Abstract
Decapentaplegic (Dpp) plays an essential role in Drosophila development, and analyses of the Dpp signaling pathway have contributed greatly to understanding of the actions of the TGF-β superfamily. Intracellular signaling of the TGF-β superfamily is mediated by Smad proteins, which are now grouped into three classes. Two Smads have been identified in Drosophila. Mothers against dpp (Mad) is a pathway-specific Smad, whereas Daughters against dpp (Dad) is an inhibitory Smad genetically shown to antagonize Dpp signaling. Here we report the identification of a common mediator Smad in Drosophila, which is closely related to human Smad4. Mad forms a heteromeric complex with Drosophila Smad4 (Medea) upon phosphorylation by Thick veins (Tkv), a type I receptor for Dpp. Dad stably associates with Tkv and thereby inhibits Tkv-induced Mad phosphorylation. Dad also blocks hetero-oligomerization and nuclear translocation of Mad. We also show that Mad exists as a monomer in the absence of Tkv stimulation. Tkv induces homo-oligomerization of Mad, and Dad inhibits this step. Finally, we propose a model for Dpp signaling by Drosophila Smad proteins.
INTRODUCTION
Members of the TGF-β superfamily regulate growth and differentiation of various cell lineages. TGF-βs, activins/inhibins, and bone morphogenetic proteins (BMPs) form three major subfamilies (Kingsley, 1994). In Drosophila, Decapentaplegic (Dpp), 60A, and Screw (Scw) have been identified as BMP-related molecules (Hogan, 1996). 60A is structurally similar to BMP-5, -6, -7, and -8, whereas Dpp is similar to BMP-2 and -4. The greater divergence in the sequence of Scw from members of the BMP family suggests that the orthologue of Scw has not been identified in vertebrates. Of the BMP-like ligands in Drosophila, Dpp is the best characterized. Dpp plays a pivotal role in Drosophila development. Dpp is required for the establishment of embryonic dorsal-ventral polarity, gut formation, and outgrowth and patterning of imaginal disks such as those of the wings and eyes (Sekelsky et al., 1995). Genetic analyses of dpp phenotypes have greatly contributed to elucidation of the mechanism of signaling by the TGF-β superfamily (Padgett et al., 1997).
TGF-β-related proteins bind to two types of transmembrane serine/threonine kinase receptors, termed types I and II (Kingsley, 1994; Derynck and Feng, 1997). The type II kinase is constitutively active. Upon ligand binding, the type I and II receptors form a heteromeric complex. Type II then transphosphorylates type I at the juxtamembrane region and activates the type I kinase (Wrana et al., 1994a; Wieser et al., 1995; Souchelnytskyi et al., 1996). Type I is the effector subunit of the receptor complex. An amino acid change in the juxtamembrane region of type I receptors results in constitutive activation of the kinases (Wieser et al., 1995; Derynck and Feng, 1997). These mutant type I receptors elicit ligand-specific responses in the absence of ligands or type II receptors, indicating that type I receptors phosphorylate downstream signaling components.
In Drosophila, Punt is the type II receptor for Dpp, and Thick veins (Tkv) and Saxophone (Sax) are the type I receptors for it (Brummel et al., 1994; Nellen et al., 1994; Penton et al., 1994; Xie et al., 1994). Another type I receptor in Drosophila, Atr-I, binds activin in the presence of Punt (Wrana et al., 1994b). Tkv and Sax have at least partial functional overlap in vivo, but null mutations of the genes result in distinct phenotypes (Brummel et al., 1994). Null tkv homozygotes die during late embryonic stages and fail to undergo dorsal closure. In contrast, sax-null homozygotes die during late larval stages and produce little or no imaginal disks. A model has been proposed in which Sax responds only to high concentrations of Dpp in the dorsal-most region of the Drosophila embryo, and Tkv responds to lower levels of Dpp throughout the region of the presumptive ectoderm normally specified by Dpp (Nellen et al., 1994). The difference between the kinase domains of the two receptors also suggests that they may target different substrates (Kawabata et al., 1998a). Recently, a short region with nine amino acids between the kinase subdomains IV and V of type I receptors, termed “L45 loop,” was shown to determine the signaling specificity of type I receptors (Feng and Derynck, 1997). This region of Tkv is similar to that of BMP type IA (BMPR-IA/ALK3) and type IB (BMPR-IB/ALK6) receptors, whereas Sax is related to ALK1 and activin type I receptor (ActR-I/ALK2).
The first of the substrates of the receptors for the TGF-β superfamily was identified through genetic screens in Drosophila (Raftery et al., 1995; Sekelsky et al., 1995; Padgett et al., 1997). Mothers against dpp (Mad) was identified as a genetic enhancer of dpp phenotypes. Subsequently three sma genes were identified in Caenorhabditis elegans as genes involved in the signaling pathway of daf-4, a type II receptor for an unidentified BMP-like ligand (Savage et al., 1996). An increasing number of vertebrate homologues of Mad and sma have been identified and are now generically denoted Smad. Nine vertebrate Smads have been reported (Heldin et al., 1997; Padgett et al., 1997) and have been grouped into three classes based on structure and function. Pathway-specific Smads are directly phosphorylated by type I receptors. Smad2 and Smad3 are substrates of the TGF-β and activin receptors, whereas Smad1, Smad5, and possibly Smad8/MADH6 propagate BMP-specific signals. In contrast, Smad4, which belongs to the second class, is a common mediator required by all pathways. Phosphorylated pathway-specific Smads form heteromeric complexes with Smad4, translocate into the nucleus, and activate a certain set of genes. In the case of the Mix.2 gene, expression of which is induced by activin in Xenopus embryos, a novel transcription factor, forkhead activin signal transducer-1, was shown to be incorporated in the Smad complex (Chen et al., 1996). The Smad–forkhead activin signal transducer-1 complex directly binds to the activin-responsive element in the Mix.2 promoter and activates its transcription.
Smads in the third class antagonize signaling by pathway-specific Smads and Smad4. Smad6 (Imamura et al., 1997; Hata et al., 1998), Smad7 (Hayashi et al., 1997; Nakao, 1997b), and Xenopus Smad8 (XSmad8) (Nakayama et al., 1998) have been shown to inhibit TGF-β/activin and/or BMP signalling. The common structure of Smads in this class diverges from that of the other Smads (Heldin et al., 1997). Pathway-specific Smads share two conserved regions, the MH1 domain in the N-terminal part and the MH2 domain in the C-terminal part, and have the Ser-Ser-X-Ser (SSXS) motif at the C-terminal end. The last two serines of the SSXS motif are sites of direct phosphorylation by the type I receptors (Abdollah et al., 1997; Souchelnytskyi et al., 1997). Smad4 contains the MH1 and MH2 domains but not the SSXS motif. Inhibitory Smads, however, share only the MH2 domain, and their N-terminal half diverges from the conserved MH1 domain. Mechanisms by which inhibitory Smads exert their antagonistic effects have been examined in the mammalian system. Smad6 and 7 stably associate with type I receptors and then inhibit phosphorylation of pathway-specific Smads (Hayashi et al., 1997; Imamura et al., 1997; Nakao et al., 1997b). In BMP signaling, Smad6 may also compete with Smad4 in association with Smad1 (Hata et al., 1998). Daughters against dpp (Dad) was identified as a gene whose expression is induced by Dpp. Dad is structurally similar to these vertebrate inhibitory Smads and was shown to antagonize Dpp signaling (Tsuneizumi et al., 1997). Expressions of Dad, Smad6, and Smad7 are regulated by ligands, and the autoregulatory feedback loop via inhibitory Smads seems to be conserved between invertebrates and vertebrates (Nakao et al., 1997b; Tsuneizumi et al., 1997; Takase et al., 1998).
The protein sequence of Mad is closely related to that of Smad1/5/8 specific to BMP signals. Consistently, Mad functions downstream of Tkv, a receptor for BMP-related Dpp (Raftery et al., 1995; Sekelsky et al., 1995; Newfeld et al., 1996; Wiersdorff et al., 1996; Maduzia and Padgett, 1997; Newfeld et al., 1997). Although phosphorylation of Mad by Tkv has not been demonstrated, BMP-2 induced phosphorylation of endogenous Mad in Drosophila cell lines (Newfeld et al., 1997). Constitutively active Tkv caused nuclear accumulation of Mad proteins (Maduzia and Padgett, 1997). Mad has also been shown to bind to the “quadrant enhancer” of the vestigial gene, expression of which is induced by Dpp (Kim et al., 1997). Mad in this case binds to DNA through its MH1 domain. The molecular basis of the regulation of Mad activity has not been fully established. Here we identified Drosophila Smad4 and show that Mad interacts with Drosophila Smad4 upon phosphorylation by Tkv. We also examined negative regulation of Mad by Dad and found that Dad stably associates with Tkv and prevents Mad from being phosphorylated by the receptor. Furthermore, we show that homo-oligomerization of Mad is induced by Tkv, and that Dad inhibits this step.
MATERIALS AND METHODS
Plasmid Construction
Construction of the Drosophila Smad4 expression plasmid was performed as follows. The full coding region was amplified by PCR with simultaneous elimination of the internal EcoRI site. An EcoRI site and an XhoI site were attached to the N-terminus and the C-terminus, respectively. The EcoRI–XhoI Drosophila Smad4 fragment was subcloned into myc-pcDNA3, which adds a myc tag N-terminally to the insert (Imamura et al., 1997). The original construction of the expression plasmids of Smad1, Smad2, Smad4, and constitutively active TGF-β type I (TβR-I) and BMP type I (BMPR-I) receptors was previously described (Imamura et al., 1997). Smad1, Smad2, and Smad4 with an epitope tag were then subcloned into another expression vector, pcDEF3 (Goldman et al., 1996), to increase expression level. The sites used for resubcloning were BamHI and XbaI for Smad1, KpnI and XbaI for Smad2, and BamHI and XbaI for Smad4. The Mad and Dad expression plasmids were prepared in a similar manner. In each case, the coding region was amplified and inserted into an appropriate epitope-tagging expression vector at EcoRI and XhoI sites. Mad was subcloned into FLAG-pcDNA3 and myc-pcDNA3. Dad was subcloned into FLAG-pcDNA3 and myc-pcDNA3 then into pcDEF3 using KpnI and XbaI. The construction of the expression plasmids for the Drosophila receptors was performed as described elsewhere (Oeda et al., 1998). All of the PCR products were sequenced.
Cloning of Drosophila Smad4
The GenBank database was searched with the human Smad4 sequence (GenBank accession number U44378) using BLAST (Altschul et al., 1990), and a Drosophila expressed sequence tag clone (LD07433) with a high degree of homology was found. The N-terminus of LD07433 corresponded to the 79th amino acid of human Smad4, and the missing N-terminal region was obtained from screening of a Drosophila cDNA library from 4-d larvae in λgt10 (Clontech, Palo Alto, CA). The probe, excised using EcoRI and EcoRV, contained a 0.35-kb fragment from the N terminus of LD07433. The screening was performed using standard procedures. Five clones of various sizes and locations were obtained and partially sequenced. Several methionines around the putative N-terminus of the coding region were found, and the methionine that corresponds to the first methionine of human Smad4 was chosen as the starting amino acid. The cDNA clone obtained from screening was combined with LD07433 to obtain the full-length Drosophila Smad4. The coding region was sequenced, and the deduced amino acid sequence was aligned with other sequences using DNASTAR (Madison, WI).
Affinity Cross-Linking, Immunoprecipitation, and Western Blotting
COS-7 cells were used in transfection experiments. Cells were maintained in DMEM containing 10% FBS. Transfection was performed using DMRIE-C (Life Technologies, Gaithersburg, MD) or FuGENE 6 (Boehringer Mannheim, Indianapolis, IN).
Cells were transfected with an appropriate combination of expression plasmids, washed, scraped, and solubilized in a buffer containing 20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1% Triton X-100, 1% Trasylol, and 1 mM PMSF. Lysates were cleared and incubated with anti-FLAG M2 (Eastman Kodak, Rochester, NY) monoclonal antibody, followed by incubation with protein G-Sepharose beads (Amersham Pharmacia Biotech, Piscataway, NJ). The beads were washed four times with the solubilization buffer. Thereafter, the immunoprecipitates were eluted by boiling for 3 min in SDS sample buffer (100 mM Tris-HCl, pH 8.8, 0.01% bromophenol blue, 36% glycerol, 4% SDS) containing 10 mM dithiothreitol and subjected to SDS-gel electrophoresis. Proteins were then electrotransferred to nitrocellulose filters, immunoblotted with anti-myc 9E10 (PharMingen, San Diego, CA) antibody or anti-hemagglutinin (HA) 3F10 antibody (Boehringer Mannheim) and detected using the enhanced chemiluminescence detection system (Amersham, Pharmacia Biotech, Piscataway, NJ).
Iodination of BMP-2 and subsequent immunoprecipitation were performed as described (Nakao et al., 1997a). Briefly, BMP-2 was iodinated using the chloramine T method, and cross-linking was performed with disuccinimidyl suberate (Pierce, Rockford, IL). Cells were lysed and directly subjected to gel electrophoresis, or immunoprecipitation with anti-HA 12CA5 (Boehringer Mannheim) or anti-FLAG antibodies and protein A- or protein G-Sepharose beads (Amersham Pharmacia Biotech) followed by gel electrophoresis. Receptor complexes were detected using a Fuji BAS 2000 bioimaging analyzer (Fuji Photo Film, Tokyo, Japan).
For in vivo phosphorylation experiments, cells were labeled with [32P]orthophosphate for 4 h, treated with the indicated amount of BMP-2 for the last 1 h of [32P]orthophosphate labeling, and subjected to immunoprecipitation, gel electrophoresis, and analyses with autoradiography. The expression level of Mad was monitored by straight Western blotting or immunoprecipitation followed by Western blotting. The intensities of bands were determined, and the ratio between [32P]orthophosphate incorporation and protein expression was calculated. Phosphorylated Smad proteins were also detected by immunoprecipitation followed by Western blotting using anti-phosphoserine antibody (Zymed Laboratories, South San Francisco, CA).
Nuclear Translocation
Subcellular localization of Mad was determined by immunostaining. Cells were grown in LAB-TEK chambers (Nunc, Naperville, IL), transfected, washed with PBS, and fixed with acetone. Cells were then incubated with 5% normal horse serum, washed, incubated with anti-FLAG antibody, washed again, incubated with biotinylated antibody against mouse immunoglobulin, washed, and incubated with FITC-labeled streptavidin. After a final wash, cells were covered with glycerin and observed by fluorescence microscopy.
RESULTS
Cloning of Drosophila Smad4 (Medea)
Smad4 is a common mediator required by both TGF-β/activin and BMP signaling pathways (Heldin et al., 1997). Loss of Smad4 correlates with loss of responses to TGF-β/activin (de Winter et al., 1997; Grau et al., 1997; Zhou et al., 1998). Identification of the Smad4 ortholog in Drosophila is thus essential to investigation of the Dpp signaling pathway. We searched the expressed sequence tag database and found a Drosophila clone (LD07433) with a high degree of homology to human Smad4. LD07433 lacked the N-terminal region of the coding sequence, and we screened a Drosophila cDNA library using LD07433 as a probe. Multiple independent clones were isolated and sequenced. The predicted amino acid sequence of the open reading frame is shown in Figure 1. It encodes a protein of 745 amino acids with a calculated molecular mass of 78.9 kDa. The protein is most similar to Smad4 among vertebrate Smads. During the preparation of this manuscript, three papers describing the cloning of the Medea gene were reported (Das et al., 1998; Hudson et al., 1998; Wisotzkey et al., 1998). Our Drosophila Smad4 was identical to Medea, and we therefore refer to our clone as Medea. Medea has both MH1 and MH2 domains but lacks the SSXS motif. This structural feature is shared by Smad4. The MH1 and MH2 domains are highly conserved between Medea and Smad4, with >80% identity for both. Medea, however, contains a much longer linker region rich in glutamines, glycines, and prolines. Other Drosophila Smads, Mad and Dad, exhibit a lower degree of similarity with Medea (Figure 1).
Figure 1.
Alignment of the predicted protein sequence of Drosophila Smad4/Medea with those of human Smad4, Mad, and Dad. The first methionine of Medea shown was chosen based on its similarity to that of human Smad4. The open reading frame encodes 745 amino acids. Dashes were inserted to maximize the alignment score. Residues identical to those of Medea are highlighted. The region located between triangles is the linker. Medea and Smad4 are highly conserved in the MH1 and MH2 regions. Note that Medea is longer than human Smad4 by ∼200 amino acids because of its long linker region. The Medea sequence has been deposited in GenBank with accession number AF057162.
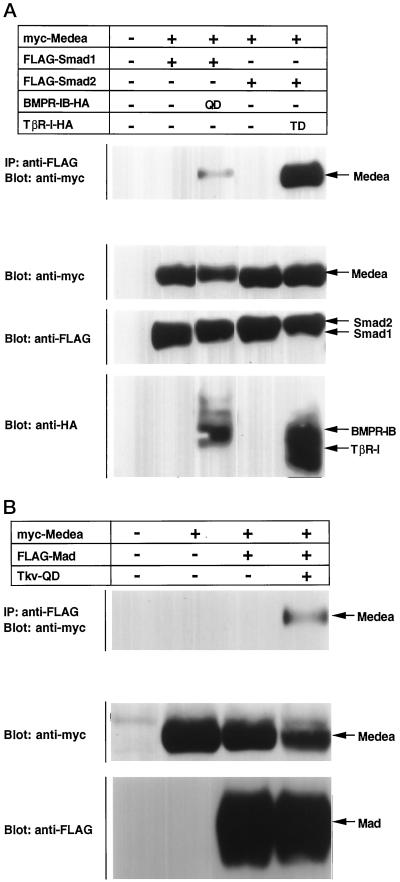
We examined whether Medea and Smad4 are functionally conserved. Smad1 interacted with Smad4 upon BMP stimulation, whereas Smad2 associated with Smad4 in a TGF-β-dependent manner. Smad1/2 was cotransfected into COS cells together with Medea in the absence or presence of constitutively active type I receptors (Figure 2A). Smad1 interacted with Medea in the presence of activated BMPR-IB/ALK6. Likewise, Smad2 bound to Medea in the presence of activated TGF-β type I receptor (TβR-I/ALK5). Medea thus has biochemical functions similar to those of Smad4, indicating that Medea is the orthologue of Smad4.
Figure 2.
Interaction of Medea with pathway-specific Smads. (A) FLAG-tagged Smad1 or FLAG-tagged Smad2 was introduced into COS cells with myc-tagged Medea in the absence or presence of the HA-tagged constitutively active type I receptor for BMP (BMPR-IB; QD) or TGF-β (TβR-I; TD). Interaction was detected by immunoprecipitation with anti-FLAG antibody followed by Western blotting with anti-myc antibody. The expression of Medea, Smad1, Smad2 and receptors was monitored by straight Western blotting with antibodies against the respective tags. (B) Association of Mad with Medea was examined in a similar experiment. FLAG-tagged Mad and myc-tagged Medea were expressed in COS cells in the absence or presence of constitutively active Tkv (Tkv-QD). The expression of each protein was monitored by straight Western blotting.
Mad Is Activated by Tkv
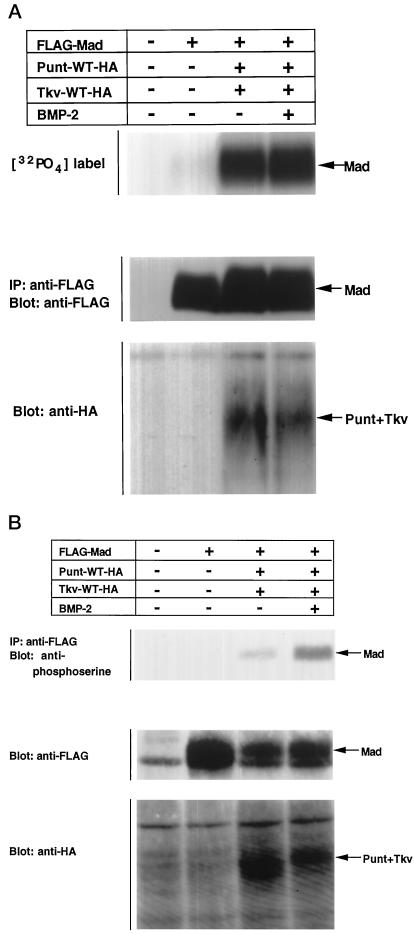
In vertebrates, pathway-specific Smads have been shown to associate with type I receptors and to undergo phosphorylation. Several lines of evidence strongly suggest that Tkv phosphorylates Mad (Maduzia and Padgett, 1997; Newfeld et al., 1997), but no direct evidence of this has been presented. We studied the phosphorylation of Mad by Tkv in two assays. Mad was introduced into COS cells with wild-type Tkv and Punt. Cells were labeled with [32P]orthophosphate, treated with BMP-2, lysed, and subjected to gel electrophoresis (Figure 3A). Coexpression of Tkv and Punt induced phosphorylation of Mad, which is probably caused by receptor activation through spontaneous association of the type I and II receptors, as has been demonstrated for TGF-β receptors (Souchelnytskyi et al., 1996). When BMP-2 was added, the phosphorylation of Mad was further enhanced.
Figure 3.
Phosphorylation of Mad by activated Tkv. (A) FLAG-tagged Mad was transfected into COS cells with or without Dpp receptors (Punt and Tkv). Cells were then labeled with [32P]orthophosphate, treated with 300 ng/ml BMP-2 for 1 h, lysed, and subjected to gel electrophoresis followed by analyses with a phosphorimager. The expression level of Mad was monitored by Western blotting. (B) Phosphorylation of Mad by Tkv was examined by Western blotting. Mad was immunoprecpitated with anti-FLAG antibody and immunoblotted with anti-phosphoserine antibody.
We next used anti-phosphoserine antibody (Figure 3B). This antibody has been shown to recognize ligand-dependent phosphorylation of Smad1, 2, 3, and 5 (our unpublished results) (Nishimura et al., 1998). COS cells were transfected with the expression plasmids for Mad and/or the BMP receptors, treated with BMP-2, lysed, and subjected to immunoprecipitation followed by Western blotting with anti-phosphoserine antibody. As in [32P]orthophosphate labeling, coexpression of Tkv and Punt induced phosphorylation of Mad, and BMP-2 treatment increased phosphorylation. This finding also indicates that phosphorylation by constitutively active Tkv (Tkv-QD) occurs at serine residues. Notably, Mad was slightly phosphorylated even in the absence of Tkv stimulation, as was found with [32P]orthophosphate labeling (Figure 3A), whereas anti-phosphoserine antibody did not recognize the basal phosphorylation of Mad (Figure 3B). Thus, the anti-phosphoserine antibody specifically detected phosphorylation of the SSXS motif by Tkv. Similar results were obtained with mammalian Smads (our unpublished results).
We next examined whether Tkv induces hetero-oligomerization of Mad with Medea. As shown in Figure 2B, Mad formed a complex with Medea in the presence of Tkv-QD, demonstrating that Mad and Medea act downstream of Tkv. Tkv-QD also caused nuclear translocation of Mad (see below) (Maduzia and Padgett, 1997).
Dad Interferes with Phosphorylation of Mad by Tkv
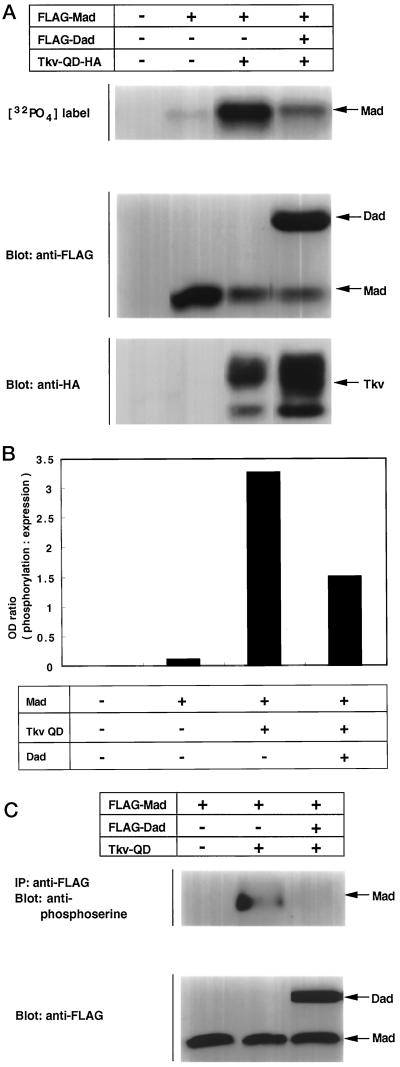
Dad has been genetically shown to inhibit Dpp signaling in vivo (Tsuneizumi et al., 1997). We examined the molecular basis of this inhibitory effect. The effect of Dad on Mad phosphorylation by Tkv was studied in the two assays described above. Various combinations of Mad, Dad, and Tkv-QD were introduced into COS cells. In the first experiment, cells were labeled with [32P]orthophosphate in vivo, and incorporation of radioactivity into Mad was detected. As in Figure 4, A and B, Dad inhibited phosphorylation of Mad by Tkv-QD. Next, anti-phosphoserine antibody was used. As in the orthophosphate labeling, phosphorylation of Mad diminished in the presence of Dad (Figure 4C).
Figure 4.
Inhibition by Dad of Tkv-induced phosphorylation of Mad. (A) Inhibition by Dad of Mad phosphorylation was examined by [32P]orthophosphate labeling. The experiment was performed as shown in Figure 3A. Expression levels of proteins were monitored by Western blotting. (B) Intensities of bands were determined with a densitometer, and the ratio between [32P]orthophosphate incorporation and protein expression level was calculated. (C) Effect of Dad on Tkv-induced Mad phosphorylation was examined. Anti-phosphoserine antibody was used as in Figure 3B. Expression levels of the proteins were monitored by Western blotting.
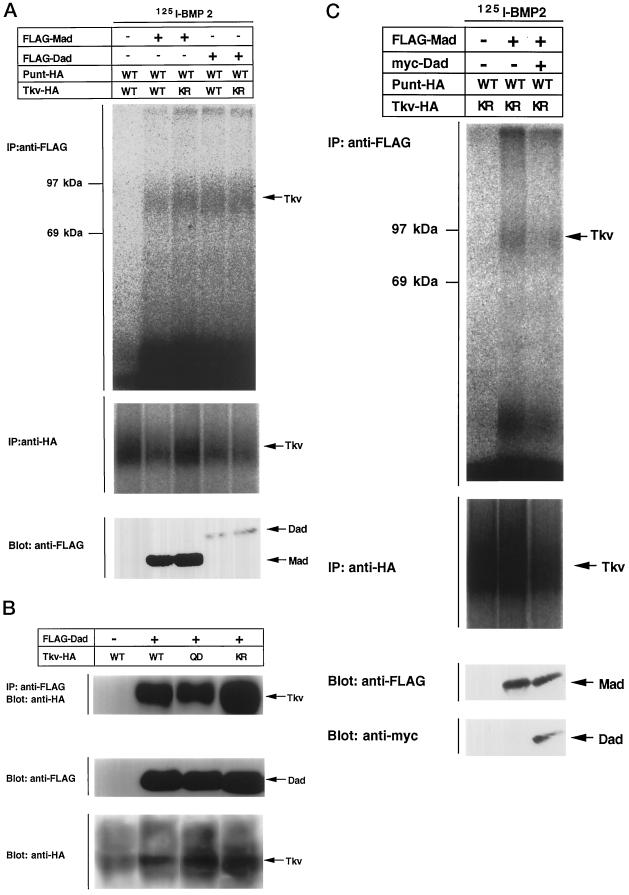
In vertebrates, inhibitory Smads such as Smad6 and Smad7 have been shown to stably associate with type I receptors (Hayashi et al., 1997; Imamura et al., 1997; Nakao et al., 1997b; Hata et al., 1998). We investigated the interaction of Mad or Dad with Tkv. Cells were transfected with an appropriate combination of expression plasmids, affinity labeled with iodinated BMP-2, and subjected to immunoprecipitation with antibodies against Mad or Dad. It was previously shown that pathway-specific Smads associate with type I receptors upon ligand stimulation, but that this interaction is too brief to detect under natural conditions (Macías-Silva et al., 1996). The interaction can be observed when the type I kinases are rendered inactive or when the C-terminal phosphorylation sites of the Smads are modified to be resistant to phosphorylation. Mad interacted with the kinase-defective form of Tkv, whereas the interaction of Mad with wild type Tkv was still detectable (Figure 5A). The interaction of Mad with Tkv might thus be more stable than that of mammalian Smads with receptors. Dad interacted with wild-type Tkv as efficiently as with the kinase-defective form of Tkv. Notably, almost the same amount of Tkv was immunoprecipitated with Mad and Dad, although the expression level of Mad was much higher than that of Dad. Thus the affinity of Dad with Tkv seems to be higher than that of Mad. Stable interaction was also observed with immunoprecipitation followed by Western blotting (Figure 5B). Finally, we found that the interaction of Mad with Tkv was hampered by expression of Dad (Figure 5C). Dad thus inhibited phosphorylation of Mad by Tkv by competing with Mad in association with the receptor.
Figure 5.
Interaction of Mad and Dad with Tkv. (A) Affinity cross-linking using 125I-BMP-2 was performed to examine the interaction of Mad and Dad with Tkv. Cells were transfected with FLAG-tagged Mad (lanes 1 and 2) or Dad (lanes 3 and 4) with combinations of wild-type (WT; lanes 1 and 3) or kinase-defective (KR; lanes 2 and 4) Tkv and Punt and affinity labeled with 125I-BMP-2. The lysates were subjected to immunoprecipitation with anti-FLAG antibody and detection by SDS-PAGE. Expression levels of proteins were monitored by Western blotting. (B) Interaction of FLAG-tagged Dad with HA-tagged Tkv was examined by immunoprecipitation with anti-FLAG antibody followed by Western blotting with anti-HA antibody. Wild-type (WT), the constitutively active form (QD), and the kinase-defective form (KR) of Tkv were used. Expression levels of proteins were monitored by Western blotting. (C) Effect of Dad expression on the interaction of Mad with Tkv was examined by affinity cross-linking followed by immunoprecipitation. Mad, Dad, and receptors were tagged with FLAG, myc, and HA, respectively. For coimmunoprecipitation, anti-FLAG antibody was used. Expression levels of proteins were monitored by Western blotting.
Dad Inhibits Oligomerization and Nuclear Translocation of Mad
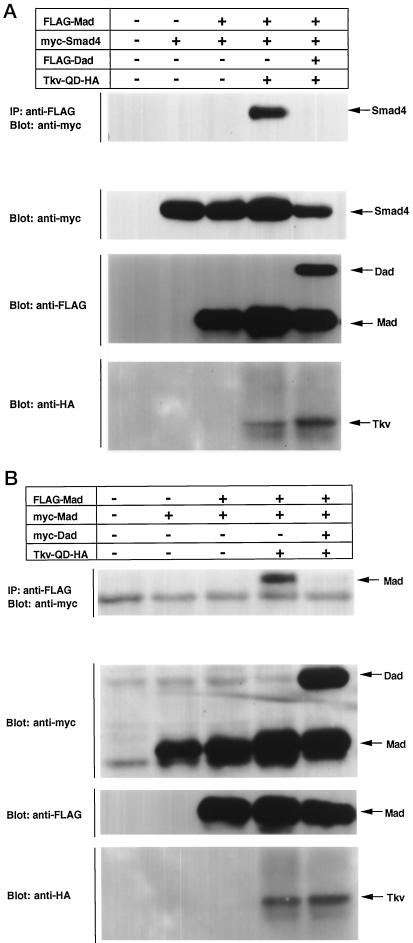
Oligomerization of the Smad proteins is a critical step in their activation. Most of the cancer-derived mutations of Smad4 or Smad2, as well as mutations of the Mad and sma genes causing developmental defects, are mapped to their MH2 domains. Based on the recently revealed crystal structure of the Smad4 MH2 domain, these mutations can be sorted into three groups: those that are located in the hydrophobic core and destabilize the overall structure, those that disrupt hetero-oligomeric interaction, and those that disrupt homo-oligomeric complex formation (Shi et al., 1997). Tkv-QD causes hetero-oligomerization of Mad with Medea (Figure 2B). The effect of Dad on the hetero-oligomerization was examined. We tested whether Dad can inhibit Tkv-QD-induced complex formation of Mad and human Smad4. As shown in Figure 6A, the hetero-oligomerization of Mad with Smad4 was efficiently blocked. Dad thus blocked a critical step in the activation of Mad.
Figure 6.
Inhibition of Tkv-induced oligomerization of Mad by Dad. (A) The effect of Dad on hetero-oligomerization of Mad with Smad4 was examined. Cells were transfected with the indicated combinations of plasmids. The lysates were subjected to immunoprecipitation with anti-FLAG antibody followed by immunoblotting with anti-myc antibody. Expression levels of proteins were monitored by Western blotting. (B) Effect of Dad on homo-oligomerization of Mad was examined. Cells were transfected with the indicated combinations of plasmids including FLAG-tagged and myc-tagged Mad. The lysates were subjected to immunoprecipitation with anti-FLAG antibody followed by immunoblotting with anti-myc antibody. Expression levels of proteins were monitored by Western blotting.
We previously observed that Smad2 and Smad3 associate with each other in a TGF-β-dependent manner (Nakao et al., 1997a). This finding suggested that pathway-specific Smads may exist as monomers in the absence of ligand stimulation and form oligomeric complexes upon phosphorylation by type I receptors. We tested this hypothesis for Mad. As shown in Figure 6B, Mad existed as a monomer in the absence of Tkv-QD, and Tkv-QD induced homo-oligomerization of Mad. The activation of Mad by Tkv-QD thus appears to consist of a sequential linkage of phosphorylation, homo-oligomerization, hetero-oligomerization, and nuclear translocation. When Dad was coexpressed, the homo-oligomer formation of Mad induced by Tkv-QD was inhibited (Figure 6B).
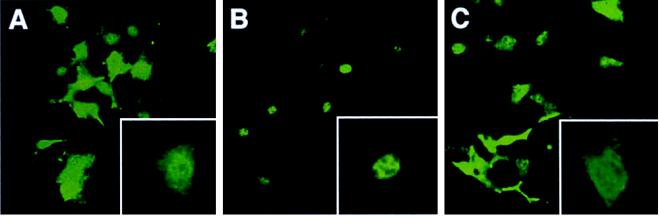
Smad proteins translocate into the nucleus after phosphorylation and oligomerization. The effect of Dad on this step was examined (Figure 7). COS cells were transfected with various combinations of Mad, Dad, and Tkv-QD, and the subcellular localization of Mad was determined by immunofluorescence microscopy. Mad was localized throughout the cell in unstimulated cells, and Tkv-QD induced nuclear accumulation of the Mad proteins. When Dad was coexpressed, nuclear translocation of Mad was blocked. The percentage of cells displaying predominant nuclear staining increased from 12 to 96% upon Tkv stimulation and decreased to 11% in the presence of Dad. An almost identical result was obtained in another experiment (our unpublished results). This finding again demonstrates that Dad inhibits Mad activation by Tkv.
Figure 7.
Inhibition of nuclear translocation of Mad by Dad. Nuclear translocation of Mad in the absence or presence of Dad was examined by immunostaining. COS cells were transfected with various combinations of Mad, Dad, and Tkv-QD. Anti-FLAG antibody was used as the first antibody. Subcellular localization was detected using FITC-labeled streptavidin, and fluorescence microscopy. (A) Mad; (B) Mad + Tkv-QD; (C) Mad + Tkv-QD + Dad. A representative cell, in each case, of higher magnification is shown in the inset.
DISCUSSION
Smad proteins propagate signals of the TGF-β superfamily (Heldin et al., 1997). Three classes of Smads have been identified: pathway-specific Smads, common mediators, and inhibitory Smads. Pathway-specific Smads undergo phosphorylation by the type I receptors, hetero-oligomerize with a common mediator, and then translocate into the nucleus where they transactivate a certain set of genes. Inhibitory Smads block the activation of pathway-specific Smads. In Drosophila, Mad and Dad have been identified as a pathway-specific Smad and an inhibitory Smad, respectively. Both Smads are involved in Dpp-Tkv signaling. If the mechanism of the signal transduction by Smads is conserved between vertebrates and invertebrates, Mad requires a partner to mediate Dpp signaling. Indeed, C. elegans has Sma-4, which is closely related to Smad4 in structure (Savage et al., 1996), although its biochemical functions are unclear. We identified Drosophila Smad4 based on its sequence similarity to human Smad4. During the preparation of this manuscript, three works describing the cloning of the Medea gene were reported (Das et al., 1998; Hudson et al., 1998; Wisotzkey et al., 1998). Our Drosophila Smad4 was identical to Medea. The three papers presented evidence that Medea acts as a common mediator Smad in Dpp signaling in vivo. Here we have presented the molecular basis of the Medea function. Medea has the MH1 and MH2 domains but lacks the SSXS motif, which is a structural feature unique to Smad4 among the Smad family proteins. Medea interacted with Smad1 or Smad2 upon stimulation by type I receptors, demonstrating that Medea and Smad4 are functionally conserved. Mad transiently interacted with Tkv and underwent phosphorylation. Tkv-QD induced association of Mad with Medea. Thus Mad and Medea together propagate signals specific to Tkv. Wisotzkey et al. (1998) showed that Mad and Medea form constitutive heteromeric complexes, which differs from our results. The authors raised the possibility that Mad is constitutively phosphorylated at the C terminus in the cells that they used.
The activity of Dpp is tightly controlled both intracellularly and extracellularly. Short gastrulation (Sog) and Tolloid (Tld) are extracellular factors that control Dpp activity (Marques et al., 1997; Piccolo et al., 1997). Dad antagonizes Dpp signaling intracellularly (Tsuneizumi et al., 1997). Dad exhibited patterns of expression similar to those of Dpp during embryonic and imaginal development, and ectopic expression of Dpp induced expression of Dad. Interestingly, Dad antagonized Dpp, as demonstrated in various assays. Expression of Dad along the wing margin caused a partial or almost complete loss of wing structure, resembling phenotypes resulting from defects in Dpp signaling. Dad also repressed expression of a Dpp-inducible gene, optomotor-blind (omb). Taken together, these findings indicate that Dpp induces expression of its own antagonist, Dad, and that Dad plays a key role in an autoregulatory circuit controlling Dpp signaling.
Dad has homology to mammalian Smad6 (Imamura et al., 1997; Hata et al., 1998), Smad7 (Hayashi et al., 1997; Nakao et al., 1997b), and Xenopus XSmad8 (Nakayama et al., 1998). These vertebrate Smads inhibit signaling of the TGF-β/activin and/or BMP signaling. Interestingly, expression of Smad6 or Smad7 was induced by ligands (Nakao et al., 1997b; Takase et al., 1998), suggesting that the autoregulatory mechanism controlling Smad signaling is conserved between invertebrates and vertebrates. In this study, we have shown that Dad blocks the activation of Mad by Tkv. Dad inhibited phosphorylation, homo- and hetero-oligomerization, and nuclear translocation of Mad. Dad associated with Tkv with a higher affinity than Mad. Dad prevented Mad from binding to the receptor, and, as a consequence, inhibited Tkv-induced phosphorylation of Mad. It was recently reported that human Smad6 competes with Smad4 in association with Smad1, thereby inhibiting BMP/Smad1 signaling (Hata et al., 1998). We examined whether Dad interacts with Mad using various conditions but have not been able to detect the interaction (our unpublished results). The reason for this discrepancy is not known at present, but different organisms may use different mechanisms to regulate signaling by the TGF-β superfamily.
Previously, a model was proposed in which a trimer of pathway-specific Smad and a trimer of Smad4 form a hetero-hexameric complex upon ligand treatment (Shi et al., 1997). Here we have shown that Mad exists as a monomer in the absence of receptor stimulation, and Tkv-QD induces homo-oligomerization. Similar results were obtained for mammalian Smads (Kawabata et al., 1998b). The number of Mad molecules incorporated in the homo-oligomeric complex was not determined in this study. This finding explains well the mechanism of complex formation of Smad2 and Smad3 after TβR-I stimulation (Nakao et al., 1997a). Phosphorylation of both Smads appears to be required for Smad2–Smad3 interaction, because Smad6 blocks TβR-I-induced phosphorylation of Smad2 but not that of Smad3 (Imamura et al., 1997). Homo-oligomerization of Mad was efficiently blocked by Dad, whereas the inhibition by Dad of Mad phosphorylation was partial. Partial inhibition of Mad phosphorylation may lead to more extensive inhibition of homo-oligomerization, because all of the Mad molecules in the homo-oligomers must be phosphorylated.
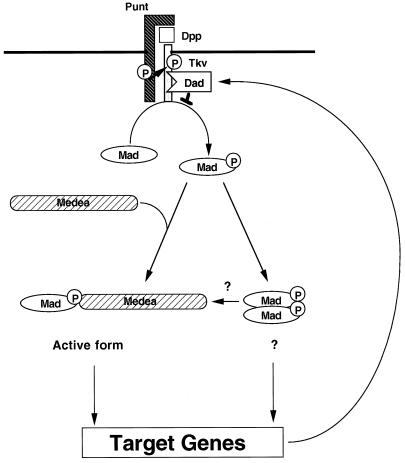
Finally, we propose the following model of Dpp signaling by Mad, Medea, and Dad (Figure 8): Dpp induces phosphorylation of Mad through Tkv and Punt. Mad then forms homo-oligomeric complexes and/or hetero-dimerizes with Medea. Oligomers of Mad and Medea translocate into the nucleus where they transactivate target genes such as vestigial. Dad is one such target, and its expression is induced by Dpp. Dad stably binds to Tkv and interrupts phosphorylation of Mad by Tkv.
Figure 8.
Model of the Dpp signaling pathway: cascades of phosphorylation and dynamic rearrangement of protein interactions. Dpp induces hetero-oligomerization of Punt and Tkv. Punt activates Tkv by transphosphorylating the juxtamembrane region. Activated Tkv phosphorylates Mad. Mad forms homo-oligomeric complexes and/or hetero-oligomeric complexes with Medea. It is not known whether Mad homo-oligomers have specific activity in vivo. Hetero-oligomers of Mad and Medea translocate into the nucleus where they transactivate target genes such as vestigial. Dad is one such target, and its expression is induced by Dpp. Dad stably binds to Tkv and interrupts phosphorylation of Mad by Tkv. The numbers of each Smad molecule in the homo- and hetero-oligomeric complexes remain to be determined.
ACKNOWLEDGMENTS
We are grateful to J. Yingling, X.-F. Wang, A. Nakao, P. ten Dijke, S. Kern, W. Gelbart, M. Hoffmann, and K. Basler for the Smad and Drosophila receptor plasmids, J.A. Langer for pcDEF3, and T.K. Sampath for BMP-2. This study was supported by grants-in-aid for scientific research from the Ministry of Education, Science, Sports, and Culture of Japan and special coordination funds for promoting science and technology from the Science and Technology Agency. K.M. was supported by the Yamanouchi Foundation for Research on Metabolic Disorders. M.K. was supported by the Takeda Science Promotion Foundation and the Cell Science Research Foundation.
REFERENCES
- Abdollah S, Macías SM, Tsukazaki T, Hayashi H, Attisano L, Wrana JL. TβRI phosphorylation of Smad2 on Ser465 and Ser467 is required for Smad2-Smad4 complex formation and signaling. J Biol Chem. 1997;272:27678–27685. doi: 10.1074/jbc.272.44.27678. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Brummel TJ, Twombly V, Marques G, Wrana JL, Newfeld SJ, Attisano L, Massagué J, O’Connor MB, Gelbart WM. Characterization and relationship of Dpp receptors encoded by the saxophone and thick veins genes in Drosophila. Cell. 1994;78:251–261. doi: 10.1016/0092-8674(94)90295-x. [DOI] [PubMed] [Google Scholar]
- Chen X, Rubock MJ, Whitman M. A transcriptional partner for MAD proteins in TGF-β signalling. Nature. 1996;383:691–696. doi: 10.1038/383691a0. [DOI] [PubMed] [Google Scholar]
- Das P, Maduzia LL, Wang H, Finelli AL, Cho S-H, Smith MM, Padgett RW. The Drosophila gene Medea demonstrates the requirement for different classes of Smads in dpp signaling. Development. 1998;125:1519–1528. doi: 10.1242/dev.125.8.1519. [DOI] [PubMed] [Google Scholar]
- de Winter JP, Roelen BAJ, ten Dijke P, van der Burg B, van den Eijnden-van Raaij AJM. DPC4 (SMAD4) mediates transforming growth factor-β1 (TGF-β1) induced growth inhibition and transcriptional response in breast tumour cells. Oncogene. 1997;14:1891–1899. doi: 10.1038/sj.onc.1201017. [DOI] [PubMed] [Google Scholar]
- Derynck R, Feng X-H. TGF-β receptor signaling. Biochim Biophys Acta. 1997;1333:F105–F150. doi: 10.1016/s0304-419x(97)00017-6. [DOI] [PubMed] [Google Scholar]
- Feng X-H, Derynck R. A kinase subdomain of transforming growth factor-β (TGF-β) type I receptor determines the TGF-β intracellular signaling specificity. EMBO J. 1997;16:3912–3923. doi: 10.1093/emboj/16.13.3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman LA, Cutrone EC, Kotenko SV, Krause CD, Langer JA. Modifications of vectors pEF-BOS, pcDNA1 and pcDNA3 result in improved convenience and expression. Biotechniques. 1996;21:1013–1015. doi: 10.2144/96216bm10. [DOI] [PubMed] [Google Scholar]
- Grau AM, Zhang L, Wang W, Ruan S, Evans DB, Abbruzzese JL, Zhang W, Chiao PJ. Induction of p21waf1 expression and growth inhibition by transforming growth factor β involve the tumor suppressor gene DPC4 in human pancreatic adenocarcinoma cells. Cancer Res. 1997;57:3929–3934. [PubMed] [Google Scholar]
- Hata A, Lagna G, Massagué J, Hemmati BA. Smad6 inhibits BMP/Smad1 signaling by specifically competing with the Smad4 tumor suppressor. Genes Dev. 1998;12:186–197. doi: 10.1101/gad.12.2.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi H, et al. The MAD-related protein Smad7 associates with the TGFβ receptor and functions as an antagonist of TGFβ signaling. Cell. 1997;89:1165–1173. doi: 10.1016/s0092-8674(00)80303-7. [DOI] [PubMed] [Google Scholar]
- Heldin C-H, Miyazono K, ten Dijke P. TGF-β signalling from cell membrane to nucleus through SMAD proteins. Nature. 1997;390:465–471. doi: 10.1038/37284. [DOI] [PubMed] [Google Scholar]
- Hogan BL. Bone morphogenetic proteins: multifunctional regulators of vertebrate development. Genes Dev. 1996;10:1580–1594. doi: 10.1101/gad.10.13.1580. [DOI] [PubMed] [Google Scholar]
- Hudson JB, Podos SD, Keith K, Simpson SL, Ferguson EL. The Drosophila Medea gene is required downstream of dpp and encodes a functional homolog of human Smad4. Development. 1998;125:1407–1420. doi: 10.1242/dev.125.8.1407. [DOI] [PubMed] [Google Scholar]
- Imamura T, Takase M, Nishihara A, Oeda E, Hanai J, Kawabata M, Miyazono K. Smad6 inhibits signalling by the TGF-β superfamily. Nature. 1997;389:622–626. doi: 10.1038/39355. [DOI] [PubMed] [Google Scholar]
- Kawabata M, Imamura T, Miyazono K. Signal transduction by bone morphogenetic proteins. Cytokine Growth Factor Rev. 1998a;9:49–61. doi: 10.1016/s1359-6101(97)00036-1. [DOI] [PubMed] [Google Scholar]
- Kawabata, M., Inoue, H., Hanyu, A., Imamura, T., and Miyazono, K. (1998b). Smad proteins exist as monomers in vivo and undergo homo- and hetero-oligomerization upon activation by serine/threonine kinase receptors. EMBO J. (in press). [DOI] [PMC free article] [PubMed]
- Kim J, Johnson K, Chen HJ, Carroll S, Laughon A. Drosophila Mad binds to DNA and directly mediates activation of vestigial by Decapentaplegic. Nature. 1997;388:304–308. doi: 10.1038/40906. [DOI] [PubMed] [Google Scholar]
- Kingsley DM. The TGF-β superfamily: new members, new receptors, and new genetic tests of function in different organisms. Genes Dev. 1994;8:133–146. doi: 10.1101/gad.8.2.133. [DOI] [PubMed] [Google Scholar]
- Macías-Silva M, Abdollah S, Hoodless PA, Pirone R, Attisano L, Wrana JL. MADR2 is a substrate of the TGFβ receptor and its phosphorylation is required for nuclear accumulation and signaling. Cell. 1996;87:1215–1224. doi: 10.1016/s0092-8674(00)81817-6. [DOI] [PubMed] [Google Scholar]
- Maduzia LL, Padgett RW. Drosophila MAD, a member of the Smad family, translocates to the nucleus upon stimulation of the dpp pathway. Biochem Biophys Res Commun. 1997;238:595–598. doi: 10.1006/bbrc.1997.7353. [DOI] [PubMed] [Google Scholar]
- Marques G, Musacchio M, Shimell MJ, Wunnenberg SK, Cho KW, O’Connor MB. Production of a DPP activity gradient in the early Drosophila embryo through the opposing actions of the SOG and TLD proteins. Cell. 1997;91:417–426. doi: 10.1016/s0092-8674(00)80425-0. [DOI] [PubMed] [Google Scholar]
- Nakao A, et al. TGF-β receptor-mediated signalling through Smad2, Smad3 and Smad4. EMBO J. 1997a;16:5353–5362. doi: 10.1093/emboj/16.17.5353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakao A, et al. Identification of Smad7, a TGFβ-inducible antagonist of TGF-β signalling. Nature. 1997b;389:631–635. doi: 10.1038/39369. [DOI] [PubMed] [Google Scholar]
- Nakayama T, Snyder M, Grewal S, Tsuneizumi K, Tabata T, Christian J. Xenopus Smad8 acts downstream of BMP-4 to modulate its activity during vertebrate embryonic patterning. Development. 1998;125:857–867. doi: 10.1242/dev.125.5.857. [DOI] [PubMed] [Google Scholar]
- Nellen D, Affolter M, Basler K. Receptor serine/threonine kinases implicated in the control of Drosophila body pattern by decapentaplegic. Cell. 1994;78:225–237. doi: 10.1016/0092-8674(94)90293-3. [DOI] [PubMed] [Google Scholar]
- Newfeld SJ, Chartoff EH, Graff JM, Melton DA, Gelbart WM. Mothers against dpp encodes a conserved cytoplasmic protein required in DPP/TGF-β responsive cells. Development. 1996;122:2099–2108. doi: 10.1242/dev.122.7.2099. [DOI] [PubMed] [Google Scholar]
- Newfeld SJ, Mehra A, Singer MA, Wrana JL, Attisano L, Gelbart WM. Mothers against dpp participates in a DPP/TGF-β responsive serine-threonine kinase signal transduction cascade. Development. 1997;124:3167–3176. doi: 10.1242/dev.124.16.3167. [DOI] [PubMed] [Google Scholar]
- Nishimura R, Kato Y, Chen D, Harris SE, Mundy GR, Yoneda T. Smad5 and DPC4 are key molecules in mediating BMP-2-induced osteoblastic differentiation of the pluripotent mesenchymal precursor cell line C2C12. J Biol Chem. 1998;273:1872–1879. doi: 10.1074/jbc.273.4.1872. [DOI] [PubMed] [Google Scholar]
- Oeda E, Oka Y, Miyazono K, Kawabata M. Interaction of Drosophila inhibitors of apoptosis with Thick veins, a type I serine/threonine kinase receptor for Decapentaplegic. J Biol Chem. 1998;273:9353–9356. doi: 10.1074/jbc.273.16.9353. [DOI] [PubMed] [Google Scholar]
- Padgett RW, Savage C, Das P. Genetic and biochemical analysis of TGF-β signal transduction. Cytokine Growth Factor Rev. 1997;8:1–9. doi: 10.1016/s1359-6101(96)00050-0. [DOI] [PubMed] [Google Scholar]
- Penton A, Chen Y, Staehling HK, Wrana JL, Attisano L, Szidonya J, Cassill JA, Massagué J, Hoffmann FM. Identification of two bone morphogenetic protein type I receptors in Drosophila and evidence that Brk25D is a decapentaplegic receptor. Cell. 1994;78:239–250. doi: 10.1016/0092-8674(94)90294-1. [DOI] [PubMed] [Google Scholar]
- Piccolo S, Agius E, Lu B, Goodman S, Dale L, De Robertis EM. Cleavage of Chordin by Xolloid metalloprotease suggests a role for proteolytic processing in the regulation of Spemann organizer activity. Cell. 1997;91:407–416. doi: 10.1016/s0092-8674(00)80424-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raftery LA, Twombly V, Wharton K, Gelbart WM. Genetic screens to identify elements of the decapentaplegic signaling pathway in Drosophila. Genetics. 1995;139:241–254. doi: 10.1093/genetics/139.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage C, Das P, Finelli AL, Townsend SR, Sun CY, Baird SE, Padgett RW. Caenorhabditis elegans genes sma-2, sma-3, and sma-4 define a conserved family of transforming growth factor β pathway components. Proc Natl Acad Sci USA. 1996;93:790–794. doi: 10.1073/pnas.93.2.790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekelsky JJ, Newfeld SJ, Raftery LA, Chartoff EH, Gelbart WM. Genetic characterization and cloning of mothers against dpp, a gene required for decapentaplegic function in Drosophila melanogaster. Genetics. 1995;139:1347–1358. doi: 10.1093/genetics/139.3.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Hata A, Lo RS, Massagué J, Pavletich NP. A structural basis for mutational inactivation of the tumour suppressor Smad4. Nature. 1997;388:87–93. doi: 10.1038/40431. [DOI] [PubMed] [Google Scholar]
- Souchelnytskyi S, Tamaki K, Engstrom U, Wernstedt C, ten Dijke P, Heldin C-H. Phosphorylation of Ser465 and Ser467 in the C terminus of Smad2 mediates interaction with Smad4 and is required for transforming growth factor-β signaling. J Biol Chem. 1997;272:28107–28115. doi: 10.1074/jbc.272.44.28107. [DOI] [PubMed] [Google Scholar]
- Souchelnytskyi S, ten Dijke P, Miyazono K, Heldin C-H. Phosphorylation of Ser165 in TGF-β type I receptor modulates TGF-β1-induced cellular responses. EMBO J. 1996;15:6231–6240. [PMC free article] [PubMed] [Google Scholar]
- Takase M, Imamura T, Sampath TK, Takeda K, Ichijo H, Miyazono K, Kawabata M. Induction of Smad6 mRNA by bone morphogenetic proteins. Biochem Biophys Res Commun. 1998;244:26–29. doi: 10.1006/bbrc.1998.8200. [DOI] [PubMed] [Google Scholar]
- Tsuneizumi K, Nakayama T, Kamoshida Y, Kornberg TB, Christian JL, Tabata T. Daughters against dpp modulates dpp organizing activity in Drosophila wing development. Nature. 1997;389:627–631. doi: 10.1038/39362. [DOI] [PubMed] [Google Scholar]
- Wiersdorff V, Lecuit T, Cohen SM, Mlodzik M. Mad acts downstream of Dpp receptors, revealing a differential requirement for dpp signaling in initiation and propagation of morphogenesis in the Drosophila eye. Development. 1996;122:2153–2162. doi: 10.1242/dev.122.7.2153. [DOI] [PubMed] [Google Scholar]
- Wieser R, Wrana JL, Massagué J. GS domain mutations that constitutively activate TβR-I, the downstream signaling component in the TGF-β receptor complex. EMBO J. 1995;14:2199–2208. doi: 10.1002/j.1460-2075.1995.tb07214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisotzkey RG, Mehra A, Sutherland DJ, Dobens LL, Liu X, Dohrmann C, Attisano L, Raftery LA. Medea is a Drosophila Smad4 homolog that is differentially required to potentiate DPP responses. Development. 1998;125:1433–1445. doi: 10.1242/dev.125.8.1433. [DOI] [PubMed] [Google Scholar]
- Wrana JL, Attisano L, Wieser R, Ventura F, Massagué J. Mechanism of activation of the TGF-β receptor. Nature. 1994a;370:341–347. doi: 10.1038/370341a0. [DOI] [PubMed] [Google Scholar]
- Wrana JL, Tran H, Attisano L, Arora K, Childs SR, Massagué J, O’Connor MB. Two distinct transmembrane serine/threonine kinases from Drosophila melanogaster form an activin receptor complex. Mol Cell Biol. 1994b;14:944–950. doi: 10.1128/mcb.14.2.944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie T, Finelli AL, Padgett RW. The Drosophila saxophone gene: a serine-threonine kinase receptor of the TGF-β superfamily. Science. 1994;263:1756–1759. doi: 10.1126/science.8134837. [DOI] [PubMed] [Google Scholar]
- Zhou S, Buckhaults P, Zawel L, Bunz F, Riggins G, Le DJ, Kern SE, Kinzler KW, Vogelstein B. Targeted deletion of smad4 shows it is required for transforming growth factor β and activin signaling in colorectal cancer cells. Proc Natl Acad Sci USA. 1998;95:2412–2416. doi: 10.1073/pnas.95.5.2412. [DOI] [PMC free article] [PubMed] [Google Scholar]