Abstract
In Listeria monocytogenes, the alternative sigma factor σB plays important roles in stress tolerance and virulence. Here, we present the identification of SbrA, a novel small noncoding RNA that is produced in a σB-dependent manner. This finding adds the σB regulon to the growing list of stress-induced regulatory circuits that include small noncoding RNAs.
The gram-positive bacterium Listeria monocytogenes is an important food-borne pathogen associated with life-threatening invasive infections in humans and animals (33). In order to establish an infection, L. monocytogenes must be able to grow and survive under a variety of stress conditions, including those encountered during food processing, in the gastrointestinal tract of the host, and, at the later stages of infection, within the intracellular environments of host cells.
The alternative sigma factor σB plays an important role in general stress response and virulence in L. monocytogenes. Mutant strains lacking functional σB are more sensitive toward a variety of stress conditions, including low pH and oxidative stress (9, 36). Furthermore, σB contributes to the ability of L. monocytogenes to invade nonphagocytic human cells and plays a critical role during the gastrointestinal stage of listeriosis in guinea pigs (10, 19). Recently, the σB regulon in L. monocytogenes has been studied by genomic strategies (1, 13, 29). Whole-genome microarray and proteome analyses of the σB regulon in L. monocytogenes strain 10403S and strain EGD-e revealed more than 100 genes to be positively controlled by σB, including genes encoding proteins with established roles in stress resistance (e.g., clpC, gadA, and ctc) and virulence (e.g., inlAB, inlD, bsh, and prfA). In addition, a large number of genes were found to be negatively influenced by σB, in particular genes encoding flagellar and ribosomal proteins. Intriguingly, many of the σB-dependent effects observed in these studies appear to be indirect, i.e., through secondary gene regulatory systems, such as the stress-related transcriptional regulators HrcA and CtsR, the key virulence regulator PrfA, and the RNA chaperone Hfq (15, 16).
In recent years, genome-wide experiment- and computer-based screens have led to the discovery of hundreds of small RNA molecules (sRNAs) in various bacterial species (6, 20, 21, 22, 27, 34, 37). Intriguingly, many of these sRNAs are induced only under specific growth conditions, such as in the stationary growth phase. In several cases, the regulatory mechanisms underlying this upregulation involve the action of alternative stress sigma factors (18, 20, 25, 31). The majority of the sRNAs studied thus far regulate gene expression by pairing with target mRNAs to change their translation and/or stability, whereas other sRNAs bind to and alter the activity of proteins (12). The RNA-binding protein Hfq is critical for the regulatory functions of multiple sRNAs. Hfq acts as an RNA chaperone to promote sRNA-mRNA duplex formation (2, 32). Furthermore, binding of Hfq increases the stability of several sRNAs, most likely by protecting them from degradation by RNases. In L. monocytogenes, Hfq plays a role in stress tolerance and contributes to pathogenesis in mice (7). We recently identified a number of sRNAs that interact with Hfq in L. monocytogenes, suggesting that Hfq-binding sRNAs are implicated in stress tolerance and virulence in this organism (6).
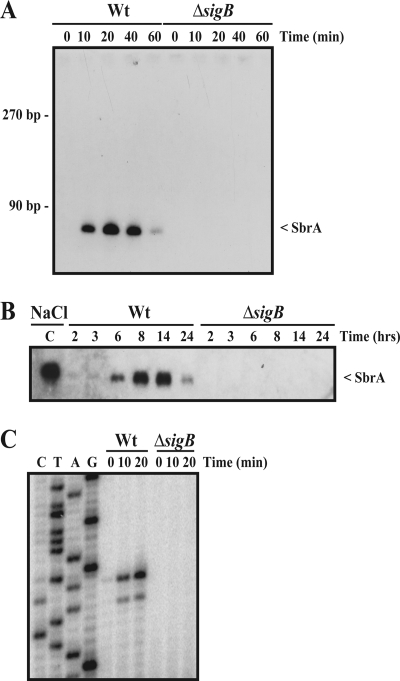
In several cases, it has been shown that the synthesis of sRNAs is tightly regulated at the level of transcription by known regulatory proteins, including alternative sigma factors, as exemplified by σE in Escherichia coli and Salmonella spp. (18, 25), and σ54 in E. coli and Vibrio cholerae (20, 31). These findings prompted us to search for σB-dependent sRNAs in L. monocytogenes. To identify σB-dependent sRNAs in L. monocytogenes EGD-e, we used a straightforward bioinformatics approach. Since bacterial sRNAs are often encoded by intergenic regions (IGRs), we searched for putative σB binding sites (GTTT-N14 to 17-GGKWWWW) and rho-independent transcriptional terminator structures within the IGRs of the L. monocytogenes EGD-e genome using the ListiList web page (http://genolist.pasteur.fr/ListiList/). Here, we defined an intrinsic terminator as an inverted repeat with a ΔG no higher than −10 kcal/mol, followed by a stretch of at least five U's. Putative σB binding sites located within 150 bases upstream from and in the same orientation as an open reading frame were discarded. Our initial screen resulted in the identification of four putative sRNA loci, named sbrA, sbrB, sbrC, and sbrD for sigmaB-dependent small RNA A to D (Table 1). These four loci all contain sequences with a high level of homology to the σB consensus sequence followed by putative rho-independent transcription terminator sequences. To examine whether σB-dependent sRNAs were expressed from these four loci, we performed Northern blotting using RNA sampled from wild-type (wt) and ΔsigB mutant strains subjected to osmotic stress (Fig. 1A). In these experiments, we observed that SbrA was induced in the wt strain in a transient fashion, reaching a maximum level around 20 min after the addition of NaCl. Importantly, SbrA was not observed in the ΔsigB strain. No σB-dependent expression of SbrB, SbrC, and SbrD could be observed under these experimental conditions (data not shown).
TABLE 1.
Genetic organization of candidate σB-regulated sRNAs in L. monocytogenes EGD-e
| Gene | Flanking genesa | Length | σB promoterb |
|---|---|---|---|
| sbrA | lmo1374 > > > lmo1375 | 70 | TAAAAATGTTTTAATCTAGGTTTAGCGGGTATTGTTTAGT |
| sbrB | lmo1251 > < < lmo1252 | 168 | TTATTTGGTTTTACTTGTTGTTGGCTTGGTTTTTGTTTTTC |
| sbrC | lmo1515 > < > lmo1516 | 73 | ATATAGCGTTTTCAATTGTTTGCTTTTGGTATTTATTTCAG |
| sbrD | atpI < < < lmo1537 | 68 | TCCGGCTGTTTTTTCCGTTCTTTTTCTTGGGAAAAGAACTTT |
The orientation of the proposed sRNA gene as well as upstream and downstream genes is indicated by arrows.
Proposed σB consensus −10 and −35 regions are shown in bold.
FIG. 1.
Identification of novel σB-regulated sRNAs. (A) Northern blot showing the expression of SbrA after NaCl stress. Overnight cultures of wild-type (Wt) and ΔsigB cells were diluted 100 times in brain heart infusion (BHI) medium and grown to early exponential phase, at which point 4% NaCl was added. The samples were drawn for RNA extraction at the indicated time points after the addition of NaCl. RNA purification and Northern blotting were performed as previously described (6) using a specific 32P-labeled RNA probe prepared by the in vitro transcription of a PCR product generated by using the primers sbrA-Fwd T7 and sbrA-Rev (the primers are listed in Table S1 in the supplemental material). (B) Northern blot showing the expression of SbrA during growth in BHI. Samples were drawn at the indicated time points. The time points of 2, 3, and 6 h correspond to the early to late exponential growth phases, whereas the 8-, 14-, and 24-h time points correspond to the early to late stationary growth phases. Lane C is a positive control corresponding to NaCl-induced SbrA. Wt, wild type. (C) Mapping of the 5′ transcriptional start site of SbrA by primer extension. Wild-type (Wt) and ΔsigB cells were grown in BHI to early exponential phase, at which point 4% NaCl was added. The samples were drawn at the indicated time points. Following RNA extraction, primer extension was performed as previously described (6) using the primer sbrA-1 (see Table S1 in the supplemental material).
In addition to osmotic stress conditions, the activity of σB in L. monocytogenes is known to increase when cells enter the stationary growth phase. By Northern blotting, we tested the expression of the four putative sRNAs in wt and ΔsigB strains during growth in rich medium. We found that SbrA was present in wt cells in a growth-phase-dependent manner, reaching a maximum level when the cells enter the stationary growth phase (Fig. 1B). Again, sRNAs corresponding to SbrB, SbrC, and SbrD were not observed under these growth conditions (data not shown). With respect to SbrB and SbrC, the lack of induction may be due to the presence of the T nucleotide instead of the G consensus nucleotide within the putative −10 region of the σB binding site (Table 1). The lack of σB-dependent expression of SbrD may be due to the rather long spacing (17 nucleotides) separating the consensus −10 and −35 sequence elements of the putative σB binding site (Table 1). To map the 5′ end of SbrA, we performed primer extension analysis with RNA samples from wt and ΔsigB strains subjected to osmotic stress (Fig. 1C). We found that the 5′ end of SbrA maps to the region located immediately downstream from the putative σB promoter element.
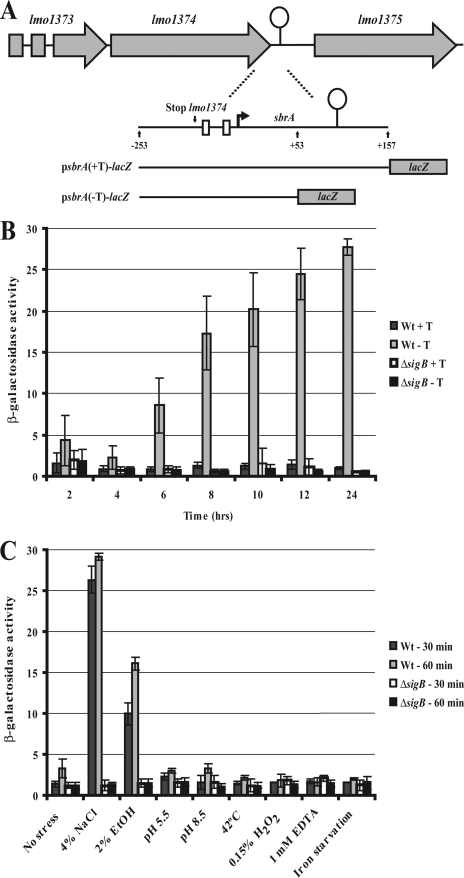
To verify the presence of a σB-dependent promoter upstream from sbrA, we constructed a transcriptional fusion of the sbrA promoter region to a promoter-less lacZ gene in the vector pTCV-lac (28). The resulting plasmid, named psbrA(-T)-lacZ (see Fig. 2A), was introduced into the wt and ΔsigB strains, and the level of β-galactosidase activity was determined under various growth conditions. During growth in rich medium, we observed an increase in sbrA(-T)-lacZ expression in the wt cells, reaching a maximum level in the stationary growth phase (Fig. 2B). In contrast, only negligible levels of β-galactosidase activity were recorded for the ΔsigB cells. Next, we examined the expression of sbrA(-T)-lacZ under various stress conditions (Fig. 2C). We observed that the level of β-galactosidase activity increased dramatically in the wt strain subjected to 4% NaCl or 2% ethyl alcohol, whereas the activity in ΔsigB cells remained constant. These results clearly indicate the presence of a σB-dependent promoter in the sbrA promoter region.
FIG. 2.
β-Galactosidase assay of transcriptional sbrA-lacZ fusions. (A) Description of transcriptional fusions of the sbrA promoter to lacZ. For the construction of psbrA(-T)-lac (containing the sbrA promoter-lacZ fusion without the intrinsic terminator [i.e., from −253 to +53 relative to the SbrA transcription start site]), primers sbrA-2 and sbrA-4 were used (see Table S1 in the supplemental material). For the construction of psbrA(+T)-lac (containing the sbrA promoter fusion, including the intrinsic terminator [i.e., from −253 to +157 relative to the SbrA transcription start site]), primers sbrA-2 and sbrA-3 were used (see Table S1 in the supplemental material). The PCR products were inserted into the vector pTCV-lac (26). (B) sbrA(+T)-lacZ and sbrA(-T)-lacZ expression in wild-type (Wt) and ΔsigB strains during growth in BHI medium. The samples were drawn at the indicated time points, and the amount of β-galactosidase activity was measured as previously described (7). (C) sbrA(-T)-lacZ expression in wild-type (Wt) and ΔsigB strains in response to various stress conditions. Cells were grown to an optical density at 600 nm of 0.3 and subjected to the indicated stress condition for 30 or 60 min. At these time points, 1-ml samples were drawn, and the amount of β-galactosidase activity was measured.
To investigate the function of the putative rho-independent transcriptional terminator located 42 to 70 nucleotides downstream from the sbrA transcription start site, we fused the entire sbrA gene, including the sbrA promoter region and the putative transcription terminator, to the lacZ gene in pTCV-lac, resulting in the plasmid construct psbrA(+T)-lacZ (Fig. 2A). The level of β-galactosidase activity in the wt cells and the ΔsigB cells harboring the sbrA(+T)-lacZ fusion plasmid remained negligible under all conditions tested, suggesting that the majority of the σB-dependent transcripts originating from the sbrA promoter region indeed ends at this transcription terminator structure (Fig. 2B).
Based on the findings described above, we infer that sbrA is a 70-nucleotide σB-dependent sRNA encoded by the IGR between lmo1374 and lmo1375. Furthermore, we note that no potential open reading frames are likely to be encoded by sbrA. We conducted a thorough BLAST and FASTA search for homologues of SbrA. We found that while SbrA is highly conserved in sequenced Listeria species (see Fig. S1 in the supplemental material), it was not identified in closely related bacterial species such as Bacillus subtilis and Staphylococcus aureus. The lmo1374 gene encodes a lipoamide acyltransferase and is part of the bkd operon which plays a crucial role in the ability of L. monocytogenes to modulate its membrane lipid composition in response to temperature and pH (11, 39). The −35 box of the σB-dependent promoter is located immediately downstream from the stop codon of lmo1374, suggesting that the SbrA transcript and the transcripts originating from the bkd operon all terminate at the same transcription terminator structure. The lmo1375 gene is located 265 bp downstream from the sbrA-bkd transcription terminator and encodes a putative aminotripeptidase.
To analyze the functional importance of SbrA, we constructed a ΔsbrA mutant strain using splicing by overlap extension PCR (14). The mutant strain lacks the −10 box of the σB-dependent promoter and 37 nucleotides at the 5′ end of the SbrA-encoding gene, thus conserving the sbrA-bkd transcription terminator. The growth or survival of wt and ΔsbrA strains was followed under a variety of conditions, including low-temperature, osmotic, acid, and alcohol stress. Under all conditions tested, no significant differences in growth or survival could be recorded for the wt and ΔsbrA strains (data not shown).
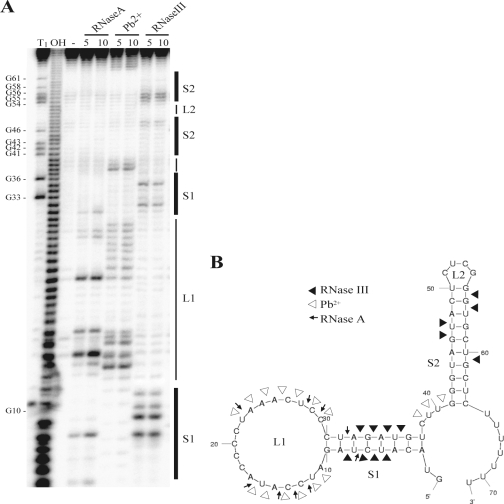
In order to predict a putative function of a sRNA, it may be helpful to know its secondary structure at the molecular level. The MFOLD program predicts a rather simple structure composed of a 5′ stem-loop and a 3′ transcription terminator stem-loop (Fig. 3B). We confirmed the structure predicted by MFOLD by probing in vitro-transcribed and 5′-end-labeled SbrA with lead(II) acetate, RNase A, and RNase III (Fig. 3A and B). Many of the sRNAs studied thus far act as antisense RNAs by base pairing with target mRNAs near the ribosome binding site and translation start site, resulting in the inhibition of translation and an increase in mRNA degradation (12). The nucleotides that participate in this antisense base pairing often lie in single-stranded regions of the sRNAs. We note that the loop of the 5′ stem-loop of SbrA (termed L1) contains 19 single-stranded nucleotides rich in CUs which could potentially base-pair with a variety of GA-rich ribosomal binding sites (Fig. 3B), a regulatory mechanism employed by several sRNAs, including Salmonella CyaR (26), S. aureus RNAIII (5), and E. coli OxyS (3).
FIG. 3.
Secondary structure of SbrA RNA. (A) The secondary structure of SbrA was mapped by RNase and lead(II) acetate cleavage. SbrA was prepared by the in vitro transcription of a PCR product generated by using the primers T7-sbrA-Fwd and sbrA-Rev-2 (see Table S1 in the supplemental material) using the MaxiScript in vitro transcription kit (Ambion). For structural probing, 50 fmol SbrA and 3 μg tRNA were incubated in structure buffer with or without (−) the specific ribonuclease RNase A (which cleaves the 3′ end of unpaired C and U residues) or RNase III (which is specific for double-stranded RNA) or with 5 mM lead(II) acetate (which cleaves single-stranded RNA) for 5 or 10 min. For a sequencing reaction of SbrA (lane T1), 50 fmol SbrA and 3 μg tRNA were incubated in sequencing buffer (Ambion) with RNase T1 (cleaves single-stranded RNA at G residues) for 5 min. An RNA ladder (lane OH) was prepared by boiling 100 fmol SbrA in hydroxide buffer (Ambion) for 5 min. After the indicated time, all enzyme reactions were stopped by the addition of 40 μl stop solution (Ambion). Lead(II) acetate cleavage reactions were stopped by the addition of EDTA to a final concentration of 10 mM. (B) Secondary structure of SbrA proposed by MFOLD (http://frontend.bioinfo.rpi.edu/applications/mfold/cgi-bin/rna-form1.cgi). The arrows indicate cleavage by ribonucleases and lead(II) acetate.
In an attempt to identify putative mRNAs targeted by SbrA, we used the program TargetRNA (http://snowwhite.wellesley.edu/targetRNA/), which identifies and scores target mRNAs based on the minimum free energy of the target interaction with an sRNA gene (30). We used a hybridization seed of at least seven consecutive base-pairing nucleotides and focused our search on genomic regions surrounding start codons. A number of the top candidates predicted by TargetRNA (kdpA, lmo2421, lmo0397, lmo0698, lmo1602, and lmo1292) were selected for further analysis. In order to test whether the translation of these mRNAs was affected by SbrA, in-frame translational fusions of the target genes to lacZ were produced in pCK-lac (see Fig. S2 and S3 in the supplemental material). Furthermore, we tested the mRNA levels of lmo0397, lmo0698, and lmo1602 by Northern blotting, semiquantitative reverse transcriptase PCR, and primer extension experiments, respectively, before and after the induction of SbrA production from an IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible promoter (see Fig. S4 in the supplemental material). Under all experimental conditions, none of the putative target genes predicted by TargetRNA appeared to be under the control of SbrA.
FIG. 4.
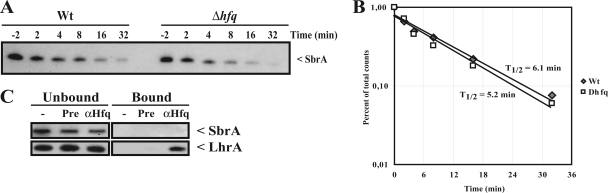
SbrA stability and interaction with Hfq. (A) Northern blot showing the stability of SbrA after the addition of rifampin. Wild-type (Wt) and Δhfq cells were grown to early exponential phase, and SbrA expression was induced by the addition of 4% NaCl. After 20 min, transcription was blocked by the addition of 15 μg/ml rifampin. (B) Quantification of the Northern blot in panel A. The approximate half-lives (T1/2) are indicated. (C) The in vivo interaction between SbrA and Hfq was tested by coimmunoprecipitation using α-Hfq antiserum. Coimmunoprecipitations and Northern blots of immunoprecipitated RNA was performed as described previously (6). Coimmunoprecipitations were performed using α-Hfq antiserum (αHfq). As negative controls, we used no serum (−) and pre-immune serum (Pre) in the coimmunoprecipitation experiments. In the left panels (Unbound), control RNA that did not bind to the Protein A-Sepharose-coupled antibodies was used for Northern blotting. In the right panels (Bound), the coimmunoprecipitated RNA was tested by Northern blotting. For the Northern blotting experiments, we used the SbrA-specific probe described in the legend to Fig. 1A. As a positive control, we used a probe directed against the Hfq-binding sRNA LhrA (6).
A large number of sRNAs depend on Hfq for function and/or stability. To test whether the stability of SbrA was affected by Hfq, we carried out stability assays of SbrA in wt and Δhfq strains (Fig. 4A). SbrA expression was first induced by an osmotic upshift. After 20 min, transcription was blocked by the addition of rifampin. In both strains, the half-life of SbrA was approximately 5 to 6 min, suggesting that SbrA does not depend on Hfq for stability (Fig. 4B). Even though SbrA does not depend on Hfq for stability, it still may interact with Hfq. To investigate whether SbrA interacts with Hfq in vivo, we conducted a coimmunoprecipitation experiment using Hfq antibodies (Fig. 4C). As a positive control, we included a known Hfq-binding sRNA, LhrA (6). In these experiments, only LhrA was retained by the Hfq-specific antibodies, suggesting that Hfq does not bind SbrA in vivo. Thus, we conclude that SbrA is an Hfq-independent sRNA.
Conclusion.
The sRNA-encoding gene sbrA is part of the σB regulon, and we find it likely that SbrA plays a role in the σB-dependent regulation of stress response, metabolism, and virulence. The lack of phenotypes for the ΔsbrA mutant strain indicates that SbrA, like many other sRNAs, may be involved in the fine-tuning of gene expression. Alternatively, a role for SbrA may be observed under very specific growth conditions only, which still remain to be defined. Regarding a mechanism of action for SbrA, our initial characterization showed that it does not interact with Hfq and most likely functions independently of this RNA chaperone. In E. coli, all sRNAs that control gene expression by base pairing with target mRNA interact with Hfq (12). In contrast to this, S. aureus RNAIII acts as an antisense RNA independently of Hfq (5, 17). Thus, although SbrA does not interact with Hfq, the possibility that SbrA could function by an antisense mechanism should not be excluded. However, an antisense-based mechanism is clearly not supported by our initial test of mRNAs predicted to be targeted by SbrA. As an alternative, SbrA may exert its function by interacting with target proteins, as exemplified by the sRNAs CsrB and CsrC (4) and 6S RNA (35). A key to the functional role and mechanism of action of SbrA may be found in the genomic studies of the σB regulon in L. monocytogenes (1, 13, 29). These studies revealed that more than 100 genes in L. monocytogenes are negatively influenced by σB. Furthermore, a large portion of the σB-activated genes are not preceded by a σB-dependent promoter. Thus, σB affects the expression of multiple genes in an indirect manner that could involve the action of regulatory sRNAs, such as SbrA.
Supplementary Material
Acknowledgments
This work was funded by the Danish Natural Science Research Council.
Footnotes
Published ahead of print on 11 July 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Abram, F., W.-L. Su, M. Wiedmann, K. J. Boor, P. Coote, C. Botting, K. A. G. Karatzas, and C. P. O'Byrne. 2008. Proteomic analyses of a Listeria monocytogenes mutant lacking σB identify new components of the σB regulon and highlight a role for σB in the utilization of glycerol. Appl. Environ. Microbiol. 74594-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aiba, H. 2007. Mechanism of RNA silencing by Hfq-binding small RNAs. Curr. Opin. Microbiol. 10134-139. [DOI] [PubMed] [Google Scholar]
- 3.Argaman, L., and S. Altuvia. 2000. flhA repression by OxyS RNA: kissing complex formation at two sites results in a stable antisense-target RNA complex. J. Mol. Biol. 3001101-1112. [DOI] [PubMed] [Google Scholar]
- 4.Babitzke, P., and T. Romeo. 2007. CsrB sRNA family: sequestration of RNA-binding regulatory proteins. Curr. Opin. Microbiol. 10156-163. [DOI] [PubMed] [Google Scholar]
- 5.Boisset, S., T. Geissmann, E. Huntzinger, P. Fechter, N. Bendridi, M. Possedko, C. Chevalier, A. C. Helfer, Y. Benito, A. Jacquier, C. Gaspin, F. Vandenesch, and P. Romby. 2007. Staphylococcus aureus RNAIII coordinately represses the synthesis of virulence factors and the transcription regulator Rot by an antisense mechanism. Genes Dev. 211353-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christiansen, J. K., J. S. Nielsen, T. Ebersbach, P. Valentin-Hansen, L. Sogaard-Andersen, and B. H. Kallipolitis. 2006. Identification of small Hfq-binding RNAs in Listeria monocytogenes. RNA 121383-1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christiansen, J. K., M. H. Larsen, H. Ingmer, L. Sogaard-Andersen, and B. H. Kallipolitis. 2004. The RNA-binding protein Hfq of Listeria monocytogenes: role in stress tolerance and virulence. J. Bacteriol. 1863355-3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reference deleted.
- 9.Ferreira, A., C. P. O'Byrne, and K. J. Boor. 2001. Role of σB in heat, ethanol, acid, and oxidative stress resistance and during carbon starvation in Listeria monocytogenes. Appl. Environ. Microbiol. 674454-4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garner, M. R., B. L. Njaa, M. Wiedmann, and K. J. Boor. 2006. Sigma B contributes to Listeria monocytogenes gastrointestinal infection but not to systemic spread in the guinea pig infection model. Infect. Immun. 74876-886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giotis, E. S., D. A. McDowell, I. S. Blair, and B. J. Wilkinson. 2007. Role of branched-chain fatty acids in pH stress tolerance in Listeria monocytogenes. Appl. Environ. Microbiol. 73997-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gottesman, S. 2004. The small RNA regulators of Escherichia coli: roles and mechanisms. Annu. Rev. Microbiol. 58303-328. [DOI] [PubMed] [Google Scholar]
- 13.Hain, T., H. Hossain, S. S. Chatterjee, S. Machata, U. Volk, S. Wagner, B. Brors, S. Haas, C. T. Kuenne, A. Billion, S. Otten, J. Pane-Farre, S. Engelmann, and T. Chakraborty. 2008. Temporal transcriptomic analysis of the Listeria monocytogenes EGD-e sigmaB regulon. BMC Microbiol. 820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horton, R. M. 1995. PCR-mediated recombination and mutagenesis. SOEing together tailor-made genes. Mol. Biotechnol. 393-99. [DOI] [PubMed] [Google Scholar]
- 15.Hu, Y., H. F. Oliver, S. Raengpradub, M. E. Palmer, R. H. Orsi, M. Wiedmann, and K. J. Boor. 2007. Transcriptomic and phenotypic analyses suggest a network between the transcriptional regulators HrcA and σB in Listeria monocytogenes. Appl. Environ. Microbiol. 737981-7991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu, Y., S. Raengpradub, U. Schwab, C. Loss, R. H. Orsi, M. Wiedmann, and K. J. Boor. 2007. Phenotypic and transcriptomic analyses demonstrate interactions between the transcriptional regulators CtsR and Sigma B in Listeria monocytogenes. Appl. Environ. Microbiol. 737967-7980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huntzinger, E., S. Boisset, C. Saveanu, Y. Benito, T. Geissmann, A. Namane, G. Lina, J. Etienne, B. Ehresmann, C. Ehresmann, A. Jacquier, F. Vandenesch, and P. Romby. 2005. Staphylococcus aureus RNAIII and the endoribonuclease III coordinately regulate spa gene expression. EMBO J. 24824-835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johansen, J., A. A. Rasmussen, M. Overgaard, and P. Valentin-Hansen. 2006. Conserved small non-coding RNAs that belong to the sigmaE regulon: role in down-regulation of outer membrane proteins. J. Mol. Biol. 3641-8. [DOI] [PubMed] [Google Scholar]
- 19.Kim, H., K. J. Boor, and H. Marquis. 2004. Listeria monocytogenes σB contributes to invasion of human intestinal epithelial cells. Infect. Immun. 727374-7378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lenz, D. H., K. C. Mok, B. N. Lilley, R. V. Kulkarni, N. S. Wingreen, and B. L. Bassler. 2004. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell 11869-82. [DOI] [PubMed] [Google Scholar]
- 21.Livny, J., A. Brencic, S. Lory, and M. K. Waldor. 2006. Identification of 17 Pseudomonas aeruginosa sRNAs and prediction of sRNA-encoding genes in 10 diverse pathogens using the bioinformatic tool sRNAPredict2. Nucleic Acids Res. 343484-3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mandin, P., F. Repoila, M. Vergassola, T. Geissmann, and P. Cossart. 2007. Identification of new noncoding RNAs in Listeria monocytogenes and prediction of mRNA targets. Nucleic Acids Res. 35962-974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reference deleted.
- 24.Reference deleted.
- 25.Papenfort, K., V. Pfeiffer, F. Mika, S. Lucchini, J. C. Hinton, and J. Vogel. 2006. SigmaE-dependent small RNAs of Salmonella respond to membrane stress by accelerating global omp mRNA decay. Mol. Microbiol. 621674-1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Papenfort, K., V. Pfeiffer, S. Lucchini, A. Sonawane, J. C. D. Hinton, and J. Vogel. 2008. Systematic deletion of Salmonella small RNA genes identifies CyaR, a conserved CRP-dependent riboregulator of OmpX synthesis. Mol. Microbiol. 68890-906. [DOI] [PubMed] [Google Scholar]
- 27.Pichon, C., and B. Felden. 2005. Small RNA genes expressed from Staphylococcus aureus genomic and pathogenicity islands with specific expression among pathogenic strains. Proc. Natl. Acad. Sci. USA 10214249-14254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poyart, C., and P. Trieu-Cuot. 1997. A broad-host-range mobilizable shuttle vector for the construction of transcriptional fusions to beta-galactosidase in gram-positive bacteria. FEMS Microbiol. Lett. 156193-198. [DOI] [PubMed] [Google Scholar]
- 29.Raengpradub, S., M. Wiedmann, and K. J. Boor. 2008. Comparative analysis of the σB-dependent stress responses in Listeria monocytogenes and Listeria innocua strains exposed to selected stress conditions. Appl. Environ. Microbiol. 74158-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tjaden, B., S. S. Goodwin, J. A. Opdyke, M. Guillier, D. X. Fu, S. Gottesman, and G. Storz. 2006. Target prediction for small, noncoding RNAs in bacteria. Nucleic Acids Res. 342791-2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Urban, J. H., K. Papenfort, J. Thomsen, R. A. Schmitz, and J. Vogel. 2007. A conserved small RNA promotes discoordinate expression of the glmUS operon mRNA to activate GlmS synthesis. J. Mol. Biol. 373521-528. [DOI] [PubMed] [Google Scholar]
- 32.Valentin-Hansen, P., M. Eriksen, and C. Udesen. 2004. The bacterial Sm-like protein Hfq: a key player in RNA transactions. Mol. Microbiol. 511525-1533. [DOI] [PubMed] [Google Scholar]
- 33.Vázquez-Boland, J. A., M. Kuhn, P. Berche, T. Chakraborty, G. Domínguez-Bernal, W. Goebel, B. González-Zorn, J. Wehland, and J. Kreft. 2001. Listeria pathogenesis and molecular virulence determinants. Clin. Microbiol. Rev. 14584-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vogel, J., and C. M. Sharma. 2005. How to find small non-coding RNAs in bacteria. Biol. Chem. 3861219-1238. [DOI] [PubMed] [Google Scholar]
- 35.Wassarman, K. M. 2007. 6S RNA: a regulator of transcription. Mol. Microbiol. 651425-1431. [DOI] [PubMed] [Google Scholar]
- 36.Wiedmann, M., T. J. Arvik, R. J. Hurley, and K. J. Boor. 1998. General stress transcription factor σB and its role in acid tolerance and virulence of Listeria monocytogenes. J. Bacteriol. 1803650-3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilderman, P. J., N. A. Sowa, D. J. FitzGerald, P. C. FitzGerald, S. Gottesman, U. A. Ochsner, and M. L. Vasil. 2004. Identification of tandem duplicate regulatory small RNAs in Pseudomonas aeruginosa involved in iron homeostasis. Proc. Natl. Acad. Sci. USA 1019792-9797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reference deleted.
- 39.Zhu, K., D. O. Bayles, A. Xiong, R. K. Jayaswal, and B. J. Wilkinson. 2005. Precursor and temperature modulation of fatty acid composition and growth of Listeria monocytogenes cold-sensitive mutants with transposon-interrupted branched-chain alpha-keto acid dehydrogenase. Microbiology 151615-623. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.