Abstract
The ultimate membrane localization and function of most of the 185 predicted Pseudomonas aeruginosa PAO1 lipoproteins remain unknown. We constructed a fluorescent lipoprotein, CSFPOmlA-ChFP, by fusing the signal peptide and the first four amino acids of the P. aeruginosa outer membrane lipoprotein OmlA to the monomeric red fluorescent protein mCherry (ChFP). When cells were plasmolyzed with 0.5 M NaCl, the inner membrane separated from the outer membrane and formed plasmolysis bays. This permits the direct observation of fluorescence in either the outer or inner membrane. CSFPOmlA-ChFP was shown to localize in the outer membrane by fluorescence microscopy and immunoblotting analysis of inner and outer membrane fractions. The site-directed substitution of the amino acids at positions +2, +3, and +4 in CSFPOmlA-ChFP was performed to test the effects on lipoprotein localization of a series of amino acid sequences selected from a panel of predicted lipoproteins. We confirmed Asp+2 and Lys+3 Ser+4 function as inner membrane retention signals and identified four novel inner membrane retention signals: CK+2 V+3 E+4, CG+2 G+3 G+4, CG+2 D+3 D+4, and CQ+2 G+3 S+4. These inner membrane retention signals are found in 5% of the 185 predicted P. aeruginosa lipoproteins. Full-length chimeras of predicted lipoproteins PA4370 and PA3262 fused to mCherry were shown to reside in the inner membrane and showed a nonuniform or patchy distribution in the membrane. The optical sectioning of cells producing PA4370CGDD-ChFP and PA3262CDSQ-ChFP by confocal microscopy improved the resolution and indicated a helix-like localization pattern in the inner membrane. The method described here permits the in situ visualization of lipoprotein localization and should work equally well for other membrane-associated proteins.
Bacterial lipoproteins are lipid anchored to the inner and outer membranes of gram-negative bacteria. Three fatty acids are attached to the N-terminal cysteine residue that immediately follows the cleavage site of lipoprotein signal peptides (13). The lipids integrate into the periplasm-facing leaflet of either membrane, leaving most lipoproteins exposed to the periplasm (13), although some lipoproteins are surface exposed (9, 16). We have previously used fluorescent microscopy to visualize red fluorescent lipoproteins (lipoRFPs) directly in both the outer and inner membranes of Escherichia coli and other Enterobacteriaceae (10). The method is based on the use of monomeric red fluorescent protein, which is stable and fluorescent in the periplasm (1). LipoRFPs were created by fusing mRFP1 to the signal peptides and the first few amino acids of a known lipoprotein. The lipoRFPs were effectively translocated across the inner membrane and remained either in the inner membrane or were transported to the outer membrane via the Lol lipoprotein-sorting machinery, according to the presence or absence of inner membrane retention signals (10).
We previously identified all exported proteins encoded by the Pseudomonas aeruginosa PAO1 genome, including all secreted proteins and proteins localized to the inner membrane, the periplasm, or outer membrane (11). Surprisingly, 38% of the 5,570 predicted open reading frames in the genome encode proteins predicted to be in the P. aeruginosa cell envelope, including 185 lipoproteins (11), most of which are annotated as hypothetical proteins with unknown functions. Most Escherichia coli lipoproteins are sorted through the Lol lipoprotein-sorting machinery, comprised of five proteins (LolABCDE), to the outer membrane (23). Lipoproteins that contain an aspartate (Asp+2) following the first cysteine residue are localized in the inner membrane (25), which accounts for about 5% of the approximately 90 predicted or known E. coli lipoproteins. Inner membrane lipoproteins are not recognized by the LolCDE complex, an ABC transporter that normally releases lipoproteins to the periplasmic chaperone LolA and ultimately to LolB and the outer membrane (13). Non-aspartate inner membrane retention signals at the +2 position subsequently were identified when it was found that tyrosine, phenylalanine, tryptophan, glycine, and proline also could function as inner membrane retention signals when followed by an asparagine residue at position +3 in an artificial lipoprotein, lipoMalE (18). Although these non-aspartate inner membrane retention signals are not found in E. coli, they are found in a small number of predicted lipoproteins in other bacteria and function similarly when fused to monomeric red fluorescent protein 1 (mRFP1) in other Enterobacteriaceae (10).
The rules governing the sorting of P. aeruginosa lipoproteins were assumed to be the same as those for other gram-negative bacteria, until it was recently reported that amino acids at positions +3 and +4 were essential for determining the inner membrane localization of P. aeruginosa lipoproteins (12). Specifically, Lys+3 and Ser+4 were shown to be required for inner membrane localization (12). Since there are only three predicted lipoproteins with this specific amino acid sequence encoded by the P. aeruginosa PAO1 genome and there are many more lipoproteins that are predicted or known to be in the inner membrane, additional P. aeruginosa inner membrane retention signals likely exist. As a first step to determining the function of the predicted lipoproteins, we wanted to determine the lipoprotein sorting signals that define their ultimate localization in the cell envelope.
MATERIALS AND METHODS
Strains and growth conditions.
All cloning and recombinant DNA manipulations were performed in E. coli DH5α or E. coli PAP105 (10). Plasmids were introduced into P. aeruginosa PAO1 by electroporation as previously described (2). All cultures were grown in Luria-Bertani medium at 37°C, unless otherwise specified, with 30 μg/ml gentamicin (P. aeruginosa) or 100 μg/ml ampicillin (E. coli) to maintain plasmids.
Plasmid construction.
DNA encoding the mCherry variant of mRFP1 was PCR amplified from pRSETb-mCherry (19) with primers Xba_mRFP_mega and ChFP_rev and was cloned as an XbaI-HindIII fragment into the Pseudomonas-E. coli shuttle vector pUCP22 (24) to construct pCHAP6655. The omlA signal peptide-encoding region and promoter was cloned as an EcoRI-XbaI fragment upstream of mCherry to create the gene fusion encoding CSFPOmlA-mCherry fluorescent protein (CSFPOmlA-ChFP) construct (pCHAP6656). The XbaI linker encodes two amino acids (Ser and Arg) between the lipoprotein sequence and mCherry; this linker previously was shown not to interfere with lipoRFP localization (10). The N terminus of mCherry was modified to include the mRFP1 N terminus (six amino acids) followed by the mCherry sequence beginning at amino acid 12.
The native promoter and full-length predicted lipoprotein PA4370 were PCR amplified with primers 4370_for and 4370_rev, and the truncated PA4370 (the promoter and first 60 amino acids) of PA4370 was PCR amplified with primers 4370_for and 4370_rev2. Full-length and truncated PA4370 genes were cloned as EcoRI-XbaI fragments upstream of the mCherry gene to construct pCHAP6672 and pCHAP6670, respectively. The native promoter and full-length predicted lipoprotein PA3262 was PCR amplified with primers 3262_for and 3262_rev and cloned as an EcoRI-XbaI fragment upstream of mCherry to construct pCHAP6688. PCR amplifications were performed in 30 cycles of 94°C for 30 s, 50°C for 39 s, and 72°C for 1 min with 10 M primers and the JumpStart REDTaq ReadyMix kit (Sigma). (See Tables 1 and 2 for a list of all primers and plasmids used in this study.)
TABLE 1.
Primers used in this study
| Primer | Linker | 5′-3′ Sequence |
|---|---|---|
| Xba_mRFP_Mega | XbaI | CAGTCTAGAATGGCCTCCTCCGAGGACGTCATCAAGGAGTTCATGCGCTTCAAGG |
| ChFP_rev | HindIII | GCAAGCTTTTACTTGTACAGCTCGTCCATG |
| omlA_for | EcoRI | GCGAATTCGCTATCGGCGAGCTCGAACAC |
| omlA_rev | XbaI | GCTCTAGAAGGAAACGAGCAACCGGCGAG |
| CX_ChFP_for1 | ATGGCCTCCTCCGAGGACGTCATC | |
| CD_ChFP_mut | TCTGGAAGGAAAGTCGCAACCGGCGAGTGCGGCAAGCCC | |
| 4370_for | EcoRI | GCGAATTCGGACGGAGCTCCGGACGAAG |
| 4370_rev | XbaI | GCTCTAGACTTGATCACCGCCTTGGCGG |
| 4370_rev2 | XbaI | GCTCTAGAGAACTCGTGGTCGGCGTTGTC |
| 4370_mut1 | GGCGCTTCGGCTTTCTTAGGAAACGAGCAGCCGGCGAGGGAG | |
| 4370_mut2 | CTCCCTCGCCGGCTGCTCGTTTCCTAAGAAAGCCGAAGCGCC | |
| 3262_for | EcoRI | GCGAATTCTCCTTCCTCAGCTGTATCTCTTG |
| 3262_rev | XbaI | GCTCTAGATGCGAATCGCAGCCGGAGAGAAC |
| FNS-F | GCACTCGCCGGTTGCTTCAACAGCGGCGCCTCCTCCGAGGACG | |
| FNS-R | CGTCCTCGGAGGAGGCGCCGCTGTTGAAGCAACCGGCGAGTGC | |
| KVE-F | GCACTCGCCGGTTGCAAGGTGGAGGGCGCCTCCTCCGAGGACG | |
| KVE-R | CGTCCTCGGAGGAGGCGCCCTCCACCTTGCAACCGGCGAGTGC | |
| DSQ-F | GCACTCGCCGGTTGCGACAGCCAGGGCGCCTCCTCCGAGGACG | |
| DSQ-R | CGTCCTCGGAGGAGGCGCCCTGGCTGTCGCAACCGGCGAGTGC | |
| GKS-F | GCACTCGCCGGTTGCGGCAAGTCGGGCGCCTCCTCCGAGGACG | |
| GKS-R | CGTCCTCGGAGGAGGCGCCCGACTTGCCGCAACCGGCGAGTGC | |
| GGC-F | GCACTCGCCGGTTGCGGCGGCGGCGGCGCCTCCTCCGAGGACG | |
| GGC-R | CGTCCTCGGAGGAGGCGCCGCCGCCGCCGCAACCGGCGAGTGC | |
| GNG-F | GCACTCGCCGGTTGCGGCAACGGCGGCGCCTCCTCCGAGGACG | |
| GNG-R | CGTCCTCGGAGGAGGCGCCGCCGTTGCCGCAACCGGCGAGTGC | |
| GDD-F | GCACTCGCCGGTTGCGGCGACGACGGCGCCTCCTCCGAGGACG | |
| GDD-R | CGTCCTCGGAGGAGGCGCCGTCGTCGCCGCAACCGGCGAGTGC | |
| AST-F | GCACTCGCCGGTTGCGCCAGCACCGGCGCCTCCTCCGAGGACG | |
| AST-R | CGTCCTCGGAGGAGGCGCCGGTGCTGGCGCAACCGGCGAGTGC | |
| QGS-F | GCACTCGCCGGTTGCCAGGGCAGCGGCGCCTCCTCCGAGGACG | |
| QGS-R | CGTCCTCGGAGGAGGCGCCGCTGCCCTGGCAACCGGCGAGTGC | |
| SSQ-F | GCACTCGCCGGTTGCAGCAGCCAGGGCGCCTCCTCCGAGGACG | |
| SSQ-R | CGTCCTCGGAGGAGGCGCCCTGGCTGCTGCAACCGGCGAGTGC |
TABLE 2.
Plasmids used in this study
| Plasmid | Description | Reference or source |
|---|---|---|
| pUCP22 | E. coli-Pseudomonas shuttle vector; Gmr, Apr | 24 |
| pRSETb-mCherry | ChFP | 19 |
| pCHAP6655 | ChFP with mRFP1 N terminus | This study |
| pCHAP6656 | omlA promoter; CSFPOmlA-ChFP | This study |
| pCHAP6660 | omlA promoter; CDFPOmlA-ChFP | This study |
| pCHAP6670 | PA4370AA1-60-ChFP | This study |
| pCHAP6672 | Full-length PA4370CGDD-ChFP | This study |
| pCHAP6673 | Full-length PA4370CSFP-ChFP | This study |
| pCHAP6688 | Full-length PA3262CDSQ-ChFP | This study |
| pCHAP6697 | CGNG-ChFP | This study |
| pCHAP6698 | CKVE-ChFP | This study |
| pCHAP6699 | CGKS-ChFP | This study |
| pCHAP6700 | CDSQ-ChFP | This study |
| pCHAP6701 | CQGS-ChFP | This study |
| pCHAP6702 | CFNS-ChFP | This study |
| pCHAP6703 | CGDD-ChFP | This study |
| pCHAP6704 | CGGG-ChFP | This study |
| pCHAP6705 | CAST-ChFP | This study |
| pCHAP6706 | CSSQ-ChFP | This study |
Site-directed mutagenesis of DNA encoding lipoprotein sorting signals.
The mutagenesis of the pCHAP6656 construct encoding CSFPOmlA-ChFP was performed to change the serine at the +2 position of the mature lipoprotein to an aspartate residue, as previously described (10). The mutagenic reverse primer was designed with the mutation engineered at the 5′ primer end. For mutagenesis, inverse PCR with nonoverlapping, phosphorylated primers CX_ChFP_for1 and CD_CFR_mut was performed to produce a linear PCR product. The mutagenized PCR product was ligated to circularize it, which resulted in a sequence change of the mature lipoprotein from CSFP to CDFP. PCR amplifications were performed in 15 cycles of 98°C for 10 s, 53°C for 20 s, and 72°C for 3 min with Phusion high-fidelity DNA polymerase (New England Biolabs).
Alternatively, overlapping extension PCR was performed to create simultaneous amino acid substitutions at positions +2, +3, and +4 as previously described (7). This method was used to change the OmlA sequence CSFP to a series of amino acids found in predicted P. aeruginosa lipoproteins at these positions and also to modify the same positions of the full-length predicted lipoprotein PA4370. First-round PCR amplifications were performed using 20 ng of pCHAP6656 template in 25 cycles of 98°C for 10 s, 56°C for 20 s, and 72°C for 1 min with Phusion high-fidelity DNA polymerase (New England Biolabs) and 5% dimethylsulfoxide. Second-round PCR amplifications were performed using round-one PCR products as the template and the original flanking primers. All plasmids and mutagenesis constructs were confirmed by DNA sequencing.
Membrane fractionation.
P. aeruginosa membranes were isolated as previously described. Briefly, 200-ml cultures were grown for 4.5 h at 30°C (optical density at 600 nm of 0.5). Membranes were prepared from cells disrupted in a French press and separated by flotation sucrose gradient centrifugation as previously described (17). Twenty fractions (250 μl) were collected from the top of the gradients, separated by sodium dodecyl sulfate-12% polyacrylamide gel electrophoresis, and transferred to nitrocellulose membranes. mCherry fluorescent lipoproteins (lipoChFPs) were detected by immunoblotting with anti-mRFP1 antibodies. The primary antibodies were purified by adsorption to disrupted P. aeruginosa PAO1 cells to reduce nonspecific reactions.
Live-cell imaging.
Overnight cultures were subcultured 1/100 and grown for 3 h (optical density at 600 nm of 0.5). To prepare cells for microscopy, 0.5 ml of culture was pelleted and resuspended in 10 μl of Luria-Bertani medium or 10 μl of plasmolysis solution (0.5 M NaCl). One microliter of control cells or plasmolyzed cells was immobilized on a thin layer of 1% agarose in water or 1% agarose in 0.5 M NaCl (to maintain plasmolysis). Live cells were visualized by epifluorescence microscopy within 15 min of slide preparation with a Zeiss Axioplan 2 microscope or a Leica DMIRE2 inverted microscope, both equipped with a Hamamatsu charge-coupled device camera. Images were collected with OpenLab software. Red fluorescence was detected with rhodamine or Cy5 filters with exposure times of 1 to 5 s. Confocal images of live bacteria were acquired using a Leica TCS SP5 confocal point-scanning laser microscope. The images then were deconvoluted using the Huygens2 (Bitplane) deconvolution, from which a theoretical point-squared function was obtained. Images were analyzed with Photoshop to obtain maximum-intensity projections.
Spheroplast preparation.
Five-milliliter cultures grown as described above were converted to spheroplasts as previously described (10). Cells from 1 ml of the culture were pelleted, resuspended in 75 μl of cold TSE buffer (0.1 M Tris acetate, 16% sucrose, 5 mM EDTA, pH 8.2) to which lysozyme (150 μg/ml) and 7.5 μl of cold water were added, and incubated on ice for 5 min. Spheroplasts were stabilized with MgSO4 at a final concentration of 15 mM and were pelleted at low speed (3,000 rpm). The spheroplast pellet was resuspended in TSM buffer (0.05 M Tris acetate, 8% sucrose, 10 mM MgSO4, pH 8.2) and viewed on agarose beds as described above.
RESULTS
LipoChFPs are membrane localized in P. aeruginosa.
LipoRFPs were used previously for the direct observation of lipoproteins in either the outer or inner membrane in the Enterobacteriaceae (10). We modified this approach to visualize lipoproteins directly in P. aeruginosa and to determine the lipoprotein sorting signals that operate in this opportunistic pathogen. We constructed lipoChFPs by fusing the signal peptide from the known P. aeruginosa outer membrane lipoprotein OmlA (15) to the mCherry variant of mRFP1. The chimeric lipoprotein CSFPOmlA-ChFP was produced from the native omlA promoter and was shown to correctly localize to the cell envelope (Fig. 1A). The plasmolysis of P. aeruginosa was performed by treating cells with 0.5 M NaCl, which results in the formation of plasmolysis bays due to the separation of the outer and inner membranes (Fig. 1). The plasmolysis of E. coli cells typically is performed with 15% sucrose treatment, but this was insufficient to plasmolyze P. aeruginosa. After high-salt plasmolysis, the fluorescence of CSFPOmlA-ChFP was observed only in the outer membrane (Fig. 1A). The outer membrane localization of lipoChFP was confirmed by the isolation of outer and inner membrane fractions by sucrose density centrifugation, separation by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and immunoblotting with antibodies against mRFP1. CSFPOmlA-ChFP was found only in outer membrane fractions that were enriched with the known P. aeruginosa outer membrane proteins OprD, OprF, OprG, OprH, and OprI (data not shown) (6). We concluded that the mCherry protein was lipid modified in this chimeric lipoChFP and correctly localized to the outer membrane.
FIG. 1.

LipoChFPs localize to either the outer or inner membrane of P. aeruginosa cells. (A) CSFPOmlA-ChFP localizes to the cell envelope in untreated cells (top) and to the outer membrane in cells plasmolyzed by 0.5 M NaCl treatment (bottom). Plasmolysis causes the inner membrane (IM) to invaginate and form periplasmic bays, as indicated by white arrows. Representative localization patterns are shown in plasmolyzed cells producing the outer membrane (OM)-localized CAST-ChFP (B) or the IM-localized CKVE-ChFP (C). Phase-contrast images are shown on the left, and fluorescent images are shown on the right.
Inner membrane retention signals in P. aeruginosa lipoproteins.
All E. coli inner membrane lipoproteins follow the canonical Asp+2 rule. Using LipoP analysis, we previously identified 185 predicted lipoproteins encoded in the P. aeruginosa PAO1 genome, but only 5 of them have Asp at the +2 position (11). We replaced the +2, +3, and +4 amino acid positions of the model lipoprotein CSFPOmlA-ChFP with sequences found in several predicted P. aeruginosa lipoproteins and tested for retention in the inner membrane. The two distinct localization patterns of outer membrane- or inner membrane-localized lipoChFPs are shown in Fig. 1B and C, respectively. For inner membrane lipoproteins, the fluorescence is strongest in the plasmolysis bays, along the contours of the inner membrane. Most of the inner membrane retention signals identified here do not contain Lys+3 Ser+4 (Table 3). CDSQ-ChFP localized to the inner membrane and CSSQ-ChFP localized to the outer membrane in plasmolyzed cells, which confirms that the Asp+2 rule also operates in P. aeruginosa (Table 3). However, CDFP-ChFP, which differs only in the +2 position of our model protein CSFPOmlA-ChFP, did not localize to the inner membrane, indicating that amino acids at positions +3 and +4 can influence the function of the canonical inner membrane retention signal Asp+2. CFNS-ChFP and CGNG-ChFP also both localized to the inner membrane, which is consistent with the non-aspartate inner membrane signals Phe+2 Asn+3 and Gly+2 Asn+3 identified in E. coli (18) and that operate in many Enterobacteriaceae (10) (Table 3). CGKS-ChFP localized to the inner membrane, consistent with the localization of MexA or lipoMalE containing the Lys+3 Ser+4 inner membrane retention signal (12). The novel inner membrane retention signals identified include CK+2 V+3 E+4, CG+2 G+3 G+4, CG+2 D+3 D+4, and CQ+2 G+3 S+4 (Table 3). In total, 9 of the 185 predicted P. aeruginosa lipoproteins contain the exact sequences demonstrated here to cause inner membrane localization (Table 3). Thus, 5% of P. aeruginosa lipoproteins have an experimentally verified inner membrane retention signal.
TABLE 3.
Summary of lipoChFP inner membrane retention signals
| Sorting signala | Lipoproteinb | Protein function | P. aeruginosa IM retention rulec | Localization in P. aeruginosa | Localization in E. coli |
|---|---|---|---|---|---|
| CSFP | PA4756, OmlA | Maintain cell envelope integrity | None | OM | OM |
| CDFP | OmlA +2 variant | None | OM | IM | |
| CAST | PA4876, OsmE | Osmotic shock response | None | OM | OM |
| CAST | PA5310 | Lipid metabolism | None | OM | OM |
| CDSQ | PA3262 | Peptidyl-prolyl cis-trans isomerase | Asp+2 | IM | IM |
| CGKS | PA0425, MexA | RND efflux membrane fusion protein | Lys+3 Ser+4 | IM | OM |
| CGKS | PA2321 | Glucokinase | Lys+3 Ser+4 | IM | OM |
| CFNS | Not present | Non-aspartate+2 | IM | IM | |
| CGNG | PA3677 | RND efflux membrane fusion protein | Non-aspartate+2 | IM | IM |
| CGGG | PA5041, PilP | Type IV pilus assembly | Novel | IM | OM |
| CGGG | PA5414 | Hypothetical protein | Novel | IM | OM |
| CKVE | PA1723, PscJ | Type III secretion machinery | Novel | IM | OM |
| CQGS | PA1812, MltD | Lytic murein transglycosylase D | Novel | IM | OM |
| CGDD | PA4370, IcmP | Metalloprotease | Novel | IM | IM |
The first four amino acids of the mature N terminus from predicted P. aeruginosa lipoproteins in the C +2 +3 +4 region.
The PA identification number and protein name, if available, which contains the lipoprotein sorting signal listed to the left.
IM, inner membrane; OM, outer membrane.
Plasmids encoding this panel of lipoChFPs were introduced into E. coli. All P. aeruginosa inner membrane-localized lipoChFPs also were inner membrane localized, except for four (CGGG, CGKS, CKVE, and CQGS) (Table 3). These four sequences are Pseudomonas-specific inner membrane retention signals.
Predicted full-length lipoproteins PA4370 and PA3262 are located in the inner membrane.
Two predicted P. aeruginosa lipoproteins were selected to construct full-length translational fusions to mCherry. PA4370 encodes an insulin-cleaving metalloproteinase (IcmP) whose activity was detected in outer membrane protein fractions; however, these authors did not report the presence of a predicted lipoprotein signal peptide (3). PA3262 encodes a probable peptidyl-prolyl cis-trans isomerase with homology to MIP (macrophage infectivity potentiator), which is a surface-exposed, outer membrane lipoprotein in Neisseria and Chlamydia (9, 14). Regions encoding the native promoters and entire open reading frames (lacking stop codons) of predicted lipoproteins PA4370 and PA3262 were cloned upstream of mCherry. Both PA4370CGDD-ChFP and PA3262CDSQ-ChFP were localized in the cell envelope but were observed unexpectedly in the inner membrane after high-salt plasmolysis (Fig. 2A, B). Since these proteins were candidates for surface-exposed lipoproteins, their inner membrane localization was unexpected. The mature N-terminal sequences of PA3262 and PA4370 were CDSQ and CGDD, respectively. Both of these four-amino-acid sequences already were shown to target lipoChFPs to the inner membrane (Table 3). This confirms that the information required for lipoprotein sorting is contained in the first four amino acids of the N terminus.
FIG. 2.

Full-length chimeras of mCherry to the proteins PA4370 and PA3262 localize to the inner membrane (IM) of P. aeruginosa. Each panel consists of untreated cells (top), plasmolyzed cells (bottom), phase-contrast images (left), and fluorescent images (right). All proteins shown localize to the cell envelope in untreated cells. In cells plasmolyzed by 0.5 M NaCl treatment, (A) PA4370CGDD-ChFP and (B) PA3262CDSQ-ChFP localize to the inner membrane, and (C) the site-directed substitution variant PA4370CSFP-ChFP is rerouted to the outer membrane (OM).
To demonstrate that the full-length mCherry fusions were not somehow impeded from reaching the outer membrane, the C+2+3+4 region of the full-length PA4370CGDD-ChFP chimera was changed to include the outer membrane-targeting signal CSFP. PA4370CSFP-ChFP was located in the outer membrane, as expected (Fig. 2C). The ability to change the sorting signal of PA4370 and reroute the full-length protein between membranes strongly supports the hypothesis that PA4370 is a genuine, lipid-modified inner membrane protein.
Spheroplast analysis of inner membrane- and outer membrane-localized lipoChFPs.
When gram-negative bacteria are converted to spheroplasts, the rod shape is converted to round due to the degradation of peptidoglycan and loss of structural integrity. Spheroplasts were prepared from P. aeruginosa that produced lipoChFPs localized in the inner or outer membrane. CSFPOmlA-ChFP localizes to the outer membrane (Fig. 1A), but fluorescence was not totally lost after spheroplast treatment (Fig. 3A). This partial staining of spheroplasts indicates that a significant portion of the outer membrane remains attached to P. aeruginosa spheroplasts. This contrasts with spheroplast formation in E. coli, in which most of the outer membrane is lost and very little lipoRFP fluorescence remains (10). PA4370CGDD-ChFP localizes to the inner membrane (Fig. 2A) and was evenly distributed over the entire perimeter of spheroplasts (Fig. 3B). The results of spheroplast analysis are consistent with the localization of lipoChFPs in plasmolyzed cells.
FIG. 3.

Spheroplasts of P. aeruginosa cells producing (A) outer membrane (OM)-localized CSFPOmlA-ChFP and (B) inner membrane (IM)-localized PA4370CGDD-ChFP. Phase-contrast images are shown on the left, and fluorescent images are shown on the right.
Optical sectioning of cells producing full-length lipoChFPs.
Images of P. aeruginosa producing either full-length PA4370CGDD-ChFP or PA3262CDSQ-ChFP using epifluorescence microscopy indicated a nonuniform fluorescence-staining pattern in the membrane. The fluorescence of PA4370CGDD-ChFP appeared stronger at the cell poles (Fig. 3A), while PA3262CDSQ-ChFP displayed a very patchy localization pattern throughout the membrane (Fig. 3B). In order to improve the resolution of these localization patterns, optical sections throughout the cell width were obtained with confocal point-scanning laser microscopy. In optical sections approaching the cell surface, a distinct localization pattern was seen that resembles a helical or coiled localization pattern for both chimeric proteins (Fig. 4). This localization structure also has been observed in the membrane-associated cytoskeletal protein MreB (8, 20), cell division proteins MinDE (20), and the surface-exposed outer membrane proteins and lipopolysaccharide (5).
FIG. 4.
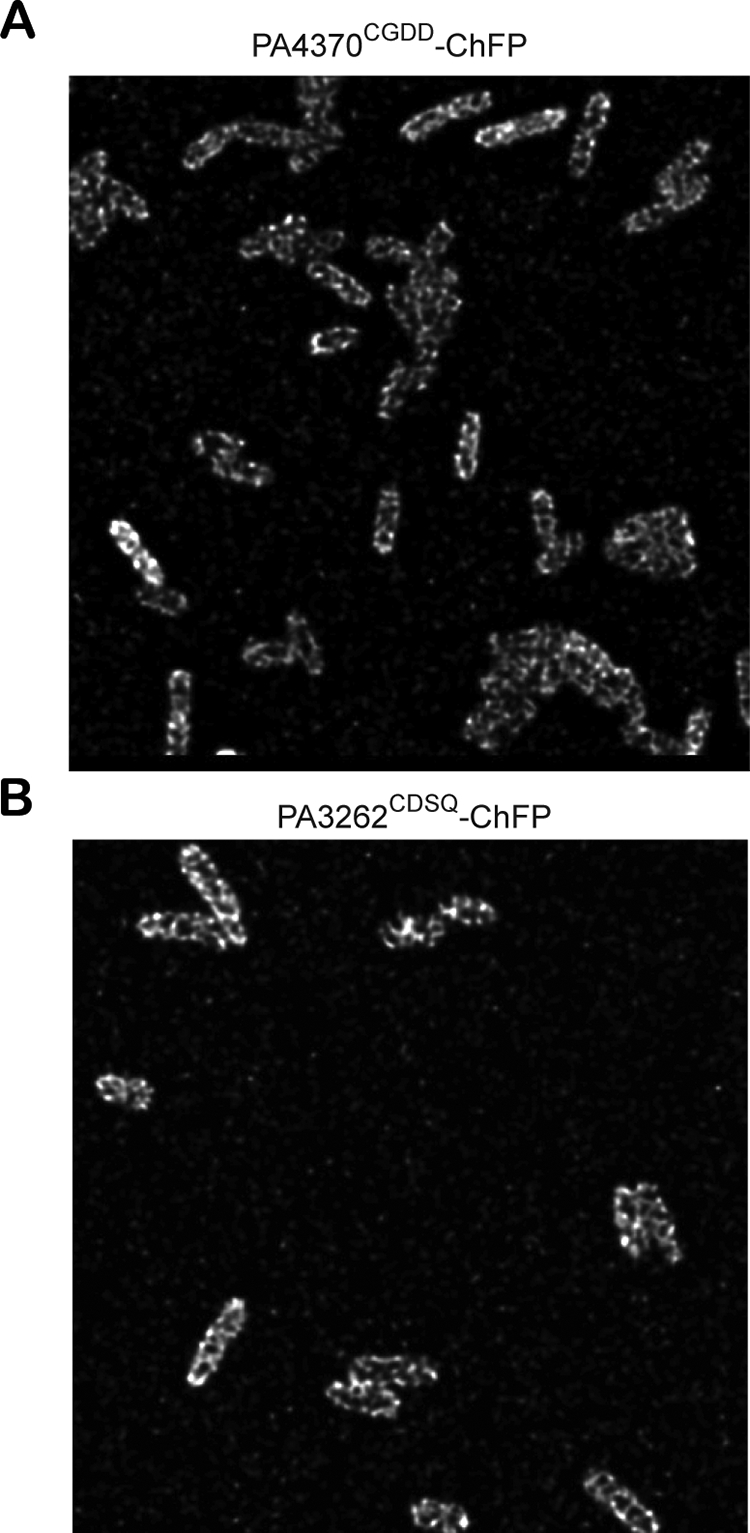
Helix or coiled localization pattern of lipoChFPs. Shown are confocal microscopy images taken from optical slices corresponding to the cell surface in P. aeruginosa producing the full-length chimeric fusions PA4370CGDD-ChFP (A) and PA3262CDSQ-ChFP (B).
DISCUSSION
We developed a novel lipoChFP for the direct observation of lipoproteins in either the outer or inner membrane of P. aeruginosa. LipoChFPs were encoded from a broad-host-range plasmid that should permit similar analyses of additional gram-negative species. We modified our previous lipoRFP strategy by replacing the monomeric red fluorescent protein with the mCherry variant, which is brighter and more stable (19), as well as using Pseudomonas-specific plasmids, promoters, and lipoprotein signal peptides. Substitutions in the first few amino acids of mature lipoproteins permit the rerouting of lipoChFPs between the inner and outer membranes, confirming that the sorting signals are within the first four amino acids. This strategy therefore might be used to demonstrate that predicted lipoproteins are in fact lipid modified and sorted between membranes via the Lol lipoprotein sorting pathway.
We have confirmed the recent findings of Narita and Tokuda (12) that the canonical Asp+2 does function as an inner membrane retention signal in P. aeruginosa; however, we also showed that Asp+2 must have the appropriate amino acids in positions +3 and +4 to function as an inner membrane retention signal in P. aeruginosa. Previous studies have shown that the ability of Asp+2 to function as an inner membrane retention signal in E. coli is modulated by adjacent amino acids (4) as well as by protein folding (17). Here, we also have confirmed that Lys+3 Ser+4 are essential for the inner membrane retention of Pseudomonas lipoproteins (12), and we identified novel amino acid sequences that act as inner membrane retention signals. In lipoproteins with CGDD N-terminal sequences, the two aspartates in positions +3 and +4 might function in a manner analogous to that of Asp+2, given that Asp+2 Asp+3 is one of the strongest inner membrane retention signals in E. coli (22). However, given the lack of conserved properties of the amino acids in the inner membrane retention signals CGGG, CQGS, and CKVE, it is not clear how these novel signals function. Of the nine inner membrane lipoproteins identified in this study, only two were predicted by PSORTb to be localized in the inner membrane, while two were predicted to localize in the outer membrane and five had an unknown localization (www.pseudomonas.com).
P. aeruginosa encodes homologs to each of the lolABCDE genes, and the LolCDE complex from P. aeruginosa recently was shown to release lipoproteins to the LolA chaperone (21). Reconstituted proteoliposomes with the LolCDE from P. aeruginosa did not release to LolA lipoproteins that contained the Lys+3 Ser+4 inner membrane retention signal. This suggests that the Lol machinery functions similarly to that of E. coli, except for the different requirements of lipoprotein recognition by LolCDE. The inner membrane retention signals identified previously (12) and here can be found in 11 of the 185 LipoP-predicted lipoproteins. At least 6% of the predicted PAO1 lipoproteins contain inner membrane retention signals that likely serve to evade recognition by LolCDE. This was not a systematic screen for inner membrane retention signals, and there probably are others. The most common amino acids in the +2 position of P. aeruginosa (103/185) and E. coli lipoproteins are alanine and serine (data not shown). Since these amino acids are found mostly in outer membrane lipoproteins, this indicates that the majority of lipoproteins in both organisms likely are localized in the outer membrane.
Surprisingly, inner membrane retention signals were identified in two lipoproteins that initially were predicted to be surface exposed. PA3262 is a homolog of a surface-localized lipoprotein in other species, but it probably resides in the inner membrane of P. aeruginosa. Our localization studies of PA4370 are in direct contrast to the initial biochemical characterization of this protease, which indicated that protease activity was detected in outer membrane fractions of sucrose gradients (3). We confirmed that the full-length protein localizes to the inner membrane, the first four amino acids, CGDD, target mCherry to the inner membrane, and the full-length protein is rerouted to the outer membrane after the introduction of a known outer membrane-targeting sequence. The latter observation indicates that the original full-length mCherry chimera was not obstructed in targeting the outer membrane and that the true localization is in the inner membrane. These contrasting results highlight the potential for incorrect localization assignments of membrane proteins that are biochemically purified in complex, multistep purifications.
The primary advantage to the lipoChFP approach to determine lipoprotein localization is to observe directly the in situ membrane localization after plasmolysis. Confocal microscopy improves the resolution of the membrane localization pattern. Despite the smaller size of P. aeruginosa compared to that of E. coli, we still were able to resolve a nonuniform, membrane localization of two separate inner membrane lipoproteins. Viewing optical sections near the cell surface, away from the midline, indicated a coiled or helix-like pattern. There are several examples of a helix-like localization in bacteria, including the membrane-associated helices of MreB and MinD that form on the cytoplasmic face of the inner membrane (8, 20) and the helical localization of surface-exposed outer membrane proteins and lipopolysaccharide (5). We report here the first example of an inner membrane-anchored lipoprotein that faces the periplasm with a similar localization pattern. The ability to label either the inner or outer membrane with fluorescent proteins should have many applications in future bacterial membrane studies.
Acknowledgments
S.L. was supported by a fellowship from the Canadian Louis Pasteur Foundation and is currently the Westaim-ASRA Chair in Bacterial Biofilm Research. This research was supported by Westaim and the Alberta Science and Research Authority (ASRA). M.M. was supported by a postdoctoral fellowship from the United States National Science Foundation.
Footnotes
Published ahead of print on 18 July 2008.
REFERENCES
- 1.Chen, J. C., P. H. Viollier, and L. Shapiro. 2005. A membrane metalloprotease participates in the sequential degradation of a Caulobacter polarity determinant. Mol. Microbiol. 551085-1103. [DOI] [PubMed] [Google Scholar]
- 2.Chuanchuen, R., C. T. Narasaki, and H. P. Schweizer. 2002. Benchtop and microcentrifuge preparation of Pseudomonas aeruginosa competent cells. BioTechniques 33760:762-763. [DOI] [PubMed] [Google Scholar]
- 3.Fricke, B., O. Parchmann, K. Kruse, P. Rucknagel, A. Schierhorn, and S. Menge. 1999. Characterization and purification of an outer membrane metalloproteinase from Pseudomonas aeruginosa with fibrinogenolytic activity. Biochim. Biophys. Acta 1454236-250. [DOI] [PubMed] [Google Scholar]
- 4.Gennity, J. M., H. Kim, and M. Inouye. 1992. Structural determinants in addition to the amino-terminal sorting sequence influence membrane localization of Escherichia coli lipoproteins. J. Bacteriol. 1742095-2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ghosh, A. S., and K. D. Young. 2005. Helical disposition of proteins and lipopolysaccharide in the outer membrane of Escherichia coli. J. Bacteriol. 1871913-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hancock, R. E., and A. M. Carey. 1979. Outer membrane of Pseudomonas aeruginosa: heat 2-mercaptoethanol-modifiable proteins. J. Bacteriol. 140902-910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horton, R. M., H. D. Hunt, S. N. Ho, J. K. Pullen, and L. R. Pease. 1989. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 7761-68. [DOI] [PubMed] [Google Scholar]
- 8.Jones, L. J., R. Carballido-Lopez, and J. Errington. 2001. Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis. Cell 104913-922. [DOI] [PubMed] [Google Scholar]
- 9.Leuzzi, R., L. Serino, M. Scarselli, S. Savino, M. R. Fontana, E. Monaci, A. Taddei, G. Fischer, R. Rappuoli, and M. Pizza. 2005. Ng-MIP, a surface-exposed lipoprotein of Neisseria gonorrhoeae, has a peptidyl-prolyl cis/trans isomerase (PPIase) activity and is involved in persistence in macrophages. Mol. Microbiol. 58669-681. [DOI] [PubMed] [Google Scholar]
- 10.Lewenza, S., D. Vidal-Ingigliardi, and A. P. Pugsley. 2006. Direct visualization of red fluorescent lipoproteins indicates conservation of the membrane sorting rules in the family Enterobacteriaceae. J. Bacteriol. 1883516-3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lewenza, S., J. L. Gardy, F. S. Brinkman, and R. E. Hancock. 2005. Genome-wide identification of Pseudomonas aeruginosa exported proteins using a consensus computational strategy combined with a laboratory-based PhoA fusion screen. Genome Res. 15321-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Narita, S., and H. Tokuda. 2007. Amino acids at positions 3 and 4 determine the membrane specificity of Pseudomonas aeruginosa lipoproteins. J. Biol. Chem. 28213372-13378. [DOI] [PubMed] [Google Scholar]
- 13.Narita, S., S. Matsuyama, and H. Tokuda. 2004. Lipoprotein trafficking in Escherichia coli. Arch. Microbiol. 1821-6. [DOI] [PubMed] [Google Scholar]
- 14.Neff, L., S. Daher, P. Muzzin, U. Spenato, F. Gulacar, C. Gabay, and S. Bas. 2007. Molecular characterization and subcellular localization of macrophage infectivity potentiator, a Chlamydia trachomatis lipoprotein. J. Bacteriol. 1894739-4748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ochsner, U. A., A. I. Vasil, Z. Johnson, and M. L. Vasil. 1999. Pseudomonas aeruginosa fur overlaps with a gene encoding a novel outer membrane lipoprotein, OmlA. J. Bacteriol. 1811099-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pugsley, A. P., M. G. Kornacker, and A. Ryter. 1990. Analysis of the subcellular location of pullulanase produced by Escherichia coli carrying the pulA gene from Klebsiella pneumoniae strain UNF5023. Mol. Microbiol. 459-72. [DOI] [PubMed] [Google Scholar]
- 17.Robichon, C., M. Bonhivers, and A. P. Pugsley. 2003. An intramolecular disulphide bond reduces the efficacy of a lipoprotein plasma membrane sorting signal. Mol. Microbiol. 491145-1154. [DOI] [PubMed] [Google Scholar]
- 18.Seydel, A., P. Gounon, and A. P. Pugsley. 1999. Testing the “+2 rule” for lipoprotein sorting in the Escherichia coli cell envelope with a new genetic selection. Mol. Microbiol. 34810-821. [DOI] [PubMed] [Google Scholar]
- 19.Shaner, N. C., R. E. Campbell, P. A. Steinbach, B. N. Giepmans, A. E. Palmer, and R. Y. Tsien. 2004. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 221567-1572. [DOI] [PubMed] [Google Scholar]
- 20.Shih, Y. L., T. Le, and L. Rothfield. 2003. Division site selection in Escherichia coli involves dynamic redistribution of Min proteins within coiled structures that extend between the two cell poles. Proc. Natl. Acad. Sci. USA 1007865-7870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tanaka, S. Y., S. Narita, and H. Tokuda. 2007. Characterization of the Pseudomonas aeruginosa Lol system as a lipoprotein sorting mechanism. J. Biol. Chem. 28213379-13384. [DOI] [PubMed] [Google Scholar]
- 22.Terada, M., T. Kuroda, S. I. Matsuyama, and H. Tokuda. 2001. Lipoprotein sorting signals evaluated as the LolA-dependent release of lipoproteins from the cytoplasmic membrane of Escherichia coli. J. Biol. Chem. 27647690-47694. [DOI] [PubMed] [Google Scholar]
- 23.Tokuda, H., and S. Matsuyama. 2004. Sorting of lipoproteins to the outer membrane in E. coli. Biochim. Biophys. Acta 1694IN1-IN9. [PubMed] [Google Scholar]
- 24.West, S. E., H. P. Schweizer, C. Dall, A. K. Sample, and L. J. Runyen-Janecky. 1994. Construction of improved Escherichia-Pseudomonas shuttle vectors derived from pUC18/19 and sequence of the region required for their replication in Pseudomonas aeruginosa. Gene 14881-86. [DOI] [PubMed] [Google Scholar]
- 25.Yamaguchi, K., F. Yu, and M. Inouye. 1988. A single amino acid determinant of the membrane localization of lipoproteins in E. coli. Cell 53423-432. [DOI] [PubMed] [Google Scholar]


