Abstract
The GerT protein of Bacillus cereus shares 74% amino acid identity with its homolog GerN. The latter is a Na+/H+-K+ antiporter that is required for normal spore germination in inosine. The germination properties of single and double mutants of B. cereus ATCC 10876 reveal that unlike GerN, which is required for all germination responses that involve the GerI germinant receptor, the GerT protein does not have a significant role in germination, although it is required for the residual GerI-mediated inosine germination response of a gerN mutant. In contrast, GerT has a significant role in outgrowth; gerT mutant spores do not outgrow efficiently under alkaline conditions and outgrow more slowly than the wild type in the presence of high NaCl concentrations. The GerT protein in B. cereus therefore contributes to the success of spore outgrowth from the germinated state during alkaline or Na+ stress.
Bacillus cereus ATCC 10876 spores germinate in inosine or in l-alanine as sole germinants (5). They require both GerI and GerQ germinant receptors for germination in inosine as the sole germinant, whereas the GerL receptor is responsible for most of the response to l-alanine as the sole germinant, with a smaller contribution from the GerI receptor (3). The GerN protein is needed for successful inosine germination, as spores of a gerN mutant of B. cereus ATCC 10876 demonstrate a slow and abnormally ordered germination response in inosine as the sole germinant (24). The residual germination was very asynchronous, and some spores appeared to become phase dark before losing heat resistance (24). This GerN protein is a homolog of a widely distributed family of cation transporters (9) and has been demonstrated to mediate Na+/H+ and Na+/H+-K+ antiport activity when expressed at a low level in Escherichia coli (21). Such GerN-mediated antiport activity is therefore apparently required for the germination response mediated by the GerI germinant receptor in B. cereus. The role of such an ion transport protein in germination remains unclear, especially as other germinant receptors in the same strain do not require a functional GerN protein; for example, the gerL-dependent alanine response is almost identical to that of the wild type in a gerN mutant (24). Both GerN and a closely related homolog are encoded in B. cereus ATCC 14579 (13) and in other members of the family, including Bacillus anthracis (17) and Bacillus thuringiensis (16). Both genes appear to be monocistronic, and their coding sequences are widely separated on the genome. We have called this second gerN-like gene gerT (19). An entirely different (spore coat) protein in B. subtilis has recently been called gerT (7), but that gene does not have a homolog in B. cereus, and as B subtilis does not carry an equivalent homolog of the B. cereus gerN or gerT gene, the nomenclature will not overlap. We describe here the spore germination and outgrowth phenotypes of mutants of B. cereus ATCC 10876 carrying an insertionally inactivated gerT gene. Resulting phenotypes suggest that the GerN and GerT proteins have distinct roles in germination and outgrowth.
MATERIALS AND METHODS
Bacterial growth, media, and strains.
Escherichia coli cloning strain TG1 and B. cereus ATCC 10876-derived strains were routinely cultured in L broth (Difco Bacto tryptone at 10 g liter−1, Difco yeast extract at 10 g liter−1, and NaCl at 5 g liter−1, pH 7.2) or on L agar containing the appropriate antibiotics (for E. coli, ampicillin at 50 μg ml−1 and chloramphenicol at 30 μg ml−1; for B. cereus, erythromycin and lincomycin at 1 and 25 μg ml−1, respectively, and kanamycin at 50 μg ml−1). Spores of B. cereus were prepared in CCY medium, harvested, and washed at least 10 times in water, as described previously (5). Oxoid nutrient broth (NB; pH 7.4) was used for outgrowth experiments.
The B. cereus strains are listed in Table 1. The E. coli K-12 strains used for attempts at complementation of ion transport defects were the Na+/H+ antiporter-deficient strain KNabc (chaA nhaA nhaB) (15) and the potassium uptake-deficient strain TK2420 (ΔkdpABC trkD1 ΔtrkA) (11), as already used in the study of gerN (21). Strain KNabc is defective in sodium efflux and consequently is unable to grow at high concentrations of Na+ (>75 mM) but will grow in a modified low-sodium medium, LBK (11), which contains 10 mM NaCl and 50 mM KCl. To test for Na+ sensitivity, colonies of KNabc/pAS1 and KNabc/pGEM3zf+ were grown for 24 h at 37°C in 5 ml of LBK plus chloramphenicol, and the culture was then diluted 250-fold into LBK, in which the NaCl concentration was varied, and grown for 16 h at 37°C.
TABLE 1.
Bacillus cereus strains used
| Strain | Relevant genotype/antibiotic resistance marker(s) | Source or reference |
|---|---|---|
| ATCC10876 UM20.1 | trp-1 Strr | 3 |
| AM1314 | Tn917-LTV1::gerIA5 Eryrtrp-1 Strr | 3 |
| AM1311 | Tn917-LTV1::gerQA2Eryrtrp-1 Strr | 3 |
| AM1316 | Tn917-LTV1::gerLA1 Eryrtrp-1 Strr | 3 |
| AM1421 | gerN17::pMUTIN4 Eryrtrp-1 Strr | 23 |
| AM1631 | gerT::pSMUT Kanrtrp-1 Strr | This work |
| AM1632 | gerN17::pMUTIN4 gerT::pSMUT Kanr Eryrtrp-1 Strr | This work |
Cloning and sequencing of gerT from B. cereus ATCC 10876.
PCR primers based on the B. anthracis genome sequence data around BA0819 (gerT) were used to amplify the regions flanking the gerT gene of B. cereus, using high-fidelity Taq Extend (Boehringer Mannheim). The resulting B. cereus-derived PCR products were sequenced, and primers were designed for cloning B. cereus gerT without its promoter region, as a 1.1-kb fragment. These were APS11 (CTGGTCGACTAAAGGAGGAGCAGATGCTAT) and APS12 (TTCGAGCTCCTATCTATACAAAATATTTC), incorporating (underlined) SalI and SacI sites, respectively, at their ends. Following restriction enzyme digestion, the PCR product was ligated into SalI- and SacI-digested pGEM3zf+, with gerT in the reverse orientation relative to the vector's lac promoter but downstream of the T7 promoter. E. coli TG1 (a strain lacking T7 polymerase) was transformed, yielding plasmid pAS1 for sequencing and complementation studies.
The monocistronic gerT locus encodes a 375-amino-acid GerT protein, with 99% amino acid identity to the equivalent protein in B. anthracis. A potential ribosome binding site is appropriately located upstream of the putative gerT start codon, and the gerT stop codon is followed by a potential rho-independent terminator.
Construction of gerT-null mutants of B. cereus.
Plasmid vector pSMUT lacks an origin of replication for B. cereus but carries a ColE1 replicon and a β-lactamase gene for amplification and selection, respectively, in E. coli. It is a derivative of pMUTIN4 (25), in which the lacZ gene has been removed and the erythromycin/lincomycin resistance gene replaced by a kanamycin resistance cassette.
Primers APS7 (CGTGAATTCGGTAAGTTAATTGTTGGTTA) and APS8 (AACGGATCCCAATAACAATCGGCTGTGAA) were used to PCR amplify a 1.0-kb internal fragment of gerT from 108 bp downstream of the start of the gerT open reading frame to 76 bp before the stop codon of gerT. This PCR fragment was digested with (underlined) EcoRI and BamHI, respectively, and ligated with EcoRI- and BamHI-digested pSMUT, yielding plasmid pAS3. B. cereus was electroporated with 2.5 μg of pAS3 DNA, and transformants were selected on nutrient agar containing kanamycin. Integration into the chromosome by a single crossover within the region of homology interrupts the gerT gene so that it encodes a protein truncated by 25 amino acids from the C terminus. The disruption of the gerT gene and expected novel junction fragments were confirmed by PCR in a transformant named AM1631 (gerT1::pSMUT).
Construction of a gerN gerT double mutant.
The gerN mutant used in this study was strain AM1421 (gerN17::pMUTIN4), in which the gerN gene was insertionally inactivated by integration of plasmid pMNAP, derived from pMUTIN4 by cloning of an internal fragment of the gerN gene, spanning bases 365 to 813 of the open reading frame; integration of this plasmid results in C-terminal truncation of GerN by 120 amino acids. The inactivated gene was checked by Southern blotting (23). This gerN17::pMUTIN4 mutation was introduced into strain AM1631 (gerT1::pSMUT) by generalized transduction with CP51ts, as described previously (5). Transductants were selected for resistance to erythromycin and lincomycin and screened for retention of the kanamycin resistance marker. Colony PCR confirmed the presence of the mutations in both genes. One transductant was retained and named AM1632 (gerN17 gerT1).
Germination experiments.
Spore germination used washed spores, heat activated for 30 min at 70°C in H2O, and conditions were as described previously (3), unless otherwise stated. Optical density at 490 nm (OD490) was measured at intervals on a Wallac Victor plate reader. For amino acid-enhanced l-alanine germination, germination was initiated by a combination of a subgerminal concentration of l-alanine (20 μM) plus histidine, proline, or tryptophan at 10 mM or tyrosine at 1 mM. The buffer conditions were those optimal for l-alanine as the sole germinant: Tris-HCl (10 mM), pH 8.9, with NH4Cl (50 mM), and germination at 30°C. Spores were preincubated in the alanine racemase inhibitor O-carbamyl-d-serine (5 μg/ml) in germination buffer for 5 min before addition of germinants to prevent any conversion of l-alanine to its competitive inhibitor d-alanine. For amino acid-enhanced inosine germination, the germination buffer was Tris-HCl (10 mM), pH 8.0, NaCl (10 mM), and spores were germinated at 37°C; inosine was used in this experiment at a just-subgerminal concentration (40 μM), and the same amino acid adjuncts were added as described above.
Nucleotide sequence accession number.
The fully overlapped sequence of the gerT region has been submitted to GenBank (accession number EU789572).
RESULTS
GerT complements the Na+ sensitivity of an E. coli mutant.
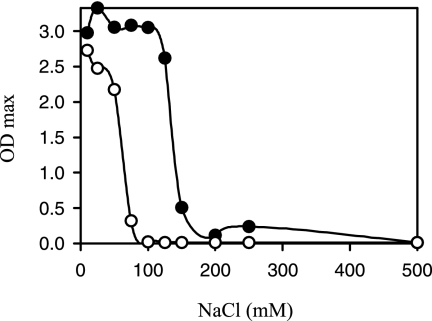
Cloning the gerN gene into E. coli strains with deficient Na+/H+ antiporter activity and K+ uptake (KNabc and TK2420, respectively) was reported to complement their respective compromised phenotypes (21). A similar test was carried out for the homologous gerT gene (Fig. 1). Introduction of the vector plasmid pGEM3zf+ did not improve the growth of the KNabc strain, which was unable to grow in concentrations of NaCl over 75 mM; in contrast, KNabc/pAS1, containing a gerT gene cloned without an efficient promoter upstream, as had been done for gerN previously, was able to grow in NaCl concentrations up to 150 mM. These data suggest that GerT provides some additional capacity for Na+ efflux. Attempts to introduce pAS1 into the K+ transport-deficient E. coli strain TK2420 (10) were unsuccessful, so no predictions can be made about the potential for K+ transport by the GerT protein.
FIG. 1.
Effect of different Na+ concentrations on the growth of E. coli strain KNabc upon introduction of the gerT gene of B. cereus. The ODs of cultures were measured after 16 h of growth at 37°C. The precise growth conditions are described in Materials and Methods. Symbols: ○, KNabc/pGEM3zf+, a strain containing the plasmid vector only; •, KNabc/pAS1, containing the gerT gene cloned in pGEM3zf+.
Spore germination in single and double mutants in response to single germinants.
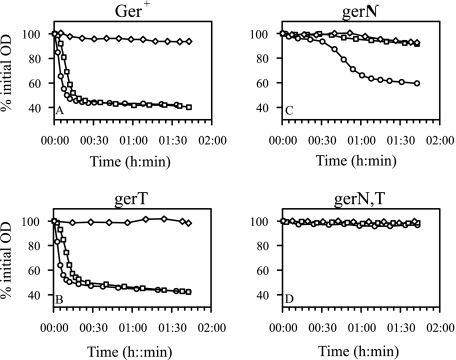
Water-washed, heat-activated spores of mutants carrying insertionally inactivated gerN and gerT genes, individually and in combination, were used in germination assays (Fig. 2). Germination was measured as the fall in OD of spore suspensions after exposure to inosine. Spores of both the wild type and the gerT mutant germinated rapidly at 0.1 mM and 1 mM inosine. As previously reported (24), the gerN mutant spores have a severe germination defect in inosine but still show some residual response in high (1 mM) inosine concentrations; this residual response is dependent on GerT, as it is absent in a gerN gerT double mutant. GerT is therefore not required for germination in inosine, provided that GerN protein is available, but it can provide a partially functional substitute for GerN if the GerN is missing from the spore. Germination in l-alanine under optimal conditions in the mutants was essentially identical to that in the wild type (data not shown); under these conditions, the receptor involved is GerL (3); therefore, neither GerN nor GerT is required for germination involving the GerL receptor.
FIG. 2.
Effect of gerN and gerT mutations on the germination response to inosine as the sole germinant, measured by fall in OD of spore suspensions. Spore suspensions were incubated in 10 mM Tris-HCl, pH 8.0, 10 mM NaCl at 37°C over a range of inosine concentrations. (A) Strain B. cereus ATCC 10876 (ger+); (B) AM1631 (gerT1::pSMUT); (C) AM1421 (gerN17::pMUTIN4); (D) AM1632 (gerN17::pMUTIN4 gerT1::pSMUT). Symbols represent different inosine concentrations as follows: ⋄, no inosine; □, 100 μM; ○, 1 mM. The data for 10 μM inosine are superimposed on the “no inosine” line.
If the GerI receptor is required for a response to combinations of germinants, GerN is also required.
Although most members of the B. cereus family germinate in l-alanine and inosine, either individually or in combination, the range of germinant receptors encoded by different strains is variable, as is consequently the detailed germination behavior. Unlike B. cereus ATCC 14579 (12), which has a slightly different complement of germinant receptors, B. cereus ATCC 10876 germinates in l-alanine plus aromatic amino acids in a fashion generally similar to that for the so-called AEA, or aromatic-enhanced alanine, response of B. anthracis (8). In ATCC 10876, a subgerminal concentration of l-alanine (20 μM) is effective in combination with tryptophan or tyrosine, although there is no response with histidine or proline (Fig. 3A). Spores of gerI, gerL, or gerN mutants all fail to germinate in the alanine-plus-tryptophan or tyrosine combinations, but gerQ and gerT mutants germinate like the wild type (reference 19 and data not shown). These data demonstrate a requirement for both GerL and GerI germinant receptors, consistent with previous observations for the equivalent response in B anthracis Sterne (8). The third receptor required in B. anthracis, GerS, is also encoded in B. cereus ATCC 10876 (2) but has not been mutated to test its function. Another germinant combination effective in B. anthracis is inosine plus amino acids. In B. cereus ATCC 10876, subgerminal (40 μM) inosine with either tryptophan or histidine proves an effective germinant combination (Fig. 3B). As is the case for inosine as the sole germinant, both GerI and GerQ receptors and GerN are required here, but there is no requirement for the GerL alanine receptor or for GerT (Fig. 3C). These data altogether suggest that in every case when the GerI receptor is required, so also is GerN; the GerT protein does not compensate functionally for GerN in these circumstances.
FIG. 3.
Germination in subgerminative concentrations of alanine or inosine in combination with amino acid adjuncts. (A) Germination of spores of B. cereus ATCC 10876, the Ger+ parental strain, in l-alanine (20 μM) plus other amino acids. Symbols: □, no addition; ○, 10 mM histidine; ▵, 10 mM proline; ▴, 1 mM tyrosine; •, 10 mM tryptophan. (B) Germination of spores of B. cereus ATCC 10876, the Ger+ parental strain, in 40 μM inosine plus amino acids. Symbols are as in panel A: □, no addition; ○, 10 mM histidine; ▵, 10 mM proline; ▴, 1 mM tyrosine; •, 10 mM tryptophan. (C) Germination of mutants in inosine (40 μM) plus tryptophan (10 mM). Symbols: ○, B. cereus ATCC 10876, the Ger+ parental strain; ⋄, gerL; ▴, gerT. These three graphs are superimposed and demonstrate complete germination. The following graphs showed little or no germination and are largely superimposed: •, gerQ; ▵, gerI; □, gerN.
GerT has a role in outgrowth in high salt concentrations and at alkaline pH.
As gerN and gerT mutants germinate normally in l-alanine, it is possible to test for a role for these genes in spore outgrowth. Spores were germinated in l-alanine in Tris-HCl buffer, harvested by centrifugation, and resuspended in NB to allow outgrowth. Spores of the parent strain and the gerN and gerT mutants all outgrew at similar rates in NB (Fig. 4B). In NB adjusted to pH 9.5 with NaOH, the wild type and gerN mutant spores outgrew at the same rate after lags of ca. 30 and 60 min, respectively, but a clear defect was observed for the gerT mutant spores, which did not outgrow significantly within the period of the experiment. The gerT mutation also reduced the ability of spores to outgrow in NB at pH 7.4 in the presence of additional 0.7 M NaCl (Fig. 4C); there was significant cell lysis and no outgrowth above the initial OD within 2.5 h. Unlike the situation in outgrowth, vegetative cells of these mutants all grew at the same rate as the wild type in NB at pH 9.5 (Fig. 4D). Sensitivity to alkaline pH in the gerT mutant is therefore limited to the outgrowing state. No difference was seen in the behavior of mutant and wild-type cells during vegetative growth following salt stress (data not shown). These data suggest that during vegetative growth, but not during outgrowth, alternative sodium efflux systems are likely to be present at a sufficient level for dealing with these environmental stresses, although the nature of such Na+ transport systems in the B. cereus family has not yet been explored.
FIG. 4.
Effect of gerT and gerN mutations on spore outgrowth and on vegetative growth. Spores were germinated in l-alanine and then resuspended in fresh medium for outgrowth. •, wild type; □, gerN; ○, gerT. (A) Outgrowth in NB adjusted to pH 9.5. (B) Outgrowth in NB. (C) Outgrowth in NB plus 0.7 M NaCl. (D) Vegetative growth in NB, pH 9.5.
DISCUSSION
Based on the phenotypes of B. cereus mutants, the related proteins GerN and GerT, both of which are likely to have Na+/H+ antiport activity, have primary roles in germination and outgrowth, respectively, and as neither protein is required for NaCl resistance in vegetative cells, their role appears to be mainly in spore biology. There is some evidence that both gerN and gerT genes are expressed during sporulation; the gerN gene of B cereus is specifically sporulation expressed (20) at 4 hours after initiation of sporulation at a very low level; according to lacZ fusion data, its expression level is approximately 1/3 the level seen for the gerI operon and 1/100 that of exsA, which encodes a spore coat morphogenetic protein (1). A lacZ fusion to the gerT gene of B. cereus is not available, but the gerT gene of B. anthracis (BA 0819) has been reported in microarray data as late-sporulation expressed (14).
Role of GerT.
Both GerN and GerT are members of the CPA2 monovalent cation:proton antiporter family of membrane transport proteins (18) and are homologs of the NapA family of proteins. The GerN protein behaves as a Na+/H+-K+ antiporter and is essential for normal GerI receptor function, possibly in the restoration of local ion balance (21). As yet, the transport capabilities of GerT have not been directly measured, but the NaCl and alkali sensitivity of the B. cereus gerT mutant during spore outgrowth and the ability of the cloned gerT gene to improve the growth of a Na+ transport-defective E. coli mutant in complementation studies both suggest a role in Na+ efflux. As described above, there is evidence for gerT expression during late sporulation, and GerT must be present in dormant spores, as it is responsible for the phenotypic difference between gerN spores and gerN gerT double mutant spores in germination at a stage before de novo protein synthesis. Whether preexisting GerT protein in the germinated spore is sufficient for outgrowth or whether in normal cells gerT is expressed again during outgrowth is not known.
The GerN and -T proteins are significantly different in sequence and separately encoded in the genome. Consequently, the different roles of GerN and GerT proteins in germination could reflect differences in their ion transport activity or specificity, association with other proteins, or relative levels of protein. There are other reports that precise functions can vary between homologs in the NapA family of transport proteins (27).
The report that led to our initial study of GerN in B. cereus was that a related protein, GrmA, is required for germination in Bacillus megaterium ATCC 12872 (22). A recent report demonstrates that, in contrast, GrmA is not required for germination in the apparently equivalent B. megaterium strain QMB1551 (4). Caution needs to be exercised when different strains of a species are considered, as genomic differences in the spectrum of encoded and functional germinant receptors may be reflected in different germination properties; for example, the germination behaviors of B. cereus strains ATCC 14579 and ATCC 10876 show significant differences (12). It is not obvious why, even within a single species, one germinant receptor (GerI) might be strictly dependent on an ion antiporter like GerN, while other receptors are not. The analysis of GerT has not clarified this issue but has highlighted a second role for proteins of this family, this time in outgrowth.
Potential significance of GerT and GerN in the biology of the B. cereus family.
Considering that it is a naive cell, encountering a new environment, the newly germinated spore will have to deal with environmental stress, for example, by salt or alkali, and it appears that the GerT protein in the B. cereus family is one protein that has been recruited to this role. For example, when spores germinate in the alkaline midgut of insects or germinate in alkaline or saline soils, the GerT protein may be an important factor contributing to the resumption of growth.
In an attempt to screen for potential germination defects in gerT mutant spores, we studied amino acid-enhanced germination in subgerminal alanine and inosine. This did not reveal any GerT function but demonstrated that GerN is important in any circumstances where the GerI receptor is implicated in germination. In B. anthracis, the GerH germinant receptor has been implicated in virulence (26). Apart from sequence differences in the long repeat region near the N termini of GerHA and GerIA, the GerH proteins are almost identical in amino acid sequence to the GerI proteins of B. cereus ATCC 10876. Unlike B. cereus, B. anthracis does not germinate in inosine as the sole germinant; the absence of gerQ in the B. anthracis genome may explain this failure, so one does not need to invoke a significant difference in function between the GerI and GerH receptors. We predict, therefore, that GerN is likely to be required in B. anthracis for GerH receptor function. This is supported by recent evidence (6) that gerH, gerN, and gerT mutants were all enriched in large-scale screens designed to detect defects in sporulation, germination, or outgrowth of B. anthracis.
Acknowledgments
This work was funded by the University of Sheffield through a University Research Studentship to A.S.
We thank Terry Krulwich and Arthur Guffanti for the gift of strains and plasmid vectors.
Footnotes
Published ahead of print on 18 July 2008.
REFERENCES
- 1.Bailey-Smith, K., S. J. Todd, T. W. Southworth, J. Proctor, and A. Moir. 2005. The ExsA protein of Bacillus cereus is required for assembly of coat and exosporium onto the spore surface. J. Bacteriol. 1873800-3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barlass, P. J. 1998. Characterisation of the L-alanine spore germination response of Bacillus cereus. Ph.D. thesis. University of Sheffield, Sheffield, United Kingdom.
- 3.Barlass, P. J., C. W. Houston, M. O. Clements, and A. Moir. 2002. Germination of Bacillus cereus spores in response to L-alanine and to inosine: the roles of gerL and gerQ operons. Microbiology 1482089-2095. [DOI] [PubMed] [Google Scholar]
- 4.Christie, G., and C. R. Lowe. 2007. Role of chromosomal and plasmid-borne receptor homologues in the response of Bacillus megaterium QM B1551 spores to germinants. J. Bacteriol. 1894375-4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clements, M. O., and A. Moir. 1998. Role of the gerI operon of Bacillus cereus 569 in the response of spores to germinants. J. Bacteriol. 1806729-6735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Day, W. A., S. L. Rasmussen, B. M. Carpenter, S. N. Peterson, and A. M. Friedlander. 2007. Microarray analysis of transposon insertion mutations in Bacillus anthracis: global identification of genes required for sporulation and germination. J. Bacteriol. 1893296-3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferguson, C. C., A. H. Camp, and R. Losick. 2007. gerT, a newly discovered germination gene under the control of the sporulation transcription factor σK in Bacillus subtilis. J. Bacteriol. 1897681-7689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fisher, N., and P. Hanna. 2005. Characterization of Bacillus anthracis germinant receptors in vitro. J. Bacteriol. 1878055-8062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fujisawa, M., M. Ito, and T. A. Krulwich. 2007. Three two-component transporters with channel-like properties have monovalent cation/proton antiport activity. Proc. Natl. Acad. Sci. USA 10413289-13294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goldberg, E. B., T. Arbel, J. Chen, R. Karpel, G. A. Mackie, S. Schuldiner, and E. Padan. 1987. Characterization of a Na+/H+ antiporter gene of Escherichia coli. Proc. Natl. Acad. Sci. USA 842615-2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guffanti, A. A., J. B. Cheng, and T. A. Krulwich. 1998. Electrogenic antiport activities of the gram-positive tet proteins include a Na+(K+)/K+ mode that mediates net K+ uptake. J. Biol. Chem. 27326447-26454. [DOI] [PubMed] [Google Scholar]
- 12.Hornstra, L. M., Y. P. de Vries, M. H. J. Wells-Bennik, W. M. de Vos, and T. Abee. 2006. Characterization of germination receptors of Bacillus cereus ATCC 14579. Appl. Environ. Microbiol. 7244-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ivanova, N., A. Sorokin, I. Anderson, N. Galleron, B. Candelon, V. Kapatral, A. Bhattacharyya, G. Reznik, N. Mikhailova, A. Lapidus, L. Chu, M. Mazur, E. Goltsman, N. Larsen, M. D'Souza, T. Walunas, Y. Grechkin, G. Pusch, R. Haselkorn, M. Fonstein, S. D. Ehrlich, R. Overbeek, and N. Kyrpides. 2003. Genome sequence of Bacillus cereus and comparative analysis with Bacillus anthracis. Nature 42387-91. [DOI] [PubMed] [Google Scholar]
- 14.Liu, H. B., N. H. Bergman, B. Thomason, S. Shallom, A. Hazen, J. Crossno, D. A. Rasko, J. Ravel, T. D. Read, S. N. Peterson, J. Yates, and P. C. Hanna. 2004. Formation and composition of the Bacillus anthracis endospore. J. Bacteriol. 186164-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nozaki, K., K. Inaba, T. Kuroda, M. Tsuda, and T. Tsuchiya. 1996. Cloning and sequencing of the gene for Na+/H+ antiporter of Vibrio parahaemolyticus. Biochem. Biophys. Res. Commun. 222774-779. [DOI] [PubMed] [Google Scholar]
- 16.Radnedge, L., P. G. Agron, K. K. Hill, P. J. Jackson, L. O. Ticknor, P. Keim, and G. L. Andersen. 2003. Genome differences that distinguish Bacillus anthracis from Bacillus cereus and Bacillus thuringiensis. Appl. Environ. Microbiol. 692755-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Read, T. D., S. N. Peterson, N. Tourasse, L. W. Baillie, I. T. Paulsen, K. E. Nelson, H. Tettelin, D. E. Fouts, J. A. Eisen, S. R. Gill, E. K. Holtzapple, O. A. Okstad, E. Helgason, J. Rilstone, M. Wu, J. F. Kolonay, M. J. Beanan, R. J. Dodson, L. M. Brinkac, M. Gwinn, R. T. DeBoy, R. Madpu, S. C. Daugherty, A. S. Durkin, D. H. Haft, W. C. Nelson, J. D. Peterson, M. Pop, H. M. Khouri, D. Radune, J. L. Benton, Y. Mahamoud, L. X. Jiang, I. R. Hance, J. F. Weidman, K. J. Berry, R. D. Plaut, A. M. Wolf, K. L. Watkins, W. C. Nierman, A. Hazen, R. Cline, C. Redmond, J. E. Thwaite, O. White, S. L. Salzberg, B. Thomason, A. M. Friedlander, T. M. Koehler, P. C. Hanna, A. B. Kolsto, and C. M. Fraser. 2003. The genome sequence of Bacillus anthracis Ames and comparison to closely related bacteria. Nature 42381-86. [DOI] [PubMed] [Google Scholar]
- 18.Saier, M. H., B. H. Eng, S. Fard, J. Garg, D. A. Haggerty, W. J. Hutchinson, D. L. Jack, E. C. Lai, H. J. Liu, D. P. Nusinew, A. M. Omar, S. S. Pao, I. T. Paulsen, J. A. Quan, M. Sliwinski, T. T. Tseng, S. Wachi, and G. B. Young. 1999. Phylogenetic characterization of novel transport protein families revealed by genome analyses. Biochim. Biophys. Acta 14221-56. [DOI] [PubMed] [Google Scholar]
- 19.Senior, A. P. 2005. GerT, an ion transporter homologue in Bacillus cereus, and its role in spore germination. Ph.D. thesis, University of Sheffield, Sheffield, United Kingdom.
- 20.Southworth, T. W. 2004. The GerN ion antiporter of Bacillus cereus and its contribution to spore germination. Ph.D. thesis. University of Sheffield, Sheffield, United Kingdom.
- 21.Southworth, T. W., A. A. Guffanti, A. Moir, and T. A. Krulwich. 2001. GerN, an endospore germination protein of Bacillus cereus, is an Na+/H+-K+ antiporter. J. Bacteriol. 1835896-5903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tani, K., T. Watanabe, H. Matsuda, M. Nasu, and M. Kondo. 1996. Cloning and sequencing of the spore germination gene of Bacillus megaterium ATCC 12872: similarities to the NaH- antiporter gene of Enterococcus hirae. Microbiol. Immunol. 4099-105. [DOI] [PubMed] [Google Scholar]
- 23.Thackray, P. D. 1999. The molecular genetics of inosine-dependent germination of Bacillus cereus spores. Ph.D. thesis. University of Sheffield, Sheffield, United Kingdom.
- 24.Thackray, P. D., J. Behravan, T. W. Southworth, and A. Moir. 2001. GerN, an antiporter homologue important in germination of Bacillus cereus endospores. J. Bacteriol. 183476-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vagner, V., E. Dervyn, and S. D. Ehrlich. 1998. A vector for systematic gene inactivation in Bacillus subtilis. Microbiology 1443097-3104. [DOI] [PubMed] [Google Scholar]
- 26.Weiner, M. A., and P. C. Hanna. 2003. Macrophage-mediated germination of Bacillus anthracis endospores requires the gerH operon. Infect. Immun. 713954-3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wutipraditkul, N., R. Waditee, A. Incharoensakdi, T. Hibino, Y. Tanaka, T. Nakamura, M. Shikata, and T. Takabe. 2005. Halotolerant cyanobacterium Aphanothece halophytica contains NapA-type Na+/H+ antiporters with novel ion specificity that are involved in salt tolerance at alkaline pH. Appl. Environ. Microbiol. 714176-4184. [DOI] [PMC free article] [PubMed] [Google Scholar]