Rogers, and R. T. Marconi. 2005. Putative coiled-coil structural elements of the BBA68 protein of the Lyme disease spirochetes are required for formation of its factor H binding site. J. Bacteriol. 1871317-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McDowell, J. V., J. Wolfgang, L. Senty, C. M. Sundy, M. J. Noto, and R. T. Marconi. 2004. Demonstration of the involvement of outer surface protein E coiled-coil structural domains and higher order structural elements in the binding of infection-induced antibody and the complement-regulatory protein, factor H. J. Immunol. 1737471-7480. [DOI] [PubMed] [Google Scholar]
- 21.McEvoy, M. M., A. C. Hausrath, G. B. Randolph, S. J. Remington, and F. W. Dahlquist. 1998. Two binding modes reveal flexibility in kinase/response regulator interactions in the bacterial chemotaxis pathway. Proc. Natl. Acad. Sci. USA 957333-7338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McGowan, S., I. S. Lucet, J. K. Cheung, M. M. Awad, J. C. Whisstock, and J. I. Rood. 2002. The FxRxHrS motif: a conserved region essential for DNA binding of the VirR response regulator from Clostridium perfringens. J. Mol. Biol. 322997-1011. [DOI] [PubMed] [Google Scholar]
- 23.McGowan, S., J. R. O'Connor, J. K. Cheung, and J. I. Rood. 2003. The SKHR motif is required for biological function of the VirR response regulator from Clostridium perfringens. J. Bacteriol. 1856205-6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mole, B. M., D. A. Baltrus, J. L. Dangl, and S. R. Grant. 2007. Global virulence regulation networks in phytopathogenic bacteria. Trends Microbiol. 15363-371. [DOI] [PubMed] [Google Scholar]
- 25.Narikiyo, M., C. Tanabe, Y. Yamada, H. Igaki, Y. Tachimori, H. Kato, M. Muto, Montensano, H. Sakamoto, Y. Nakajima, and H. Sasaki. 2004. Frequent and preferential infection of Treponema denticola, Streptococcus mitis, and Streptococcus anginosus in esophageal cancers. Cancer Sci. 95569-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nikolskaya, A. N., and M. Y. Galperin. 2002. A novel type of conserved DNA-binding domain in the transcriptional regulators of the AlgR/AgrA/LytR family. Nucleic Acids Res. 302453-2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nishino, K., and A. Yamaguchi. 2002. EvgA of the two-component signal transduction system modulates production of the YhiUV multidrug transporter in Escherichia coli. J. Bacteriol. 1842319-2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Offenbacher, S., H. L. Jared, P. G. O'Reilly, S. R. Wells, G. E. Salvi, H. P. Lawrence, S. S. Socransky, and J. D. Beck. 1998. Potential pathogenic mechanisms of periodontitis associated pregnancy complications. Ann. Periodontol. 3233-250. [DOI] [PubMed] [Google Scholar]
- 29.Ohta, N., and A. Newton. 2003. The core dimerization domains of histidine kinases contain recognition specificity for the cognate response regulator. J. Bacteriol. 1854424-4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ohtani, K., H. I. Kawsar, K. Okumura, H. Hayashi, and T. Shimizu. 2003. The VirR/VirS regulatory cascade affects transcription of plasmid-encoded putative virulence genes in Clostridium perfringens strain 13. FEMS Microbiol. Lett. 222137-141. [DOI] [PubMed] [Google Scholar]
- 31.Ojaimi, C., C. Brooks, D. Akins, S. Casjens, P. Rosa, A. Elias, A. Barbour, A. Jasinskas, J. L. Benach, L. Katona, J. D. Radolf, M. Caimano, J. Skare, K. Swingle, S. Sims, and I. Schwartz. 2002. Borrelia burgdorferi gene expression profiling with membrane-based arrays. Methods Enzymol. 358165-177. [DOI] [PubMed] [Google Scholar]
- 32.Ojaimi, C., V. Mulay, D. Liveris, R. Iyer, and I. Schwartz. 2005. Comparative transcriptional profiling of Borrelia burgdorferi clinical isolates differing in capacities for hematogenous dissemination. Infect. Immun. 736791-6802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parkinson, J. S., and E. C. Kofoid. 1992. Communication modules in bacterial signaling proteins. Annu. Rev. Genet. 2671-112. [DOI] [PubMed] [Google Scholar]
- 34.Paterson, G. K., C. E. Blue, and T. J. Mitchell. 2006. Role of two-component systems in the virulence of Streptococcus pneumoniae. J. Med. Microbiol. 55355-363. [DOI] [PubMed] [Google Scholar]
- 35.Qian, Q., C. Y. Lee, J. D. Helmann, and M. A. Strauch. 2002. AbrB is a regulator of the sigma(W) regulon in Bacillus subtilis. FEMS Microbiol. Lett. 211219-223. [DOI] [PubMed] [Google Scholar]
- 36.Revel, A. T., A. M. Talaat, and M. V. Norgard. 2002. DNA microarray analysis of differential gene expression in Borrelia burgdorferi, the Lyme disease spirochete. Proc. Natl. Acad. Sci. USA 991562-1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rogers, E. A., and R. T. Marconi. 2007. Delineation of species-specific binding properties of the CspZ protein (BBH06) of Lyme disease spirochetes: evidence for new contributions to the pathogenesis of Borrelia spp. Infect. Immun. 755272-5281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sanders, D. A., B. L. Gillece-Castro, A. M. Stock, A. L. Burlingame, and D. E. Koshland, Jr. 1989. Identification of the site of phosphorylation of the chemotaxis response regulator protein, CheY. J. Biol. Chem. 26421770-21778. [PubMed] [Google Scholar]
- 39.Scotcher, M. C., F. B. Rudolph, and G. N. Bennett. 2005. Expression of abrB310 and SinR, and effects of decreased abrB310 expression on the transition from acidogenesis to solventogenesis, in Clostridium acetobutylicum ATCC 824. Appl. Environ. Microbiol. 711987-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seshadri, R., G. S. A. Myers, H. Tettelin, J. A. Eisen, J. F. Heidelberg, R. J. Dodson, T. M. Davidsen, R. T. DeBoy, D. E. Fouts, D. H. Haft, J. Selengut, Q. Ren, L. M. Brinkac, R. Madupu, J. Kolonay, S. A. Durkin, S. C. Daugherty, J. Shetty, A. Shvartsbeyn, E. Gebregeorgis, K. Geer, G. Tsegaye, J. Malek, B. Ayodeji, S. Shatsman, M. P. McLeod, D. Smajs, J. K. Howell, S. Pal, A. Amin, P. Vashisth, T. Z. McNeill, Q. Xiang, E. Sodergren, E. Baca, G. M. Weinstock, S. J. Norris, C. M. Fraser, and I. T. Paulsen. 2004. Comparison of the genome of the oral pathogen Treponema denticola with other spirochete genomes. Proc. Natl. Acad. Sci. USA 1015646-5651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shafikhani, S. H., and T. Leighton. 2004. AbrB and Spo0E control the proper timing of sporulation in Bacillus subtilis. Curr. Microbiol. 48262-269. [DOI] [PubMed] [Google Scholar]
- 42.Socransky, S., A. Haffajee, M. Cugini, C. Smith, and R. J. Kent. 1998. Microbial complexes in subgingival plaque. J. Clin. Periodontol. 25134-144. [DOI] [PubMed] [Google Scholar]
- 43.Stock, A. M., V. L. Robinson, and P. N. Goudreau. 2000. Two-component signal transduction. Annu. Rev. Biochem. 69183-215. [DOI] [PubMed] [Google Scholar]
- 44.Strauch, M. A. 1995. Delineation of AbrB-binding sites on the Bacillus subtilis spo0H, kinB, ftsAZ, and pbpE promoters and use of a derived homology to identify a previously unsuspected binding site in the bsuB1 methylase promoter. J. Bacteriol. 1776999-7002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Strauch, M. A., B. G. Bobay, J. Cavanagh, F. Yao, A. Wilson, and Y. Le Breton. 2007. Abh and AbrB control of Bacillus subtilis antimicrobial gene expression. J. Bacteriol. 1897720-7732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Strauch, M. A., M. Perego, D. Burbulys, and J. A. Hoch. 1989. The transition state transcription regulator AbrB of Bacillus subtilis is autoregulated during vegetative growth. Mol. Microbiol. 31203-1209. [DOI] [PubMed] [Google Scholar]
- 47.Strauch, M. A., G. B. Spiegelman, M. Perego, W. C. Johnson, D. Burbulys, and J. A. Hoch. 1989. The transition state transcription regulator abrB of Bacillus subtilis is a DNA binding protein. EMBO J. 81615-1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tzeng, Y. L., X. Zhou, S. Bao, S. Zhao, C. Noble, and D. S. Stephens. 2006. Autoregulation of the MisR/MisS two-component signal transduction system in Neisseria meningitidis. J. Bacteriol. 1885055-5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wolanin, P. M., P. A. Thomason, and J. B. Stock. 2002. Histidine protein kinases: key signal transducers outside the animal kingdom. Genome Biol. 3S3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wright, J. S., III, and R. J. Kadner. 2001. The phosphoryl transfer domain of UhpB interacts with the response regulator UhpA. J. Bacteriol. 1833149-3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yamamoto, H., M. Murata, and J. Sekiguchi. 2000. The CitST two-component system regulates the expression of the Mg-citrate transporter in Bacillus subtilis. Mol. Microbiol. 37898-912. [DOI] [PubMed] [Google Scholar]
- 52.Yang, C. K., J. H. Kim, and M. R. Stallcup. 2006. Role of the N-terminal activation domain of the coiled-coil coactivator in mediating transcriptional activation by beta-catenin. Mol. Endocrinol. 203251-3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang, H., A. Raji, M. Theisen, P. R. Hansen, and R. T. Marconi. 2005. bdrF2 of the Lyme disease spirochetes is coexpressed with a series of cytoplasmic proteins and is produced specifically during early infection. J. Bacteriol. 187175-184. [DOI] [PMC free article] [PubMed] [Google Scholar]