Abstract
In Anabaena sp. strain PCC 7120, differentiation of heterocysts takes place in response to the external cue of combined nitrogen deprivation, allowing the organism to fix atmospheric nitrogen in oxic environments. NtcA, a global transcriptional regulator of cyanobacteria, is required for activation of the expression of multiple genes involved in heterocyst differentiation, including key regulators that are specific to the process. We have set up a fully defined in vitro system, which includes the purified Anabaena RNA polymerase, and have studied the effects of NtcA and its signaling effector 2-oxoglutarate on RNA polymerase binding, open complex formation, and transcript production from promoters of the hetC, nrrA, and devB genes that are activated by NtcA at different stages of heterocyst differentiation. Both RNA polymerase and NtcA could specifically bind to the target DNA in the absence of any effector. 2-Oxoglutarate had a moderate positive effect on NtcA binding, and NtcA had a limited positive effect on RNA polymerase recruitment at the promoters. However, a stringent requirement of both NtcA and 2-oxoglutarate was observed for the detection of open complexes and transcript production at the three investigated promoters. These results support a key role for 2-oxoglutarate in transcription activation in the developing heterocyst.
Cyanobacteria are phototrophic prokaryotes that, being the organisms that developed oxygenic photosynthesis, have played a crucial role in the evolution of our planet and of life on it (27). Nowadays, cyanobacteria contribute to an important fraction of the primary production in vast oligotrophic areas of oceans (14, 20). Their contribution to N2 fixation in oceans and other ecosystems is also remarkable and results in a net input of combined nitrogen into the biosphere (43, 56). To fix atmospheric nitrogen in oxic environments, cyanobacteria have to cope not only with external oxygen but also with that produced intracellularly as a result of water-splitting photosynthesis. To meet this challenge, some filamentous strains differentiate specialized cells called heterocysts when combined nitrogen is not available. Heterocysts exhibit many structural and metabolic differences with regard to vegetative cells (52), which arise during the course of a developmental process that involves activation of the expression of many genes and repression of others (9, 12, 19, 54). At the molecular level, NtcA, the cyanobacterial N control factor, plays a principal role in transcriptional regulation for the triggering and progress of heterocyst differentiation as well as in the mature heterocyst (reviewed in reference 19).
NtcA is a ca. 222-amino-acid protein of universal distribution in cyanobacteria, including strains from very different habitats and various taxonomic groups (18). NtcA belongs to the CRP (cyclic AMP receptor protein)/FNR (fumarate and nitrate reduction regulator) family of transcriptional regulators and has been shown to bind DNA in specific sites bearing the critical sequence element GTAN8TAC (32, 47). In most cases studied to date, NtcA acts as a transcriptional activator on promoters constituted by a −10 determinant with the consensus sequence TAN3T and an NtcA binding box in place of the −35 determinant (i.e., centered at ca. −41.5 nucleotides with regard to the transcription start point [tsp]). This arrangement has been considered the “canonical NtcA-activated promoter” (18) and conforms to the general structure of bacterial class II-activated promoters (7).
NtcA-dependent transcription activation can take place when cells are in the absence of ammonium, the preferred N source. In addition to being autoregulated at the level of gene expression, NtcA is modulated at the activity level as a function of the C-to-N balance of the cells. Thus, NtcA-dependent genes are activated only under conditions that lead to high C-to-N ratios, even in mutant strains that constitutively overexpress the NtcA protein from synthetic promoters (34, 40). 2-Oxoglutarate (2-OG), a metabolite that reflects the C-to-N balance of cyanobacterial cells (33, 39), has a positive effect on the expression of NtcA-activated genes (49). In vitro, 2-OG has been shown to increase NtcA binding to, and transcription from, the promoters of the glnA and ntcA genes of the unicellular cyanobacterium Synechococcus sp. strain PCC 7942 (45, 48) and to facilitate NtcA binding to the promoters of ntcA (29) and nrrA (35) of Anabaena sp. strain PCC 7120.
Previously (42), RNA polymerase (RNAP) was isolated from vegetative cells of the heterocyst-forming cyanobacterium Anabaena sp. strain PCC 7120, and in vitro assays have shown this protein to initiate transcription at a number of vegetative cell promoters but not at a promoter of glnA that is used in heterocysts. In this work, we have developed procedures for high-yield purification of the RNAP from Anabaena and have set up an in vitro system in which we have studied the effect of NtcA and its metabolic effector 2-OG on RNAP binding, open complex formation, and transcription at three regulated promoters activated during heterocyst differentiation.
MATERIALS AND METHODS
Plasmid construction.
Total DNA from Anabaena sp. strain PCC 7120 was isolated from cells grown in BG11 medium as described previously (8). DNA constructions, sequencing by the dideoxy chain termination method, and PCR were performed by standard procedures (2). The DNA nucleotide sequences of all the cloned genes in all the final constructs were verified.
The 1,321-bp DNA fragment from the Anabaena sp. strain PCC 7120 sigA region contained in plasmid pCSAV143 was amplified by PCR with oligonucleotides RD1 and RD2 (Table 1) and genomic DNA from strain PCC 7120 as the template, digested with BamHI and PstI, and cloned into BamHI- and PstI-digested expression vector pQE10 (Qiagen). The 1,024-bp DNA fragment from the rpoA (encoding the RNAP α subunit) region of Anabaena contained in plasmid pCSV25 was amplified by PCR with oligonucleotides RA1 and RA2 (Table 1) and genomic DNA from strain PCC 7120 as the template, digested with BamHI and PvuII, and cloned into BamHI- and HincII-digested expression vector pQE9 (Qiagen). The 3,527-bp DNA fragment from the rpoB (encoding the RNAP β subunit) region of Anabaena sp. strain PCC 7120 included in plasmid pCSAV155 was amplified in three segments by PCR with oligonucleotides RB5 and RB2, RB3 and RB7, and RB6 and RB4 (Table 1) and genomic DNA from strain PCC 7120 as the template. These fragments were cloned in pGEM-T Easy vector (Promega), producing plasmids pCSAV150, pCSAV151, and pCSAV152, respectively. An EcoNI-ScaI fragment from pCSAV151 was ligated to the EcoNI-ScaI fragment from pCSAV152, generating plasmid pCSAV153, which was cut with ClaI and SalI enzymes and ligated to a ClaI-SalI fragment from pCSAV150, producing plasmid pCSAV154, which contains the complete rpoB gene in vector pGEM-T Easy. The NcoI fragment from pCSAV154 was finally cloned into NcoI-digested expression vector pTrc99A, rendering pCSAV155. The 1,924-bp DNA fragment from the rpoC1 (encoding the RNAP γ subunit) region of Anabaena sp. strain PCC 7120 included in plasmid pCSAV158 was amplified by PCR with oligonucleotides RC110 and RC111 (Table 1) and genomic DNA from strain PCC 7120 as the template, digested with NcoI and SmaI, and cloned into NcoI-SmaI-digested expression vector pTrc99A. Plasmid pCSAV157 contains a 4,164-bp DNA fragment from the rpoC2 (encoding the β′ subunit) region of Anabaena sp. strain PCC 7120. This gene was amplified in three segments by PCR with oligonucleotides RC25 and RC26, RC27 and RC28, and RC29 and RC210 (Table 1) and genomic DNA from strain PCC 7120 as the template. The DNA product amplified with oligonucleotides RC25 and RC26 was digested with EcoRI and BamHI and cloned in the expression vector pTrc99A cut with the same restriction enzymes, producing plasmid pCSAV156. The DNA fragment amplified with oligonucleotides RC27 and RC28 was first cloned in pGEM-T Easy and then excised with DraIII and SalI and cloned in the same restriction sites of pCSAV156, producing pCSAV156B. Finally, the product of the amplification with oligonucleotides RC29 and RC210 was cut with Eco47III and SalI and inserted into pCSAV156B digested with Eco47III and SalI, producing pCSAV157.
TABLE 1.
Oligodeoxynucleotides used in this study
| Oligodeoxynucleotide | Sequence (5′-3′) | Positions relative to the translation start of the corresponding gene |
|---|---|---|
| RD1 | GGAGCATGAGGATCCCGGCATGAACCAG | −19 to +9 (rpoD) |
| RD2 | CCTACATTTCTGGGGCATTC | +1318 to +1299 (rpoD) |
| RA1 | AAGGGAGGCGGATCCGTGGCGCAGTTTC | −15 to +13 (rpoA) |
| RA2 | GACCATGACGGATTAGCTCAG | +1080 to +1060 (rpoA) |
| RB5 | CGGCTAATGACACCCATGGTAGTGTCATC | −21 to +8 (rpoB) |
| RB2 | GGGATGAGACTGGCTGAATATG | +500 to +479 (rpoB) |
| RB3 | GTGCGATCGCCTGGAGTTTATTAC | +424 to +447 (rpoB) |
| RB7 | CATCTTGTACCAGTCGTTCGG | +2134 to +2114 (rpoB) |
| RB6 | GTAGTTGCTGGTCAGGTGC | +1987 to +2005 (rpoB) |
| RB4 | GTGCTGAGTTCTGAGTAAAG | +3524 to +3505 (rpoB) |
| RC110 | CATTAGTGAATAGTCCCGGGTT | +1939 to +1918 (rpoC1) |
| RC111 | CAGCCCATGGGTATGAGACCCGCCCAAAC | −12 to +17 (rpoC1) |
| RC25 | GACTAACGAATTCATGATTTTTCGC | −13 to +12 (rpoC2) |
| RC26 | GTTTCGACGGGATCCCCATTTTTC | +1517 to +1494 (rpoC2) |
| RC27 | GCCAGAGGTGGTTTGATTTG | +1422 to +1437 (rpoC2) |
| RC28 | GATGTCACGACGCAAAACTAAGG | +2532 to +2510 (rpoC2) |
| RC29 | GACCTGACCTTGAGGATGCAG | +2461 to +2476 (rpoC2) |
| RC210 | GATATAGGATGTTGTCGACGTTGTG | +4173 to +4249 (rpoC2) |
| DB1 | CCCCCTACTCCCTTTCC | −825 to −809 (devB) |
| DB3 | CCTGCTTCTTCTGGTCAGCG | −807 to −788 (devB) |
| DB4 | CATAACATTTCCCCAAGTC | −580 to −598 (devB) |
| DB13 | TCCGTCACCCTTGACAT | −3 to +17 (devB) |
| HC1 | TAGTACATCGGTGAGGGGTG | −693 to −674 (hetC) |
| HC2 | TGTGAGCAACATCGACATCTG | −411 to −431 (hetC) |
| PA1 | CAGAGCAGCCGATTGTCTGTTG | +97 to +76 (psbA) |
| PA2 | CTGATAAGTGAGCTATTCAC | −145 to −126 (psbA) |
| NA1 | CTGGCGGCTTGATGCACACGG | +104 to +84 (nrrA) |
| NA2 | ATTCAGGCTGAAATGTGAAGGG | −498 to −477 (nrrA) |
| NA14 | CGTAGAGGTAATTGTGGC | −96 to −79 (nrrA) |
Cloning and purification of recombinant Anabaena RNAP.
Overproduction and purification of recombinant Anabaena sp. strain PCC 7120 RNAP was performed as described for the E. coli enzyme (44), with modifications. Histidine-tagged α and SigA subunits were purified from isopropyl-β-d-thiogalactopyranoside (IPTG)-induced cultures of E. coli containing plasmids pCSV25 and pCSAV143, respectively. Extracts from the inclusion bodies of those cultures solubilized in buffer A (50 mM Tris-HCl [pH 8.0], 6 M guanidine hydrochloride, 0.3 M NaCl, 10% glycerol) were chromatographed through a 1.25-ml Ni2+-charged His-Select cartridge (Sigma) by using a fast protein liquid chromatography system (Pharmacia). Samples containing α or SigA were supplemented with 50% (final concentration) of glycerol and stored at −20°C.
β, β′, and γ subunits were purified from inclusion bodies of IPTG-induced E. coli strains containing plasmids pCSAV155, pCSAV157, and pCSAV158, respectively. Inclusion bodies were solubilized in buffer B (50 mM Tris-HCl [pH 8.0], 6 M guanidine hydrochloride, 10 mM MgCl2, 10 μM ZnCl2, 1 mM EDTA, 10 mM dithiothreitol [DTT], 10% glycerol) and used as crude preparations.
For preparation of RNAP, a mixture of approximately 80 μg of hexahistidine-tagged α, 300 μg of crude preparation of β, 600 μg of β′, and 600 μg of γ and, separately, approximately 300 μg of hexahistidine-tagged SigA were dialyzed against buffer C (50 mM Tris-HCl [pH 8.0], 200 mM KCl, 10 mM MgCl2, 1 mM DTT, 5 mM 2-mercaptoethanol, 20% glycerol), containing decreasing amounts of urea (1 h with 6 M, 1 h with 5 M, 1 h with 4 M, 1.5 h with 3 M, 1.5 h with 2 M, and 3 to 6 h with 1 M), and finally against buffer D (50 mM Tris-HCl [pH 8.0], 200 mM KCl, 20% glycerol). Following dialysis, the two preparations were combined, incubated for 45 min at 30°C, cleared by centrifugation (16,000 × g, 15 min at 4°C), and chromatographed through a 1.25-ml Ni2+-charged His-Select cartridge (Sigma). Contaminating proteins were eluted by washing the column with 25 volumes of buffer D plus 3.5 mM of imidazole. Bound RNAP was eluted with a linear gradient of imidazole (3.5 mM to 460 mM) in buffer D. Samples containing the complete RNAP holoenzyme were concentrated by centrifugal ultrafiltration (Amicon 100000 MWCO; 2,000 × g at 4°C), mixed with glycerol (50% final concentration), and stored at −20°C. Just before use, the RNAP preparation was supplemented with an ∼3-fold molar excess of the purified and reconstituted SigA protein and incubated for 15 min at 32°C.
Proteins were analyzed by standard electrophoresis in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels, followed by staining with Coomassie brilliant blue R. Protein concentration was estimated by a dye-binding assay (Bio-Rad) based on that of Bradford (5).
Overproduction and purification of recombinant Anabaena NtcA protein.
NtcA was purified from crude extracts of E. coli containing plasmid pCSAM61 (37), which were chromatographed through a 1-ml HiTrap heparin HP column (Pharmacia Biotech). Bound NtcA was eluted with a linear gradient of NaCl (500 mM to 2 M) in 20 mM sodium phosphate buffer (pH 7.0) supplemented with 10% glycerol. Samples containing NtcA were desalted by centrifugal ultrafiltration (Amicon 10000 MWCO; 7,000 × g at 4°C) and stored in 20 mM sodium phosphate buffer (pH 7.0) containing 250 mM NaCl and 10% glycerol at −20°C.
Preparation of DNA fragments.
The double-stranded DNA fragments used in electrophoretic mobility shift assays (EMSA) or potassium permanganate footprinting and in vitro transcription experiments were obtained by PCR amplification, followed by purification of the corresponding products with a GFX gel band purification kit (Pharmacia Biotech). The fragment of the hetC region was obtained with primers HC1 and HC2 (positions −121 to +162 with regard to the tsp) (Table 1) and plasmid pCSAM83 (36) as the template. The devB fragment (positions −103 to +125 with regard to the tsp) was amplified using primers DB3 and DB4 (Table 1) and plasmid pCSAM155 (which contains the devB promoter region, amplified with oligonucleotides DB1 and DB13 and cloned in pMBL-T vector) (A. M. Muro-Pastor, unpublished data) as the template. The nrrA fragment used in in vitro transcription and potassium permanganate footprinting assays was amplified using oligonucleotides NA1 and NA2 (positions −471 to +131) (Table 1), and the fragment used in EMSA was amplified with primers NA14 and NA1 (positions −69 to +131) (Table 1) and plasmid pCSAM113 (35) as the template. The psbA fragment encompasses positions −145 to +97 with regard to the translation start of the gene, amplified using primers PA1 and PA2 (Table 1) and plasmid pRL278 (4) as the template.
EMSA.
EMSA was carried out using 5 to 10 fmol of the specific DNA fragment end labeled with T4 polynucleotide kinase (Boehringer) and [γ-32P]ATP, and 0.07 mg/ml poly(dI-dC) as nonspecific competitor DNA, in a final volume of 20 μl. The reaction mixtures with the corresponding promoter fragment were incubated with purified NtcA for 5 min at 32°C and then with the reconstituted RNAP holoenzyme supplemented with purified SigA (see above). The protein-DNA complexes were separated on native 5% polyacrylamide gels in low-ionic-strength buffer (40 mM Tris [pH 8.0], 40 mM boric acid, 0.8 mM EDTA) at 200 V for 3 to 5 h at 4°C with a prerun of 1 h. Images of radioactive gels were obtained and quantified with a Cyclone storage phosphor system (Packard).
DNase I footprint assays.
Protein-DNA complexes were formed in a final volume of 70 μl of buffer [10 mM Tris-HCl (pH 8.0), 30 mM KCl, 10 mM MgCl2, 5 mM CaCl2, 2 mM DTT, 0.07 mg/ml bovine serum albumin, 0.07 mg/ml poly(dI-dC)], with 10 fmol of 32P-end-labeled (with T4 polynucleotide kinase and [γ-32P]ATP) DNA fragment, 1 mM ATP, and the reconstituted RNAP holoenzyme. The reaction mixtures were incubated for 30 min at 32°C. Then, DNase I (1 U; Roche) was added in a buffer containing 50 mM sodium acetate and 6 mM MgSO4, and the reaction was immediately stopped with 320 μl of stop solution (0.5 mM EDTA, 0.125% SDS, 12.5 mM ammonium acetate [pH 5.2]). The products were resolved in 6% polyacrylamide-4.68 M urea gels next to the corresponding sequencing ladder. Images of radioactive gels were obtained as described above.
Potassium permanganate footprinting assays.
Potassium permanganate footprinting was performed with minor modifications of the procedure described by Jiang and Gralla (22). Complexes were formed under the same conditions as those described for EMSA, with 5 to 10 fmol of nonlabeled DNA template and 1 mM ATP (psbA and devB) or 1 mM ATP plus 1 mM GTP (hetC). After 30 min of incubation, 4 mM potassium permanganate was added and then incubated for 1 min at 31°C. Potassium permanganate-hypersensitive sites were detected by asymmetric PCR using end-labeled primers (HC1, NA1, DB4, and PA1) (Table 1) and EcoTaq polymerase (Ecogen). PCR products were analyzed on 6% polyacrylamide-4.68 M urea gels next to the corresponding sequencing ladder. Images of radioactive gels were obtained as described above.
In vitro transcription assays.
The runoff transcription assays were performed in a total volume of 30 μl of buffer (40 mM Tris-HCl [pH 8.0], 50 mM KCl, 10 mM MgCl2, 0.1 mM EDTA, 0.5 mg/ml bovine serum albumin, 5% glycerol). Complexes were formed under the same conditions as those described for EMSA. Transcription was started by the addition of 3 μl of a substrate solution of 0.15 mM each of ATP, GTP, and UTP, 20 μM CTP, and 3 μCi [α-32P]CTP. After 30 min of incubation at 37°C, the reaction was terminated by the addition of 30 μl phenol, 10 mM EDTA, and 3 μl of 200-ng/μl glycogen prepared in 3 M sodium acetate (pH 5.2). The products were precipitated with ethanol and fractionated by electrophoresis on 6% polyacrylamide-4.68 M urea gels. Images of radioactive gels were obtained as described above.
RESULTS
Anabaena RNAP.
As a first step to study the relations of NtcA and the cyanobacterial RNAP at the regulated promoters, purification of RNAP of Anabaena sp. strain PCC 7120 was addressed in order to set up a fully defined in vitro assay system and to avoid artifacts that could arise from the use of a heterologous RNAP. The core cyanobacterial RNAP has a subunit composition different from that of the typical proteobacterial α2ββ′, as found in, e.g., E. coli. The cyanobacterial core is constituted of α2ββ′γ subunits, in which the two domains found in the E. coli β′ subunit are split into two subunits in cyanobacteria: the γ subunit corresponds to the amino-terminal half of the E. coli β′ subunit, and the β′ subunit corresponds to the carboxy-terminal half of the E. coli β′ subunit (3, 42).
Besides the principal, group 1 σ factor SigA, multiple alternative group 2 σ factors have been identified in the several cyanobacteria investigated to date (6, 26, 55). In the unicellular cyanobacterium Synechocystis sp. strain PCC 6803, the expression of some NtcA-dependent genes has been shown to take place principally with SigA, whereas the group 2 factors SigB and SigC could contribute to expression under particular cellular conditions (21). Because NtcA-activated promoters encompass a standard −10 determinant (18, 32), we first chose to set up in vitro assays with RNAP bearing the principal Anabaena type 1 σ factor, SigA.
Purification of Anabaena RNAP was accomplished through two different approaches. First, it was isolated, by chelating affinity chromatography, from a derivative of strain PCC 7120 that expresses a modified version of the β′ subunit carrying a polyhistidine tail fused to its carboxy terminus (E. Olmedo-Verd, A. M. Muro-Pastor, E. Flores, and A. Herrero, unpublished data). Second, it was reconstituted from the single subunits cloned and expressed in E. coli, as described in Materials and Methods (Fig. 1). The two preparations of Anabaena RNAP were tested in the different types of in vitro assays used throughout this study, rendering similar results. Thus, because higher purification yields were obtained with the enzyme expressed in E. coli, this was the source of the enzyme that was used subsequently and in the experiments described below. Figure 2 shows the performance of the cloned Anabaena RNAP in a consensus-type promoter, that of the psbA gene from Amaranthus hybridus, which includes standard −10 and −35 determinants and is utilized in Anabaena sp. strain PCC 7120 (51). Binding of RNAP to the promoter region leading to footprinting signals from ca. −55 to +20 bp with respect to the tsp of the gene (Fig. 2A; see also reference 10), melting of the promoter exposing T's to the reaction with potassium permanganate (Fig. 2B; see below), and production of full transcripts for the DNA fragment used (Fig. 2C) took place efficiently. These results show an adequate performance of the obtained RNAP preparations.
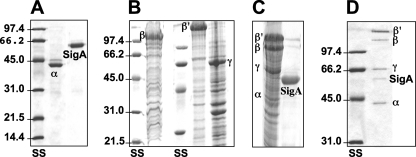
FIG. 1.
Preparation of recombinant RNAP from Anabaena sp. strain PCC 7120. Preparations of His-tagged α and σ (SigA) subunits of Anabaena RNAP expressed in E. coli and purified (A) and of solubilized inclusion bodies from E. coli containing the β, β′, and γ subunits (B), all of them obtained under denaturing conditions, were subjected to SDS-PAGE and stained with Coomassie brilliant blue R. Portions of the preparations of His-tagged α, β, β′, and γ subunits were mixed and renatured together. The resulting preparation was mixed with a preparation of renatured His-SigA, and the obtained reconstituted RNAP holoenzyme was purified by chromatography through chelating Sepharose. Preparations obtained after renaturation (C) and holoenzyme purification (D) were also analyzed by SDS-PAGE. See Materials and Methods for further details. Size standards (SS) in kDa are shown.
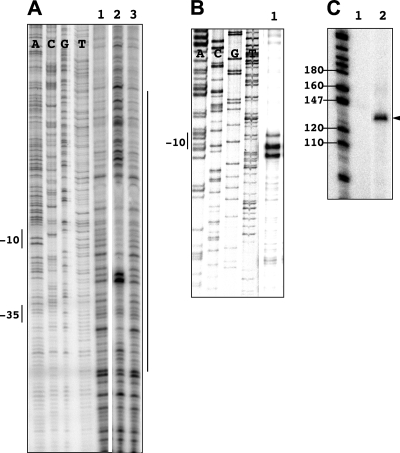
FIG. 2.
Activity of the cloned Anabaena RNAP on the promoter of the psbA gene. (A) Results of DNase I footprinting. The DNA fragment of the promoter region of the psbA gene was incubated without (lanes 1 and 3) or with (lane 2) purified RNAP holoenzyme (11 nM plus 40 nM SigA) before DNase I treatment (noncoding strand represented). The positions of the −10 and −35 determinants as well as the footprinted region are indicated by vertical lines. (B) Results of potassium permanganate footprinting. The DNA fragment encompassing the promoter of the psbA gene was incubated with purified RNAP holoenzyme (60 nM plus 100 nM SigA) before potassium permanganate treatment (lane 1, the coding strand subjected to primer extension). The −10 promoter element is indicated in the sequence ladder. (C) Results of the in vitro runoff transcription assay. The psbA DNA fragment was incubated without (lane 1) or with (lane 2) RNAP (15 nM plus 48 nM SigA) before the addition of nucleoside triphosphates (see Materials and Methods for details). The arrowhead indicates the full transcript corresponding to the DNA fragment used. Size markers (in nucleotides) are also indicated.
Binding of Anabaena RNAP to promoters activated by NtcA during heterocyst differentiation.
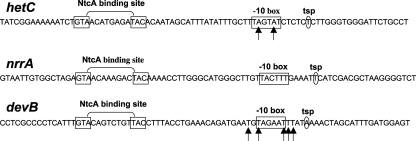
Binding of purified RNAP and the effect of NtcA and 2-OG were tested with DNA fragments containing a promoter of hetC, nrrA, or devB. hetC encodes an ABC-type exporter required at an early stage of heterocyst development (25), nrrA encodes a response regulator proposed to mediate NtcA-dependent activation of hetR, the first-acting regulatory gene specific for heterocyst differentiation (11, 12), and the devBCA operon encodes an ABC-type exporter involved in the maturation of the heterocyst envelope (15). The expression of hetC (36), nrrA (35), and devBCA (16) is activated upon combined nitrogen deprivation from NtcA-dependent promoters. Binding of RNAP and NtcA to the promoters of these genes was studied by EMSA. As described previously, NtcA is able to bind to the three tested promoters in the absence of any effector (16, 35, 36), although, as in other regulated promoters, binding affinity might increase to some extent in the presence of 2-OG (35; our unpublished results).
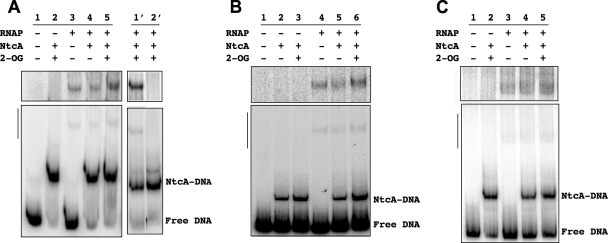
Figure 3 shows that RNAP was able to bind to the three tested promoters both in the presence and in the absence of NtcA, alone or together with 2-OG (upper parts of Fig. 3). The addition of NtcA, preferably with 2-OG, prior to the addition of RNAP had some positive effect on the amount of RNAP-containing complexes detected. Quantification of the results shown in Fig. 3 gave the following relative amounts of DNA bound to RNAP for the assays containing RNAP, RNAP plus NtcA, and RNAP plus NtcA and 2-OG, respectively: hetC, 1:1.1:2.0 (Fig. 3A, lanes 3 to 5); nrrA, 1:1.5:1.9 (Fig. 3B, lanes 4 to 6); and devB, 1:1.4:1.7 (Fig. 3C, lanes 3 to 5). Although unspecific interactions of RNAP with the ends of DNA fragments can lead to misleading interpretations of binding features in EMSA, we are confident that the observed binding was specific because site-directed mutation of the hetC promoter fragment changing the −10 determinant from TAGTAT to GGGTAG completely abolished RNAP binding but not NtcA binding (Fig. 3A, lanes 1′ to 2′).
FIG. 3.
Results of EMSA of the binding of Anabaena RNAP and NtcA to promoters of the hetC, nrrA, and devB genes. The DNA fragment of the hetC (A), nrrA (B), or devB (C) promoter region was incubated, as indicated, with purified NtcA without or with 2-OG (0.6 mM), then supplemented with purified RNAP holoenzyme, and further incubated before being loaded onto PAGE gels (protein concentrations: NtcA, 142 nM for hetC and nrrA and 130 nM for devB; RNAP, 48 nM for hetC and 60 nM for nrrA and devB, plus 110 nM SigA in the three cases [see Materials and Methods for details]). Lanes 1′ and 2′ of panel A show the results of an independent assay in which a DNA fragment bearing a mutated −10 box with the sequence GGGTAG instead of the native TAGTAT one was used in lane 2′ (protein concentrations: NtcA, 130 nM; RNAP, 60 nM plus 110 nM SigA). The upper panels are overexposed images of the top portions (indicated with vertical lines) of the bottom gels showing the RNAP-containing complexes. The positions of the binary NtcA-DNA complexes and of the free DNA are indicated.
NtcA-dependent transcription.
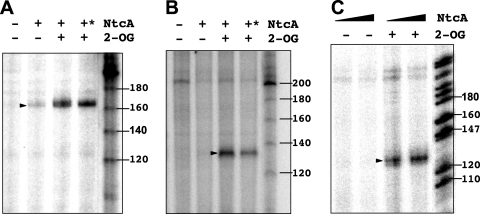
Given that both RNAP and NtcA were able to independently bind to the DNA of the tested Anabaena-regulated promoters, we wanted to assess the requirement of NtcA and 2-OG for transcription from those promoters. Runoff in vitro transcription was tested with Anabaena RNAP in the presence and absence of NtcA and 2-OG. The bands corresponding to the hetC (Fig. 4A), nrrA (Fig. 4B), and devB (Fig. 4C) transcripts (162, 131, and 125 nucleotides, respectively [distance from the tsp to the end of the corresponding fragment; see Materials and Methods]) were readily produced in the presence of NtcA plus 2-OG but not when RNAP was tested alone or in the presence of NtcA without 2-OG. With the three promoters, the requirement of NtcA and 2-OG was observed irrespective of the order of the addition of RNAP and NtcA (not shown). No effect of NtcA or 2-OG in transcript production from the constitutive psbA promoter described above was observed (not shown). Remarkably, these results show that, in spite of independent binding of NtcA and RNAP to DNA in the presence and absence of 2-OG (Fig. 3), transcription from the tested hetC, nrrA, and devB promoters requires both NtcA and 2-OG.
FIG. 4.
Effect of NtcA and 2-OG on transcription from promoters of the hetC, nrrA, and devB genes. In vitro runoff transcription assays were performed with DNA fragments of the hetC (A), nrrA (B), or devB (C) genomic region in the presence or absence of NtcA and 2-OG as indicated. NtcA (130 nM [A], 170 nM [B], and 41 or 100 nM [C]), supplemented with 2-OG (0.6 mM) when indicated, was incubated with the DNA fragment before the addition of RNAP (27 nM plus 88 nM SigA [A and B] and 29 nM plus 96 nM SigA [C]; see Materials and Methods for details). Where indicated with *, RNAP was first added to the DNA preparation and, after incubation, supplemented with NtcA and 2-OG. Arrowheads indicate full transcripts encompassing positions from the corresponding tsp to the end of the Anabaena fragment used. Size markers (in nucleotides) are also indicated.
Open complex formation.
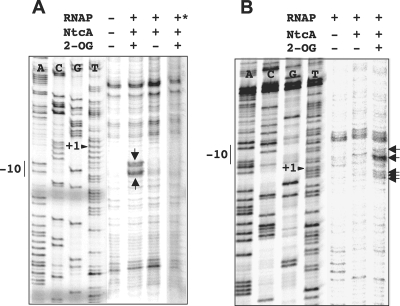
To trace back the requirement of NtcA and 2-OG in the sequence of events leading to transcription activation at the tested promoters, permanganate footprinting assays were performed to detect open complex formation in the presence and absence of NtcA and 2-OG. Opening of the DNA strands, which during transcription initiation takes place at the promoter, can render T's in the melted region exposed to potassium permanganate so that the detection of reactive T's in the region from ca. −12 to +2 is indicative of the presence of promoter open complexes (for details, see reference 10). Reactive T's can be detected as points of termination in primer extension reactions performed with the treated DNA. In the hetC promoter (Fig. 5A), extension product ends mapping at the T's in the −10 box of the coding strand were specifically detected when the promoter DNA was incubated with RNAP and NtcA plus 2-OG but not when 2-OG was omitted (Fig. 5A and 6). Note that when the DNA was incubated in the absence of RNAP (Fig. 5A), or simply subjected to primer extension without previous treatment (not shown), all the other extension products shown in Fig. 5A were produced. These products should therefore correspond to nonspecific stops during the extension reaction. No specific ends were detected when the DNA fragment of the hetC promoter bearing a mutated −10 box (Fig. 3A) was used (Fig. 5A). Specific reactivity to potassium permanganate was also apparent for T's between positions −12 and −3 in the noncoding strand of the devB promoter (Fig. 5B and 6). Although only weakly labeled, T's at the −10 box in the noncoding strand of the nrrA promoter could also be detected (not shown). Remarkably, in all three studied promoters, reactive T's were detected only when both NtcA and 2-OG were present in the assay. These results indicate that the concourse of the transcriptional activator NtcA and its effector 2-OG is required for an open complex to be formed in the assayed promoters.
FIG. 5.
Effect of NtcA and 2-OG on permanganate footprinting in promoters of the hetC and devB genes. The DNA fragments of the genomic regions of hetC (the fragment with the mutated −10 box in the lane marked with *; see the text for details) (A) and devB (B) genes were incubated, as indicated, with purified NtcA (188 and 130 nM, respectively) without or with 2-OG (0.6 mM) and then with purified RNAP holoenzyme (100 nM plus 390 nM SigA and 60 nM plus 110 nM SigA, respectively) and further incubated before potassium permanganate treatment (see Materials and Methods for details). Arrows point to the reactive T's (the noncoding strand of hetC and the coding strand of devB were subjected to primer extention). The −10 promoter element and the tsp (+1) are indicated in the corresponding sequence ladder.
FIG. 6.
Features of the NtcA-regulated hetC, nrrA, and devB promoters used in this study. The depicted sequences (noncoding strands) are included in the DNA fragments of the corresponding promoters used throughout this study (see Materials and Methods for details). The locations of the sequence signature of the NtcA binding site, the −10 box, and the tsp are as described by Muro-Pastor et al. (36) for hetC, Muro-Pastor et al. (35) for nrrA, and Fiedler et al. (16) for devB. Arrows point to the positions marked in potassium permanganate footprinting in the promoters of hetC and devB (Fig. 5).
DISCUSSION
NtcA, a cyanobacterial transcription factor exerting N control, is essential for the developmental process leading to heterocyst-containing filaments. Thus, ntcA mutant strains are unable to initiate heterocyst differentiation (17, 46, 50, 53). Besides that, a substantial number of genes are expressed from NtcA-dependent promoters in differentiating cells or in mature heterocysts, among them the hetC, nrrA, and devB genes (19, 35). Here, we have investigated the requirement of NtcA and the metabolite 2-OG during transcription activation of hetC, nrrA, and devB promoters from the cyanobacterium Anabaena sp. strain PCC 7120, using a defined in vitro system that includes the RNAP of the same organism. In the three cases, a stringent requirement of NtcA and 2-OG is observed for transcription. However, RNAP is able to bind the DNA of the three promoters in the absence of NtcA, with only a ≤2-fold positive effect of NtcA plus 2-OG on the amount of DNA-RNAP-containing complexes formed. Thus, the strong requirement of the regulator at the level of transcript production does not seem to respond only to an effect on RNAP recruitment at these promoters.
Concerning the effect of 2-OG, this metabolite has been shown to exert a moderate positive effect on the affinity of NtcA for the NtcA-regulated promoters, whose magnitudes vary from one to another (see above). However, because NtcA can bind DNA to a substantial degree in the absence of 2-OG and because the amounts of DNA-bound RNAP do not vary significantly when different ratios of free DNA versus NtcA-bound DNA are used (not shown), it is unlikely that the observed requirement of 2-OG for transcription activation responds solely to an effect on NtcA binding. Instead, NtcA and 2-OG would be required at a later step during the formation of the active transcriptional complex. Because DNA melting is detected only in the presence of NtcA and 2-OG at the three investigated promoters, an action of the regulator, which would require 2-OG, at the step of open complex formation could be considered. In this respect, the role of NtcA as a transcriptional activator resembles that of CRP, which at class II-activated promoters increases both the binding of RNAP to DNA for the formation of the RNAP-promoter closed complex and the rate of isomerization of the closed to the open complex (30). However, whereas CRP, as well as its homologous regulators FNR (24) and CooA (23), is unable to specifically bind its DNA targets in the absence of its effector (28), NtcA is capable of specific binding to DNA in the absence of additional factors.
As mentioned above, a positive effect of 2-OG on in vitro binding of NtcA to some NtcA-regulated genes of Anabaena has already been shown, and 2-OG added externally to a derivative of strain PCC 7120 expressing a cloned 2-OG transporter has been reported to promote differentiation of (likely nonfunctional) heterocysts in the presence of nitrate but not of ammonium (31). However, our observations of a stringent requirement of 2-OG for the expression of the Anabaena hetC, nrrA, and devB genes provide strong evidence in support of a key regulatory role of this metabolite at transcription activation in the developing heterocyst. Unfortunately, little is known about the metabolic features of the Anabaena cells that are undergoing differentiation, including information about metabolite levels. At the whole-filament level, the icd gene, which encodes the isocitrate dehydrogenase that synthesizes 2-OG, has been shown to be expressed at higher levels in diazotrophic cultures of Anabaena sp. strain PCC 7120 than in cultures using nitrate or ammonium as the nitrogen source (38). On the other hand, in the unicellular cyanobacterium Synechocystis sp. strain PCC 6803, the expression of the icd gene is activated by NtcA (38). Given that the ntcA gene is induced in proheterocysts (41), activation by NtcA of the icd gene during heterocyst differentiation could be a mechanism contributing to high 2-OG levels in the differentiating cell.
Finally, it is worth pointing out that open complex formation was more readily detected with the psbA promoter than with the hetC, nrrA, or devB promoter used here, even in the presence of NtcA and 2-OG, consistent with higher levels of in vitro transcription of the psbA gene (compare Fig. 2 with Fig. 4 and 5). This might reflect the in vivo situation, where low levels of expression have been detected at least for the hetC (36) and devB (16) genes. However, once the requirement of NtcA activation has been met, the expression of these regulated genes could be further modulated in vivo. A number of other protein factors have been identified, mainly by genetic studies, as regulators of heterocyst differentiation (for details, see references 19 and 54), although in most cases neither their precise mechanisms of action nor their molecular targets have been identified. Also, the protein PipX has been described as a coactivator of NtcA in Synechococcus sp. (13). Possible roles of many candidates as transcriptional regulators can be tested in the in vitro system that we have set up here, as can the promoter specificities of the different alternative sigma factors found to be upregulated in the differentiating cells (1). Additionally, this system should permit the study of the regulation of other promoters that, being activated during heterocyst differentiation in an NtcA-dependent manner, do not exhibit NtcA binding sites and are thus apt to be activated by NtcA indirectly (11, 19).
Acknowledgments
We thank Virginia Rodríguez for her contribution to the cloning of the RNAP α subunit, Silvia Picossi for experimental advice, Rocío Rodríguez for skillful technical assistance, and Ignacio Luque and Alicia M. Muro-Pastor for critical reading of the manuscript.
This work was supported by grants BFU2004-00872 and BFU2007-60457 from the Ministerio de Educación y Ciencia (Spain).
Footnotes
Published ahead of print on 25 July 2008.
REFERENCES
- 1.Aldea, M. R., R. A. Mella-Herrera, and J. W. Golden. 2007. Sigma factor genes sigC, sigE, and sigG are upregulated in heterocysts of the cyanobacterium Anabaena sp. strain PCC 7120. J. Bacteriol. 1898392-8396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 2008. Current protocols in molecular biology. Greene Publishing and Wiley-Interscience, New York, NY.
- 3.Bergsland, K. J., and R. Haselkorn. 1991. Evolutionary relationships among eubacteria, cyanobacteria, and chloroplasts: evidence from the rpoC1 gene of Anabaena sp. strain PCC 7120. J. Bacteriol. 1733446-3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Black, T. A., Y. Cai, and C. P. Wolk. 1993. Spatial expression and autoregulation of hetR, a gene envolved in the control of heterocyst development in Anabaena. Mol. Microbiol. 977-84. [DOI] [PubMed] [Google Scholar]
- 5.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72248-254. [DOI] [PubMed] [Google Scholar]
- 6.Brahamsha, B., and R. Haselkorn. 1992. Identification of multiple RNA polymerase sigma factor homologs in the cyanobacterium Anabaena sp. strain PCC 7120: cloning, expression, and inactivation of the sigB and sigC genes. J. Bacteriol. 1747273-7282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Browning, D. F., and S. J. Busby. 2004. The regulation of bacterial transcription initiation. Nat. Rev. Microbiol. 257-65. [DOI] [PubMed] [Google Scholar]
- 8.Cai, Y., and C. P. Wolk. 1990. Use of a conditionally lethal gene in Anabaena sp. strain PCC 7120 to select for double recombinants and to entrap insertion sequences. J. Bacteriol. 1723138-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campbell, E. L., M. L. Summers, H. Christman, M. E. Martin, and J. C. Meeks. 2007. Global gene expression patterns of Nostoc punctiforme in steady-state dinitrogen-grown heterocyst-containing cultures and at single time points during the differentiation of akinetes and hormogonia. J. Bacteriol. 1895247-5256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Haseth, P. L., M. L. Zupancic, and M. T. Record. 1998. RNA polymerase-promoter interactions: the comings and goings of RNA polymerase. J. Bacteriol. 1803019-3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ehira, S., and M. Ohmori. 2006. NrrA directly regulates expression of hetR during heterocyst differentiation in the cyanobacterium Anabaena sp. strain PCC 7120. J. Bacteriol. 1888520-8525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ehira, S., and M. Ohmori. 2006. NrrA, a nitrogen-responsive response regulator facilitates heterocyst development in the cyanobacterium Anabaena sp. strain PCC 7120. Mol. Microbiol. 591692-1703. [DOI] [PubMed] [Google Scholar]
- 13.Espinosa, J., K. Forchhammer, S. Burillo, and A. Contreras. 2006. Interaction network in cyanobacterial nitrogen regulation: PipX, a protein that interacts in a 2-oxoglutarate dependent manner with PII and NtcA. Mol. Microbiol. 61457-469. [DOI] [PubMed] [Google Scholar]
- 14.Falkowski, P. G., and A. H. Knoll. 2008. An introduction to primary producers in the sea: who they are, what they do, and when they evolved, p. 1-6. In P. G. Falkowski and A. H. Knoll (ed.), Evolution of primary producers in the sea. Elsevier Academic Press, Burlington, MA.
- 15.Fiedler, G., M. Arnold, S. Hannus, and I. Maldener. 1998. The DevBCA exporter is essential for envelope formation in heterocysts of the cyanobacterium Anabaena sp. strain PCC 7120. Mol. Microbiol. 271193-1202. [DOI] [PubMed] [Google Scholar]
- 16.Fiedler, G., A. M. Muro-Pastor, E. Flores, and I. Maldener. 2001. NtcA-dependent expression of the devBCA operon, encoding a heterocyst-specific ATP-binding cassette transporter in Anabaena spp. J. Bacteriol. 1833795-3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frías, J. E., E. Flores, and A. Herrero. 1994. Requirement of the regulatory protein NtcA for the expression of nitrogen assimilation and heterocyst development genes in the cyanobacterium Anabaena sp. PCC 7120. Mol. Microbiol. 14823-832. [DOI] [PubMed] [Google Scholar]
- 18.Herrero, A., A. M. Muro-Pastor, and E. Flores. 2001. Nitrogen control in cyanobacteria. J. Bacteriol. 183411-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herrero, A., A. M. Muro-Pastor, A. Valladares, and E. Flores. 2004. Cellular differentiation and the NtcA transcription factor in filamentous cyanobacteria. FEMS Microbiol. Rev. 28469-487. [DOI] [PubMed] [Google Scholar]
- 20.Hess, W. R. 2008. Comparative genomics of marine cyanobacteria and their phages, p. 89-116. In A. Herrero and E. Flores (ed.), The cyanobacteria: molecular biology, genomics and evolution. Caister Academic Press, Norfolk, United Kingdom.
- 21.Imamura, S., K. Tanaka, M. Shirai, and M. Asayama. 2006. Growth phase-dependent activation of nitrogen-related genes by a control network of group 1 and group 2 sigma factors in a cyanobacterium. J. Biol. Chem. 2812668-2675. [DOI] [PubMed] [Google Scholar]
- 22.Jiang, Y., and J. D. Gralla. 1995. Nucleotide requirements for activated RNA polymerase II open complex formation in vitro. J. Biol. Chem. 2701277-1281. [DOI] [PubMed] [Google Scholar]
- 23.Kerby, R. L., H. Youn, M. V. Thorsteinsson, and G. P. Roberts. 2003. Repositioning about the dimer interface of the transcription regulator CooA: a major signal transduction pathway between the effector and DNA-binding domains. J. Mol. Biol. 325809-823. [DOI] [PubMed] [Google Scholar]
- 24.Khoroshilova, N., H. Beinert, and P. J. Kiley. 1995. Association of a polynuclear iron-sulfur center with a mutant FNR protein enhances DNA binding. Proc. Natl. Acad. Sci. USA 922499-2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khudyakov, I., and C. P. Wolk. 1997. hetC, a gene coding for a protein similar to bacterial ABC protein exporters, is involved in early regulation of heterocyst differentiation in Anabaena sp. strain PCC 7120. J. Bacteriol. 1796971-6978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khudyakov, I. Y., and J. W. Golden. 2001. Identification and inactivation of three group 2 sigma factor genes in Anabaena sp. strain PCC 7120. J. Bacteriol. 1836667-6675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Knoll A. H. 2008. Cyanobacteria and Earth history, p. 1-19. In A. Herrero and E. Flores (ed.), The cyanobacteria: molecular biology, genomics and evolution. Caister Academic Press, Norfolk, United Kingdom.
- 28.Kolb, A., S. Busby, H. Buc, S. Garges, and S. Adhya. 1993. Transcriptional regulation by cAMP and its receptor protein. Annu. Rev. Biochem. 62749-795. [DOI] [PubMed] [Google Scholar]
- 29.Laurent, S., H. Chen, S. Bedu, F. Ziarelli, L. Peng, and C.-C. Zhang. 2005. Nonmetabolizable analogue of 2-oxoglutarate elicits heterocyst differentiation under repressive conditions in Anabaena sp. PCC 7120. Proc. Natl. Acad. Sci. USA 1029907-9912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lawson, C. L., D. Swigon, K. S. Murakami, S. A. Darst, H. M. Berman, and R. H. Ebright. 2004. Catabolite activator protein: DNA binding and transcription activation. Curr. Opin. Struct. Biol. 1410-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li, J.-H., S. Laurent, V. Konde, S. Bédu, and C.-C. Zhang. 2003. An increase in the level of 2-oxoglutarate promotes heterocyst development in the cyanobacterium Anabaena sp. PCC 7120. Microbiology 1493257-3263. [DOI] [PubMed] [Google Scholar]
- 32.Luque, I., E. Flores, and A. Herrero. 1994. Molecular mechanism for the operation of nitrogen control in cyanobacteria. EMBO J. 132862-2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luque, I., and K. Forchhammer. 2008. Nitrogen assimilation and C/N balance sensing, p. 335-382. In A. Herrero and E. Flores (ed.), The cyanobacteria: molecular biology, genomics and evolution. Caister Academic Press, Norfolk, United Kingdom.
- 34.Luque, I., M. F. Vázquez-Bermúdez, J. Paz-Yepes, E. Flores, and A. Herrero. 2004. In vivo activity of the nitrogen control transcription factor NtcA is subjected to metabolic regulation in Synechococcus sp. strain PCC 7942. FEMS Microbiol. Lett. 23647-52. [DOI] [PubMed] [Google Scholar]
- 35.Muro-Pastor, A. M., E. Olmedo-Verd, and E. Flores. 2006. All4312, an NtcA-regulated two-component response regulator in Anabaena sp. strain PCC 7120. FEMS Microbiol. Lett. 256171-177. [DOI] [PubMed] [Google Scholar]
- 36.Muro-Pastor, A. M., A. Valladares, E. Flores, and A. Herrero. 1999. The hetC gene is a direct target of the NtcA transcriptional regulator in cyanobacterial heterocyst development. J. Bacteriol. 1816664-6669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muro-Pastor, A. M., A. Valladares, E. Flores, and A. Herrero. 2002. Mutual dependence of the expression of the cell differentiation regulatory protein HetR and the global nitrogen regulator NtcA during heterocyst development. Mol. Microbiol. 441377-1385. [DOI] [PubMed] [Google Scholar]
- 38.Muro-Pastor, M. I., J. C. Reyes, and F. J. Florencio. 1996. The NADP+-isocitrate dehydrogenase gene (icd) is nitrogen regulated in cyanobacteria. J. Bacteriol. 1784070-4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Muro-Pastor, M. I., J. C. Reyes, and F. J. Florencio. 2001. Cyanobacteria perceive nitrogen status by sensing intracellular 2-oxoglutarate levels. J. Biol. Chem. 27638320-38328. [DOI] [PubMed] [Google Scholar]
- 40.Olmedo-Verd, E., E. Flores, A. Herrero, and A. M. Muro-Pastor. 2005. HetR-dependent and -independent expression of heterocyst-related genes in an Anabaena strain overproducing the NtcA transcription factor. J. Bacteriol. 1871985-1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Olmedo-Verd, E., A. M. Muro-Pastor, E. Flores, and A. Herrero. 2006. Localized induction of the ntcA regulatory gene in developing heterocysts of Anabaena sp. strain PCC 7120. J. Bacteriol. 1886694-6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schneider, G. J., M. E. Tumer, C. Richard, G. Borbely, and R. Haselkorn. 1987. Purification and characterization of RNA polymerase from the cyanobacterium Anabaena 7120. J. Biol. Chem. 2621433-1439. [PubMed] [Google Scholar]
- 43.Stal, L. J., and J. P. Zehr. 2008. Cyanobacterial nitrogen fixation in the ocean: diversity, regulation and ecology, p. 423-446. In A. Herrero and E. Flores (ed.), The cyanobacteria: molecular biology, genomics and evolution. Caister Academic Press, Norfolk, United Kingdom.
- 44.Tang, H., K. Severinov, A. Goldfarb, and R. H. Ebright. 1995. Rapid RNA polymerase genetics: one-day, no-column preparation of reconstituted recombinant Escherichia coli RNA polymerase. Proc. Natl. Acad. Sci. USA 924902-4906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tanigawa, R., M. Shirokane, S.-I. Maeda, T. Omata, K. Tanaka, and H. Takahashi. 2002. Transcriptional activation of NtcA-dependent promoters of Synechococcus sp. PCC 7942 by 2-oxoglutarate in vitro. Proc. Natl. Acad. Sci. USA 994251-4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thiel, T., and B. Pratte. 2001. Effect on heterocyst differentiation of nitrogen fixation in vegetative cells of the cyanobacterium Anabaena variabilis ATCC 29413. J. Bacteriol. 183280-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vázquez-Bermúdez, M. F., E. Flores, and A. Herrero. 2002. Analysis of binding sites for the nitrogen-control transcription factor NtcA in the promoters of Synechococcus nitrogen-regulated genes. Biochim. Biophys. Acta 157895-98. [DOI] [PubMed] [Google Scholar]
- 48.Vázquez-Bermúdez, M. F., A. Herrero, and E. Flores. 2002. 2-Oxoglutarate increases the binding affinity of the NtcA (nitrogen control) transcription factor for the Synechococcus glnA promoter. FEBS Lett. 51271-74. [DOI] [PubMed] [Google Scholar]
- 49.Vázquez-Bermúdez, M. F., A. Herrero, and E. Flores. 2003. Carbon supply and 2-oxoglutarate effects on expression of nitrate reductase and nitrogen-regulated genes in Synechococcus sp. strain PCC 7942. FEMS Microbiol. Lett. 221155-159. [DOI] [PubMed] [Google Scholar]
- 50.Wei, T.-F., T. S. Ramasubramanian, and J. W. Golden. 1994. Anabaena sp. strain PCC 7120 ntcA gene required for growth on nitrate and heterocyst development. J. Bacteriol. 1764473-4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wolk, C. P., Y. Cai, and J.-M. Panoff. 1991. Use of a transposon with luciferase as a reporter to identify environmentally responsive genes in a cyanobacterium. Proc. Natl. Acad. Sci. USA 885355-5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wolk, C. P., A. Ernst, and J. Elhai. 1994. Heterocyst metabolism and development, p. 769-823. In D. A. Bryant (ed.), The molecular biology of cyanobacteria. Kluwer, Dordrecht, Germany.
- 53.Wong, F. C. Y., and J. C. Meeks. 2002. Establishment of a functional symbiosis between the cyanobacterium Nostoc punctiforme and the bryophyte Anthoceros punctatus requires genes involved in nitrogen control and initiation of heterocyst differentiation. Microbiology 148315-323. [DOI] [PubMed] [Google Scholar]
- 54.Xu, X., J. Elhai, and C. P. Wolk. 2008. Transcriptional and developmental responses by Anabaena to deprivation of fixed nitrogen, p. 383-422. In A. Herrero and E. Flores (ed.), The cyanobacteria: molecular biology, genomics and evolution. Caister Academic Press, Norfolk, United Kingdom.
- 55.Yoshimura, H., S. Okamoto, Y. Tsumuraya, and M. Ohmori. 2007. Group 3 sigma factor gene, sigJ, a key regulator of desiccation tolerance, regulates the synthesis of extracellular polysaccharide in cyanobacterium Anabaena sp. strain PCC 7120. DNA Res. 1413-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zehr, J. P., J. B. Waterbury, P. J. Turner, J. P. Montoya, E. Omoregie, G. F. Steward, A. Hansen, and D. M. Kart. 2001. Unicellular cyanobacteria fix N2 in the subtropical North Pacific Ocean. Nature 412635-638. [DOI] [PubMed] [Google Scholar]