Abstract
Chromatin immunoprecipitation and microarray (ChIP-chip) analysis showed that the nitric oxide (NO)-sensitive repressor NsrR from Escherichia coli binds in vivo to the promoters of the tynA and feaB genes. These genes encode the first two enzymes of a pathway that is required for the catabolism of phenylethylamine (PEA) and its hydroxylated derivatives tyramine and dopamine. Deletion of nsrR caused small increases in the activities of the tynA and feaB promoters in cultures grown on PEA. Overexpression of nsrR severely retarded growth on PEA and caused a marked repression of the tynA and feaB promoters. Both the growth defect and the promoter repression were reversed in the presence of a source of NO. These results are consistent with NsrR mediating repression of the tynA and feaB genes by binding (in an NO-sensitive fashion) to the sites identified by ChIP-chip. E. coli was shown to use 3-nitrotyramine as a nitrogen source for growth, conditions which partially induce the tynA and feaB promoters. Mutation of tynA (but not feaB) prevented growth on 3-nitrotyramine. Growth yields, mutant phenotypes, and analyses of culture supernatants suggested that 3-nitrotyramine is oxidized to 4-hydroxy-3-nitrophenylacetate, with growth occurring at the expense of the amino group of 3-nitrotyramine. Accordingly, enzyme assays showed that 3-nitrotyramine and its oxidation product (4-hydroxy-3-nitrophenylacetaldehyde) could be oxidized by the enzymes encoded by tynA and feaB, respectively. The results suggest that an additional physiological role of the PEA catabolic pathway is to metabolize nitroaromatic compounds that may accumulate in cells exposed to NO.
Phagocytic cells of the mammalian innate immune system synthesize both superoxide and nitric oxide (NO) in response to infection (12). Superoxide and NO react to form peroxynitrite (ONOO−), which nitrates tyrosine residues in proteins, forming 3-nitrotyrosine (3-NTyr). Immunological detection of 3-NTyr suggests that bacterial proteins can be nitrated in vivo within phagocytic cells (7, 11). Under some pathological conditions, host proteins may also become nitrated on tyrosine residues (27). Tyrosine nitration can lead to the loss or perturbation of enzyme activity. For example, nitration of the Escherichia coli glutamine synthetase mimics the regulatory modification (adenylylation), while nitration of the adenylylated form of the enzyme causes complete inactivation (3). It seems likely that the nitration of tyrosine residues by peroxynitrite in phagocytic cells contributes to their killing activity, though this has not been demonstrated conclusively. Several bacterial pathogens express a peroxynitrite reductase activity, which may be an important virulence determinant (6). It is possible that the ability to repair the damage caused by peroxynitrite is also an important contributor to the ability of some pathogens to escape phagocyte killing.
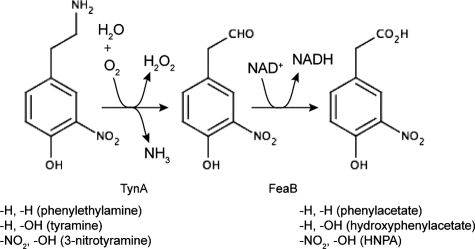
Protein tyrosine nitration has been described as irreversible (3), and E. coli cell extracts show no evidence of an ability to repair nitrated proteins (23). On the other hand, there is evidence for a protein tyrosine “denitrase” activity in rat tissues and in mitochondria (21, 22), and nitrated proteins may be subject to more rapid turnover than their native counterparts (15, 36). Despite these observations, the fate of nitrated proteins remains poorly understood. The degradation of nitrated proteins (whether or not it is selective) would liberate free 3-NTyr, and so there is some interest in the biochemical fate of this molecule in both host cells and invading pathogens. In rat PC12 cells, 3-NTyr can be converted to 4-hydroxy-3-nitrophenylacetate by the sequential action of an aromatic amino acid decarboxylase, an amine oxidase, and a NAD-linked dehydrogenase (4). The intermediates in this pathway are 3-nitrotyramine and 4-hydroxy-3-nitrophenylacetaldehyde (Fig. 1). Bacteria isolated on the basis of their ability to use 3-NTyr as a carbon and energy source convert 3-NTyr to 4-hydroxy-3-nitrophenylacetate via 4-hydroxy-3-nitrophenylpyruvate through the sequential action of a deaminase and a decarboxylase. The nitro group is then removed from 4-hydroxy-3-nitrophenylacetate by a novel denitrase activity (29).
FIG. 1.
Pathways for the catabolism of phenylethylamine and tyramine and a proposed pathway for the catabolism of 3-nitrotyramine. The first reaction is catalyzed by the amine oxidase (TynA) and the second by a NAD-linked dehydrogenase (FeaB). Substituents at the 3 and 4 positions are shown below the structures along with the names of the corresponding compounds. HNPA, 4-hydroxy-3-nitrophenylacetate.
The responses of bacteria to oxygen and nitrogen radicals attract considerable interest, in part because of their roles in the innate immune response (12). In the case of NO, diverse bacteria express several different NO detoxification enzymes, and there are numerous regulatory systems that have been reported to respond to NO (12, 38). In the context of the current work, the regulator of interest is NsrR, a transcriptional repressor from the Rrf2 family, which probably contains an iron-sulfur cluster and is sensitive to sources of NO. NsrR has been shown to act as an NO-sensitive regulator of gene expression in several organisms (1, 2, 5, 14, 17, 19, 28, 31, 33), and targets for NsrR regulation have been predicted (34). In E. coli, NsrR regulates expression of the NO-detoxifying flavohemoglobin, along with several other genes and operons, some of which are of unknown or poorly understood function (5, 13, 24, 40).
In this report, we show that 3-nitrotyramine can be used as a nitrogen source by cultures of E. coli, supporting growth at slow rates. We present evidence that the pathway of 3-nitrotyramine degradation to 4-hydroxy-3-nitrophenylacetate is similar to that found in rat cells (4), involving a periplasmic amine oxidase (TynA, also known as MaoA) and a cytosolic NAD-linked dehydrogenase (FeaB, also known as PadA). The tynA and feaB promoters are bound by NsrR in vivo, and NsrR exerts a weak, though significant, degree of control on both promoters. Overexpression of NsrR represses the tynA and feaB promoters and severely retards growth on phenylethylamine (PEA), catabolism of which requires TynA and FeaB activities. Expression of the tynA and feaB genes is upregulated by growth on PEA and 3-nitrotyramine, regulation that requires an AraC-type regulator encoded by the feaR gene. We speculate that one physiological function of TynA and FeaB is to metabolize nitrated aromatic compounds that may accumulate in cells exposed to NO and superoxide.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
The strains and plasmids used in this work are listed in Table 1. Transposon insertions in the tynA and feaB genes of E. coli MG1655 were obtained by P1 transduction, using strains JD22473 (tynA::Tn10) and JD22470 (feaB::Tn10) from the National BioResource Project (Japan) as the donors. To construct an unmarked deletion in the lacZ gene of MG1655, we first introduced a lacZ::kan mutation from strain VJS8363 (a gift from Valley Stewart) and then removed the kanamycin resistance cartridge by site-specific recombination with pCP20 (8). The nsrR gene was disrupted by replacing the coding region with a kanamycin resistance cassette, using the λred recombinase method, with pKD4 as the template and primers designed to generate a nonpolar mutation (8). The mutation was transferred to other strains by P1 transduction. To convert the insertion mutation to an unmarked nonpolar deletion, we transformed the strains with pCP20, and kanamycin/ampicillin-sensitive transformants were identified after colony purification at 43°C (8). Reporter strains with feaR::kan mutations were constructed by P1 transduction using JW1379 (from the National BioResource Project, Japan) as the donor. The structures of all insertion and deletion mutants were confirmed at each step by PCR.
TABLE 1.
E. coli strains and plasmids used in this study
| Strain or plasmid | Genotype | Source or reference |
|---|---|---|
| E. coli strains | ||
| MG1655 | rph-1 | E. coli Genetic Stock Center |
| JOEY216 | MG1655 feaB::Tn10 | This work |
| JOEY217 | MG1655 tynA::Tn10 | This work |
| JOEY238 | MG1655 ΔnsrR | This work |
| JOEY270 | MG1655 ΔlacZ | This work |
| JOEY271 | JOEY270 λtynA-lacZ | This work |
| JOEY272 | JOEY270 λfeaB-lacZ | This work |
| JOEY335 | JOEY270 ΔnsrR | This work |
| JOEY336 | JOEY335 λfeaB-lacZ | This work |
| JOEY337 | JOEY335 λtynA-lacZ | This work |
| JOEY448 | JOEY270 λtynA-lacZ feaR::kan | This work |
| JOEY449 | JOEY335 λtynA-lacZ feaR::kan | This work |
| JOEY450 | JOEY270 λfeaB-lacZ feaR::kan | This work |
| JOEY451 | JOEY335 λfeaB-lacZ feaR::kan | This work |
| Plasmids | ||
| pRS415 | 35 | |
| pSTBlue-1 | Novagen | |
| p2795 | 18 | |
| pJP07 | nsrR, epitope tagged at the 3′ end, cloned in p2795 | This work |
The rich medium for routine propagation of E. coli strains was L broth (tryptone, 10 g liter−1; yeast extract, 5 g liter−1; NaCl, 5 g liter−1). For growth tests and enzyme determinations, a defined medium (37) was used, supplemented with the indicated carbon and nitrogen sources and with Casamino acids (0.05% [wt/vol]), as needed. For growth with alternative nitrogen sources, the ammonium sulfate in this medium was substituted with sodium sulfate. PEA has limited solubility in water, so it was added directly to the bulk medium, which was then sterilized by filtration. Growth on PEA is temperature sensitive (32) and is significantly improved by the addition of Casamino acids to growth media. Therefore, all PEA cultures were grown at 30°C in the presence of 0.05% (wt/vol) Casamino acids (in 250-ml flasks shaken at 250 rpm) and were inoculated with precultures grown in glucose minimal medium. For cultures grown on glucose with 3-nitrotyramine as the nitrogen source, we found that growth was improved by restricting the oxygen supply (which perhaps alleviated the oxidative stress associated with the production of hydrogen peroxide by the amine oxidase). Precultures were grown at 30°C in 5 ml of medium in 16-mm culture tubes rotated at ∼50 rpm. Experimental cultures were grown at 30°C in 20 to 50 ml of medium, in 250-ml flasks shaken at 60 to 70 rpm.
Genetic manipulations.
The tynA and feaB promoter regions (on 279- and 247-bp fragments, respectively) were amplified by PCR (primer sequences for these and other procedures are listed in Table S1 in the supplemental material) and cloned into pSTBlue-1, using methods similar to those described previously (5). Promoter fusions to lacZ were then constructed in pRS415, transferred to λRS45, and integrated into the chromosome as described previously (5, 35). The plasmid pJP07 contains the nsrR gene (with its own promoter) modified at the 3′ end by the addition of sequences encoding the 3XFlag epitope tag (41). The modified nsrR gene was amplified from the chromosome of strain JOEY135 (10) and cloned into p2795, a high-copy number plasmid derived from pBluescript (18). The C-terminal epitope tag has no detectable effect on the activity of NsrR, either in vivo or in vitro (unpublished work). The same modification was used to identify NsrR binding sites by chromatin immunoprecipitation and microarray analysis (ChIP-chip). For ChIP-chip experiments, published procedures were followed for strain constructions, growth of cultures, chromatin extraction, DNA labeling, array hybridization, and data analysis (10).
Enzyme assays.
Extracts for the TynA and FeaB activity assays were prepared from 50-ml cultures grown to late exponential phase. Cell pellets were washed three times and resuspended in 1 ml of basal minimal medium (with no carbon or nitrogen source). Cells were disrupted by sonication and then centrifuged at 16,000 × g at 4°C for 20 min. To remove membrane fragments, extracts were centrifuged at 100,000 × g at 4°C for 1 h.
To assay the amine oxidase TynA, we measured oxygen uptake rates at 30°C, using a Clark-type electrode (Hansatech Instruments, King's Lynn, Norfolk, England) in a 0.1 M phosphate buffer (pH 7.0), 1.5 mM Na2SO4. Reactions were started by the addition of 100 μM substrate, a concentration chosen to avoid substrate inhibition by PEA. FeaB activities were assayed at 30°C in 50 mM potassium phosphate (pH 7.0) containing 2 mM NAD+. Reactions were initiated by the addition of 50 μM PEA or 100 μM 3-nitrotyramine, and the absorbance at 340 nm was followed with a Cary 50 spectrophotometer (Varian, Palo Alto, CA).
Enzyme kinetic data were analyzed by direct curve fitting using Kaleidagraph (Synergy Software, Reading, PA) software. Where substrate inhibition was evident, data were fitted to the Haldane equation (equation 1); otherwise data were fitted to the Michaelis-Menten equation.
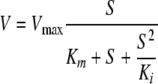 |
(1) |
β-Galactosidase activities were measured according to published protocols (25). All enzyme activities were measured in duplicate with samples from three independently grown cultures.
Chemicals and analytical methods.
3-Nitrotyramine was purchased from Apin Chemicals (Abingdon, United Kingdom). Concentrations of stock solutions of 3-nitrotyramine were determined spectrophotometrically. The molar extinction coefficient of 3-nitrotyramine (422 nm) at pH 7.5 is 2,800 M−1 cm−1 (26). Using this value, we determined the extinction coefficient to be 1,973 M−1 cm−1 at pH 7.0 and used this latter value for measuring the concentrations of stock solutions. Diethylenetriamine (DETA)-NONOate was purchased from Cayman Chemicals (Ann Arbor, MI). This compound decomposes at pH 7.4, with a half-life (t0.5) of 20 h at 37°C and 56 h at 22 to 25°C, and releases two equivalents of NO (Cayman Chemicals). The half-life of DETA-NONOate under the conditions of our experiments (pH 7.0; 30°C) is not known, but we assume that it is between 20 and 56 h, and the interpretation of results is not affected by the exact half-life of the compound. DETA-NONOate was added to cultures at the time of inoculation and was present throughout growth.
3-Nitrotyramine and 4-hydroxy-3-nitrophenylacetate concentrations were measured in filtered culture supernatants, using previously published methods (29), except that a 100-mm column was used for high-performance liquid chromatography (HPLC).
RESULTS
Regulation of tynA and feaB by NsrR.
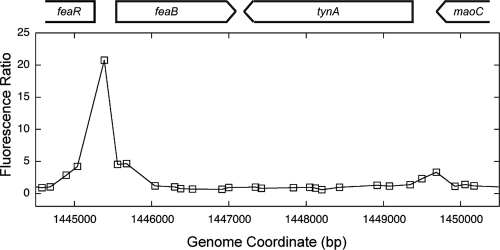
We have recently used transcriptomics (13) and ChIP-chip analysis (unpublished data) to identify E. coli genes regulated by NsrR. The ChIP-chip approach identified binding sites for NsrR in the intergenic region between feaB and the regulatory gene feaR and in the promoter region of the adjacent tynA gene (Fig. 2). Full results of the genome-wide mapping of NsrR binding sites using ChIP-chip will be published elsewhere. The tynA and feaB genes encode the first two enzymes of a pathway (Fig. 1) that is required for the utilization of PEA as a carbon and energy source or utilization of tyramine or dopamine as a nitrogen source (9).
FIG. 2.
ChIP-chip data for the feaR-feaB-tynA region of the E. coli chromosome. For the ChIP-chip experiments, cultures were grown anaerobically in the presence and absence of nitrate. In the absence of nitrate, NsrR binding sites should be occupied. In the presence of nitrate, some NO is generated as a by-product of nitrite respiration, and the NsrR regulon is derepressed (5), indicating that NsrR binding sites are vacant under these conditions. After ChIP, the two samples were labeled (with Cy5 and Cy3) and hybridized together to a high-density microarray. High fluorescence ratios therefore indicate the presence of NsrR binding sites, where binding is sensitive to the presence of nitrate in culture medium. Full technical details of this experiment have been published previously (10). The data shown are the mean fluorescence ratios from three experiments, with each of the three data sets centered on a mean ratio of 1 prior to averaging. The coordinates are for a noncurrent version of the E. coli MG1655 genome sequence (http://genolist.pasteur.fr/Colibri/).
To determine whether there is any regulation of these genes by NsrR, we fused the feaR, feaB, and tynA promoters to lacZ and transferred the reporter fusions in single copies to the chromosome. We found no evidence for NsrR regulation of the feaR promoter (data not shown). In exponential-phase cultures growing in complex medium (not shown) or in defined medium with glycerol as the carbon source, the tynA and feaB promoters had low activities in both a wild-type strain and an nsrR mutant (Table 2). However, in cultures grown on PEA as the sole source of carbon and energy, the activities of both promoters increased substantially (Table 2). Regulation of the tynA and feaB genes by PEA (and tyramine) has been observed previously (16, 42) and is presumed to involve FeaR, a predicted regulatory protein with an AraC-type DNA binding domain, though this regulation has not been confirmed biochemically (9). Accordingly, feaR mutants grew very poorly on PEA, and the tynA and feaB promoters were not induced by PEA in a feaR mutant (Table 2). In cultures of the nsrR mutant grown on PEA, we consistently observed small, though significant (20 to 50%), increases in feaB and tynA promoter activities (representative data are shown in Table 2). The low feaB promoter activities observed in a feaR mutant were also derepressed to a small extent in the feaR nsrR double mutant. Thus, the magnitude of the repression exerted by NsrR on the feaB promoter is similar in both the presence and the absence of FeaR.
TABLE 2.
Activities of reporter fusions to the tynA and feaB promoters in nsrR and feaR mutantsa
| Genotype | β-Galactosidase activity (Miller Units)b
|
|||||
|---|---|---|---|---|---|---|
| Glycerol
|
Phenylethylamine
|
Nitrotyramine
|
||||
| tynA-lacZ | feaB-lacZ | tynA-lacZ | feaB-lacZ | tynA-lacZ | feaB-lacZ | |
| Wild type | ND | 147 (9) | 849 (30) | 1,055 (51) | 333 (14) | 420 (37) |
| ΔnsrR | ND | 182 (10) | 1024 (81) | 1,570 (61) | 435 (5) | 487 (15) |
| feaR::kan | ND | 153 (15) | NDc | 87 (1) | NG | NG |
| ΔnsrR feaR::kan | ND | 195 (3) | ND | 104 (7) | NG | NG |
Cultures were grown at 30°C in defined medium containing 5 mM phenylethylamine or 40 mM glycerol as the carbon source or containing 11.1 mM glucose as the carbon source with 1.31 mM nitrotyramine as the nitrogen source.
Numbers in parentheses are 1 standard deviation. Units are as defined by Miller (25). ND, not detectable (<2 units μmol/min/mg protein). NG, no growth.
Growth of the feaR mutant on phenylethylamine was severely impaired.
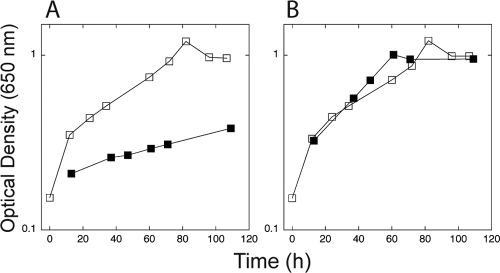
In contrast to the small effects of the nsrR mutation, we found that overexpression of nsrR (by increasing the gene copy number) had large effects. Transformation with a high-copy number plasmid carrying a cloned nsrR gene resulted in severely impaired growth on PEA (Fig. 3) and 17- and 6-fold reductions in the activities of the tynA and feaB promoters, respectively (Table 3). Addition of a compound (DETA-NONOate) that releases NO very slowly (t0.5 = 20 to 56 h) in these slow-growing cultures restored both growth on PEA and maximal promoter activities (Fig. 3 and Table 3). This suggests that increasing the copy number of the nsrR gene causes repression of the tynA and feaB promoters and, therefore, impaired growth on PEA. Under these conditions, the activities of both promoters can be regulated by NO. This repression by NsrR that can be alleviated by NO presumably involves NsrR binding to the sites identified by ChIP-chip analysis (Fig. 2). The cellular concentration of NsrR (under the conditions used for these experiments) seems to be poised such that its removal (by mutation) has small effects on these promoters, but overexpression causes severe repression. We were interested in determining the physiological properties of the feaB and tynA gene products that might provide a rationale for the inclusion of these genes in the NsrR regulon.
FIG. 3.
Overexpression of nsrR inhibits growth on PEA. (A) Cultures of JOEY272 transformed with pJP07 (which expresses nsrR, closed symbols) or with p2795 (vector control, open symbols) were cultured in media containing PEA as the sole source of carbon and energy. (B) The experiment shown in panel A was repeated, except that the culture medium were amended with 100 μM DETA-NONOate, a source of NO. The data shown are representative of multiple experiments. β-Galactosidase activities measured in cultures grown under these conditions are shown in Table 3.
TABLE 3.
Activities of the tynA and feaB promoters exposed to NOa
| Plasmid | β-Galactosidase activity (Miller Units)b
|
|||
|---|---|---|---|---|
|
tynA
|
feaB
|
|||
| −NO | +NO | −NO | +NO | |
| pJP07(nsrR) | 28 (3)b | 705 (39) | 188 (6) | 1491 (21) |
| p2795 (vector control) | 465 (21) | 662 (15) | 1136 (61) | 1331 (19) |
Activities are shown for the tynA and feaB promoters in reporter strains containing multiple copies of the nsrR gene and exposed (+) or not (−) to NO. Cultures grown with PEA as the carbon source were supplemented with 100 μM DETA-NONOate, which decomposes with a t0.5 of between 20 and 56 h under the conditions of this experiment, to release 2 equivalents of NO.
Numbers in parentheses are 1 standard deviation. Units are as defined by Miller (25).
Utilization of 3-nitrotyramine as a nitrogen source.
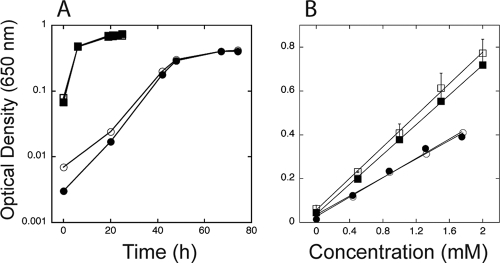
TynA and FeaB have broad substrate specificities and, besides PEA, can also oxidize tyramine to 4-hydroxyphenylacetate. E. coli K-12 strains cannot further oxidize 4-hydroxyphenylacetate and so use tyramine only as a nitrogen source. Dopamine may also be used as a substrate by this pathway and is oxidized to dihydroxyphenylacetate (9). Decarboxylation of 3-NTyr yields 3-nitrotyramine (4), and so we wondered whether 3-nitrotyramine might be a substrate for TynA and 4-hydroxy-3-nitrophenylacetaldehyde a substrate for FeaB. Assuming the presence of an as-yet-unidentified 3-NTyr decarboxylase, TynA and FeaB would provide a pathway for the conversion of 3-NTyr to 4-hydroxy-3-nitrophenylacetate (Fig. 1), a pathway similar to that described for rat PC12 cells (4). E. coli MG1655 grew slowly in a defined medium containing 3-nitrotyramine as the sole source of nitrogen (Fig. 4). The growth yield (as estimated by the final culture density per mole of substrate) of cultures grown on 3-nitrotyramine was ∼59% of that of cultures grown on (NH4)2SO4 (Fig. 4). Although we cannot exclude other physiological explanations for the reduced growth yield, it is at least consistent with the notion that only one nitrogen of 3-nitrotyramine can be assimilated. A tynA mutant of MG1655 failed to grow on 3-nitrotyramine (data not shown), whereas a feaB mutant grew with the same yield as that of the wild-type strain (Fig. 4). The phenotypes of the tynA and feaB mutants are consistent with the pathway shown in Fig. 1, and, together with the growth yield data suggest that growth on 3-nitrotyramine is at the expense of the amino group.
FIG. 4.
Utilization of 3-nitrotyramine as a nitrogen source for growth. (A) Growth of wild-type (open symbols) and nsrR mutant (closed symbols) cultures on glucose plus 2 mM ammonium sulfate (squares) or glucose plus 2 mM 3-nitrotyramine (circles). (B) Growth yields of E. coli MG1655 (open symbols) and a feaB mutant (closed symbols) for cultures using ammonia or 3-nitrotyramine as the nitrogen source. Cultures were grown in defined media containing (NH4)2SO4 (squares) or 3-nitrotyramine (circles) as the sole nitrogen source at the indicated concentrations. Optical densities (650 nm) of cultures were measured at the end of growth.
Growth on 3-nitrotyramine as the sole source of nitrogen induced the tynA and feaB promoters in a wild-type strain and in an nsrR mutant, the β-galactosidase activities being about 40% of those observed for cultures grown on PEA (Table 2). FeaR is thought to mediate substrate inducibility of the tynA and feaB genes (9), and a feaR mutant cannot grow on 3-nitrotyramine. Thus, upregulation of the two promoters by 3-nitrotyramine is independent of NsrR and probably requires FeaR. The identity of the ligand for FeaR has not been established; it may not be PEA (or tyramine), given that TynA is located in the periplasm and that its substrate is, therefore, presumably not transported into the cell. In any case, the simplest explanation for our results is that 3-nitrotyramine, or a molecule related to 3-nitrotyramine (perhaps 4-hydroxy-3-nitro-phenylacetaldehyde), can function with FeaR to control the activity of the tynA and feaB promoters. In nsrR mutants grown on 3-nitrotyramine, the tynA and feaB promoters showed activities that were modestly increased compared to those of the wild-type strains, as was the case for cultures grown on PEA (Table 2).
The pathway for 3-nitrotyramine catabolism.
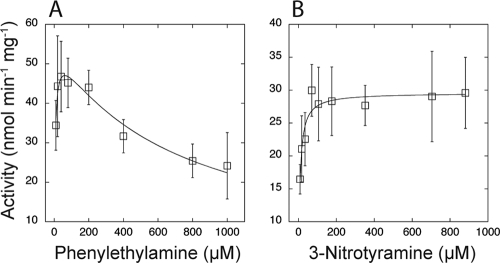
To test the prediction (Fig. 1) that 3-nitrotyramine is a substrate for TynA, we assayed substrate-dependent amine oxidase activity by following oxygen uptake by cell extracts in a Clark-type oxygen electrode. Using PEA as the substrate, we found evidence for substrate inhibition (Fig. 5), as has been reported previously (39). The activity data fitted well to the Haldane equation (equation 1) for substrate inhibition, with estimates of apparent Km = 5.5 ± 1.4 μM and Ki = 690 ± 109 μM. With 3-nitrotyramine as the substrate, oxygen uptake rates in the same cell extracts were somewhat lower (Vmax = 29.6 ± 0.7 versus 55.5 ± 2.9 nmol/min/mg protein for PEA) but followed Michaelis-Menten kinetics, with an estimated apparent Km value of 7.2 ± 1.3 μM (Fig. 5). The tynA mutant of E. coli MG1655 does not grow on PEA (conditions which are required to induce activity); therefore, we were unable to assay 3-nitrotyramine-dependent oxygen uptake in a tynA mutant (to provide direct proof that TynA is responsible for the measured activity). Nevertheless, other data we present in this paper lend confidence to the idea that TynA is the enzyme responsible for oxidizing 3-nitrotyramine.
FIG. 5.
Phenylethylamine and 3-nitrotyramine oxidation by cell extracts of E. coli MG1655. Enzyme activities were assayed by following oxygen uptake in a Clark-type electrode, with extracts of cells grown on phenylethylamine as the carbon and energy source. At each substrate concentration, activities were measured in duplicate, in extracts of three independently grown cultures. Each data point is therefore the mean of six determinations, and the error bars are 1 standard deviation. (A) With phenylethylamine as the substrate, there is evidence of substrate inhibition, as reported previously (39). Data were fitted to the Haldane equation (equation 1) for substrate inhibition, providing estimates of the apparent Km (5.5 ± 1.4 μM) and inhibition constant, Ki (680 ± 109 μM) values. (B) With 3-nitrotyramine as the substrate, there is no evidence of substrate inhibition. The data were therefore fitted to the Michaelis-Menten equation, with an estimated apparent Km value of 7.2 ± 1.3 μM.
We measured substrate-dependent oxygen uptake activities in cultures of MG1655 and in an nsrR mutant culture grown under a range of conditions that were similar to those used for assays of reporter fusions. The enzyme had extremely low activity or was undetectable in cells grown on glycerol (Table 4), which is consistent with the assays of tynA promoter activity (Table 2). TynA activity was detected in cells grown on PEA or 3-nitrotyramine (Table 4), which is again consistent with the reporter fusion assays. TynA activities (measured with either substrate) were ∼10-fold higher in PEA- versus 3-nitrotyramine-grown cells (Table 4), whereas the tynA promoter was only ∼2.5-fold more active in PEA-grown cells (Table 2). This discrepancy may be indicative of some posttranscriptional control of the tynA gene. Importantly, the activity assays provide additional confirmation of the suggestion that 3-nitrotyramine acts as an inducer of the catabolic pathway.
TABLE 4.
Phenylethylamine- and 3-nitrotyramine-dependent O2 uptakea
| Growth substrate | O2 uptake (nmol min−1 mg protein−1)b
|
|||
|---|---|---|---|---|
| MG1655
|
MG1655 ΔnsrR
|
|||
| Phenylethyl- amine | 3-Nitroty- ramine | Phenylethyl- amine | 3-Nitroty- ramine | |
| Phenylethylamine | 66 (5) | 41 (3) | 73 (11) | 44 (7) |
| Glycerol | ND | ND | ND | 3 (1) |
| 3-Nitrotyramine | 6 (3) | 4 (2) | 7 (2) | 4 (1) |
Phenylethylamine- and 3-nitrotyramine-dependent O2 uptake in extracts of cells grown on phenylethylamine or glycerol (as the carbon source) or on 3-nitrotyramine (as the nitrogen source). Cultures were grown at 30°C in defined medium containing 5 mM phenylethylamine or 40 mM glycerol as the carbon source or containing 11.1 mM glucose as the carbon source with 2.56 mM nitrotyramine as the nitrogen source.
Numbers in parentheses are 1 standard deviation (SD). ND, not detectable (<2 nmol min−1 mg protein−1).
We were unable to test the role of FeaB in 3-nitrotyramine catabolism directly, since the postulated substrate (4-hydroxy-3-nitrophenylacetaldehyde) is not commercially available. Therefore, we developed a coupled assay in which FeaB activity can be measured in cell extracts in the physiological direction by adding the substrate for TynA, which is oxidized in situ to generate the FeaB substrate. FeaB activity was measured by following the reduction of NAD+ to NADH. A feaB mutant can utilize both PEA (32) and 3-nitrotyramine (Fig. 4) as nitrogen sources, presumably because the mutant can liberate the amino group of PEA and 3-nitrotyramine through the activity of TynA. The feaB mutant grown on PEA as a nitrogen source showed no PEA- or 3-nitrotyramine-dependent reduction of NAD+ with the FeaB assay, confirming that FeaB is responsible for the measured activity.
Using the coupled NAD+-linked assay, we could detect FeaB activity in cell extracts with PEA as the substrate for the assay (Table 5), activities that did not differ significantly from those measured with the known FeaB substrate phenylacetaldehyde (data not shown). FeaB activity was low or undetectable in cells grown on glycerol, though this may be a reflection of the absence of TynA activity under these conditions. In cells grown on 3-nitrotyramine as the nitrogen source, FeaB activity was detectable at levels similar to those seen with cells grown on PEA (Table 5). The major conclusion that can be drawn from the results is that oxidation of 3-nitrotyramine by TynA generates an intermediate (presumably 4-hydroxy-3-nitrophenylacetaldehyde) that can be further oxidized by FeaB. Thus, the predicted product of the pathway is 4-hydroxy-3-nitrophenylacetate (Fig. 1). This hypothesis was tested by determination of 3-nitrotyramine and 4-hydroxy-3-nitrophenylacetate in culture supernatants, using HPLC. After the growth (of MG1655 and its nsrR mutant) on a limiting concentration (∼1 mM; Fig. 4), 3-nitrotyramine was undetectable in culture supernatants, and there was almost stoichiometric (88 to 90%) accumulation of 4-hydroxy-3-nitrophenylacetate (data not shown). Thus, 4-hydroxy-3-nitrophenylacetate is the likely end product of 3-nitrotyramine metabolism in E. coli.
TABLE 5.
Phenylethylamine- and 3-nitrotyramine-dependent NAD+ reductiona
| Growth substrate | NAD+ reduction (nmol min−1 mg protein−1)b
|
|||
|---|---|---|---|---|
| MG1655
|
MG1655 ΔnsrR
|
|||
| Phenylethyl- amine | 3-Nitroty- ramine | Phenylethyl- amine | 3-Nitroty- ramine | |
| Phenylethylamine | 38 (2.3) | 18 (1.6) | 35 (2.8) | 15 (2.2) |
| Glycerol | ND | 2.4 (1.0) | 1.0 (0.7) | 1.3 (0.3) |
| 3-Nitrotyramine | 54 (2.5) | 20 (6.4) | 42 (4.4) | 14 (4.0) |
Data show phenylethylamine- and 3-nitrotyramine-dependent NAD+ reduction in extracts of cells grown on phenylethylamine or glycerol (as the carbon source) or on nitrotyramine (as the nitrogen source). Cultures were grown at 30°C in defined medium containing 5 mM phenylethylamine or 40 mM glycerol as the carbon source or containing 11.1 mM glucose as the carbon source with 1.3 mM nitrotyramine as the nitrogen source.
Numbers in parentheses are 1 standard deviation (SD). ND, not detectable (<1 nmol min−1 mg protein−1).
DISCUSSION
The starting point for the work described in this paper was the discovery, using ChIP-chip analysis, of NsrR binding sites in the tynA and feaB promoter regions. We went on to show that NsrR can function as a regulator of tynA and feaB expression and that the enzymes encoded by these genes can oxidize 3-nitrotyramine, in addition to the previously described substrates. These findings illustrate one advantage of the ChIP-chip approach as a means of identifying regulon members. We could not have discovered NsrR regulation of tynA and feaB by comparing the transcriptomes (or proteomes) of a wild-type strain and an nsrR mutant, because (i) regulation requires cultures to be grown on PEA, and there would be no a priori reason to choose those growth conditions for a transcriptomics experiment; and (ii) revealing the full extent of NsrR regulation requires overexpression rather than deletion of nsrR; again, it is unlikely we would have chosen to use those conditions in a transcriptomics experiment. The tynA and feaB promoters have not been well characterized, and the nature of the DNA sequence recognized by NsrR is incompletely understood, although a consensus sequence has been proposed, which is a long inverted repeat (5, 34). Analysis of the ChIP-chip targets (unpublished work) suggests that NsrR can bind to half of the inverted repeat sequence, but, in the absence of in vitro data, we cannot reach firm conclusions about the locations of the NsrR binding sites in the tynA and feaB promoters.
Deletion of nsrR has very small effects on the tynA and feaB promoters, while overexpression of nsrR causes severe repression. We have observed similar effects at some other NsrR-regulated promoters (unpublished work) and believe that the concentration of NsrR is typically poised in a range that is insufficient to repress some promoters that are potentially controlled by NsrR. In this case, understanding the factors that regulate expression of the nsrR gene becomes especially important, since conditions that lead to the upregulation of nsrR would potentially lead to the regulation of promoters (such as tynA and feaB) that may otherwise escape repression. In this context, we have found that the nsrR promoter is ∼twofold more active in cultures grown in minimal medium than in medium supplemented with amino acids (unpublished data). This effect, albeit small, may provide an explanation for our observation that good growth on PEA requires the addition of amino acids to growth media.
The amine oxidase (TynA) and phenylacetaldehyde dehydrogenase (FeaB) enzymes of E. coli K-12 strains have been viewed as providing a straightforward pathway for the catabolism of PEA, tyramine, and dopamine (9). Two recently published observations suggest that these enzymes might have alternative and/or additional physiological roles. First, tynA mutants express the SOS response constitutively, which has been interpreted as indicating that the amine oxidase is responsible for removing an endogenously generated genotoxic compound (30). Second, feaB was identified in a screen for genes important for survival under planktonic (versus biofilm) growth conditions (20). This observation implies that a substrate for FeaB was present in the minimal growth medium used or can be generated endogenously. Thus, there is circumstantial evidence from independent studies to suggest that TynA and FeaB might have roles in catabolizing endogenously generated substrates. The substrate inhibition of TynA (Fig. 5) indicates that the enzyme is significantly inhibited by the concentrations of PEA typically used in growth media (1 mM). Since TynA is located in the periplasm (32), it is exposed to the medium concentration of PEA. The inhibition of TynA by physiologically relevant concentrations of PEA suggests that the enzyme is not optimally suited to a major role in PEA catabolism, which may account for the very slow growth on PEA (Fig. 3). Our results clearly show that TynA and FeaB also provide a pathway for the metabolism of 3-nitrotyramine (which does not exert substrate inhibition on TynA) and that the corresponding genes are regulated by NsrR and by NO. A rationale for this regulatory pattern may be provided if 3-nitrotyramine accumulates, and must be disposed of, in cells exposed to NO. 3-Nitrotyramine can be generated by the decarboxylation of 3-NTyr (4), which may accumulate in cells exposed to NO and superoxide (12). However, E. coli strains are not known to express or encode an aromatic amino acid decarboxylase, and there is therefore no known pathway from 3-NTyr to 3-nitrotyramine. Accordingly, 3-NTyr cannot be used as a nitrogen source by E. coli MG1655, and there is no 3-NTyr-mediated stimulation of oxygen uptake or NAD+ reduction in cell extracts (L. D. Rankin, and S. Spiro, unpublished observations). It remains to be seen whether 3-nitrotyramine is a physiologically significant substrate for TynA, either endogenously generated or encountered in natural environments. The wider significance of these observations also remains to be established. Homologs of tynA and feaB have restricted distributions in sequenced genomes and are found in the same organism quite infrequently. The feaR-feaB-tynA region of E. coli strain MG1655 is absent from several other E. coli strains, including some clinical isolates. Thus, the metabolism we have identified may not be ubiquitously important. On the other hand, we would predict that organisms capable of expressing tyrosine decarboxylase along with homologs of tynA and feaB are potentially capable of degrading 3-nitrotyrosine.
Supplementary Material
Acknowledgments
We thank Valley Stewart, Barry Wanner, Michael Hensel, and the National Bioresource Project (Japan) for gifts of strains and plasmids.
This work was supported by grant MCB-0702858 from the National Science Foundation (to S.S.) and by grant AB07CBT002 from the Army Research Office and the Defense Threat Reduction Agency (to J.C.S.).
Footnotes
Published ahead of print on 25 July 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Bang, I. S., L. Liu, A. Vazquez-Torres, M. L. Crouch, J. S. Stamler, and F. C. Fang. 2006. Maintenance of nitric oxide and redox homeostasis by the Salmonella flavohemoglobin Hmp. J. Biol. Chem. 28128039-28047. [DOI] [PubMed] [Google Scholar]
- 2.Beaumont, H. J. E., S. I. Lens, W. N. M. Reijnders, H. V. Westerhoff, and R. J. M. van Spanning. 2004. Expression of nitrite reductase in Nitrosomonas europaea involves NsrR, a novel nitrite-sensitive transcription repressor. Mol. Microbiol. 54148-158. [DOI] [PubMed] [Google Scholar]
- 3.Berlett, B. S., B. Friguet, M. B. Yim, P. B. Chock, and E. R. Stadtman. 1996. Peroxynitrite-mediated nitration of tyrosine residues in Escherichia coli glutamine synthetase mimics adenylylation: relevance to signal transduction. Proc. Natl. Acad. Sci. USA 931776-1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blanchard-Fillion, B., D. Prou, M. Polydoro, D. Spielberg, E. Tsika, Z. Wang, S. L. Hazen, M. Koval, S. Przedborski, and H. Ischiropoulos. 2006. Metabolism of 3-nitrotyrosine induces apoptotic death in dopaminergic cells. J. Neurosci. 266124-6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bodenmiller, D. M., and S. Spiro. 2006. The yjeB (nsrR) gene of Escherichia coli encodes a nitric oxide-sensitive transcriptional regulator. J. Bacteriol. 188874-881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bryk, R., P. Griffin, and C. Nathan. 2000. Peroxynitrite reductase activity of bacterial peroxiredoxins. Nature 407211-215. [DOI] [PubMed] [Google Scholar]
- 7.Chakravortty, D., I. Hansen-Wester, and M. Hensel. 2002. Salmonella Pathogenicity island 2 mediates protection of intracellular Salmonella from reactive nitrogen intermediates. J. Exp. Med. 1951155-1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 976640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diaz, E., A. Ferrandez, M. A. Prieto, and J. L. Garcia. 2001. Biodegradation of aromatic compounds by Escherichia coli. Microbiol. Mol. Biol. Rev. 65523-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Efromovich, S., D. Grainger, D. Bodenmiller, and S. Spiro. 2008. Genome-wide identification of binding sites for the nitric oxide sensitive transcriptional regulator NsrR. Methods Enzymol. 437211-233. [DOI] [PubMed] [Google Scholar]
- 11.Evans, T. J., L. D. K. Buttery, A. Carpenter, D. R. Springall, J. M. Polak, and J. Cohen. 1996. Cytokine-treated human neutrophils contain inducible nitric oxide synthase that produces nitration of ingested bacteria. Proc. Natl. Acad. Sci. USA 939553-9558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fang, F. C. 2004. Antimicrobial reactive oxygen and nitrogen species: concepts and controversies. Nat. Rev. Microbiol. 2820-832. [DOI] [PubMed] [Google Scholar]
- 13.Filenko, N., S. Spiro, D. F. Browning, D. Squire, T. W. Overton, J. Cole, and C. Constantinidou. 2007. The NsrR regulon of Escherichia coli K-12 includes genes encoding the hybrid cluster protein and the periplasmic, respiratory nitrite reductase. J. Bacteriol. 1894410-4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilberthorpe, N. J., M. E. Lee, T. M. Stevanin, R. C. Read, and R. K. Poole. 2007. NsrR: a key regulator circumventing Salmonella enterica serovar Typhimurium oxidative and nitrosative stress in vitro and in IFN-γ-stimulated J774.2 macrophages. Microbiology 1531756-1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gow, A. J., D. Duran, S. Malcolm, and H. Ischiropoulos. 1996. Effects of peroxynitrite-induced protein modifications on tyrosine phosphorylation and degradation. FEBS Lett. 38563-66. [DOI] [PubMed] [Google Scholar]
- 16.Hanlon, S. P., T. K. Hill, M. A. Flavell, J. M. Stringfellow, and R. A. Cooper. 1997. 2-Phenylethylamine catabolism by Escherichia coli K-12: gene organization and expression. Microbiology 143513-518. [DOI] [PubMed] [Google Scholar]
- 17.Heurlier, K., M. J. Thomson, N. Aziz, and J. W. B. Moir. 2008. The nitric oxide (NO)-sensing repressor NsrR of Neisseria meningitidis has a compact regulon of genes involved in NO synthesis and detoxification. J. Bacteriol. 1902488-2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Husseiny, M. I., and M. Hensel. 2005. Rapid method for the construction of Salmonella enterica serovar Typhimurium vaccine carrier strains. Infect. Immun. 731598-1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Isabella, V., L. F. Wright, K. Barth, J. M. Spence, S. Grogan, C. A. Genco, and V. L. Clark. 2008. cis- and trans-acting elements involved in regulation of norB (norZ), the gene encoding nitric oxide reductase in Neisseria gonorrhoeae. Microbiology 154226-239. [DOI] [PubMed] [Google Scholar]
- 20.Junker, L. M., J. E. Peters, and A. G. Hay. 2006. Global analysis of candidate genes important for fitness in a competitive biofilm using DNA-array-based transposon mapping. Microbiology 1522233-2245. [DOI] [PubMed] [Google Scholar]
- 21.Kamisaki, Y., K. Wada, K. Bian, B. Balabanli, K. Davis, E. Martin, F. Behbod, Y.-C. Lee, and F. Murad. 1998. An activity in rat tissues that modifies nitrotyrosine-containing proteins. Proc. Natl. Acad. Sci. USA 9511584-11589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koeck, T., X. Fu, S. L. Hazen, J. W. Crabb, D. J. Stuehr, and K. S. Aulak. 2004. Rapid and selective oxygen-regulated protein tyrosine denitration and nitration in mitochondria. J. Biol. Chem. 27927257-27262. [DOI] [PubMed] [Google Scholar]
- 23.Lightfoot, R. T., D. Shuman, and H. Ischiropoulos. 2000. Oxygen-insensitive nitroreductases of Escherichia coli do not reduce 3-nitrotyrosine. Free Radic. Biol. Med. 281132-1136. [DOI] [PubMed] [Google Scholar]
- 24.Lin, H.-Y., P. J. Bledsoe, and V. Stewart. 2007. Activation of yeaR-yoaG operon transcription by the nitrate-responsive regulator NarL is independent of oxygen- responsive regulator Fnr in Escherichia coli K-12. J. Bacteriol. 1897539-7548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller, J. H. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 26.Monzani, E., R. Roncone, M. Galliano, W. H. Koppenol, and L. Casella. 2004. Mechanistic insight into the peroxidase catalyzed nitration of tyrosine derivatives by nitrite and hydrogen peroxide. Eur. J. Biochem. 271895-906. [DOI] [PubMed] [Google Scholar]
- 27.Morrissey, B. M., K. Schilling, J. V. Weil, P. E. Silkoff, and D. M. Rodman. 2002. Nitric oxide and protein nitration in the cystic fibrosis airway. Arch. Biochem. Biophys. 40633-39. [DOI] [PubMed] [Google Scholar]
- 28.Nakano, M. M., H. Geng, S. Nakano, and K. Kobayashi. 2006. The nitric oxide-responsive regulator NsrR controls ResDE-dependent gene expression. J. Bacteriol. 1885878-5887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nishino, S. F., and J. C. Spain. 2006. Biodegradation of 3-nitrotyrosine by Burkholderia sp. strain JS165 and Variovorax paradoxus JS171. Appl. Environ. Microbiol. 721040-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O'Reilly, E. K., and K. N. Kreuzer. 2004. Isolation of SOS constitutive mutants of Escherichia coli. J. Bacteriol. 1867149-7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Overton, T. W., R. Whitehead, Y. Li, L. A. S. Snyder, N. J. Saunders, H. Smith, and J. A. Cole. 2006. Coordinated regulation of the Neisseria gonorrhoeae truncated denitrification pathway by the nitric oxide-sensitive repressor, NsrR, and nitrite-insensitive NarQ-NarP. J. Biol. Chem. 28133115-33126. [DOI] [PubMed] [Google Scholar]
- 32.Parrott, S., S. Jones, and R. A. Cooper. 1987. 2-Phenylethylamine catabolism by Escherichia coli K12. J. Gen. Microbiol. 133347-351. [DOI] [PubMed] [Google Scholar]
- 33.Rock, J. D., M. J. Thomson, R. C. Read, and J. W. B. Moir. 2007. Regulation of denitrification genes in Neisseria meningitidis by nitric oxide and the repressor NsrR. J. Bacteriol. 1891138-1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rodionov, D. A., I. L. Dubchak, A. P. Arkin, E. J. Alm, and M. S. Gelfand. 2005. Dissimilatory metabolism of nitrogen oxides in bacteria: comparative reconstruction of transcriptional networks. PLOS Comp. Biol. 1e55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simons, R. W., F. Houman, and N. Kleckner. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 5385-96. [DOI] [PubMed] [Google Scholar]
- 36.Souza, J. M., I. Choi, Q. Chen, M. Weisse, E. Daikhin, M. Yudkoff, J. Obin, J. Ara, J. Horwitz, and H. Ischiropoulos. 2000. Proteolytic degradation of tyrosine nitrated proteins. Arch. Biochem. Biophys. 380360-366. [DOI] [PubMed] [Google Scholar]
- 37.Spencer, M. E., and J. R. Guest. 1973. Isolation and properties of fumarate reductase mutants of Escherichia coli. J. Bacteriol. 114563-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spiro, S. 2007. Regulators of bacterial responses to nitric oxide. FEMS Microbiol. Rev. 31193-211. [DOI] [PubMed] [Google Scholar]
- 39.Steinebach, V., J. A. Benen, R. Bader, P. W. Postma, S. de Vries, and J. A. Duine. 1996. Cloning of the maoA gene that encodes aromatic amine oxidase of Escherichia coli W3350 and characterization of the overexpressed enzyme. Eur. J. Biochem. 237584-591. [DOI] [PubMed] [Google Scholar]
- 40.Thompson, K. M., V. Rhodius, and S. Gottesman. 2007. σE regulates and is regulated by a small RNA in Escherichia coli. J. Bacteriol. 1894243-4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Uzzau, S., N. Figueroa-Bossi, S. Rubino, and L. Bossi. 2001. Epitope tagging of chromosomal genes in Salmonella. Proc. Natl. Acad. Sci. USA 9815264-15269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamashita, M., H. Azakami, N. Yokoro, J. H. Roh, H. Suzuki, H. Kumagai, and Y. Murooka. 1996. maoB, a gene that encodes a positive regulator of the monoamine oxidase gene (maoA) in Escherichia coli. J. Bacteriol. 1782941-2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.