Abstract
Bacterial cell division is mediated by a set of proteins that assemble to form a large multiprotein complex called the divisome. Recent studies in Bacillus subtilis and Escherichia coli indicate that cell division proteins are involved in multiple cooperative binding interactions, thus presenting a technical challenge to the analysis of these interactions. We report here the use of an E. coli artificial septal targeting system for examining the interactions between the B. subtilis cell division proteins DivIB, FtsL, DivIC, and PBP 2B. This technique involves the fusion of one of the proteins (the “bait”) to ZapA, an E. coli protein targeted to mid-cell, and the fusion of a second potentially interacting partner (the “prey”) to green fluorescent protein (GFP). A positive interaction between two test proteins in E. coli leads to septal localization of the GFP fusion construct, which can be detected by fluorescence microscopy. Using this system, we present evidence for two sets of strong protein-protein interactions between B. subtilis divisomal proteins in E. coli, namely, DivIC with FtsL and DivIB with PBP 2B, that are independent of other B. subtilis cell division proteins and that do not disturb the cytokinesis process in the host cell. Our studies based on the coexpression of three or four of these B. subtilis cell division proteins suggest that interactions among these four proteins are not strong enough to allow the formation of a stable four-protein complex in E. coli in contrast to previous suggestions. Finally, our results demonstrate that E. coli artificial septal targeting is an efficient and alternative approach for detecting and characterizing stable protein-protein interactions within multiprotein complexes from other microorganisms. A salient feature of our approach is that it probably only detects the strongest interactions, thus giving an indication of whether some interactions suggested by other techniques may either be considerably weaker or due to false positives.
The process of cell division in bacteria requires the assembly of a large number of proteins at mid-cell. These proteins catalyze the separation of nucleoids, the formation of a septum, and the division of the cell into two daughter cells. In Escherichia coli, at least 15 proteins are involved in this process, 10 of which are essential for septation and for viability. The assembly of these proteins at the septal region is initiated by localization of FtsZ, followed by its interaction with the stabilizing proteins ZipA, ZapA, and FtsA, leading to the formation of the Z-ring. The presence at mid-cell of these proteins is necessary for subsequent recruitment of the cell division proteins FtsE/X, FtsK, FtsQ, FtsB, FtsL, FtsW, FtsI, FtsN, PBP 1B, AmiC, and EnvC (23, 45). In E. coli, the recruitment of Fts proteins during this dynamic process occurs largely via a dependency pathway, in which a given protein requires the presence of an “upstream” protein to be localized to mid-cell (Fig. 1A) (6). However, this representation of an apparently simple linear pathway masks a much wider array of cooperative binding interactions between Fts proteins. This more complex picture of divisome assembly is indicated by a variety of findings. First, bacterial two-hybrid assays suggest that many Fts proteins interact with multiple others in the formation of the septal ring assembly (16, 18, 30, 37). Further, genetic studies showed that the essentiality for the recruitment role of some proteins can be avoided by suppressor mutations in, or overexpression of, other divisomal proteins (1, 20, 21, 24, 26, 27, 39, 42). Finally, biochemical approaches revealed that at least some Fts proteins assemble as a complex independently of their hierarchical recruitment to the septum (5, 24).
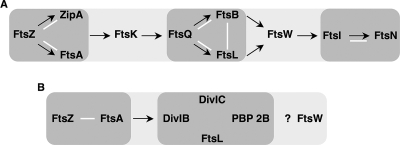
FIG. 1.
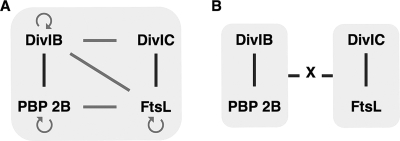
Schema of the sequential recruitment of essential cell division proteins to the septum in E. coli (A) and B. subtilis (B). Proteins that form a subcomplex independent of other divisomal proteins are surrounded by gray halos. Arrows point to the recruited downstream protein or downstream subcomplex. White bars correspond to direct protein-protein interactions demonstrated by genetic and biochemical approaches. The question mark indicates that the recruitment time of FtsW is unknown in B. subtilis.
Among these divisomal protein-protein interactions, the most well established is that between FtsQ, FtsL, and FtsB. Although FtsQ can localize to the mid-cell in the absence of FtsB and FtsL, the three proteins form a subcomplex that exists, in vivo, independently of their recruitment to the mid-cell (5). These three proteins share a similar membrane topology, with one transmembrane domain bordered by a short cytoplasmic N-terminal domain and a periplasmic C-terminal domain of varying length (40). Both FtsL and FtsB, which exhibit a leucine zipper-like motif in their periplasmic domains (8), interact strongly, although physical contact via the leucine zipper-like motifs has not been demonstrated (5, 24).
FtsQ, FtsL, and FtsB are well conserved in most bacteria, and the orthologs of these proteins in gram-positive bacteria are generally referred to as DivIB, FtsL, and DivIC, respectively (19). Even though the sequence similarities of these three proteins between gram-positive and gram-negative bacteria are weak, the three proteins from the two classes of organisms share similar size and membrane topology, and the divIB and ftsL genes show conserved chromosome positions. The function of FtsQ, FtsL, and FtsB remains enigmatic, with suggestions that FtsQ may play a role in peptidoglycan synthesis or as a regulator of divisome assembly (4, 45). In B. subtilis, FtsL (named here FtsLB) was proposed to play a direct or indirect role in Z-ring disassembly and constriction of membranes (15, 33). According to bacterial and yeast two- and three-hybrid assays, the three proteins appear to associate to form a complex; however, these interactions are not fully understood, and there are conflicting findings concerning possible interactions between DivIC and FtsLB (15, 41, 43). In Streptococcus pneumoniae, biochemical methods using copurification suggest the formation of a transient trimeric complex between DivIB and artificially restrained heterodimer of the periplasmic domains of FtsL and DivIC (38). These data overall suggest that FtsQ (DivIB), FtsL, and FtsB (DivIC) form a conserved subcomplex.
However, some properties of FtsQ (DivIB), FtsL (FtsLB), and FtsB (DivIC) might indicate differences in their functions in different organisms. First, the E. coli proteins are in much lower abundance (ca. 25 to 50 copies/cell for FtsQ and 50 to 200 copies/cell for FtsL) compared to their orthologs in B. subtilis (ca. 5,000 to 13,000 copies/cell for DivIB and ca. 50,000 copies/cell for DivIC) (46). In B. subtilis, these levels of DivIB, DivIC, and FtsLB are required for two processes: cell division and sporulation (14, 29, 34). Second, the assembly pathway and the protein stability of FtsQ (DivIB), FtsL (FtsLB), and FtsB (DivIC) differ between the two organisms (Fig. 1). In E. coli, FtsL and FtsB are codependent for their localization and their stability, and they both require the presence of FtsQ for their septal recruitment. However, FtsQ can be recruited to the septum independently of FtsL and FtsB (5). In B. subtilis, DivIB, FtsLB, and DivIC appear to cooperate with a fourth protein, PBP 2B, the B. subtilis ortholog of E. coli FtsI, a bitopic membrane protein, which possesses a transpeptidase activity in its periplasmic domain. Several lines of evidence indicate that the assembly and stability of these four proteins is interdependent (19). (i) DivIB is not essential for cell division at low temperature, but its depletion leads to cell division blockage at high temperature. This is probably due to rapid degradation of FtsLB, suggesting that DivIB stabilizes FtsLB under these conditions. (ii) DivIC is dependent on FtsLB for its stability in the presence of DivIB. (iii) Finally, FtsLB and DivIC are destabilized when PBP 2B is depleted (13-15).
The complexity of divisome formation and the essential nature of this process present a technical challenge to the genetic characterization of the network of protein-protein interactions. This is the case, in particular, for defining the specific regions of contact between two individual proteins in a system where proteins are involved in multiple interactions. Furthermore, because the localization of any one cell division protein to mid-cell is dependent on the presence of others that have already localized, specifying such contacts is complicated by the multiplicity of proteins already present in the complex. Thus, little progress has been made in defining the domains of proteins that interact with each other and in assessing the strength of these interactions. While two-hybrid systems reveal interactions, in the case of the divisome, they do not indicate whether the strength of those interactions is sufficient to allow stable complex formation between a pair of proteins.
In previous work, we described an approach that partially overcomes these problems by directing a protein to mid-cell in the absence of the upstream proteins that are normally required for its septal localization. This approach, called premature targeting, involves the fusion of a cell division protein to ZapA, a cytoplasmic protein which can localize to mid-cell by binding to FtsZ independently of all other cell division proteins (25). Thus, by fusing a protein such as FtsQ to ZapA, we were able to direct FtsQ to mid-cell in the absence of upstream proteins such as FtsA or FtsK that are normally required for its recruitment (see Fig. 1). The direct interaction between FtsQ and a particular protein is assessed using a green fluorescent protein (GFP) fusion to that protein, thus providing an easily observable phenotype: either GFP septal localization in the case of a positive interaction or a membrane subcellular location if there is no interaction or only a weak one. Using this approach, we found that FtsQ is able to promote the recruitment of the downstream proteins FtsL, FtsB, FtsW, FtsI, but not FtsN, and that it can “back-recruit” FtsK in the absence of FtsA (24).
However, while this approach helps narrow down the problems described above, it does not eliminate them, since one is looking potentially at protein-protein interactions that could involve many of the proteins still present at the septum. We recognized that a further refinement of this technique might allow us to examine the protein-protein interactions between smaller numbers of proteins and perhaps even between pairs of cell division proteins. This refinement involves utilizing the ZapA fusions in E. coli, not as a tool for studying E. coli divisome assembly, but rather for analyzing the assembly of proteins from an organism not closely related to it. Specifically, we have examined the interactions between a subset of B. subtilis cell division proteins, namely, DivIB, FtsLB, DivIC, and PBP 2B. Each of these proteins was fused to E. coli ZapA on the one hand and to GFP on the other hand. We then studied interactions of these hybrid proteins in a pairwise fashion by assessing the ability of the ZapA fusion to recruit the GFP fusion to the E. coli septum. Our results demonstrate the formation of two stable complexes: one between FtsLB and DivIC and the other between DivIB and PBP 2B. However, we did not detect the formation of a stable tetrameric complex involving all four proteins. Expression of the four B. subtilis proteins did not interfere with the E. coli cell division process. The “E. coli artificial septal targeting system” represents a novel means of dissecting protein-protein interactions among protein components of multiprotein complexes from other microorganisms that yields new types of information about these interactions.
MATERIALS AND METHODS
Media, bacterial strains, and plasmids.
The bacterial strains and plasmids used in the present study are listed in Table 1. All strains were grown in rich medium NZY liquid broth or agar (9). Rich medium was supplemented with appropriate antibiotics as follows: ampicillin at 200 μg/ml (for bla on plasmid) or at 25 μg/ml (for bla on chromosome), chloramphenicol at 10 μg/ml, kanamycin at 40 μg/ml, and spectinomycin at 100 μg/ml. l-Arabinose or d-glucose were added at 0.2% to induce or repress, respectively, the expression of genes under the control of the pBAD promoter (28). In some cases, 20 or 50 μM IPTG (isopropyl-β-d-thiogalactopyranoside) were used to induce expression of genes under the control of the pTrc promoter.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype or features | Source or reference |
|---|---|---|
| Strains | ||
| MC4100 | F−araD139 ΔlacU169 relA1 rpsL150 thi mot flb5301 deoC7 ptsF25 rbsR | Laboratory collection |
| JOE309 | MC4100 ara+ | 11 |
| KS272 | F− ΔlacX74 galE galK thi rpsL ΔphoA (ΔPvuII) | Laboratory collection |
| CR14 | JOE309 ΔftsB(Kanr)/pBAD42-ftsB | This study |
| JC60 | KS272 ftsI::TnphoAI137ΔIS50R(Kanr)/pBAD33-ftsI | Laboratory collection |
| JOE417 | JOE309 ftsQE14::kan/pJC10 | 10 |
| NB988 | JOE307 ftsL::TnphoAL81ΔIS50R(Kanr)/pJMG197 | N. Buddelmeijer |
| CR166 | CR14/pCR31 | This study |
| CR168 | NB988/pCR34 | This study |
| CR198 | JOE309 Δ(λattL-lom)::bla lacIq pDSW204-gfp-divICflag3 | This study |
| CR200 | JOE309 Δ(λattL-lom)::bla lacIq pDSW204-gfp-ftsLBflag3 | This study |
| CR202 | JOE309 Δ(λattL-lom)::bla lacIq pDSW204-gfp-divIBmyc3 | This study |
| CR208 | CR198/pMDG1 | This study |
| CR209 | CR198/pCR39 | This study |
| CR210 | CR198/pCR40 | This study |
| CR211 | CR198/pCR41 | This study |
| CR212 | CR200/pMDG1 | This study |
| CR213 | CR200/pCR39 | This study |
| CR214 | CR200/pCR40 | This study |
| CR215 | CR200/pCR41 | This study |
| CR216 | CR202/pMDG1 | This study |
| CR217 | CR202/pCR39 | This study |
| CR218 | CR202/pCR40 | This study |
| CR219 | CR202/pCR41 | This study |
| CR251 | CR208 ftsQE14::kan/pJC10 | This study |
| CR253 | CR211 ftsQE14::kan/pJC10 | This study |
| CR257 | CR213 ftsQE14::kan/pJC10 | This study |
| CR275 | CR198/pNG162 | This study |
| CR278 | CR198/pCR44 | This study |
| CR279 | CR200/pNG162 | This study |
| CR280 | CR200/pCR42 | This study |
| CR315 | JOE417/pCR38 | This study |
| CR362 | JOE309 Δ(λattL-lom)::bla lacIq pDSW204-gfp-pbpBmyc3 | This study |
| CR368 | JC60/pCR67 | This study |
| CR373 | CR362/pMDG1 | This study |
| CR374 | CR362/pCR39 | This study |
| CR375 | CR362/pCR40 | This study |
| CR376 | CR362/pCR41 | This study |
| Plasmids | ||
| pBAD33 | Arabinose-regulated promoter, pACYC184 origin, Cmr | 28 |
| pBAD42 | Arabinose-regulated promoter, pSC101 origin, Specr | 28 |
| pDSW204 | IPTG-regulated promoter, pBR322 origin, Ampr | 47 |
| pDSW207 | pDSW204-gfp-MCS (fusion vector) | 47 |
| pBAD42-ftsB | pBAD42-ftsB | N. Buddelmeijer |
| pJC10 | pBAD33-ftsQ | 11 |
| pJMG197 | pBAD33-ftsL | 22 |
| pMDG1 | pNG162-zapA-Fcyto-ftsB | M. D. Gonzalez |
| pNG162 | IPTG regulated promoter (pDSW204), pSC101 origin, Specr | 24 |
| pCR23 | pBAD33-myc3 | This study |
| pCR25 | pDSW207-flag3 | This study |
| pCR27 | pDSW207-myc3 | This study |
| pCR31 | pDSW204-gfp-divICflag3 | This study |
| pCR34 | pDSW204-gfp-ftsLBflag3 | This study |
| pCR38 | pDSW204-gfp-divIBmyc3 | This study |
| pCR39 | pNG162-zapA-Fcyto-divICflag3 | This study |
| pCR40 | pNG162-zapA-Fcyto-divIBmyc3 | This study |
| pCR41 | pNG162-zapA-Fcyto-ftsLBflag3 | This study |
| pCR42 | pNG162-divICflag3 | This study |
| pCR44 | pNG162-ftsLBflag3 | This study |
| pCR67 | pDSW204-gfp-pbpBmyc3 | This study |
| pCR52 | pBAD33-divICflag3 | This study |
| pCR54 | pBAD33-divIBmyc3 | This study |
| pCR60 | pBAD33-pbpBmyc3 | This study |
| pCR63 | pBAD33-ftsLBflag3 | This study |
| pCR68 | pBAD33-pbpBmyc3-divICflag3 | This study |
aCmr, chloramphenicol resistance; Specr, spectinomycin resistance; Ampr, ampicillin resistance; Kanr, kanamycin resistance.
Strains CR198, CR200, CR202, and CR362 were obtained by integration of plasmids pCR31, pCR34, pCR38, and pCR67, respectively, on the chromosome of JOE309 using the λInCh procedure (2). Transduction of ftsQE14::kan from JOE417 was performed to construct strains CR251, CR253, and CR257 using phage P1 (36).
Plasmids pCR25 and pCR27 were obtained by insertion of a XbaI-HindIII fragment, encoding either the triple Flag epitope or the triple myc epitope, from pNB13 (5) and pNG210 (26), respectively, into pDSW207. The four plasmids pCR31, pCR34, pCR38 and pCR67, encoding DivICflag3, FtsLBflag3, DivIBmyc3, and PBP 2Bmyc3 fused to GFP, were constructed by ligating a DNA fragment, obtained by PCR amplification from DNA of a B. subtilis strain derived from the prototrophic strain 168 (PY79) (48), using primers listed in Table S1 in the supplemental material, into the EcoRI or KpnI and the XbaI sites of pCR25 or pCR27. pMDG1 is a pSC plasmid containing zapA-Fcyto-ftsB, in which the ftsB gene is cloned between the EcoRI and HindIII sites (Mark D. Gonzalez, unpublished data). Plasmids pCR39, pCR40, and pCR41, encoding ZapA-DivICflag3, ZapA-DivIBmyc3, and ZapA-FtsLBflag3, respectively, were obtained by replacing the EcoRI-HindIII fragment of pMDG1 with the EcoRI-HindIII fragment of pCR31, pCR34, and pCR38, corresponding to divICflag3, ftsLBflag3, and divIBmyc3. The same procedure was used to construct pCR42 and pCR44, encoding DivICflag3 and FtsLBflag3, respectively, into the EcoRI and HindIII sites in pNG162. Plasmids pCR52 and pCR54, encoding DivICflag3 and DivIBmyc3, were constructed by inserting PCR fragments obtained from pCR42 and pCR38, respectively, using the primers 33divIC 5′ or 33divIB5′ and universal pTrc3′, into the SalI and the HindIII sites of pBAD33. Plasmids pCR60 and pCR63, encoding PBP 2Bmyc3 and FtsLBmyc3, were constructed by inserting PCR fragments, obtained from DNA of a B. subtilis 168 (PY79) (48) and pCR34, respectively, using the primers 33pbp5′ and 33pbp3′ or the primers 33ftsLB5′ and universal pTrc3′, into the KpnI and the XbaI sites of pCR23. This pCR23 plasmid corresponds to a pBAD33 (28) containing the triple myc epitope from pNG210 (26), inserted into XbaI-HindIII cloning sites. The plasmid pCR68 was constructed by insertion of a PCR fragment, encoding PBP 2Bmyc3 and obtained from pCR60 with primers 33-pbp-IC5′ and 33-pbp-IC3′, into the KpnI and the SalI sites of pCR52. All amplified DNA fragments were verified by sequencing analysis, and we noticed the same point mutation generating the substitution L150Q in all pbpB cloned genes. Since the mutation was found in all independent amplified DNA fragments of this gene, we conclude that this mutation is present in the original strain. All primers used in the present study are listed in Table S1 in the supplemental material.
Growth conditions.
For standard growth and preparation of cell extracts, cells were grown overnight at 37°C in NZY media containing the necessary antibiotics and diluted 1:100 into the same NZY media. Cells were then grown at 30°C to an optical density at 600 nm (OD600) of ∼0.3 and then incubated with 20 or 50 μM IPTG for 30 min or 1 h.
To deplete cells of FtsB, FtsL, FtsQ, or FtsI, cells were grown overnight in NZY media at 37°C with arabinose and the necessary antibiotics. Overnight cultures were diluted 1:100 in the same NZY media and incubated at 30°C for 90 min and then diluted 1:100 in NZY media with either arabinose or glucose and antibiotics. Cells were grown at 30°C until the OD600 reached ∼0.3 and then incubated with 20 μM IPTG for 30 min.
Microscopy analysis.
After growth at 30°C and IPTG induction, cells were harvested, fixed (11), and then laid down on a 1% agarose layer as described previously (44). Samples were examined with an Eclipse 80i (Nikon) or Axioskop II (Zeiss) microscope equipped with a 100× plan-Apochromat oil immersion objective and 100-W mercury lamp. Phase-contrast and fluorescence images were captured by using a digital Sight DS-UI camera (Nikon) or an Orca-100 charge-coupled device camera (Hamamatsu Photonics) and analyzed with the software ACT-2U (Nikon) or Openlab (Improvision).
Membrane preparation.
Overnight cultures grown at 37°C in NZY media supplemented with antibiotics were diluted 1:100 into 50 ml of fresh NZY media with antibiotics and grown during 3 h at 30°C and supplemented with 20 μM IPTG during 30 min (OD600 ∼ 0.4). A total extract of each strain was prepared from 1 ml of culture by resuspending the pellet in 1× sodium dodecyl sulfate (SDS) sample buffer; the remaining 45 ml of each culture was harvested by centrifugation at 8,000 × g for 10 min at 4°C and resuspended in 300 μl of buffer I (10 mM Tris-HCl, 150 mM NaCl [pH 7.4]). Cells were disrupted by sonication and centrifuged for 10 min at 8,000 × g to eliminate cell debris. Membranes were collected by ultracentrifugation at 100,000 × g in a Beckman TL100 for 1 h at 4°C and then resuspended in 300 μl of buffer I and 30 μl of 10% n-dodecyl-β-d-maltopyranoside (Anatrace). The supernatant corresponding to the soluble fraction was stored at −20°C. The resuspended pellet of membrane proteins was incubated at 4°C for 30 min with gentle shaking for solubilization, and then solubilized membrane proteins were separated from insoluble material by ultracentrifugation at 100,000 × g for 30 min at 4°C. Insoluble fraction pellets were then resuspended in 300 μl of buffer I. The same volume of soluble, solubilized membrane and insoluble fractions were resuspended in 2× SDS sample buffer and analyzed by SDS-polyacrylamide gel electrophoresis (PAGE).
SDS-PAGE and immunoblotting.
Total cell lysates and samples containing the soluble cell fraction, the solubilized membrane fraction, and the insoluble fraction were incubated at 100°C for 3 min and separated on SDS-PAGE gels containing 12% acrylamide. Proteins were detected after transfer onto a polyvinylidene difluoride membrane (Millipore), followed by incubation with primary monoclonal antibody to GFP (Santa Cruz Biotechnology) and then horseradish peroxidase-conjugated secondary antiserum (ECL-Plus; Amersham Biosciences). Bound horseradish peroxidase-labeled antibodies were detected by enhanced chemiluminescence (Amersham).
RESULTS
To assess interactions between the B. subtilis DivIB, FtsLB, DivIC, and PBP 2B proteins using E. coli artificial septal targeting, we constructed two sets of protein fusions: GFP and ZapA fusion proteins. The GFP fusions were expressed from a pBR origin plasmid (pCR25 or pCR27, see Table 1) in which gfp is fused in frame to the 5′ extremity of divIB, ftsLB, divIC, or pbpB through a three-asparagine linker (3×Asn). The second set of protein fusions was constructed in a derivative of the low-copy plasmid pSC101 (pMDG1; Gonzalez, unpublished [see Table 1]). These plasmids expressed ZapA connected to the N terminus of DivIB, FtsLB, DivIC, or PBP 2B by a linker that includes the first cytoplasmic domain of the membrane protein MalF plus 3×Asn (24). All constructs were expressed from IPTG-inducible promoters. To facilitate the detection of proteins, we tagged DivIB and PBP 2B fusions with a Myc epitope and DivIC and FtsLB fusions with a Flag epitope, in all cases at the C-terminal ends of the cell division proteins. The expression of the fusion proteins was analyzed from whole-cell samples by Western blotting, which shows detectable levels of proteins for all of the GFP and ZapA fusions, with some degradation products, signifying some degree of instability (data not shown). Some of these proteins were tested without a C-terminal tag, and they gave the same results presented here and in the following sections.
B. subtilis DivIB, FtsLB, DivIC, and PBP 2B do not function in E. coli cell division.
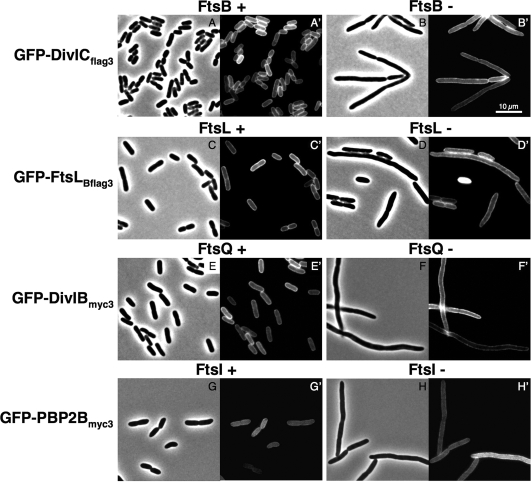
We initially considered the possibility that, despite the relatively high sequence divergence of these proteins, the B. subtilis proteins might substitute for the E. coli one in the cell division process. In that case, we would not be able to study interactions between B. subtilis proteins independently of other possible E. coli interacting proteins. In order to eliminate this possibility, we performed complementation tests in E. coli strains depleted of the E. coli ortholog of the B. subtilis protein present in the fusion tested. For example, GFP-DivIBmyc3 and ZapA-DivIBmyc3 each were tested for complementation of an E. coli FtsQ depletion strain (JOE417) carrying an ftsQ-null allele. This strain contained a wild-type ftsQ expressed from an arabinose-inducible plasmid and thus could be maintained in arabinose-containing medium but could not grow in the presence of glucose. Complementation tests with FtsLBflag3, DivICflag3, and PBP 2Bmyc3 fused to GFP or ZapA were performed in equivalent strains: the E. coli ftsL depletion strain NB988, the E. coli ftsB depletion strain CR14, and the E. coli ftsI depletion strain JC60, each one complemented by the wild-type genes ftsL, ftsB, and ftsI respectively, expressed from an arabinose-inducible promoter. On solid media containing glucose and 20 μM IPTG to induce expression of the heterologous protein fusions, these strains failed to grow, indicating that DivIBmyc3, FtsLBflag3, DivICflag3, and PBP 2Bmyc3 fusions do not complement (data not shown). No growth was observed on plates without or with IPTG (0.1 to 1 mM). The failure to restore cell division is clearly seen by microscopic examination of filament formation in liquid cultures grown with glucose (Fig. 2B, D, F, and H for the GFP fusion [data not shown for the ZapA fusion]). In contrast, cells grown in the presence of arabinose displayed a normal morphology (Fig. 2A, C, E, and G).
FIG. 2.
DivICflag3, FtsLBflag3, DivIBmyc3, and PBP 2Bmyc3 GFP fusions do not localize to the cell division site in strains depleted of their respective E. coli orthologs. E. coli strains were deleted of ftsB, ftsL, ftsQ, or ftsI at their native loci and complemented by the wild-type ftsB, ftsL, ftsQ, or ftsI gene, respectively, carried on a pBAD plasmid and induced in the presence of arabinose (FtsB+, FtsL+, FtsQ+, and FtsI+) or repressed in the presence of glucose (FtsB-, FtsL-, FtsQ-, and FtsI-). Cells expressed the GFP fusion from a pDSW204 plasmid after induction with 20 μM IPTG during 1 h at 30°C. Samples were prepared for microscopy as described in Materials and Methods. Phase-contrast and fluorescence microscopy images are shown for each set of strain and conditions. (A to B′) GFP-DivICflag3 expressed in an E. coli FtsB depletion strain (CR166); (C to D′) GFP-FtsLBflag3 expressed in an E. coli FtsL depletion strain (CR168); (E to F′) GFP-DivIBmyc3 expressed in an E. coli FtsQ depletion strain (CR315); (G to H′) GFP-PBP 2Bmyc3 expressed in an E. coli FtsI depletion strain (CR368). Bar scale, 10 μm.
It should be noted that all ZapA fusions cause slight defects in growth, yielding somewhat elongated cells at 30°C. However, cells expressing GFP fusions display a normal wild-type morphology, indicating that these B. subtilis cell division proteins do not interfere with the E. coli cell division process.
Finally, we also determined the subcellular localization of these B. subtilis GFP fusions proteins in the depletion strains described above. Cultures were grown in the presence of arabinose or glucose, and 20 μM IPTG was added during 30 min to induce expression of the GFP fusions. When strains were grown in the presence of arabinose or glucose, the fluorescent signal is distributed over the entire cell envelope, suggesting that, as expected for these bitopic membrane proteins, GFP-DivIBmyc3, GFP-FtsLBflag3, GFP-DivICflag3, and GFP-PBP 2Bmyc3 are inserted into the cytoplasmic membrane in wild-type and depletion conditions (Fig. 2A′ to H′). The inability to detect septal localization in both conditions suggests that these fusion proteins are not targeted to the E. coli septum, a finding consistent with their inability to complement for the function of their E. coli orthologs.
The lack of complementation and septal localization of these B. subtilis fusions can either be because these fusions are unable to interact with the E. coli divisomal complex and are nonfunctional or because they are unstable. However, since interaction and stability are connected in many cases, such as for E. coli FtsB and FtsL (see the introduction above), the two explanations may be connected. These possibilities should be kept in mind in evaluating the remaining experiments when negative results are obtained and are discussed below.
E. coli artificial septal targeting reveals two sets of direct protein-protein interactions: DivIC with FtsLB and DivIB with PBP 2B.
The constructs expressing GFP fusion proteins in single copy were integrated into the chromosome at the λ attachment site of the wild-type strain JOE309 (see Table 1 and Materials and Methods) (7, 35). The four resulting strains—CR198, CR200, CR202, and CR362—were then transformed with pCR39, pCR40, or pCR41, each one expressing a ZapA fusion protein (see Table 1).
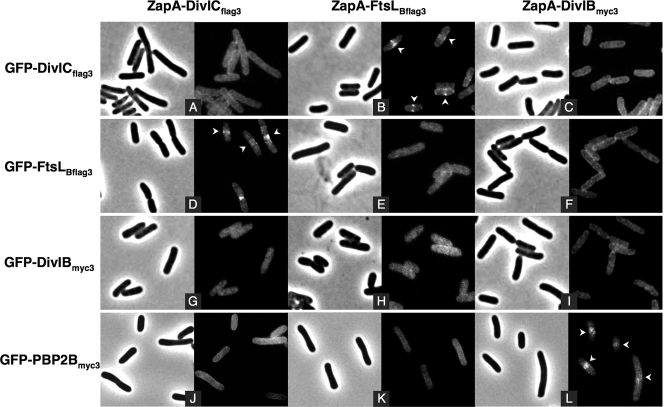
To assess protein-protein interactions, we examined the localization of each GFP fusion when coexpressed, in a pairwise fashion, with each ZapA fusion. Fluorescent dots or bands were observed at mid-cell in strains coexpressing FtsLBflag3 and DivICflag3, the former to GFP and the latter to ZapA, or vice versa, and in strains coexpressing GFP-PBP 2Bmyc3 with ZapA-DivIBmyc3 (Fig. 3B, D, and L). These results indicate that the interaction between FtsLBflag3 and DivICflag3 and between DivIBmyc3 and PBP 2Bmyc3 is strong enough to allow them to stably colocalize to the mid-cell. In contrast, all of the other pairs of proteins (i.e., FtsLBflag3/DivIBmyc3, DivICflag3/DivIBmyc3, FtsLBflag3/PBP 2Bmyc3, and DivICflag/PBP 2Bmyc3; see Fig. 3C, F to H, and J to K), as well as the three tested homodimeric combinations (i.e., FtsLBflag3/FtsLBflag3, DivICflag3/DivICflag3, and DivIBmyc3/DivIBmyc3; see Fig. 3A, E, and I) showed a diffuse fluorescent signal. These observations indicate that, in the absence of other B. subtilis cell division proteins, DivICflag3 interacts strongly with FtsLBflag3 and DivIBmyc3 interacts strongly with PBP 2Bmyc3. However, other combinations of protein fusions do not interact at all or at least not strongly enough to visualize a fluorescent signal at mid-cell.
FIG. 3.
Interaction assays with DivICflag3, FtsLBflag3, DivIBmyc3, and PBP 2Bmyc3 fused to GFP and ZapA using the E. coli artificial septal targeting. Fluorescence microscopy images show subcellular localization of DivICflag3, FtsLBflag3, DivIBmyc3, and PBP 2Bmyc3 GFP fusions when coexpressed with DivICflag3, FtsLBflag3, or DivIBmyc3 ZapA fusions. The phase-contrast images corresponding to the same field as the fluorescence images are shown. Strains (from CR209 to CR211 [A to C], CR213 to CR215 [D to F], CR217 to CR219 [G to I], and CR374 to CR376 [J to L], Table 1) were grown at 30°C, and fusions were induced during 30 min with 20 μM IPTG. Samples were then prepared for microscopy as described in Materials and Methods. Arrows indicate mid-cell fluorescence.
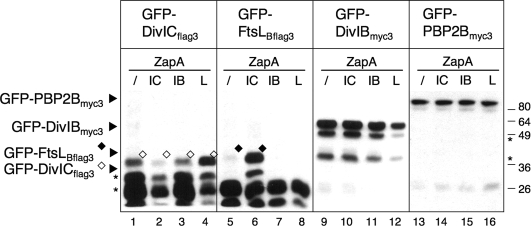
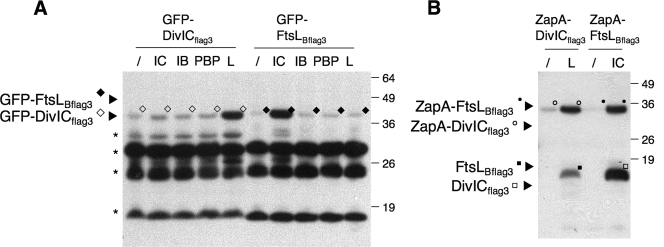
When we examined the expression level of ZapA and GFP fusions individually by immunoblotting, both series of fusions showed degradation products, indicating some degree of instability. In fact, the GFP-FtsLBflag3 and GFP-DivICflag3 fusions were barely detected when expressed at these low levels (Fig. 4, lanes 1 to 8). However, interestingly, both fusion proteins were more stable when coexpressed with ZapA-DivICflag3 or ZapA-FtsLBflag3, respectively (Fig. 4, lanes 4 and 6), suggesting that their association in a complex is sufficient to protect them from degradation. The same stability pattern was observed for ZapA fusions of FtsLBflag3 and DivICflag3 coexpressed with GFP-DivICflag3 and GFP-FtsLBflag3, respectively (data not shown). Using fractionation experiments, we confirmed that all full-length GFP fusions were present in the solubilized membrane fraction (M), whereas the smallest degradation products, likely corresponding to the unfused GFP domain, were soluble (S) (see Fig. S1 in the supplemental material).
FIG. 4.
Steady-state levels of GFP fusion proteins coexpressed with ZapA fusions. Total cell extracts from strains (CR208 to CR219 and CR373 to CR376, Table 1) coexpressing GFP-DivICflag3, GFP-FtsLBflag3, GFP-DivIBmyc3, or GFP-PBP 2Bmyc3 with ZapA-DivICflag3 (IC), ZapA-FtsLBflag3 (L), ZapA-DivIBmyc3 (IB), or ZapA-FtsB (/) as a negative control were analyzed by SDS-PAGE and immunoblotting with anti-GFP antibodies. The molecular masses of the standards are indicated to the right of the blot in kilodaltons. The “★” symbol indicates degradation products.
The results showing that the B. subtilis proteins, DivICflag3 and FtsLBflag3, are unstable when not complexed with their respective partner are relevant to our finding that the B. subtilis proteins are unable to complement E. coli strains depleted of FtsB and FtsL. Indeed, successful complementation would require, first, that Bacillus proteins be functional in E. coli and, second, an interaction strong enough, between DivICflag3 and E. coli FtsL or between FtsLBflag3 and E. coli FtsB, to stabilize both proteins and thus allow septal localization. Thus, we believe that the failure to complement is not due to the instability of DivICflag3 and FtsLBflag3 but, to the contrary, their instability is a consequence of the inability of these B. subtilis protein to bind tightly to a partner protein in the E. coli divisome.
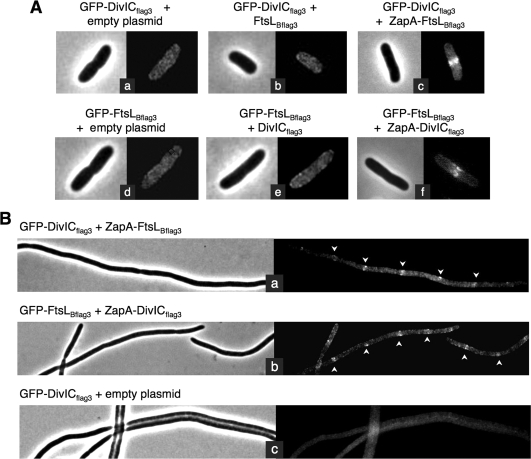
In order to confirm that the complexes observed in cases where the GFP and ZapA fusions are well recruited to the mid-cell actually do depend on the fusion of one of these proteins to ZapA, we performed the following experiments. We analyzed the cellular location of GFP-DivICflag3 or GFP-FtsLBflag3 fusions when coexpressed, respectively, with FtsLBflag3 or DivICflag3 not fused to ZapA. Strains CR198 and CR200 harboring GFP fusion genes integrated into their chromosomes were transformed with a low-copy plasmid expressing under IPTG regulation either FtsLBflag3 or DivICflag3 (see Table 1). Fluorescence microscopy revealed a diffuse fluorescence signal when GFP-DivICflag3 was coexpressed with FtsLBflag3 (Fig. 5Ab) and when GFP-FtsLBflag3 was coexpressed with DivICflag3 (Fig. 5Ae), indicating that these GFP fusions are not targeted to the mid-cell. These results support our conclusion that the DivICflag3/FtsLBflag3 complex cannot be targeted to the septum independently of one being fused to ZapA. Moreover, these results strongly indicate that, when DivICflag3 and FtsLBflag3 are costabilized, they do not localize on their own to the E. coli septum, which is consistent with subcellular localization assays presented previously.
FIG. 5.
E. coli septal targeting of DivICflag3/FtsLBflag3 complex, tested in the absence of ZapA, fused to one of them or tested in an FtsQ depletion strain. (A) Fluorescence microscopy images showing subcellular localization of DivICflag3 and FtsLBflag3 GFP fusions when coexpressed with DivICflag3, FtsLBflag3, ZapA-DivICflag3, or ZapA-FtsLBflag3 or in cells carrying an empty plasmid. Strains CR275 (a), CR278 (b), CR211 (c), CR279 (d), CR280 (e), and CR213 (f) were grown at 30°C, and tested proteins were induced for 30 min with 20 μM IPTG. Samples were then prepared for microscopy as described in Materials and Methods. (B) Subcellular localization of GFP-DivICfllag3 and GFP-FtsLBflag3 when coexpressed with ZapA-FtsLBflag3 (a) and ZapA-DivICflag3 (b), respectively, or with ZapA-FtsB as a negative control (c) in an E. coli ftsQ-depleted strain. Strains CR253, CR257, and CR251 were depleted of FtsQ and complemented by the wild-type ftsQ gene carried on a pBAD plasmid and repressed in the presence of glucose. Cells expressed the GFP fusion from a pDSW204 plasmid, integrated on the chromosome, and ZapA fusions on a plasmid, after induction with 20 μM IPTG during 30 min at 30°C. Samples were prepared for microscopy as described in Materials and Methods, and phase-contrast and fluorescence microscopy images are shown for each strain. Arrows indicate some septal localizations in filaments.
Altogether, our results indicate that DivICflag3/FtsLBflag3 and DivIBmyc3/PBP 2Bmyc3 form two independent stable complexes in E. coli, in which the septal localization is dependent on a fusion of one of the proteins to ZapA.
The interaction between DivIC and FtsLB in E. coli does not require E. coli FtsQ.
Despite the generally weak homology between B. subtilis and E. coli cell division proteins, we sought to determine whether the observed interaction and costabilization of DivICflag3 and FtsLBflag3 might be dependent on an E. coli cell division protein and not a result of the ability of the two proteins to interact independently of any other proteins. A likely candidate for such a protein is FtsQ, the E. coli ortholog of DivIB, which interacts and stabilizes E. coli FtsB and FtsL (5). To determine whether FtsQ plays a role in the observed interaction of DivICflag3 and FtsLBflag3, septal targeting analysis was performed with DivICflag3 and FtsLBflag3, fused to GFP and ZapA, in an ftsQ depletion strain. Strains CR198, CR200, and CR202 were transformed with pBAD33 carrying the wild-type ftsQ gene under the control of an arabinose-inducible promoter and then transduced with a phage P1 carrying ftsQE14::kan. The resulting strains therefore lack chromosomally expressed FtsQ but are complemented with plasmid-encoded FtsQ and express the GFP fusion protein from the chromosome and the ZapA fusion gene from a plasmid in the presence of IPTG (see Table 1). Strains are dependent on arabinose for growth and exhibit a normal morphology. When grown in the presence of glucose, however, FtsQ depletion leads to the formation of filaments (Fig. 5Bc) (11). In these cells deficient for septation, GFP-DivICflag3 and GFP-FtsLBflag3 are localized at equivalent intervals in filaments when coexpressed with ZapA-FtsLBflag3 and with ZapA-DivICflag3, respectively (Fig. 5Ba and Bb). This septal localization indicates that FtsQ does not participate in the interaction and targeting of the DivICflag3/FtsLBflag3 complex.
B. subtilis proteins DivIB, FtsLB, DivIC, and PBP 2B do not form stable trimeric or tetrameric protein complexes in E. coli.
We further used the E. coli artificial septal targeting approach to test the effect of the coexpression of a third or fourth protein on the interaction between the GFP and ZapA fusion proteins. In particular, we assumed that, in a three-hybrid assay, the test FtsLB (or DivIC) fusions would be stabilized in the presence of an unfused DivIC (or FtsLB) partner and consequently promote an interaction with one of the two other DivIB or PBP 2B fusion proteins. To perform these assays, the strains coexpressing the GFP and ZapA fusions (Fig. 3) were cotransformed with a second plasmid, pBAD33, expressing either DivIBmyc3, FtsLBflag3, DivICflag3, PBP 2Bmyc3 or the two proteins PBP 2Bmyc3 and DivICflag3. Cells were grown at 30°C and incubated for 1 h with IPTG and arabinose to induce all of the constructs. DivICflag3/FtsLBflag3 fusions and DivIBmyc3/PBP 2Bmyc3 fusions show the same positive signal at mid-cell as described previously, no matter which additional B. subtilis cell division proteins were coexpressed (Table 2). None of the other three- or four-hybrid experiments gave any significant fluorescent signal at the mid-cell, suggesting that, in these cases, the presence of the additional protein did not promote interactions between the GFP and ZapA fusion proteins (Table 2). Importantly, Western blot analyses indicate that GFP-DivICflag3 and GFP-FtsLBflag3, and similarly ZapA-DivICflag3 and ZapA-FtsLBflag3, were both stabilized when coexpressed with FtsLBflag3 and DivICflag3, respectively, as described above (Fig. 6). Hence, this costabilization of DivICflag3 and FtsLBflag3 was not sufficient to favor their interaction with the DivIBmyc3 or PBP 2Bmyc3 fusion proteins. These data suggest that the four B. subtilis cell division proteins, DivIBmyc3, FtsLBflag3, DivICflag3 and PBP 2Bmyc3, do not form a ternary or quaternary protein complex that is stable enough on its own to be localized to the septum in E. coli.
TABLE 2.
E. coli artificial septal targeting assays performed with the coexpression of three or four B. subtilis proteinsa
| Third protein | ZapA fusions
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GFP-DivICflag3
|
GFP-FtsLBflag3
|
GFP-DivIBmyc3
|
GFP-PBP 2Bmyc3
|
|||||||||
| IC | LB | IB | IC | LB | IB | IC | LB | IB | IC | LB | IB | |
| Empty pBAD33 plasmid | - | + | - | + | - | - | - | - | - | - | - | + |
| DivICflag3 | ND | + | - | + | - | - | - | - | - | - | - | + |
| FtsLBmyc3 | - | + | - | + | ND | - | - | ND | - | - | - | + |
| DivIBmyc3 | - | + | - | + | - | - | - | - | - | - | - | + |
| PBP2Bmyc3 | - | + | - | + | - | - | - | - | - | - | - | + |
| PBP2Bmyc3 + DivICflag3 | ND | ND | ND | + | ND | - | - | - | - | ND | ND | ND |
Subcellular localization of DivICflag3, FtsLBflag3, DivIBmyc3, and PBP 2Bmyc3 GFP fusions has been observed in strains coexpressing them with DivICflag3 (IC), FtsLBflag3 (LB), or DivIBmyc3 (IB) ZapA fusions and with an empty pBAD33 plasmid or DivICflag3, FtsLBmyc3, DivIBmyc3, PBP 2Bmyc3 or (PBP 2Bmyc3+ DivICflag3) expressed from plasmid pBAD33. Strains (from CR209 to CR211, CR213 to CR215, CR217 to CR219, and CR374 to CR376; Table 1) transformed with pBAD33, pCR52, pCR54, pCR60, pCR63, or pCR68 (Table 1) were grown at 30°C, and tested proteins were induced for 1 h with 50 μM IPTG and 0.2% arabinose. Samples were then prepared for microscopy as described in Materials and Methods. +, Septal localization of the GFP fusion; -, diffuse GFP signal; ND, not determined.
FIG. 6.
Steady-state levels of DivICflag3 and FtsLBflag3 GFP or ZapA fusions coexpressed with DivICflag3, DivIBmyc3, PBP 2Bmyc3, or FtsLBflag3. (A) Total cell extracts from strains (CR198 and CR200 carrying the plasmids pBAD33, pCR52, pCR54, pCR60, or pCR63 [Table 1]), coexpressing GFP-DivICflag3 or GFP-FtsLBflag3, with DivICflag3 (IC), DivIBmyc3 (IB), PBP 2B myc3 (PBP), FtsLBflag3 (L) or carrying an empty plasmid (/) as a negative control were analyzed by SDS-PAGE and immunoblotting with anti-GFP antibodies. (B) Total cell extracts from strains JOE309 coexpressing ZapA-DivICflag3 (from pCR39) or ZapA-FtsLBflag3 (from pCR41) with DivICflag3 (IC) (from pCR52) or FtsLBflag3 (L) (from pCR63) or carrying an empty plasmid (/) as a negative control were analyzed by SDS-PAGE and immunoblotting with anti-Flag antibodies. Molecular masses of the standards are indicated to the right of the blot in kDa. The “★” symbol indicates degradation products.
DISCUSSION
The process of assembly of multiprotein complexes presents a challenging problem for those seeking to understand the individual protein-protein interactions involved. For example, it has been difficult to study in vivo the interactions between specific pairs of cell division proteins and to distinguish those interactions from ones that require additional components of the larger divisomal protein complex. The dependency pathway and interdependency of septal recruitment of Bacillus cell division proteins, shown in Fig. 1, raises such problems for the study of protein-protein interactions that direct formation of the E. coli or other bacterial divisomes. The E. coli artificial septal targeting system, presented here, represents an approach that, in contrast to other approaches, likely detects only strong interactions between subsets of proteins in a bacterial multiprotein complex, those interactions that do not require other components of the complex. Specifically, this approach, in its general sense, requires expression of subsets of proteins from a multiprotein complex of one bacterium in another unrelated bacterium. The unrelated bacterium (in this case, E. coli) should not contain a comparable complex or the components of that complex should be sufficiently different from the organism under study, so that no interactions between components of the two occur. Thus, to begin with, we recruit a given foreign protein to the septal region of E. coli by fusing it to ZapA, an FtsZ-binding protein, and assess the septal localization of a second foreign protein by expressing it fused to GFP. The interaction between the two tested proteins is reflected in the detection of a septal fluorescence signal. To test this approach, we have examined the interactions between four proteins involved in the cell division process in B. subtilis by expressing them simultaneously in E. coli. Our results indicate that this fluorescence-based visual detection system is a useful tool that allows deciphering of the most robust protein-protein interactions directing formation of bacterial divisomal complexes. This system would help in the identification of new protein-protein interactions, in this case, for example, detecting proteins that promote interactions between the two binary complexes we have detected. This approach also adds to the methodologies for validating of data from other genetic and biochemical approaches. Finally, this approach may also find general use in the study of multiprotein complexes unrelated to cell division in E. coli and protein complex assembly in heterologous species.
The four B. subtilis cell division proteins we have studied—DivIB, FtsLB, DivIC, and PBP 2B—are thought to interact in their native organism, although many features of these interactions are not well established (13-15, 31, 46). We present several lines of evidence that these four B. subtilis divisomal proteins do not interfere with the E. coli cell division process. First, although these proteins are orthologs of E. coli FtsQ, FtsL, FtsB, and FtsI, respectively, they generally show weak homology. Second, E. coli strains depleted of the E. coli proteins cannot be complemented by expression of the corresponding ortholog from B. subtilis. Third, we demonstrate that GFP fusions to each of the proteins do not localize to the E. coli divisome. This result indicates that there are no interactions between these GFP fusions and host division proteins, or at least no interactions that are strong enough to recruit the B. subtilis proteins to the mid-cell. Fourth, expression of the B. subtilis proteins does not interfere with the E. coli cell division process, nor does it induce any obvious phenotype in this host.
Our results show that among the four B. subtilis proteins there are two pairs, (i) DivIC and FtsLB and (ii) DivIB and PBP 2B, that interact strongly enough so that when one of a pair is recruited to the divisome via a ZapA fusion, the other, fused to GFP, is also recruited (Fig. 7A). Some degree of interaction among the proteins of these two pairs has been previously reported in bacterial two-hybrid studies (15, 43). It is important to note that the various two-hybrid studies described to date involve expression of fusion proteins at a high level that probably increases the probability of detecting weak or transitory interactions between two proteins. While providing a means of detecting potential interactions among proteins, these studies do not indicate whether the observed interactions are strong enough to maintain proteins in a stable complex. In contrast, by physically observing the localization of the proteins to the bacterial septum, rather than by indirect assessment via expression of reporter proteins, we can, using our system, identify such strong interactions. Furthermore, the fact that low levels of protein are sufficient to detect a positive interaction between ZapA and GFP fusions is a significant advantage of this system. It is likely that weaker interactions that do play a role in divisome assembly and which are observed in other two-hybrid systems would not be detected. Distinguishing strong interactions from weak interactions (as well as from false positives) should be important in designing experiments for the detection of subcomplexes of such proteins.
FIG. 7.
(A) Positive protein-protein interactions between B. subtilis cell division proteins DivIC, FtsL, DivIB, and PBP 2B detected by E. coli artificial septal targeting and bacterial two-hybrid assays. (B) Schema of the two stable subcomplexes identified in the present study. The two subcomplexes may be strongly bound in a complex only when another protein(s) X is present. Strong interactions between proteins are indicated by dark bars, while weaker interactions are indicated by light gray bars and arrows. Circular arrows reflect proposed homodimeric interactions.
In the case of homodimer formation, we did not observe any interactions (e.g., between ZapA-DivIBmyc3 and GFP-DivIBmyc3) sufficient to localize the GFP fusion to the mid-cell. Other than the two binary complexes described above, we observed, by our technique, no other binary heterodimeric interactions. Singly, neither DivIC nor FtsLB showed an interaction with DivIB or PBP 2B in our binary assays, whereas DivIB and PBP 2B have been shown to participate in the stabilization of DivIC and FtsLB in B. subtilis (i.e., when other proteins of the divisome are present) (13, 15, 32). In addition, previous bacterial two-hybrid studies have suggested positive reciprocal interactions among pairs of the three B. subtilis proteins: PBP 2B, DivIB, and FtsLB (15). In our study, the formation of the DivIC/FtsLB complex stabilizes both members, indicating that, at least in the E. coli periplasm, DivIB and PBP 2B are not required for stabilization of this protein complex. Thus, the assembly of the FtsLB/DivIC cell division subcomplex in E. coli appears remarkably similar to the one between FtsB and FtsL. Interestingly, previous biochemical studies performed to examine the putative physical association between the extracellular domains of DivIC and FtsLB failed to find an interaction between these proteins (41). These contrasting results suggest that the full-length of the proteins (or at least “membrane-anchored” versions of these proteins) are necessary for a stable interaction between FtsLB and DivIC.
One intriguing aspect of our results is the finding that the two complexes, FtsLB/DivIC and PBP 2B/DivIB, which individually stably localize to the septum, do not interact strongly enough with each other to colocalize. These results suggest that other components of the divisomal complex are necessary to connect these two subcomplexes, despite the indications of interactions between components of the two complexes seen in previous two-hybrid assays (Fig. 7B). Thus, our results point to additional experiments to identify other protein components required for the necessary stabilizing interactions. Overall, our results indicate further the complex nature of the assembly of this multiprotein complex and the need to sort out the significance of weak and strong interactions.
The stability of individual proteins, or protein complexes, in E. coli cannot probably be extended to B. subtilis due to differences in proteases or other factors. For example, Brankamp et al. recently described a B. subtilis protease (YluC), specific for FtsLB, and for which no homolog has been reported in E. coli (3). Indeed, the fact that the two complexes, FtsLB/DivIC and PBP 2B/DivIB, are individually stabilized in E. coli, contrasts with the situation in their native environment, in which the four proteins localize interdependently, thus making biochemical studies of the individual proteins, or pairs of proteins, rather difficult (41, 43). In addition, overexpression and purification of FtsLB or DivIC, from E. coli strains, for in vitro analyses, remains unsuccessful, likely because the two proteins need to interact with each other to costabilize in E. coli. Finally, the enhanced stability of FtsLB and DivIC proteins in E. coli opens up the possibility of studying, more precisely, the nature of the interaction at the dimer interface, such as the role of the extracellular leucine-zipper-like motifs.
Previously, others groups tested similar approaches to E. coli septal targeting based on a visual detection with GFP: one group used E. coli FtsZ and GFP fusions to study complex formation by components of the Agrobacterium tumefaciens type IV secretion system (17), while another group used the B. subtilis protein DivIVA, naturally targeted to mature cell poles, and GFP fusions (12). Indeed, these approaches showed variable efficiency due to perturbation of the cell division cycle through FtsZ fusions in the first example, or difficulties in discriminating a positive polar interaction from a polar inclusion body in the second case, as overexpressed GFP fusion proteins tend to aggregates and accumulate at the cell poles (35).
In conclusion, we show that the E. coli artificial septal targeting approach represents an efficient means to detect, in E. coli, protein-protein interactions among B. subtilis cell division proteins. The enhanced stability of heterologous proteins in E. coli may, in many cases, be a major advantage of this system for performing further studies to characterize, at the molecular level, protein-protein interactions that drive complex formation. Moreover, the direct observation of the localization of proteins to the bacterial septum through fluorescence and the low level of protein fusion expression make the septal targeting approach useful in focusing on what interactions are strong enough to allow stable complex formation. Finally, this system should find a general use in the identification of new protein-protein interactions and in the study of protein complex assembly in heterologous species.
Supplementary Material
Acknowledgments
We thank M. Gonzalez for advice and helpful technical assistance, all of the members of the Beckwith laboratory for support, D. Rudner for B. subtilis genomic DNA, and D. Ladant for helpful comments on the manuscript.
This study was supported by grants from the National Institute of General Medical Sciences (GMO38922 to J.B.) and the National Health and Medical Research Council (Project Grant 519735 to G.F.K.). C.R. holds a Marie Curie Outgoing International Fellowship of the European community under contract number MOIF-CT-2005-008977. N.W.G. was an HHMI Predoctoral Fellow.
Footnotes
Published ahead of print on 11 July 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Bernard, C. S., M. Sadasivam, D. Shiomi, and W. Margolin. 2007. An altered FtsA can compensate for the loss of essential cell division protein FtsN in Escherichia coli. Mol. Microbiol. 641289-1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boyd, D., D. S. Weiss, J. C. Chen, and J. Beckwith. 2000. Towards single-copy gene expression systems making gene cloning physiologically relevant: lambda InCh, a simple Escherichia coli plasmid-chromosome shuttle system. J. Bacteriol. 182842-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bramkamp, M., L. Weston, R. A. Daniel, and J. Errington. 2006. Regulated intramembrane proteolysis of FtsL protein and the control of cell division in Bacillus subtilis. Mol. Microbiol. 62580-591. [DOI] [PubMed] [Google Scholar]
- 4.Buddelmeijer, N., M. E. Aarsman, A. H. Kolk, M. Vicente, and N. Nanninga. 1998. Localization of cell division protein FtsQ by immunofluorescence microscopy in dividing and nondividing cells of Escherichia coli. J. Bacteriol. 1806107-6116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buddelmeijer, N., and J. Beckwith. 2004. A complex of the Escherichia coli cell division proteins FtsL, FtsB, and FtsQ forms independently of its localization to the septal region. Mol. Microbiol. 521315-1327. [DOI] [PubMed] [Google Scholar]
- 6.Buddelmeijer, N., and J. Beckwith. 2002. Assembly of cell division proteins at the Escherichia coli cell center. Curr. Opin. Microbiol. 5553-557. [DOI] [PubMed] [Google Scholar]
- 7.Buddelmeijer, N., O. Francetic, and A. P. Pugsley. 2006. Green fluorescent chimeras indicate nonpolar localization of pullulanase secreton components PulL and PulM. J. Bacteriol. 1882928-2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buddelmeijer, N., N. Judson, D. Boyd, J. J. Mekalanos, and J. Beckwith. 2002. YgbQ, a cell division protein in Escherichia coli and Vibrio cholerae, localizes in codependent fashion with FtsL to the division site. Proc. Natl. Acad. Sci. USA 996316-6321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, J. C., and J. Beckwith. 2001. FtsQ, FtsL and FtsI require FtsK, but not FtsN, for colocalization with FtsZ during Escherichia coli cell division. Mol. Microbiol. 42395-413. [DOI] [PubMed] [Google Scholar]
- 10.Chen, J. C., M. Minev, and J. Beckwith. 2002. Analysis of ftsQ mutant alleles in Escherichia coli: complementation, septal localization, and recruitment of downstream cell division proteins. J. Bacteriol. 184695-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, J. C., D. S. Weiss, J. M. Ghigo, and J. Beckwith. 1999. Septal localization of FtsQ, an essential cell division protein in Escherichia coli. J. Bacteriol. 181521-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Corbin, B. D., B. Geissler, M. Sadasivam, and W. Margolin. 2004. Z-ring-independent interaction between a subdomain of FtsA and late septation proteins as revealed by a polar recruitment assay. J. Bacteriol. 1867736-7744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Daniel, R. A., and J. Errington. 2000. Intrinsic instability of the essential cell division protein FtsL of Bacillus subtilis and a role for DivIB protein in FtsL turnover. Mol. Microbiol. 36278-289. [DOI] [PubMed] [Google Scholar]
- 14.Daniel, R. A., E. J. Harry, V. L. Katis, R. G. Wake, and J. Errington. 1998. Characterization of the essential cell division gene ftsL(yIID) of Bacillus subtilis and its role in the assembly of the division apparatus. Mol. Microbiol. 29593-604. [DOI] [PubMed] [Google Scholar]
- 15.Daniel, R. A., M. F. Noirot-Gros, P. Noirot, and J. Errington. 2006. Multiple interactions between the transmembrane division proteins of Bacillus subtilis and the role of FtsL instability in divisome assembly. J. Bacteriol. 1887396-7404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Di Lallo, G., M. Fagioli, D. Barionovi, P. Ghelardini, and L. Paolozzi. 2003. Use of a two-hybrid assay to study the assembly of a complex multicomponent protein machinery: bacterial septosome differentiation. Microbiology 1493353-3359. [DOI] [PubMed] [Google Scholar]
- 17.Ding, Z., Z. Zhao, S. J. Jakubowski, A. Krishnamohan, W. Margolin, and P. J. Christie. 2002. A novel cytology-based, two-hybrid screen for bacteria applied to protein-protein interaction studies of a type IV secretion system. J. Bacteriol. 1845572-5582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.D'Ulisse, V., M. Fagioli, P. Ghelardini, and L. Paolozzi. 2007. Three functional subdomains of the Escherichia coli FtsQ protein are involved in its interaction with the other division proteins. Microbiology 153124-138. [DOI] [PubMed] [Google Scholar]
- 19.Errington, J., R. A. Daniel, and D. J. Scheffers. 2003. Cytokinesis in bacteria. Microbiol. Mol. Biol. Rev. 6752-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geissler, B., D. Elraheb, and W. Margolin. 2003. A gain-of-function mutation in ftsA bypasses the requirement for the essential cell division gene zipA in Escherichia coli. Proc. Natl. Acad. Sci. USA 1004197-4202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geissler, B., and W. Margolin. 2005. Evidence for functional overlap among multiple bacterial cell division proteins: compensating for the loss of FtsK. Mol. Microbiol. 58596-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghigo, J. M., D. S. Weiss, J. C. Chen, J. C. Yarrow, and J. Beckwith. 1999. Localization of FtsL to the Escherichia coli septal ring. Mol. Microbiol. 31725-737. [DOI] [PubMed] [Google Scholar]
- 23.Goehring, N. W., and J. Beckwith. 2005. Diverse paths to midcell: assembly of the bacterial cell division machinery. Curr. Biol. 15R514-R526. [DOI] [PubMed] [Google Scholar]
- 24.Goehring, N. W., M. D. Gonzalez, and J. Beckwith. 2006. Premature targeting of cell division proteins to midcell reveals hierarchies of protein interactions involved in divisome assembly. Mol. Microbiol. 6133-45. [DOI] [PubMed] [Google Scholar]
- 25.Goehring, N. W., F. Gueiros-Filho, and J. Beckwith. 2005. Premature targeting of a cell division protein to midcell allows dissection of divisome assembly in Escherichia coli. Genes Dev. 19127-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goehring, N. W., I. Petrovska, D. Boyd, and J. Beckwith. 2007. Mutants, suppressors, and wrinkled colonies: mutant alleles of the cell division gene ftsQ point to functional domains in FtsQ and a role for domain 1C of FtsA in divisome assembly. J. Bacteriol. 189633-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goehring, N. W., C. Robichon, and J. Beckwith. 2007. Role for the nonessential N terminus of FtsN in divisome assembly. J. Bacteriol. 189646-649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 1774121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harry, E. J., S. L. Rowland, M. S. Malo, and R. G. Wake. 1994. Expression of divIB of Bacillus subtilis during vegetative growth. J. Bacteriol. 1761172-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karimova, G., N. Dautin, and D. Ladant. 2005. Interaction network among Escherichia coli membrane proteins involved in cell division as revealed by bacterial two-hybrid analysis. J. Bacteriol. 1872233-2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Katis, V. L., and R. G. Wake. 1999. Membrane-bound division proteins DivIB and DivIC of Bacillus subtilis function solely through their external domains in both vegetative and sporulation division. J. Bacteriol. 1812710-2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Katis, V. L., R. G. Wake, and E. J. Harry. 2000. Septal localization of the membrane-bound division proteins of Bacillus subtilis DivIB and DivIC is codependent only at high temperatures and requires FtsZ. J. Bacteriol. 1823607-3611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kawai, Y., and N. Ogasawara. 2006. Bacillus subtilis EzrA and FtsL synergistically regulate FtsZ ring dynamics during cell division. Microbiology 1521129-1141. [DOI] [PubMed] [Google Scholar]
- 34.Levin, P. A., and R. Losick. 1994. Characterization of a cell division gene from Bacillus subtilis that is required for vegetative and sporulation septum formation. J. Bacteriol. 1761451-1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Margolin, W. 2000. Green fluorescent protein as a reporter for macromolecular localization in bacterial cells. Methods 2062-72. [DOI] [PubMed] [Google Scholar]
- 36.Miller, J. H. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 37.Muller, P., C. Ewers, U. Bertsche, M. Anstett, T. Kallis, E. Breukink, C. Fraipont, M. Terrak, M. Nguyen-Disteche, and W. Vollmer. 2007. The essential cell division protein FtsN interacts with the murein (peptidoglycan) synthase PBP1B in Escherichia coli. J. Biol. Chem. 28236394-36402. [DOI] [PubMed] [Google Scholar]
- 38.Noirclerc-Savoye, M., A. Le Gouellec, C. Morlot, O. Dideberg, T. Vernet, and A. Zapun. 2005. In vitro reconstitution of a trimeric complex of DivIB, DivIC, and FtsL, and their transient colocalization at the division site in Streptococcus pneumoniae. Mol. Microbiol. 55413-424. [DOI] [PubMed] [Google Scholar]
- 39.Reddy, M. 2007. Role of FtsEX in cell division of Escherichia coli: viability of ftsEX mutants is dependent on functional SufI or high osmotic strength. J. Bacteriol. 18998-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robson, S. A., and G. F. King. 2006. Domain architecture and structure of the bacterial cell division protein DivIB. Proc. Natl. Acad. Sci. USA 1036700-6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robson, S. A., K. A. Michie, J. P. Mackay, E. Harry, and G. F. King. 2002. The Bacillus subtilis cell division proteins FtsL and DivIC are intrinsically unstable and do not interact with one another in the absence of other septasomal components. Mol. Microbiol. 44663-674. [DOI] [PubMed] [Google Scholar]
- 42.Samaluru, H., L. SaiSree, and M. Reddy. 2007. Role of SufI (FtsP) in cell division of Escherichia coli: evidence for its involvement in stabilizing the assembly of the divisome. J. Bacteriol. 1898044-8052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sievers, J., and J. Errington. 2000. The Bacillus subtilis cell division protein FtsL localizes to sites of septation and interacts with DivIC. Mol. Microbiol. 36846-855. [DOI] [PubMed] [Google Scholar]
- 44.van Helvoort, J. M., and C. L. Woldringh. 1994. Nucleoid partitioning in Escherichia coli during steady-state growth and upon recovery from chloramphenicol treatment. Mol. Microbiol. 13577-583. [DOI] [PubMed] [Google Scholar]
- 45.Vicente, M., A. I. Rico, R. Martinez-Arteaga, and J. Mingorance. 2006. Septum enlightenment: assembly of bacterial division proteins. J. Bacteriol. 18819-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wadsworth, K. D., S. L. Rowland, E. J. Harry, and G. F. King. 2008. The divisomal protein DivIB contains multiple epitopes that mediate its recruitment to incipient division sites. Mol. Microbiol. 671143-1155. [DOI] [PubMed] [Google Scholar]
- 47.Weiss, D. S., J. C. Chen, J. M. Ghigo, D. Boyd, and J. Beckwith. 1999. Localization of FtsI (PBP3) to the septal ring requires its membrane anchor, the Z ring, FtsA, FtsQ, and FtsL. J. Bacteriol. 181508-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Youngman, P. J., J. B. Perkins, and R. Losick. 1983. Genetic transposition and insertional mutagenesis in Bacillus subtilis with Streptococcus faecalis transposon Tn917. Proc. Natl. Acad. Sci. USA 802305-2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.