Abstract
From the photosynthetic green sulfur bacterium Chlorobium tepidum (pro synon. Chlorobaculum tepidum), we have purified three factors indispensable for the thiosulfate-dependent reduction of the small, monoheme cytochrome c554. These are homologues of sulfur-oxidizing (Sox) system factors found in various thiosulfate-oxidizing bacteria. The first factor is SoxYZ that serves as the acceptor for the reaction intermediates. The second factor is monomeric SoxB that is proposed to catalyze the hydrolytic cleavage of sulfate from the SoxYZ-bound oxidized product of thiosulfate. The third factor is the trimeric cytochrome c551, composed of the monoheme cytochrome SoxA, the monoheme cytochrome SoxX, and the product of the hypothetical open reading frame CT1020. The last three components were expressed separately in Escherichia coli cells and purified to homogeneity. In the presence of the other two Sox factors, the recombinant SoxA and SoxX showed a low but discernible thiosulfate-dependent cytochrome c554 reduction activity. The further addition of the recombinant CT1020 protein greatly increased the activity, and the total activity was as high as that of the native SoxAX-CT1020 protein complex. The recombinant CT1020 protein participated in the formation of a tight complex with SoxA and SoxX and will be referred to as SAXB (SoxAX binding protein). Homologues of the SAXB gene are found in many strains, comprising roughly about one-third of the thiosulfate-oxidizing bacteria whose sox gene cluster sequences have been deposited so far and ranging over the Chlorobiaciae, Chromatiaceae, Hydrogenophilaceae, Oceanospirillaceae, etc. Each of the deduced SoxA and SoxX proteins of these bacteria constitute groups that are distinct from those found in bacteria that apparently lack SAXB gene homologues.
Thiosulfate is one of the most-abundant forms of reduced sulfur in nature, and the ability to oxidize this compound is distributed over many microorganisms across different phyla (6, 7, 14, 19, 21, 43). It sometimes occurs that some members within a phylum can utilize thiosulfate while others cannot. This is the case with green sulfur bacteria; Chlorobium tepidum (pro synon. Chlorobaculum tepidum [29]) and Chlorobium limicola f. sp. thiosulfatophilum utilize thiosulfate, but Chlorobium limicola DSM 245 and Prosthecochloris estuarii do not (23, 24, 29, 30, 46).
Almost all of the green sulfur bacteria, with one known exception of Chlorobium ferrooxidans, carry out anoxygenic photosynthesis with reduced sulfur compounds, such as sulfide and elemental sulfur, and some of them also use thiosulfate as the electron donor for the assimilation of various elements for growth (6, 7, 14, 23, 24, 43). Their reaction center (RC) is like photosystem I of oxygenic photosynthetic organisms, called type I or iron-sulfur type RC with ferredoxin and flavodoxin as immediate electron acceptors (27, 52, 56). The primary donor of the RC is a special pair of bacteriochlorophylls called P840, and its immediate electron donor is the RC-bound cytochrome (cyt) c551. In the moderately thermophilic C. tepidum, there seem to be multiple pathways for the reduction of the RC-bound cyt c551 (the CT1639 protein, encoded by CT1639 of C. tepidum TLS). Itoh et al. (31) showed that a soluble monoheme cyt, c554, of about 10 kDa (the CT0075 protein) donates electrons to the bound cyt c551 rather than directly to oxidized P840. A similar small, monoheme cyt, c555, is an electron acceptor in thiosulfate oxidation in C. limicola f. sp. thiosulfatophilum (39). C. tepidum seems to have an alternative electron transfer pathway, because mutant cells with the soluble cyt c554 gene (CT0075) disrupted can grow phototrophically in a medium containing sulfide and thiosulfate, although at a lower rate than the wild type (59). The alternative pathway is proposed to be sulfide → membrane-bound sulfide-quinone reductase (57) → membrane-bound quinol oxidoreductase → RC-bound cyt c551 → P840.
Two different biochemical pathways for thiosulfate oxidation are distinguishable among bacteria (22, 26, 36). In one type of pathway found in, e.g., Starkeya novella (34) and Allochromatium vinosum (28), thiosulfate is oxidized to sulfate through the cooperation of thiosulfate dehydrogenase, tetrathionate hydrolase, and trithionate hydrolase with the formation of tetrathionate as the intermediate and sulfate as the final product. In the other type of pathway, found in Paracoccus pantotrophus and some green sulfur bacteria, thiosulfate is oxidized by a sulfur oxidizing system (Sox), which is also called thiosulfate-oxidizing multienzyme system (TOMES) (36). The Sox (or TOMES) pathway consists of several proteins and does not result in the formation of tetrathionate (22). Some bacteria, e.g., S. novella (34) and A. vinosum, have both pathways (14, 26, 28).
The biochemical pathway of thiosulfate oxidation by the Sox system has been intensively studied in facultative lithotrophic bacteria, such as Paracoccus pantotrophus (22) and Paracoccus versutus (formerly Thiobacillus versutus) (36). In these bacteria, Sox proteins are localized in the periplasm, and SoxYZ, SoxAX, and SoxB are essential components of thiosulfate oxidation, with a small, soluble, monoheme c-type cyt of about 10 kDa as the electron acceptor. The SoxYZ complex contains no prosthetic group and serves as the acceptor for the reaction intermediates (47). SoxX is a monoheme cyt c, and SoxA is either a mono- or diheme cyt c depending on the bacterial species (1, 19, 32). SoxA and SoxX occur as a heterodimeric complex in P. pantotrophus (20), Rhodovulum sulfidophilum (1), and S. novella (32). The initial oxidative reaction of the pathway is proposed to be the oxidative formation of a disulfide linkage between the sulfane sulfur of thiosulfate and the cysteinyl-SH of SoxY (SoxY-SH) to yield -(S)-SSO3−, accompanied by the reduction of the small, monoheme cyt c catalyzed by SoxAX as follows (22): (SoxY-SH) + −SSO3− + 2 cyt cox → (SoxY-S)-SSO3− + 2 cyt cred + H+. SoxB contains two manganese atoms (8, 17) and is thought to catalyze the hydrolysis of (SoxY-S)-SSO3− to yield sulfate and (SoxY-S)-SH.
The fate of the -(S)-SH on SoxY seems to be different with microorganisms. In bacteria such as P. pantotrophus that have SoxCD, it is further oxidized to (-S)-SO3− on SoxY, which is again hydrolyzed by SoxB to regenerate (SoxY-SH), accompanied by the reduction of the small, monoheme cyt c (20, 22, 51). Complete oxidation of a thiosulfate generates eight electrons and two sulfate molecules.
In bacteria such as Thiothrix strains and green sulfur bacteria that have no SoxCD, (SoxY-S)-SH presumably reacts with new −SSO3− to yield SoxY that binds one more sulfur atom (sulfane sulfur, or S0), accompanied by the donation of two electrons to external cyt c, and this reaction cycle is repeated several times, yielding polysulfide groups on SoxY (54).
From the direct sequencing of the sox genes, as well as from the results of current whole-genome-sequencing projects, genes predicted to be involved in inorganic sulfur metabolism have been identified in a variety of microorganisms, including many of the genes encoding Sox proteins (soxA, soxB, soxF, soxX, soxY, and soxW) that occur in a cluster as in P. pantotrophus (19, 21, 22). In the green sulfur bacterium C. limicola f. thiosulfatophilum, Verté et al. (60) reported that sox genes occur in a similar cluster. The entire genomic sequence of C. tepidum was elucidated for the first time, and its comparison to sequences of other green sulfur bacteria revealed a fairly large number of homologous genes involved in sulfur metabolism, including sox genes as found in other bacteria (16, 25). With the increasing availability of genomic sequences from other green sulfur bacterial strains, genes possibly involved in sulfur metabolism may be compiled and compared (23, 24). The sox gene clusters were found in green sulfur bacteria that utilize thiosulfate, but generally not in those that do not utilize it, although some exceptions to the latter may exist (23, 24). Moreover, soxC and soxD have not been found in green sulfur bacteria. When fed with thiosulfate, thiosulfate-utilizing green sulfur bacteria form elemental sulfur globules as intermediates in the periplasmic space which are subsequently oxidized to sulfate (7, 9, 10, 11). The pathways for transport of elemental sulfur (or sulfane sulfur equivalent) and its subsequent metabolism in green sulfur bacteria are unknown. In the metabolic pathway, the involvement of a dissimilatory sulfur reductase that resides in cytoplasm is proposed (16, 23, 24, 25, 45). The possible pathway for transport of elemental sulfur (sulfane sulfur equivalent) or persulfide (23, 24) across the plasma membrane has been discussed based on genomic sequence analysis (9, 10, 23, 24). Recently, Chan et al. (11) generated a mutant from C. tepidum in which the region between CT0868 and CT0876 was replaced by a transposon insertion and subsequently found that the mutant was completely defective for growth on thiosulfate as the sole electron donor, suggesting that the protein(s) encoded by one or some of the genes in this region might be involved in the transport of sulfur (or a sulfane sulfur equivalent) across the plasma membrane.
The results of biochemical studies revealed the presence of several components involved in thiosulfate oxidation in green sulfur bacteria. A small, monoheme cyt c called Chlorobium cyt c555, a homolog of conventional cyt c in mitochondria, was isolated from C. limicola f. sp. thiosulfatophilum and found to be an electron acceptor of the thiosulfate-oxidizing enzyme system (38, 39, 44). In C. tepidum, cyt c554 (a homolog of C. limicola cyt c555) seems to mediate electron transport between the thiosulfate oxidation system and RC-bound cyt c551 as described above (31). Meyer et al. (44) isolated the multiheme cyt c551 from C. limicola f. sp. thiosulfatophilum, which appears to correspond to a homologue of SoxAX found in various thiosulfate-oxidizing bacteria. Kusai and Yamanaka (39) isolated thiosulfate-cyt c (multiheme cyt c551) reductase from the same strain that binds no flavin or heme, with a molecular mass of 80 kDa as estimated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis in the presence of mercaptoethanol. Brief summaries of the results of biochemical studies of thiosulfate oxidation in green sulfur bacteria have been presented in the literature (43, 62). Although the results of genome sequence analyses indicate the presence of many sox genes in green sulfur bacteria that are homologous to those found in other groups of bacteria (16, 23, 24, 25), biochemical characterization of the products encoded by sox genes in green sulfur bacteria is largely lacking.
We have purified three components necessary for thiosulfate oxidation in C. tepidum and characterized some of their biochemical properties. We found that the SoxAX complex binds a novel colorless protein factor, encoded by the hypothetical open reading frame CT1020. The gene product of CT1020, which will be referred to as SAXB (SoxAX binding protein), stimulates the thiosulfate oxidation activity of the Sox system in this bacterium.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
C. tepidum strain TLS (obtained from M. T. Madigan) was grown for 12 to 17 h at 40°C, essentially according to the methods described in reference 61.
Purification of thiosulfate-oxidizing proteins.
The cells were harvested by centrifugation (8,000 × g) at 4°C. The collected cells were resuspended in 50 mM Tris-HCl (pH 7.8), 100 mM NaCl, 10 mM EDTA, 5 mM sodium ascorbate, 5 mM dithiothreithol, 1 mM 6-amino-n-caproic acid, 1 mM phenylmethylsulfonyl fluoride and 1 mM p-aminobenzamidine·2HCl. The cell pellets were sonicated briefly and then disrupted with a French pressure cell at 140 MPa. The cell lysate was centrifuged at 20,000 × g for 20 min, and unbroken cells were removed as precipitates. The supernatant was further centrifuged at 160,000 × g for 2 h, and the resultant supernatant was fractionated by ammonium sulfate, yielding thiosulfate-oxidizing activity as the precipitate between 40% and 80% ammonium sulfate saturation. Precipitated proteins were collected by centrifugation, and the pellet was resuspended in 20 mM Tris-HCl (pH 7.8) and dialyzed against the same buffer with several changes. The dialyzed preparation was applied to a DEAE-Toyopearl 650 M column (2.5 by 25 cm; Tosoh) equilibrated with 20 mM Tris-HCl (pH 7.8), and the flowthrough fractions that contained thiosulfate-oxidizing activity with cyt c554 as the electron acceptor were collected. The buffer of the flowthrough fractions was changed to 10 mM 2-morpholinoethanesulfonic acid (MES)-NaOH buffer (pH 6.0) by ultrafiltration (YM-3; Millipore) and applied to a Hitrap SP column (two 5-ml columns in tandem; GE Healthcare). Protein was eluted with 100 ml of a linear gradient of 0 to 250 mM NaCl. The fractions (5 ml each) that had a thiosulfate-dependent cyt c554 reduction activity were combined (total volume, 20 ml) and desalted by ultrafiltration (YM-3; Millipore), and the concentrate was applied to a Hitrap Q column (two 5-ml columns in tandem). Protein was eluted with 100 ml of a linear gradient of 0 to 300 mM NaCl in 10 mM MES-NaOH buffer (pH 6.0), and fractions (5 ml each) were collected.
At this stage, thiosulfate-oxidizing activity in the presence of externally added cyt c554 was not detectable in any of the single fractions. Because it has often been shown that multiple factors are required for thiosulfate oxidation in other bacteria (20, 22, 41, 42, 51), we tried to reconstitute the activity and found that the combination of three fractions (tentatively referred to as fractions I, II, and III) restored the activity. We subsequently purified the active components separately. Fraction I (eluted from the Hitrap Q column at about 20 mM NaCl) was desalted by ultrafiltration (YM-3; Millipore) with the buffer changed to 10 mM MES-NaOH (pH 6.0) and applied to a MonoS column (bed volume, 1 ml; GE Healthcare) equilibrated with the same buffer. Proteins were eluted with 20 ml of a linear gradient of 0 to 100 mM NaCl, and purified SoxYZ was eluted at about 20 mM NaCl. The fractions eluted at about 60 mM NaCl contained a complex of SoxYZ and SoxB, but we did not study the latter fractions in detail. Fraction II (eluted from the Hitrap Q column at about 80 mM NaCl) was desalted by ultrafiltration with the buffer changed to 10 mM Tris-HCl (pH 8.7) and applied to a MonoQ column (bed volume, 1 ml; GE Healthcare) equilibrated with the same buffer. SoxB was eluted at about 60 mM NaCl with a 30-ml linear gradient of 0 to 300 mM NaCl yielding purified SoxB. Fraction III (eluted from the Hitrap Q column at about 100 mM NaCl) was desalted by ultrafiltration with the buffer changed to 10 mM Tris-HCl (pH 8.7) and applied to a MonoQ column (bed volume, 1 ml; GE Healthcare) equilibrated with 10 mM Tris-HCl (pH 8.7). Protein was eluted at about 80 mM NaCl with a 30-ml linear gradient of 0 to 300 mM NaCl, yielding purified SoxAX-CT1020 protein.
Purification of cyt c554.
Cyt c554 was purified to homogeneity from cell extracts as described previously (31).
Expression of rSoxA, rSoxX, and rCT1020 in Escherichia coli cells.
All molecular manipulations were carried out according to standard DNA techniques (53), and E. coli strains were grown in Luria-Bertani medium at 37°C. The final concentrations of antibiotics, when used, were 30 μg ml−1 for chloramphenicol and 100 μg ml−1 for ampicillin. Recombinant SoxA (rSoxA) and recombinant SoxX (rSoxX) were produced separately in E. coli strain BL21(DE3) harboring, in addition to the plasmid pEC86 (2), either pET23c::soxA or pET23c::soxX. Recombinant CT1020 protein (rCT1020) was produced in E. coli strain BL21(DE3) harboring the plasmid pET23c::CT1020. Expression of the recombinant genes was induced by adding 0.5 mM isopropyl-β-d-thiogalacto-pyranoside to the culture, followed by incubation for 16 to 18 h.
More precisely, these genes were amplified by PCR with KOD Dash (Toyobo) using the pair of oligonucleotides soxAfw (5′-CATATGAAAAAAACAATTCAGCGGGG-3′) and soxArv (5′-GAATTCTTATTTTCTTGATGCCGGG-3′) for soxA, soxXfw (5′-GAATTCGTGGCGCGTGGTTTT-3′) and soxXrv (5′-AAGCTTTCAGAGCGTGTAGAGATAATCGAC-3′) for soxX, and ct1020fw (5′-CATATGAAAAAAGTGTTATCGCTCT-3′) and ct1020rv (5′-AAGCTTTCAGTTTTTAGGAATCATC-3′) for CT1020. These PCR-generated fragments were ligated into pCR2.1 to produce the plasmids pCR2.1::soxA, pCR2.1::soxX, and pCR2.1::CT1020, respectively. pCR2.1::soxA was digested with NdeI and EcoRI, and the released insert was ligated into NdeI/EcoRI-digested pET23c to produce the plasmid pET23c::soxA. pCR2.1::soxX was digested with EcoRI and HindIII, and the released insert was ligated into EcoRI/HindIII-digested pET23c to produce the plasmid pET23c::soxX. pCR2.1::CT1020 was digested with NdeI and HindIII, and the released insert was ligated into NdeI/HindIII-digested pET23c to produce the plasmid pET23c::CT1020. These plasmids were checked for correct sequences by automated DNA sequencing using an ABI Prism 310 (Applied Biosystems).
Preparation of periplasmic extracts from overexpressing E. coli cells and purification of rSoxA, rSoxX, and rCT1020.
Periplasmic fractions were isolated by osmotic shock treatment of E. coli cells according to the method specified by Qiagen (13). Briefly, cells were harvested by centrifugation (10,000 × g for 10 min), and the pellets from 4 liters of cultures were washed once with 30 mM Tris-HCl (pH 7.8) and resuspended in 10 ml of 30 mM Tris-HCl (pH 7.8), 1 mM EDTA·2Na, and 20% sucrose. The cell suspension was stirred at room temperature for 10 min and centrifuged at 10,000 × g, 4°C for 10 min, and the supernatant was retained. The pellets were resuspended in 30 mM Tris-HCl (pH 7.8), and after being stirred for 10 min in an ice bath, the cells were again separated by centrifugation as described above. The combined supernatants were clarified by centrifugation at 184,000 × g for 1 h and used for purification of the respective recombinant proteins.
Briefly, purification of rSoxX was carried out as follows: a DEAE-Toyopearl 650 M column (10 by 2.5 cm; Tosoh) with a linear gradient of 0 to 300 mM NaCl in 20 mM Tris-HCl (pH 7.8), brought to 2 M ammonium sulfate, was used, and the supernatant, after the precipitate was removed, was applied to a HiTrap PHE column (bed volume, 5 ml; GE Healthcare), equilibrated with 2 M ammonium sulfate in the same buffer, and eluted by using a decreasing linear gradient of ammonium sulfate from 2 to 0 M; concentration and desalting were performed by ultrafiltration (Amicon ultra; Millipore) and HiTrapQ column chromatography with elution on a linear gradient of 0 to 200 mM NaCl in the same buffer.
Purification of rCT1020 and rSoxA was carried out as follows: the active fractions were eluted in the flowthrough fractions from a DEAE-Toyopearl 650 M column (10 by 2.5 cm; Tosoh) equilibrated with 20 mM Tris-HCl (pH 7.8), brought to 2 M ammonium sulfate, and the supernatant, after the precipitate was removed, was applied to a HiTrap PHE column (GE Healthcare), equilibrated with 2 M ammonium sulfate, and eluted by using a decreasing linear gradient of ammonium sulfate from 2 to 0 M in the same buffer; concentration and desalting were performed by ultrafiltration (Amicon Ultra; Millipore).
Enzyme assays.
Thiosulfate-dependent cyt c554 reduction activity was measured by using a spectrophotometer (UV2500PC; Shimadzu) at 25°C in a volume of 0.1 ml reaction mixture. The standard reaction mixture contained 20 mM MES-NaOH (pH 6.0), 50 μM cyt c554, 2 mM sodium thiosulfate, and 0.5 μM purified thiosulfate-oxidizing components (SoxYZ, SoxB, and SoxAX-CT1020 protein) unless otherwise indicated. Cyt c reduction rates were calculated by using the redox difference ɛ554 = 23.8 mM−1·cm−1 (55). The kinetic constants of enzymatic reactions were obtained by linear regression analyses.
Redox titrations.
Redox titrations were carried out at 25°C under nitrogen atmosphere with a potentiometer (HM26S; Toa) equipped with a redox electrode (PTS-5011C; Toa). The solutions contained 5 μM of proteins, 10 μM 2,3,5,6-tetramethyl-1,4-phenylenediamine, 10 μM duroquinone, and 10 μM 1-methoxy-5-methylphenazinium methyl sulfate, in either 100 mM potassium phosphate (pH 7.0) or 100 mM glycine-NaOH (pH 10). Reductive titrations were started at a high Eh by successive additions of 50 mM sodium dithionite and were followed by oxidative titrations (for pH 7.0 only) using successive additions of 10 mM potassium ferricyanide.
Analytical methods.
SDS-PAGE analysis was performed (40), and proteins were stained with Coomassie brilliant blue R 250. Heme was stained with 3,3′-dimethoxy benzidine dihydrochloride (DMBZ) (18).
N-terminal amino acid sequences were determined by the Edman degradation method using a Procise cLC 494 sequencer (Applied Biosystems).
The molecular masses of SoxAX-CT1020 protein were determined by matrix-assisted laser ionization (MALDI)-time-of-flight mass spectrometry using a Shimadzu AXIMA-CFR mass spectrometer with sinapinic acid as the matrix.
Protein was quantified by the method of Bradford (5) unless otherwise indicated.
The total heme content of SoxAX-CT1020 protein, rSoxA, and rSoxX were determined by using the alkaline pyridine hemochrome method (4). The protein concentrations of SoxA and SoxX were calculated from the heme concentration by assuming that each contains one heme per subunit and the SoxAX-CT1020 protein complex a total of two hemes per molecule.
The molecular masses of proteins were estimated by gel permeation chromatography using either (i) a fast protein liquid chromatography system (AKTA; GE Healthcare) equipped with a Superdex 200 10- by 300-mm GL column (GE Healthcare) with a flow rate of 1 ml·min−1 or (ii) an LC10-AD high-performance liquid chromatography (Shimadzu) TSK-Gel G3000PWXL 7.8- by 300-mm column with a flow rate of 0.5 ml·min−1 using a low-molecular-weight gel filtration kit (GE Healthcare) as the standard. The buffer was 50 mM Tris-HCl (pH 7.8) containing 150 mM NaCl.
RESULTS
Proteins essential for thiosulfate oxidation and their polypeptide composition.
We have purified the three components, namely, SoxYZ, SoxAX-CT1020 protein, and SoxB, to homogeneity (Fig. 1) as essential proteins for thiosulfate oxidation in vitro with the small, monoheme cyt c554 as the electron acceptor.
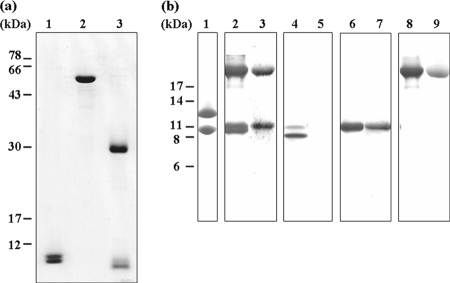
FIG. 1.
SDS-PAGE profiles of purified Sox proteins and purified recombinant proteins. (a) Lanes: 1, SoxYZ; 2, SoxB; 3, SoxAX-CT1020 protein. (b) Lanes: 1, SoxYZ; 2 and 3, SoxAX-CT1020 protein; 4 and 5, rCT1020; 6 and 7, rSoxX; 8 and 9, rSoxA. Lanes 1, 2, 4, 6, and 8 were stained with Coomassie brilliant blue, and lanes 3, 5, 7, and 9 were stained with DMBZ.
The active protein purified from fraction I was colorless, with apparent molecular masses of about 40 and 26 kDa on gel permeation chromatography (10- by 300-mm Superdex 200) analysis (data not shown). With SDS-PAGE analysis, the polypeptide composition was the same, composed of two kinds of polypeptide of about 13 kDa and 9 kDa (Fig. 1b, lane 1). The N-terminal amino acid sequence of the 13-kDa polypeptide was SWNEKAFSAS, which agrees with the one deduced from the soxY gene (CT1017) in the C. tepidum genomic database (http://www.tigr.org) that begins with Ser32, indicating that a signal peptide targeting the periplasmic space is cleaved between 31Ala and 32Ser. The N-terminal amino acid sequence of the 9-kDa polypeptide was determined to be MKIKAVVQNN, which agrees with the one deduced from the soxZ gene (CT1018) that begins with the initial methionine without cleavage of a signal peptide. When the purified preparation containing both the 26- and 40-kDa proteins was incubated with 1 mM dithiothreitol for 30 min at room temperature, the results of the subsequent chromatography revealed that the preparation contained only 26-kDa proteins. The same results were obtained when the preparation was incubated with only 1 mM thiosulfate, suggesting that the 40-kDa protein was formed during purification by the oxidation of the product-binding cysteine on SoxY between the two SoxYZ complexes. The SoxYZ activity was essentially unchanged by prior incubation of the preparation with either dithiothreitol or thiosulfate. SoxY and SoxZ form a tight complex that is difficult to separate into active components by ion-exchange chromatography, hydrophobic interaction chromatography, or gel permeation chromatography.
The active protein purified from fraction II had an apparent molecular mass of 63 kDa by gel permeation chromatography analysis (7.8- by 300-mm TSK-Gel G3000PWXL) (data not shown) and about 60 kDa by SDS-PAGE analysis (Fig. 1a, lane 2). The N-terminal amino acid sequence of TKASSDLYDF, which agrees with the one deduced from the soxB gene (CT1021) that begins with 60Thr, indicates that a signal peptide targeting the periplasmic space is cleaved between 59Ala and 60Thr. Assuming that SoxB contains two Mn per molecule, the molecular mass of SoxB is calculated to be 61,655 Da, indicating that the purified SoxB exists as a monomer.
The active protein purified from fraction III was reddish brown in color, with an apparent molecular mass of 42 kDa by gel permeation chromatography (10- by 300-mm Superdex 200) analysis (data not shown), and composed of three kinds of polypeptide of about 30 kDa, 10 kDa, and 9 kDa by SDS-PAGE analysis (Fig. 1b, lane 2). The 30-kDa and the 10-kDa bands were found to bind heme from DMBZ staining, but the 9-kDa band did not (Fig. 1b, lane 3). The N-terminal amino acid sequence of the 30-kDa band was EVNYQALVDADV, which agrees with the one deduced from the soxA gene (CT1019) that begins with 28Glu, indicating that a signal peptide targeting the periplasmic space is cleaved between 27Ala and 28Glu. The N-terminal amino acid sequence of the 10-kDa band was AAPAAVDSSV, which agrees with the one deduced from soxX gene (CT1016) that begins with 47Ala, indicating that a signal peptide targeting the periplasmic space is cleaved between 46Ala and 47Ala. The N-terminal amino acid sequence of the 9-kDa band was EPAPAAPAAS, which agrees with the one deduced from the hypothetical open reading frame CT1020 of previously unknown function that begins with 22Glu, indicating that a signal peptide targeting the periplasmic space is cleaved between 21Ala and 22Glu. The MALDI-time-of-flight MS measurements of the SoxAX-CT1020 protein complex yielded masses in agreement with the above conclusions: 29,911 Da (SoxA, calculated mass of 29,934 Da for a monoheme form; heme, 618 Da; and persulfide, 32 Da [3, 12, 15, 35]), 11,128 Da (SoxX, calculated mass of 11,130 Da for a monoheme form), and 9,376 Da (CT1020 protein, calculated mass of 9,373 Da) were the major peaks (data not shown).
Expression of rSoxA, rSoxX, and rCT1020 in E. coli cells.
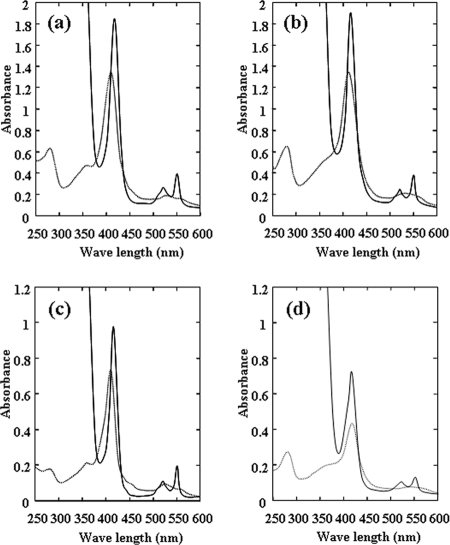
The SoxAX-CT1020 protein formed a tight complex, making it difficult to prepare each component as a separate fraction retaining activity. We expressed each component separately in E. coli cells and purified rSoxA, rSoxX, and rCT1020 (Table 1 and Fig. 1b). The dithionite-reduced form of the native SoxAX-CT1020 protein exhibited absorption peaks at 551, 522, and 417 nm (Fig. 2a). The dithionite-reduced absorption spectrum of rSoxAX-rCT1020 (Fig. 2b) was essentially identical with that of the native SoxAX-CT1020 protein. rSoxA has absorption peaks at 551, 524, and 418 nm and rSoxX at 551, 522, and 416 nm when reduced with dithionite, both typical for cyt c (Fig. 2c and d).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| E. coli strains | ||
| XL-1 Blue MR | Δ(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 recA1 endA1 gyrA96 thi-1 supE44 relA1 λ−lac | Stratagene |
| BL21(DE3) | F−ompT hsdSB(rB− mB−) gal dcm (DE3) | Novagen |
| Plasmids | ||
| pCR2.1 | Apr KmrlacZ | Invitrogen |
| pET23c | Apr T7 promoter | Novagen |
| pEC86 | CmrccmABCDEFGH in pACYC184 | 2 |
| pCR2.1::soxA | 870-bp soxA PCR fragment in pCR2.1 | This study |
| pCR2.1::soxX | 447-bp soxX PCR fragment in pCR2.1 | This study |
| pCR2.1::CT1020 | 336-bp CT1020 PCR fragment in pCR2.1 | This study |
| pET23c::soxA | 863-bp NdeI/HindIII fragment from pCR2.1::soxA in NdeI/HindIII of pET23c | This study |
| pET23c::soxX | 441-bp EcoRI/HindIII fragment from pCR2.1::soxX in EcoRI/HindIII of pET23c | This study |
| pET23c::CT1020 | 329-bp NdeI/HindIII fragment from pCR2.1::CT1020 in NdeI/HindIII of pET23c | This study |
FIG. 2.
Redox absorption spectra of the SoxAX-CT1020 protein and its components. (a) SoxAX-CT1020 protein. (b) Purified complex obtained by gel permeation chromatography from the mixture containing rSoxA, rSoxX, and rCT1020. (c) Purified rSoxX. (d) Purified rSoxA. The concentration of each protein was 5 μM based on heme determination. Dithionite-reduced spectra (solid lines) were obtained by the addition of sodium dithionite. Oxidized spectra (dotted lines) are those of the preparations as obtained.
The absorption changes of the redox titration curves of native SoxAX-CT1020 protein, rSoxA, and rSoxX are shown in Fig. 3. At pH 10.0 (Fig. 3a), the midpoint redox potential of rSoxX was +135 mV and that of rSoxA less than −550 mV. SoxAX-CT1020 protein exhibited two midpoint potentials, with the higher one at +138 mV and the lower one less than −550 mV. From these results, the former is assigned to the heme on SoxX and the latter to the one on SoxA. At pH 7.0, only reduction of one heme with a midpoint potential of +161 mV was observed, and the reduction-oxidation cycle was completely reversible (Fig. 3b).
FIG. 3.
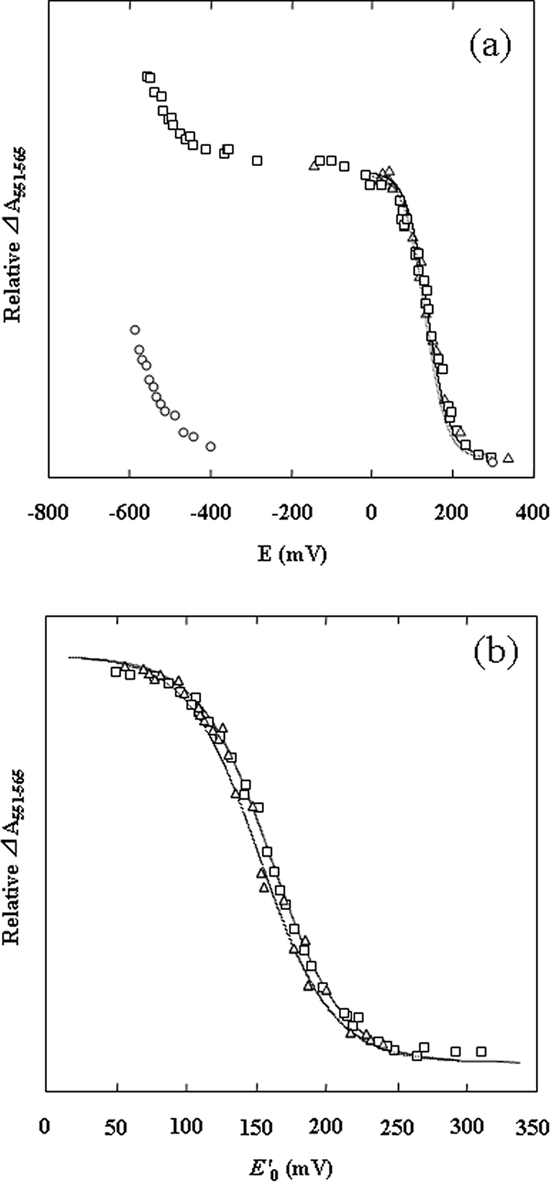
Redox titrations of SoxAX-CT1020 protein, rSoxA, and rSoxX. The absorbance was normalized based on heme determination. (a) Titrations at pH 10.0. Open squares, SoxAX-CT1020 protein; open triangles, rSoxX; open circles, rSoxA. (b) Titrations at pH 7.0. Open squares, SoxAX-CT1020 protein; open triangles, rSoxX. When Eh was stabilized, typically 2 to 3 min after the addition of dithionite or ferricyanide, the visible spectrum was recorded, and the differential absorption changes of A551 minus A565 were plotted. In panel b, the results of both reductive and oxidative titrations are plotted in the same figure. The solid line is fitted to the Nernst's n = 1 equation with Em values of +161 mV (SoxAX-CT1020 protein, pH 7.0), +153 mV (rSoxX, pH 7.0), +138 mV (SoxAX-CT1020 protein, pH 10), and +135 mV (rSoxX, pH 10), respectively.
By SDS-PAGE analysis, the apparent molecular masses of purified rSoxA (30 kDa) and rSoxX (10 kDa) were indistinguishable from those of the corresponding subunits of the native C. tepidum SoxAX-CT1020 protein complex (Fig. 1b). However, by SDS-PAGE analysis, rCT1020 showed two bands, with the major band smaller than the band found in the native SoxAX-CT1020 protein complex and the minor band indistinguishable from the band in the native complex (Fig. 1b, lane 4). The N-terminal amino acid sequences of rSoxA and rSoxX were EVNYQALVDA and AAPAAVDSSV, respectively, in complete agreement with those of SoxA and SoxX in the purified native C. tepidum SoxAX-CT1020 protein complex. The results of MALDI mass spectrometry showed a single mass peak of 11,112 Da for rSoxX and a major peak of 29,918 Da for rSoxA. The entire profiles of the MALDI mass spectrograms in the regions of these polypeptides were very similar to those of the native C. tepidum SoxAX-CT1020 protein complex (data not shown). The N-terminal amino acid sequences of the rCT1020 preparation were found to be composed of a mixture of two populations, with the major one starting at 27Ala and the minor one at 23Glu, the latter being in agreement with the native CT1020 protein in the purified C. tepidum complex. The MALDI mass spectrogram of rCT1020 showed two mass peaks of 8,923 Da (major) and 9,390 Da (minor) (data not shown), in agreement with assignments of starting points at 27Ala (8,908 Da) and 23Glu (9,373 kDa), respectively. The rCT1020 preparation used in this study is a mixture of these two populations.
Thiosulfate-dependent cyt c554 reduction kinetics and the effects of CT1020 protein.
The combination of SoxAX-CT1020 protein, SoxYZ, and SoxB was absolutely necessary for the thiosulfate-dependent reduction of the small, monoheme cyt c554. The omission of any one of the components results in no activity (Table 2), as found in other sulfur-oxidizing bacteria (20, 38, 41, 42, 51).
TABLE 2.
Kinetics parameters in thiosulfate-dependent cyt c554 reduction activitiesa
| Component | V (μmol cyt c554 reduction · μmol Sox protein−1 · min−1) | Km (mM) |
|---|---|---|
| SoxYZ, SoxB, and SoxAX-CT1020 protein | 7.9 ± 0.2 | 0.17 ± 0.02 |
| SoxYZ, SoxB, and rSoxAX-rCT1020 | 7.8 ± 0.1 | 0.18 ± 0.02 |
| SoxYZ, SoxB, rSoxA, and rSoxX | 0.24 ± 0.08 | 0.16 ± 0.02 |
| SoxYZ and SoxB | ND | |
| SoxYZ and SoxAX-CT1020 protein | ND | |
| SoxB and SoxAX-CT1020 protein | ND |
Thiosulfate-dependent cyt c554 reduction activities and Km values were determined from the [S]0/v-versus-[S]0 plot in the thiosulfate concentration range of 0 to 2 mM. The values are the means ± standard deviations of the results of three independent measurements similar to the ones whose results are shown in Fig. 4a and b. ND, not detectable (<0.01 μmol cyt c554 reduced · μmol Sox protein−1 · min−1).
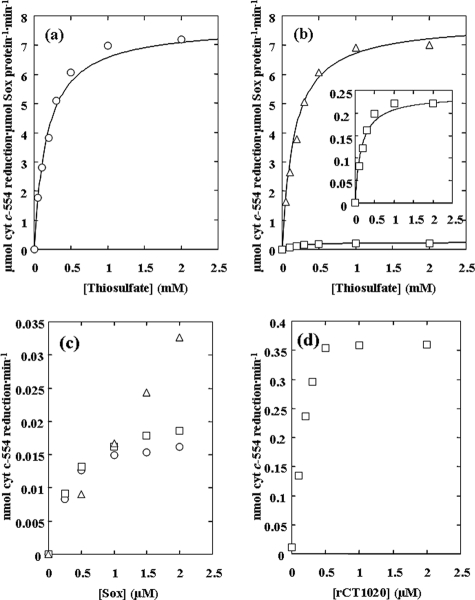
The relationship between the initial rate of cyt c554 reduction and the thiosulfate concentration (in the range of 0 to 2 mM) can be fitted to Michaelis-Menten-type kinetics with a Km value for thiosulfate of 0.17 mM (Fig. 4a and Table 2). At higher thiosulfate concentrations, inhibition of reactions was noted (data not shown), as with the Sox system of A. vinosum (28). The optimum pH for the reaction was found at about pH 6.0 to 6.5 (data not shown). In this assay system, 2.1 mol of cyt c554 was reduced per mole of thiosulfate oxidized and the reaction proceeded almost linearly for at least two minutes, during which time about 8 mol of thiosulfate was oxidized per mole of SoxYZ (data not shown). These results indicate that the product of sulfane sulfur from thiosulfate is zero-valence sulfur, possibly remaining as polysulfane bound on SoxY or as the elemental sulfur subsequently formed (54). The profile of the temperature-activity curve of the complete system was similar to those of ordinary enzyme reactions, with the optimum temperature at about 55 to 60°C. The activities determined at 2 mM thiosulfate for 1 min were 6.7 (25°C), 17.2 (35°C), 24.6 (40°C), 29.7 (45°C), 33.3 (50°C), 36.4 (55°C), 36.9 (60°C), and 28.7 (65°C) μmol cyt c554 reduced·μmol Sox protein−1·min−1.
FIG. 4.
Thiosulfate-dependent cyt c554 reduction activities. (a) Effects of thiosulfate concentration on cyt c554 reduction rates. The reaction mixture contained 0.5 μM SoxYZ, 0.5 μM SoxB, 0.5 μM SoxAX-CT1020 protein, 50 μM cyt c554, 20 mM MES-NaOH (pH 6.0), and the indicated concentrations of thiosulfate. (b) As above, except that in place of 0.5 μM SoxAX-CT1020 protein (the basic assay mixture), the following were added: open triangles, 0.5 μM each of rSoxA, rSoxX, and rCT1020; open squares, 0.5 μM each of rSoxA and rSoxX. An enlargement of the kinetics of the last combination is shown in the inset. Cyt c554 reduction was monitored by the absorbance change at 554 nm. (c) Effects of rSoxA and rSoxX concentrations in the absence of rCT1020. The basic assay mixtures contained 1.0 μM rSoxA and indicated concentrations of rSoxX (open squares), 1.0 μM rSoxX and indicated concentrations of rSoxA (open circles), or equal concentrations of both rSoxA and rSoxX as indicated (open triangles). (d) Effects of rCT1020 concentration. The basic assay mixture contained 0.5 μM rSoxA, 0.5 μM rSoxX, and the indicated concentrations of rCT1020.
The functions of purified rSoxA, rSoxX, and rCT1020 were examined in an assay system containing SoxYZ, SoxB, and cyt c554 from C. tepidum, but not SoxAX-CT1020 protein (the basic assay system). The addition of any one of the single components rSoxA, rSoxX, or rCT1020 to the basic assay system could not support thiosulfate oxidation. The same was true with the two-component mixtures of rSoxA plus rCT1020 and rSoxX plus rCT1020 protein (data not shown). However, the mixture of rSoxA and rSoxX, each at 0.5 μM, showed a low but discernible cyt c554 reduction activity when added to the basic assay system (Fig. 4b and Table 2). When the rSoxX concentration was fixed at 1.0 μM, the rate of thiosulfate-dependent cyt c554 reduction initially increased with an increase in rSoxA concentration but apparently was saturated at about 1.0 μM of rSoxA (Fig. 4c). Similar results were obtained when the rSoxX concentration was changed in the presence of a fixed concentration of rSoxA of 1.0 μM. When rSoxA and rSoxX were added simultaneously at the same concentration, the reaction rate increased with increasing concentrations of rSoxA and rSoxX and with no indication of saturation in the concentration range shown in Fig. 4c. Furthermore, the simultaneous addition of increasing concentrations of rSoxA and rSoxX exceeded the maximum rates attained when the concentration of one of the components was fixed at 1 μM (Fig. 4c).
When 0.5 μM rCT1020 was added to the basic assay system containing 0.5 μM rSoxA and 0.5 μM rSoxX, the activity increased dramatically (Fig. 4d). The activity initially increased with increasing rCT1020 concentrations and, at around 0.5 μM, reached a plateau at a level about 30-fold higher than in the presence of rSoxA and rSoxX without rCT1020. The specific activity and Km for thiosulfate of the assay system containing a mixture of rSoxA, rSoxX, and rCT1020 are shown in Table 2. The maximum activity of the assay mixture containing the three recombinant proteins was comparable to that of the mixture containing the native SoxAX-CT1020 protein complex, indicating that the recombinant proteins were overexpressed in E. coli cells in their active forms. The temperature-activity curve of the basic assay system containing all three recombinant proteins was similar to that containing native SoxAX-CT1020 protein until the temperature reached 55°C. The activity of the basic assay system containing the recombinant proteins was slightly lower than that containing the native SoxAX-CT1020 protein after 60°C, declining slightly faster than the latter with further increases in temperature (data not shown). Although the maximum activity of the system containing rSoxA and rSoxX was much lower than that of the complete system containing all three recombinant proteins, the Km value for thiosulfate was almost unchanged. The kinetics values obtained by various assays are summarized in Table 2.
rCT1020 and complex formation.
In order to study the complex formation properties of rSoxA, rSoxX, and rCT1020, the recombinant proteins were incubated in various combinations and analyzed by gel permeation chromatography (Fig. 5). When the mixture of all three proteins, rSoxA, rSoxX, and rCT1020, was incubated and applied to the column, there appeared one major peak accompanied by a minor one, with the retention time of the major one almost identical with that of the native SoxAX-CT1020 protein complex (Fig. 5, chromatogram b). The occurrence of the minor peaks might be caused by the addition of an amount of rCT1020 slightly in excess of the stoichiometric amounts of rSoxA and rSoxX. When rSoxA and rCT1020 were mixed, the major peak was eluted earlier than for rSoxA alone (chromatogram d), indicating complex formation between the two. The mixture of rSoxA and rSoxX was eluted in two independent peaks (chromatogram c), and their retention times were indistinguishable from those of the respective components applied to the column separately. The mixture of rSoxX and rCT1020 also showed no indication of complex formation in gel permeation chromatography analysis (chromatogram e).
FIG. 5.
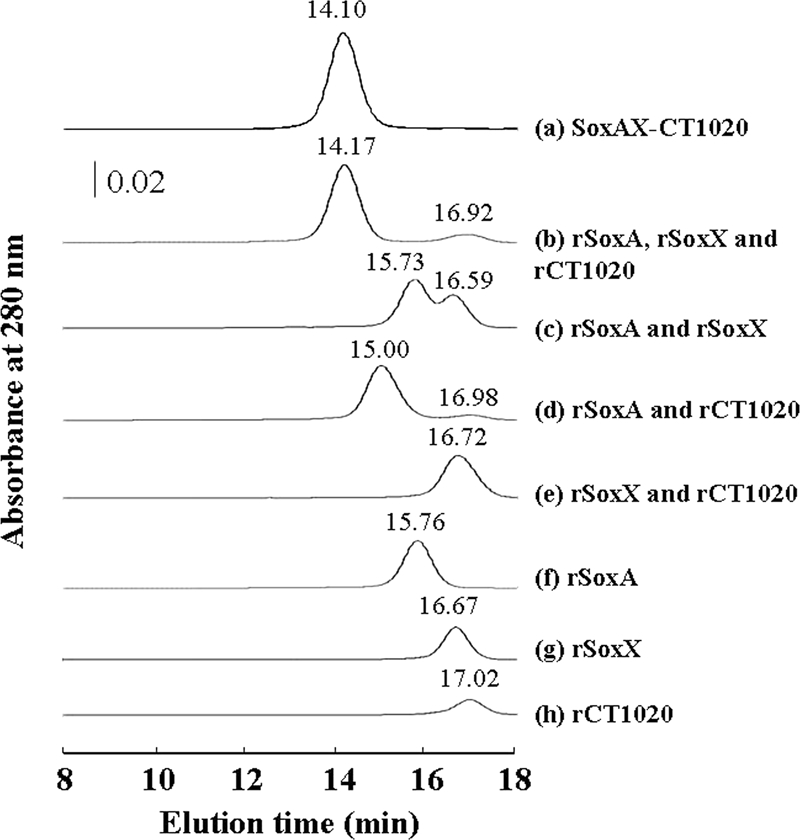
Gel permeation chromatograms of rSoxAX-rCT1020 components and mixtures of the components. Samples contained 10 μM of each protein in 100 μl.
DISCUSSION
Factors required for thiosulfate oxidation in C. tepidum.
In C. tepidum, sox gene homologues found in other thiosulfate-oxidizing bacteria are expressed under ordinary conditions of growth in a medium containing thiosulfate. In this paper, we describe the purification of three components that are indispensable for in vitro thiosulfate oxidation with the small, monoheme cyt c554 as the electron acceptor (Table 2). On the whole, the properties of the three components were similar to those found in P. pantotrophus (20, 51) and P. versutus (41, 42) but with some new characteristics that are described below. The stoichiometry of electrons donated to cyt c554 per molecule of thiosulfate oxidized was found to be 2.1, in agreement with the reaction model proposed for the P. pantotrophus Sox system where two electrons are generated in the absence of SoxCD (22). SoxYZ serves as the acceptor for the reaction intermediates (47, 58). In P. pantotrophus, SoxYZ is reported to occur as a heterodimeric or heterotetrameric protein (48). Although our preparation also contained the heterotetrameric form, this appears to be an oxidized product of the dimeric form linked by an intermolecular disulfide bridge during purification steps under aerobic conditions, because the tetrameric form disappeared upon incubation of the preparation with thiosulfate. Thus, we conclude that SoxY (13 kDa) and SoxZ (9 kDa) are a heterodimeric complex in the presence of thiosulfate. The second component is SoxB that has been proposed to catalyze the hydrolytic cleavage of the thiosulfate oxidation product bound to SoxYZ to yield sulfate, and we find that SoxB is a monomer (60 kDa).
In several bacteria, the third component occurs as heterodimeric SoxAX protein, and the three-dimensional structure of the proteins from R. sulfidophilum (3) and P. pantotrophus (15) have been determined by X-ray crystallography. SoxX from various bacteria has been reported to be a monoheme protein, while SoxA is either a diheme protein in P. pantotrophus and R. sulfidophilum or a monoheme protein in Starkeya novella (32). For this study, we prepared a complex containing SoxAX and found that the complex binds the CT1020-encoded protein of about 9 kDa as the third component and that the complex exists as a heterotrimer (Fig. 1 and Fig. 5).
Biochemical characterization of the CT1020 protein (SAXB).
Rother and Friedrich (50) report having succeeded in coexpressing soxA and soxX of P. pantotrophus in E. coli cells and having subsequently obtained the SoxAX complex in an active form. They also overexpressed soxX, but the protein alone had no detectable activity. We have succeeded in separately expressing soxA, soxX, and CT1020 of C. tepidum in E. coli cells (Fig. 1b) and subsequently obtained each protein in an active purified form (Fig. 4 and Table 2). rCT1020 is a colorless protein that does not appear to bind any prosthetic group or heavy metals. The mixture of rSoxA and rSoxX had low but definite thiosulfate-oxidizing activity in the presence of SoxYZ and SoxB, and the activity was greatly accelerated by the addition of rCT1020 (Fig. 4d). When rCT1020, rSoxA, and rSoxX were mixed in various combinations, complex formation was demonstrated by the results of gel permeation chromatography for the combinations of rCT1020 and rSoxA and rCT1020, rSoxA, and rSoxX, but not rCT1020 and rSoxX or rSoxA and rSoxX (Fig. 5). These results indicate that the CT1020 protein strengthens the association of SoxA and SoxX by binding to SoxA and, possibly, also to SoxX. Accordingly, the CT1020 protein will be referred to as SAXB (SoxAX binding protein). In addition to the structural role proposed above, SAXB might have a role in accelerating the catalytic activity by inducing a conformational change in SoxA and/or SoxX.
In the enzyme kinetics studies, when either rSoxA or rSoxX was present at a fixed concentration, the reaction rate was saturated when the concentration of the other component was increased (Fig. 4c). When the concentrations of both components were increased simultaneously, the oxidizing activity increased with the increase in the concentrations and exceeded the saturation rates attained when the concentration of either rSoxA or rSoxX was fixed. These results indicate that rSoxA and rSoxX do not work by the catalysis mechanism of random collision of the two components, because in such a mechanism, saturation of the rate (Fig. 4c) would not be observed in both instances when the concentration of each component was fixed. The results of the kinetics experiments suggest that association of the two components is required for the catalytic reaction, although a complex formation between the two components was not directly demonstrated by the results of gel permeation chromatography (Fig. 5). The association between the two components does not appear to be great enough to withstand gel permeation chromatography in the absence of SAXB.
Redox potentials of hemes of SoxAX-SAXB.
The midpoint redox potential at pH 10.0 of rSoxX was +135 mV, but that of rSoxA was unusually low, with a value of less than −550 mV. Previously, the midpoint redox potentials of all the hemes of SoxAX, irrespective of whether they were the diheme type (S. novella [33] and C. limicola f. sp. thiosulfatophilum [44]) or triheme type (P. pantotrophus [49] and R. sulfidophilum [3]) were reported to be in the range of about +135 to 200 mV. More recently, it was reported that one of the hemes has an unusually low midpoint redox potential of −432 mV in the triheme SoxAX complex from P. pantotrophus (49) and −479 mV in the diheme protein from Starkeya novella (33), respectively. Our results show that one of the hemes in the diheme SoxAX-SAXB complex from C. tepidum has a very low redox potential and that rSoxA contains this low-potential heme (Fig. 3a).
Distribution of genes homologous to the SAXB gene.
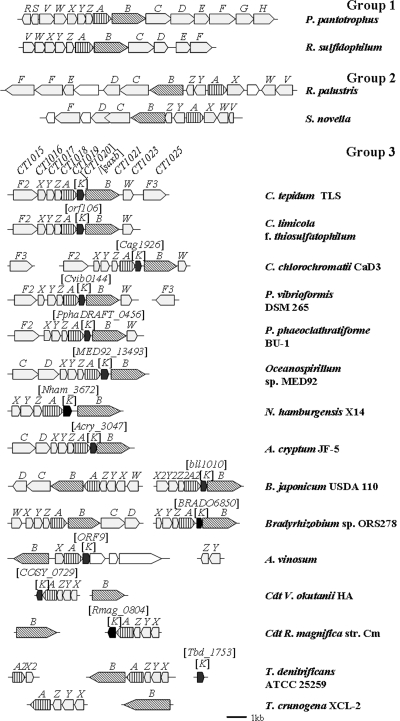
A search of the GenBank database reveals that homologues of the SAXB gene are found in a fairly large number of strains, comprising roughly one-third of the thiosulfate-oxidizing bacteria whose sox gene cluster sequences have been deposited so far and ranging over many genera, including members of the Chlorobiaceae, Chromatiaceae, Hydrogenophilaceae, Oceanospirillaceae, etc. In the Chlorobium limicola f. sp. thiosulfatophilum sox cluster, Verté et al. (60) noted the presence of an open reading frame, a homologue of CT1020 (the SAXB gene), but they considered it not to be related to thiosulfate oxidation because the homologue was absent in the sox clusters of several bacteria, including P. pantotrophus (20). Hensen et al. (28) noted the presence of genes homologous to the SAXB gene immediately downstream of soxA in several thiosulfate-oxidizing bacteria, including Allochromatium vinosum. Frigaard and Bryant (23, 24) noted that the cluster of sox genes in C. tepidum TLS, CT1015-soxXYZA-CT1020-soxBW, is conserved in the genomes of three other thiosulfate-utilizing green sulfur bacterial strains in addition to one (Chlorobium chlorochromatii CaD3) that has not been reported to grow on thiosulfate. They designated CT1020 as soxK and speculated as to its possible involvement in regenerating SoxYZ by mobilizing bound polysulfane. We have demonstrated here that the gene encodes SAXB that is required for tight binding of SoxA and SoxX. The gene is apparently absent in many other thiosulfate-oxidizing bacteria, including P. pantotrophus, S. novella, R. sulfidophilum, etc. Amino acid sequence comparison of the deduced mature SAXB homologues from various sources indicates sequence identities of from 20 to 79% to that of C. tepidum SAXB (SoxK). Some conserved amino acid sequence motifs were identified (see Fig. 8a), implying a conserved functional identity.
FIG. 8.
Proteins and amino acid sequence alignments of component of SoxAX-SAXB. The alignment was performed using the program CLUSTALW, version 1.83. Amino acid sequence alignments of predicted SAXB (SoxK) (a), SoxA (b), and SoxX (c) proteins of some bacteria representative of those in Fig. 7 are shown. (a) Amino acid sequence alignments of predicted SAXB (SoxK) proteins and the sequences of loop extension regions of type I SoxXs which are absent in group III SoxXs are compared. In panel c, the sequences of this loop extension region are underlined. (b and c) The sequences in squares have been shown by X-ray crystallography to be important for the formation of complexes between SoxA and SoxX in R. sulfidophilum and P. pantotrophus (3, 15). Asterisks show identical residues; colons show conserved substitutions; dots show semiconserved substitutions.
The SAXB gene homologue is localized to an sox gene cluster immediately downstream of soxA in all the bacteria that are found to contain SAXB gene (soxK) homologues thus far, except for Thiobacillus denitrificans and Thiomicrospira crunogena XCL-2 (Fig. 6). T. denitrificans has two sox gene clusters but only one putative SAXB gene homologue, which is found at about 1.3 Mbp and about 0.87 Mbp away from soxA1 and soxA2, respectively. We have failed to find an SAXB gene homologue in the genome of T. crunogena. In green sulfur bacteria, the SAXB gene (soxK) homologue is invariably found immediately upstream of soxB, as noted in references 23 and 24. It is also found immediately upstream of soxB in some of the bacteria that belong to the other groups, but not in others. Interestingly, Bradyrhizobium japonicum USAD 110 contains two sox gene clusters, in one of which the SAXB gene (soxK) is present and in the other of which it is absent. Moreover, the predicted SoxAXs encoded by the two gene clusters belong to different groups of proteins which are correlated with the presence and absence of the SAXB gene (soxK) (see below).
FIG. 6.
Map of the sox gene cluster. SAXB gene (soxK) homologs are indicated in black. Other hypothetical sox-related genes are shown in light gray with soxA vertically and soxB obliquely striped. The sources used were (organism, GenBank nucleotide sequence accession number): Acidiphilium cryptum JF-5, CP000697; Allochromatium vinosum, DQ441405; Bradyrhizobium japonicum USDA 110, BA000040; Bradyrhizobium sp. strain ORS278, CU234118; Chlorobium chlorochromatii CaD3, CP000108; Chlorobium limicola f. sp. thiosulfatophilum, AY074395; Chlorobium tepidum TLS, AE006470; “Candidatus Ruthia magnifica” strain Cm, CP000488; “Candidatus Vesicomyosocius okutanii” HA, AP009247; Nitrobacter hamburgensis X14, CP000319; Oceanospirillum sp. strain MED92, AAOW00000000; Paracoccus pantotrophus GB17, X79242 (EMBL nucleotide sequence database accession number); Pelodictyon phaeoclathratiforme BU-1, AAIK00000000; Prosthecochloris vibrioformis DSM 265, CP000607; Rhodopseudomonas palustris CGA009, BX571963; Rhodovulum sulfidophilum, AY005800; Starkeya novella DSMZ 506T, AF139113; Thiobacillus denitrificans ATCC 25259, CP000116; and Thiomicrospira crunogena XCL-2, CP000109.
Some amino acid sequence characteristics of SoxA and SoxX in bacteria in both the absence and presence of SAXB homologues.
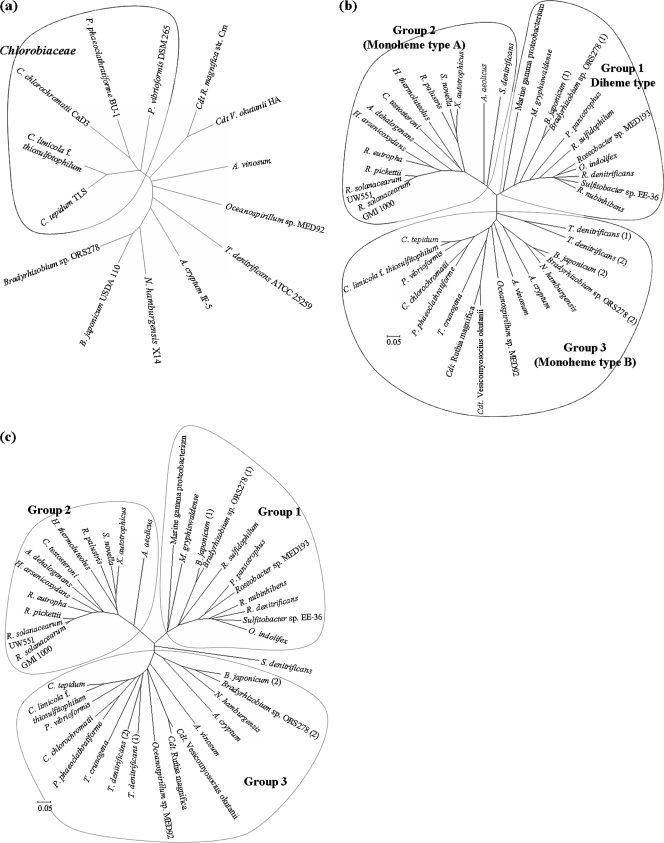
The results of phylogenetic-tree analysis of bacteria with deduced SAXB (SoxK), SoxA, and SoxX proteins are shown in Fig. 7. The green sulfur bacteria with deduced SAXB (SoxK) proteins form a cluster away from other bacteria (Fig. 7a).
FIG. 7.
Unrooted phylogenetic tree of bacteria predicted to contain SAXB (a), SoxA (b), and SoxX (c). Phylogenetic relationships were generated using Molecular Evolutionary Genetic Analysis (MEGA) software version 4.0. The sequences used were [organism, protein (GenBank accession number)]: Acidiphilium cryptum JF-5, SoxA (YP_001236154), SoxX (YP_001236151), and SAXB (YP_001236155); Allochromatium vinosum, SoxA (ABE01361), SoxX (ABE01360), and SAXB (ABE01362); Anaeromyxobacter dehalogenans 2CP-1, SoxA (ZP_02323507) and SoxX (ZP_02323506); Aquifex aeolicus VF5, SoxA (NP_214239) and SoxX (NP_214238); Bradyrhizobium japonicum USDA 110, SoxA1 (NP_770154), SoxA2 (NP_767651), SoxX1 (NP_770151), SoxX2 (NP_767654), and SAXB (NP_767650); Bradyrhizobium sp. strain ORS278, SoxA1 (YP_001205214), SoxA2 (YP_001208659), SoxX1 (YP_001205211), SoxX2 (YP_001208656), and SAXB (YP_001208656); Chlorobium chlorochromatii CaD3, SoxA (YP_380216), SoxX (YP_380213), and SAXB (YP_380217); Chlorobium limicola f. sp. thiosulfatophilum, SoxA (AAL68886), SoxX (AAL68883), and SAXB (AAL68887); Chlorobium tepidum TLS, SoxA (NP_661911), SoxX (NP_661908), and SAXB (NP_661912,); Comamonas testosteroni KF-1, SoxA (ZP_01521176) and SoxX (ZP_01521177); Candidatus Ruthia magnifica” strain Cm, SoxA (YP_903997), SoxX (YP_904000), and SAXB (YP_903996); “Candidatus Vesicomyosocius okutanii” HA, SoxA (YP_001219566), SoxX (YP_001219569), and SAXB (YP_001219565); Herminiimonas arsenicoxydans, SoxA (YP_001099497) and SoxX (YP_001099498); Hydrogenophilus thermoluteolus, SoxA (BAF34123) and SoxX (BAF34124); Magnetospirillum gryphiswaldense MSR-1, SoxA (CAM76243) and SoxX (CAM76246); Marine gammaproteobacterium strain HTCC2143, SoxA (ZP_01615028) and SoxX (ZP_01615032); Nitrobacter hamburgensis X14, SoxA (YP_578861) and SoxX (YP_578864); Oceanibulbus indolifex HEL-45, SoxA (ZP_02154073) and SoxX (ZP_02154077); Oceanospirillum sp. strain MED92, SoxA (ZP_01167150), SoxX (ZP_01167154), and SAXB (ZP_01167149); Paracoccus pantotrophus GB17, SoxA (CAA55827) and SoxX (CAB94379); Pelodictyon phaeoclathratiforme BU-1, SoxA (ZP_00588640), SoxX (ZP_00588637), and SAXB (ZP_00588641); Prosthecochloris vibrioformis DSM 265, SoxA (YP_001129671), SoxX (YP_001129674), and SAXB (YP_001129670); Ralstonia eutropha H16, SoxA (YP_727992) and SoxX (YP_727991); Ralstonia pickettii 12J, SoxA (ZP_01661484) and SoxX (ZP_01661485); Ralstonia solanacearum GMI1000, SoxA (NP_521375) and SoxX (NP_521374); Ralstonia solanacearum UW551, SoxA (ZP_00944483) and SoxX (ZP_00944484); Rhodopseudomonas palustris CGA009, SoxA (NP_949804) and SoxX (NP_949805); Rhodovulum sulfidophilum, SoxA (AAF99434) and SoxX (AAF99431); Roseobacter sp. strain MED193, SoxA (ZP_01055913) and SoxX (ZP_01055916); Roseobacter denitrificans OCh 114, SoxA (YP_681833) and SoxX (YP_681830); Roseovarius nubinhibens ISM, SoxA (ZP_00961295) and SoxX (ZP_00961298); Starkeya novella DSMZ 506T, SoxA (AAR98727) and SoxX (AAR98728); Sulfitobacter sp. strain EE-36, SoxA (ZP_00956138) and SoxX (ZP_00956135); Sulfurimonas denitrificans DSM 1251, SoxA (YP_392779) and SoxX (YP_392776); Thiobacillus denitrificans ATCC 25259, SoxA1 (YP_314322), SoxA2 (YP_314676), SoxX1 (YP_314325), SoxX2 (YP_314675), and SAXB (YP_315511); Thiomicrospira crunogena XCL-2, SoxA (YP_390871) and SoxX (YP_390874); and Xanthobacter autotrophicus Py2, SoxA (YP_001416427) and SoxX (YP_001416428).
The SoxXs of various bacteria invariably contain one heme, and the SoxAs either one or two heme groups (1, 19, 32). For example, the SoxAs from P. pantotrophus and R. sulfidophilum each have two c-type hemes and two conserved heme-binding motifs (CXXCH) (1, 20). By contrast, the SoxAs from S. novella and C. limicola f. sp. thiosulfatophilum each have only one heme-binding motif, due to a difference in amino acid sequence located within the N-terminal side (32, 37). From DNA sequence data, the predicted SoxA protein from C. tepidum belongs to the monoheme type. From an amino acid sequence homology search using the MEGA 4 program based on the neighbor-joining method, we found that SoxA sequences may be classified into three groups (Fig. 7b): group 1 (diheme), represented by the SoxAs of Paracoccus and Rhodovulum bacteria; group 2 (monoheme, type A), represented by the SoxAs of S. novella and R. palustris; and group 3 (monoheme, type B), represented by the SoxAs of members of the Chlorobiacieae (32). SoxXs may also be classified into three groups (Fig. 7c) that completely correspond to the grouping of SoxAs with respect to strains, with the exception of SoxX from Sulfurimonas denitrificans, which constitutes an outlier in our analysis despite the fact that its SoxA appears to belong to group 2 (Fig. 7b). SoxAs and SoxXs from those bacteria that have SAXB invariably belong to group 3. However, in the genome of Thiomicrospira crunogena, whose SoxA and SoxX belong to group 3 in the above analysis, we could not find a gene encoding an SAXB (SoxK) homologue using a BLAST search with any SAXB (SoxK) homologue protein as the query sequence. As stated above, Thiobacillus denitrificans contains two sox gene clusters with the SAXB gene (soxK) located a distance away from the clusters, and both of the predicted SoxAXs belong to the groups that are correlated with the presence of the SAXB gene (soxK).
Although predicted SoxAs show considerable overall amino acid sequence identity across strains, each group of SoxAs and SoxXs has its own characteristics (Fig. 7 and 8). Compared with group 1 and group 3 predicted SoxA sequences, the group 2 predicted SoxA sequence has a shorter N terminus by about 20 to 30 amino acids and insertions of 9 and 7 residues in the middle and in the vicinity of the C terminus, respectively. Each of the predicted sequences is assumed to have a signal peptide, and there may be some uncertainty in the sequence of the mature protein on the N-terminal side. We have found that C. tepidum SoxA begins with the 28th glutamic acid sequence, and it seems that each group 2 predicted mature SoxA is actually shorter than the group 3 protein by about 20 to 30 amino acids on the N-terminal side. The group 1 predicted SoxA sequence and its group 3 counterpart seem to be rather similar to each other, with only minor differences of short insertions/deletions of less than 6 residues. (Fig. 8b).
Although all of the predicted SoxX sequences show considerable amino acid sequence identity with each other, the overall profile across the groups shows greater sequence diversity than that predicted for the SoxA sequences. The predicted group 2 SoxX has a notable N-terminal extension of about 40 to 90 amino acids as compared with the group 1 and group 3 SoxX sequences (Fig. 8c). Due to the presence of a periplasmic targeting signal sequence on the N-terminal end, there may be some ambiguity in the prediction of mature proteins from DNA sequences. In a comparison of SoxX proteins whose N-terminal sequences have been biochemically determined, the S. novella group 2 SoxX has a longer N terminus than the SoxXs from the following: group 1, 47 residues (P. pantotrophus) and 45 residues (R. sulfidophilum), and group 3, 86 residues (C. tepidum). Compared with the group 1 SoxX, group 2 and group 3 SoxX proteins have a significant 20-amino-acid deletion (from residue 122 to 141 of R. sulfidophilum SoxX) (3) (Fig. 8c). The group 2 SoxX has a 12-residue insertion between residues 60 and 61 of R. sulfidophilum. The group 3 SoxX has no such insertion. The strings of 20-amino-acid sequences of group 1 SoxXs corresponding to the deleted sequence in group 3 and group 2 SoxXs were compared with SAXBs from several group 3 bacteria by using CLUSTALW, and the results of the amino acid sequence alignment indicate that there exists significant homology between them (Fig. 8a). From the previously reported X-ray crystallographic results for the three-dimensional structure of the group 1 SoxAX complexes of R. sulfidophilum (3) and P. pantotrophus (15), the 20-amino-acid sequence, which is absent in group 3 and group 2 SoxXs, constitutes a loop that seems to be important for complex formation with SoxA (3). From the results of the above-described sequence analyses (Fig. 8) and the gel permeation chromatography results (Fig. 5), we conclude that SAXB contributes to the tight complex formation between group 3 SoxA and SoxX. Group 2 bacteria do not seem to have the SAXB gene, although their SoxX protein has a 20-residue deletion similar to that in group 3 SoxX. In group 2 bacteria, it may be that the 12-residue insertion described above or the long N-terminal extension in SoxX makes SAXB unnecessary for the association between SoxA and SoxX.
Several research groups have reported the preparation of cyt c551s from group 3 bacteria that were essential for thiosulfate oxidation, including a 45- to 60-kDa cyt c551 from Chlorobium limicola f. sp. thiosulfatophilum strain NCIB 8346 (44); a dimeric, 30-kDa cyt c551 from Chlorobium limicola strain Tassajara (37); and a heterodimeric, 40-kDa cyt c551 from A. vinosum (28). Thus far, the presence of SAXB has not been reported for any of these strains, and the presence or absence of SAXB in the SoxAX complex among group 3 bacteria will be interesting to study in the future.
SoxB and thiosulfate-oxidizing enzyme.
From C. tepidum, we have prepared SoxB as a monomer of about 60 kDa and SoxYZ either as a heterodimer or a tetramer of about 26- and 40-kDa proteins (data not shown). Kusai and Yamanaka (39) purified a protein they called thiosulfate-oxidizing enzyme that catalyzed the thiosulfate-dependent reduction of cyt c551 (SoxAX homologue), but the requirement of SoxYZ for catalysis was not described. The enzyme was reported to be a colorless protein of 80 kDa in molecular mass as estimated by SDS-PAGE analysis in the presence of 0.5% mercaptoethanol. We found that SoxB and SoxAX-SAXB are not sufficient for thiosulfate-dependent cyt c554 reduction and that the participation of SoxYZ is also necessary (Table 2). We purified SoxYZ and SoxB as separate proteins. However, as described in Materials and Methods, C. tepidum cell extracts contained a complex composed of SoxYZ and SoxB, and the partially purified complex tended to dissociate into SoxB and SoxYZ in the subsequent gel permeation chromatography in the presence of a high salt concentration of 150 mM NaCl in 50 mM Tris-HCl (pH 7.8). From the above observations, it is tempting to speculate that the “thiosulfate oxidase” preparation of Kusai and Yamanaka (39) could be a complex of SoxYZ and SoxB, although they estimated the molecular mass of the “thiosulfate oxidase” as 80 kDa by SDS-PAGE analysis in the presence of 2-mercaptoethanol.
In addition to the well-recognized soxXYZAB-encoded proteins in other bacteria, the CT1020-encoded protein (SAXB [SoxK]) is also required for efficient in vitro oxidation of thiosulfate in C. tepidum. SAXB gene (soxK) homologues are distributed over fairly large groups of thiosulfate-oxidizing bacteria, and the SoxX and SoxA proteins of such bacteria constitute a group that is distinct from those of bacteria that do not contain SAXB gene (soxK) homologues.
Acknowledgments
We thank Linda Thöny-Meyer for the plasmid pEC86.
This work was supported in part by the Global COE program (integrative life science based on the study of biosignaling mechanisms), MEXT, Japan. This work was also supported by a high-tech research center project, MEXT, Japan.
Footnotes
Published ahead of print on 18 July 2008.
REFERENCES
- 1.Appia-Ayme, C., P. J. Little, Y. Matsumoto, A. P. Leech, and B. C. Berks. 2001. Cytochrome complex essential for photosynthetic oxidation of both thiosulfate and sulfide in Rhodovulum sulfidophilum. J. Bacteriol. 1836107-6118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arslan, E., H. Schulz, R. Zufferey, P. Künzler, and L. Thöny-Meyer. 1998. Overproduction of the Bradyrhizobium japonicum c-type cytochrome subunits of the cbb3 oxidase in Escherichia coli. Biochem. Biophys. Res. Commun. 251744-747. [DOI] [PubMed] [Google Scholar]
- 3.Bamford, V. A., S. Bruno, T. Rasmussen, C. Appia-Ayme, M. R. Cheesman, B. C. Berks, and A. M. Hemmings. 2002. Structural basis for the oxidation of thiosulfate by a sulfur cycle enzyme. EMBO J. 215599-5610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berry, E. A., and B. L. Trumpower. 1987. Simultaneous determination of hemes a, b, and c from pyridine hemochrome spectra. Anal. Biochem. 1611-15. [DOI] [PubMed] [Google Scholar]
- 5.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72248-254. [DOI] [PubMed] [Google Scholar]
- 6.Brune, D. C. 1989. Sulfur oxidation by phototrophic bacteria. Biochim. Biophys. Acta 975189-221. [DOI] [PubMed] [Google Scholar]
- 7.Brune, D. C. 1995. Sulfur compounds as photosynthetic electron donors, p. 847-870. In R. E. Blankenship, M. T. Madigan, and C. E. Bauer (ed.), Anoxygenic photosynthetic bacteria. Kluwer, Amsterdam, The Netherlands.
- 8.Cammack, R., A. Chapman, W.-P. Lu, A. Karagouni, and D. P. Kelly. 1989. Evidence that protein B of the thiosulphate-oxidizing system of Thiobacillus versutus contains a binuclear manganese cluster. FEBS Lett. 253239-243. [Google Scholar]
- 9.Chan, L.-K., R. Morgan-Kiss, and T. E. Hanson. 2008. Genetic and proteomic studies of sulfur oxidation in Chlorobium tepidum (syn. Chlorobaculum tepidum), p. 357-371. In R. Hell, C. Dahl, D. B. Knaff, and T. Leustek (ed.), Sulfur metabolism in phototrophic organisms. Springer, Dordrecht, The Netherlands.
- 10.Chan, L.-K., R. Morgan-Kiss, and T. E. Hanson. 2008. Sulfur oxidation in Chlorobium tepidum (syn. Chlorobaculum tepidum): genetic and proteomic analyses, p. 117-126. In C. Dahl and C. G. Friedrich (ed.), Microbial sulfur metabolism. Springer, Berlin, Germany.
- 11.Chan, L.-K., T. S. Weber, R. M. Morgan-Kiss, and T. E. Hanson. 2008. A genomic region required for phototrophic thiosulfate oxidation in the green sulfur bacterium Chlorobium tepidum (syn. Chlorobaculum tepidum). Microbiology 154818-829. [DOI] [PubMed] [Google Scholar]
- 12.Cheesman, M. R., P. J. Little, and B. C. Berks. 2001. Novel heme ligation in a c-type cytochrome involved in thiosulfate oxidation: EPR and MCD of SoxAX from Rhodovulum sulfidophilum. Biochemistry 4010562-10569. [DOI] [PubMed] [Google Scholar]
- 13.Crowe, J., and K. Henco. 1992. The QIAexpressionist, 2nd ed. Qiagen, Inc., Chatsworth, CA.
- 14.Dahl, C. 2008. Inorganic sulfur compounds as electron donors in purple sulfur bacteria, p. 289-317. In R. Hell, C. Dahl, D. B. Knaff, and T. Leustek (ed.), Sulfur metabolism in phototrophic organisms. Springer, Dordrecht, The Netherlands.
- 15.Dambe, T., A. Quentmeier, D. Rother, C. Friedrich, and A. J. Scheidig. 2005. Structure of the cytochrome complex SoxXA of Paracoccus pantotrophus, a heme enzyme initiating chemotrophic sulfur oxidation. J. Struct. Biol. 152229-234. [DOI] [PubMed] [Google Scholar]
- 16.Eisen, J. A., K. E. Nelson, I. T. Paulsen, J. F. Heidelberg, M. Wu, R. J. Dodson, R. Deboy, M. L. Gwinn, W. C. Nelson, D. H. Haft, E. K. Hickey, J. D. Peterson, A. S. Durkin, J. L. Kolonay, F. Yang, I. Holt, L. A. Umayam, T. Mason, M. Brenner, T. P. Shea, D. Parksey, W. C. Nierman, T. V. Feldblyum, C. L. Hansen, M. B. Craven, D. Radune, J. Vamathevan, H. Khouri, O. White, T. M. Gruber, K. A. Ketchum, J. C. Venter, H. Tettelin, D. A. Bryant, and C. M. Fraser. 2002. The complete genome sequence of Chlorobium tepidum TLS, a photosynthetic, anaerobic, green-sulfur bacterium. Proc. Natl. Acad. Sci. USA 999509-9514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Epel, B., K.-O. Schäfer, A. Quentmeier, C. Friedrich, and W. Lubitz. 2005. Multifrequency EPR analysis of the dimanganese cluster of the putative sulfate thiohydrolase SoxB of Paracoccus pantotrophus. J. Biol. Inorg. Chem. 10636-642. [DOI] [PubMed] [Google Scholar]
- 18.Francis, R. T., Jr., and R. R. Becker. 1984. Specific indication of hemoproteins in polyacrylamide gels using a double-staining process. Anal. Biochem. 136509-514. [DOI] [PubMed] [Google Scholar]
- 19.Friedrich, C. G., F. Bardischewsky, D. Rother, A. Quentmeier, and J. Fischer. 2005. Prokaryotic sulfur oxidation. Curr. Opin. Microbiol. 8253-259. [DOI] [PubMed] [Google Scholar]
- 20.Friedrich, C. G., A. Quentmeier, F. Bardischewsky, D. Rother, R. Kraft, S. Kostka, and H. Prinz. 2000. Novel genes for lithotrophic sulfur oxidation of Paracoccus pantotrophus GB17. J. Bacteriol. 1824677-4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Friedrich, C. G., A. Quentmeier, F. Bardischewsky, D. Rother, G. Orawski, P. Hellwig, and J. Fischer. 2008. Redox control of chemotrophic sulfur oxidation of Paracoccus pantotrophus, p. 139-150. In C. Dahl and C. G. Friedrich (ed.), Microbial sulfur metabolism. Springer, Berlin, Germany.
- 22.Friedrich, C. G., D. Rother, F. Bardischewsky, A. Quentmeier, and J. Fischer. 2001. Oxidation of reduced inorganic sulfur compounds by bacteria: emergence of a common mechanism? Appl. Environ. Microbiol. 672873-2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frigaard, N.-U., and D. A. Bryant. 2008. Genomic and evolutionary perspectives on sulfur metabolism in green sulfur bacteria, p. 60-76. In C. Dahl and C. G. Friedrich (ed.), Microbial sulfur metabolism. Springer, Berlin, Germany.
- 24.Frigaard, N.-U., and D. A. Bryant. 2008. Genomic insights into the sulfur metabolism of phototrophic green sulfur bacteria, p. 337-355. In R. Hell, C. Dahl, D. B. Knaff, and T. Leustek (ed.), Sulfur metabolism in phototrophic organisms. Springer, Dordrecht, The Netherlands.
- 25.Frigaard, N.-U., A. G. M. Chew, H. Li, J. A. Maresca, and D. A. Bryant. 2003. Chlorobium tepidum: insight into the structure, physiology, and metabolism of a green sulfur bacterium derived from the complete genome sequence. Photosynth. Res. 7893-117. [DOI] [PubMed] [Google Scholar]
- 26.Grimm, F., F. Bettina, and C. Dahl. 2008. Thiosulfate and sulfur oxidation in purple sulfur bacteria, p. 101-116. In C. Dahl and C. G. Friedrich (ed.), Microbial sulfur metabolism. Springer, Berlin, Germany.
- 27.Hauska, G., T. Schoedl, H. Remigy, and G. Tsiotis. 2001. The reaction center of green sulfur bacteria. Biophys. Biochim. Acta 1507260-277. [DOI] [PubMed] [Google Scholar]
- 28.Hensen, D., D. Sperling, H. G. Trüper, D. C. Brune, and C. Dahl. 2006. Thiosulphate oxidation in the phototrophic sulphur bacterium Allochromatium vinosum. Mol. Microbiol. 62794-810. [DOI] [PubMed] [Google Scholar]
- 29.Imhoff, J. F. 2003. Phylogenetic taxonomy of the family Chlorobiaceae on the basis of 16S rRNA and fmo (Fenna-Matthews-Olson protein) gene sequences. Int. J. Syst. Evol. Microbiol. 53941-951. [DOI] [PubMed] [Google Scholar]
- 30.Imhoff, J. F. 2008. Systematics of anoxygenic phototrophic bacteria, p. 269-287. In R. Hell, C. Dahl, D. B. Knaff, and T. Leustek (ed.), Sulfur metabolism in phototrophic organisms. Springer, Dordrecht, The Netherlands.
- 31.Itoh, M., D. Seo, H. Sakurai, and P. Sétif. 2002. Kinetics of electron transfer between soluble cytochrome c554 and purified reaction center complex from the green sulfur bacterium Chlorobium tepidum. Photosynth. Res. 71125-135. [DOI] [PubMed] [Google Scholar]
- 32.Kappler, U., K.-F. Aguey-Zinsou, G. R. Hanson, P. V. Bernhardt, and A. G. McEwan. 2004. Cytochrome c551 from Starkeya novella. Characterization, spectroscopic properties, and phylogeny of a diheme protein of the SoxAX family. J. Biol. Chem. 2796252-6260. [DOI] [PubMed] [Google Scholar]
- 33.Kappler, U., P. V. Bernhardt, J. Kilmartin, M. J. Riley, J. Teschner, K. J. McKenzie, and G. R. Hanson. SoxAX cytochromes: a new type of heme copper protein involved in bacterial energy generation from sulfur compounds. J. Biol. Chem., in press. [DOI] [PubMed]
- 34.Kappler, U., C. G. Friedrich, H. G. Trüper, and C. Dahl. 2001. Evidence for two pathways of thiosulfate oxidation in Starkeya novella (formerly Thiobacillus novellus). Arch. Microbiol. 175102-111. [DOI] [PubMed] [Google Scholar]
- 35.Kappler, U., G. R. Hanson, A. Jones, and A. G. McEwan. 2005. A recombinant diheme SoxAX cytochrome: implications for the relationship between EPR signals and modified heme-ligands. FEBS Lett. 5792491-2498. [DOI] [PubMed] [Google Scholar]
- 36.Kelly, D. P., J. K. Shergill, W.-P. Lu, and A. P. Wood. 1997. Oxidative metabolism of inorganic sulfur compounds by bacteria. Antonie van Leeuwenhoek 7195-107. [DOI] [PubMed] [Google Scholar]
- 37.Klarskov, K., F. Verté, G. V. Driessche, T. E. Meyer, M. A. Cusanovich, and J. V. Beeumen. 1998. The primary structure cytochrome c551 from the photosynthetic green sulfur bacterium Chlorobium limicola, strain Tassajara, reveals a novel c-type cytochrome. Biochemistry 3710555-10562. [DOI] [PubMed] [Google Scholar]
- 38.Kusai, A., and T. Yamanaka. 1973. A novel function of cytochrome c (555, Chlorobium thiosulfatophilum) in oxidation of thiosulphate. Biochem. Biophys. Res. Commun. 51107-112. [DOI] [PubMed] [Google Scholar]
- 39.Kusai, K., and T. Yamanaka. 1973. The oxidation mechanisms of thiosulphate and sulphide in Chlorobium thiosulphatophilum: roles of cytochrome c551 and cytochrome c553. Biochim. Biophys. Acta 325304-314. [DOI] [PubMed] [Google Scholar]
- 40.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227680-685. [DOI] [PubMed] [Google Scholar]
- 41.Lu, W.-P., B. E. P. Swoboda, and D. P. Kelly. 1985. Properties of the thiosulphate-oxidizing multi-enzyme system from Thiobacillus versutus. Biochim. Biophys. Acta 828116-122. [Google Scholar]
- 42.Lu, W.-P., and D. P. Kelly. 1983. Purification and some properties of two principal enzymes of the thiosulphate-oxidizing multi-enzyme system from Thiobacillus A2. J. Gen. Microbiol. 1293549-3564. [Google Scholar]
- 43.Meyer, B., J. F. Imhoff, and J. Kuever. 2007. Molecular analysis of the distribution and phylogeny of the soxB gene among sulfur-oxidizing bacteria: evolution of the Sox sulfur oxidation enzyme system. Environ. Microbiol. 92957-2977. [DOI] [PubMed] [Google Scholar]
- 44.Meyer, T. E., R. G. Bartsch, M. A. Cusanovich, and J. H. Mathewson. 1968. The cytochromes of Chlorobium thiosulfatophilum. Biochim. Biophys. Acta 153854-861. [DOI] [PubMed] [Google Scholar]
- 45.Meyer, T. E., and M. A. Cusanovich. 2003. Discovery and characterization of electron transfer proteins in the photosynthetic bacteria. Photosynth. Res. 76111-126. [DOI] [PubMed] [Google Scholar]
- 46.Petri, R., L. Podgotsek, and J. F. Imhoff. 2001. Phylogeny and distribution of the soxB gene among thiosulfate-oxidizing bacteria. FEMS Microbiol. Lett. 197171-178. [DOI] [PubMed] [Google Scholar]
- 47.Quentmeier, A., and C. G. Friedrich. 2001. The cysteine residue of the SoxY protein as the active site of protein-bound sulfur oxidation of Paracoccus pantotrophus GB17. FEBS Lett. 503168-172. [DOI] [PubMed] [Google Scholar]
- 48.Quentmeier, A., P. Hellwig, F. Bardischewsky, G. Grelle, R. Kraft, and C. G. Friedrich. 2003. Sulfur oxidation in Paracoccus pantotrophus: interaction of the sulfur-binding protein SoxYZ with the dimanganese SoxB protein. Biochem. Biophys. Res. Commun. 3121011-1018. [DOI] [PubMed] [Google Scholar]
- 49.Reijerse, E., M. Sommerhalter, P. Hellwig, A. Quentmeier, D. Rother, C. Laurich, E. Bothe, W. Lubitz, and C. G. Friedrich. 2007. The unusual redox centers of SoxXA, a novel c-type heme-enzyme essential for chemotrophic sulfur-oxidation of Paracoccus pantotrophus. Biochemistry 467804-7810. [DOI] [PubMed] [Google Scholar]
- 50.Rother, D., and C. G. Friedrich. 2002. The cytochrome complex SoxXA of Paracoccus pantotrophus is produced in Escherichia coli and functional in the reconstituted sulfur-oxidizing enzyme system. Biochim. Biophys. Acta 159865-73. [DOI] [PubMed] [Google Scholar]
- 51.Rother, D., H.-J. Henrich, A. Quentmeier, F. Bardischewsky, and C. G. Friedrich. 2001. Novel genes of the sox gene cluster, mutagenesis of the flavoprotein SoxF, and evidence for a general sulfur-oxidizing system in Paracoccus pantotrophus GB17. J. Bacteriol. 1834499-4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sakurai, H., N. Kusumoto, and K. Inoue. 1996. Function of the reaction center of green sulfur bacteria. Photochem. Photobiol. 645-13. [Google Scholar]
- 53.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 54.Sauvĕ, V., S. Bruno, B. C. Berks, and A. M. Hemmings. 2007. The SoxYZ complex carries sulfur cycle intermediates on a peptide swinging arm. J. Biol. Chem. 28223194-23204. [DOI] [PubMed] [Google Scholar]
- 55.Selvaraj, F., D. Devine, W. Zhou, D. C. Brune, M. T. Lince, and R. E. Blankenship. 1998. Purification and properties of cytochrome c553 from the green sulfur bacterium Chlorobium tepidum, p. 1593-1596. In G. Garab (ed.), Photosynthesis: mechanisms and effects, vol. III. Kluwer Academic Publishers, Dordrecht, The Netherlands. [Google Scholar]
- 56.Seo, D., A. Tomioka, N. Kusumoto, M. Kamo, I. Enami, and H. Sakurai. 2001. Purification of ferredoxins and their reaction with purified reaction center complex from the green sulfur bacterium Chlorobium tepidum. Biochim. Biophys. Acta 1503377-384. [DOI] [PubMed] [Google Scholar]
- 57.Shahak, Y., B. Arieli, E. Padan, and G. Hauska. 1992. Sulfide quinine reductase (SQR) activity in Chlorobium. FEBS Lett. 299127-130. [DOI] [PubMed] [Google Scholar]
- 58.Stout, J., L. D. Smet, B. Vergauwen, S. Savvides, and J. van Beeumen. 2008. Structural insights into component SoxY of the thiosulfate-oxidizing multienzyme system of Chlorobaculum thiosulfatophilim, p. 127-138. In C. Dahl and C. G. Friedrich (ed.), Microbial sulfur metabolism. Springer, Berlin, Germany.
- 59.Tsukatani, Y., R. Miyamoto, S. Itoh, and H. Oh-oka. 2006. Soluble cytochrome c554, CycA, is not essential for photosynthetic electron transfer in Chlorobium tepidum. FEBS Lett. 5802191-2194. [DOI] [PubMed] [Google Scholar]
- 60.Verté, F., V. Kostanjevecki, L. D. Smet, T. E. Meyer, M. A. Cusanovich, and J. J. Van Beeumen. 2002. Identification of a thiosulfate utilization gene cluster from the green phototrophic bacterium Chlorobium limicola. Biochemistry 412932-2945. [DOI] [PubMed] [Google Scholar]
- 61.Wahlund, T. M., C. R. Woese, R. W. Castenholz, and M. T. Madigan. 1991. A thermophilic green sulfur bacterium from New Zealand hot springs, Chlorobium tepidum sp. nov. Arch. Microbiol. 15681-90. [Google Scholar]
- 62.Yamanaka, T. 1996. Mechanisms of oxidation of inorganic electron donors in autotrophic bacteria. Plant Cell Physiol. 37569-574. [Google Scholar]