Abstract
As obligate intracellular, vector-borne bacteria, rickettsiae must adapt to both mammalian and arthropod host cell environments. Deciphering the molecular mechanisms of the interactions between rickettsiae and their host cells has largely been hindered by the genetic intractability of these organisms; however, research in other gram-negative pathogens has demonstrated that many bacterial determinants of attachment, entry, and pathogenesis are extracytoplasmic proteins. The annotations of several rickettsial genomes indicate the presence of homologs of the Sec translocon, the major route for bacterial protein secretion from the cytoplasm. For Rickettsia typhi, the etiologic agent of murine typhus, homologs of the Sec-translocon-associated proteins LepB, SecA, and LspA have been functionally characterized; therefore, the R. typhi Sec apparatus represents a mechanism for the secretion of rickettsial proteins, including virulence factors, into the extracytoplasmic environment. Our objective was to characterize such Sec-dependent R. typhi proteins in the context of a mammalian host cell infection. By using the web-based programs LipoP, SignalP, and Phobius, a total of 191 R. typhi proteins were predicted to contain signal peptides targeting them to the Sec translocon. Of these putative signal peptides, 102 were tested in an Escherichia coli-based alkaline phosphatase (PhoA) gene fusion system. Eighty-four of these candidates exhibited signal peptide activity in E. coli, and transcriptional analysis indicated that at least 54 of the R. typhi extracytoplasmic proteins undergo active gene expression during infections of HeLa cells. This work highlights a number of interesting proteins possibly involved in rickettsial growth and virulence in mammalian cells.
Epidemic typhus, murine typhus, and Rocky Mountain and other spotted fever rickettsioses are some of the devastating human diseases caused by pathogenic rickettsiae. Yet, despite more than a century of research, rickettsial virulence factors have not been precisely defined. The genetic intractability of these obligate intracellular, arthropod-borne bacteria has been a fundamental impediment in defining the critical mechanisms behind rickettsial pathogenesis. However, the use of genomic information and heterologous experimental systems have opened new platforms for rickettsial research and have been responsible for most of the current knowledge regarding the functional roles of rickettsial proteins, including putative virulence factors.
Although the mechanisms of rickettsial attachment, entry, phagosome escape, and infection-induced cellular injury are poorly understood, research in Escherichia coli and other gram-negative pathogens has demonstrated that a majority of the bacterial determinants involved in these processes are surface and secreted proteins, many of which are exported out of the bacterial cytoplasm through the Sec secretion system (1, 9, 15, 38, 41, 42). In E. coli, the Sec apparatus maintains a protein translocation channel in the bacterial inner membrane. The core of this channel is formed by the integral membrane proteins SecY, SecE, and SecG, and the Sec complex is completed by at least three other membrane proteins (SecD, SecF, and YajC) and a peripherally associated ATPase (SecA) (13, 38). Bacterial proteins secreted via the Sec system are synthesized in the cytoplasm as protein precursors containing an amino-terminal signal peptide. Molecular chaperones direct these unfolded preproteins to the SecYEG channel, and after translocation into the periplasm, the signal peptide is cleaved by the appropriate membrane-bound signal peptidase: signal peptidase I (LepB) for nonlipoproteins and signal peptidase II (LspA) for lipoproteins (37). The translocated protein then folds into its mature conformation and either remains in the periplasm or is further translocated through the outer membrane into the extracellular environment as a membrane-bound or free protein (37).
Genomic analyses indicate that the rickettsiae contain all of the functional components of the Sec system (12). For Rickettsia typhi, the causative agent of murine typhus, Dreher-Lesnick et al. reported the transcription of secA-B, secD-G, secY, lspA, and lepB during in vitro infections in mammalian (L929) cells at both 25°C and 37°C (10). In addition, Rahman et al. found that the R. typhi homologs of the two signal peptidases lepB and lspA, as well as the secA-encoded ATPase, were able to complement E. coli mutants lacking each of these genes (32-34). Therefore, based on these results and the known importance of the Sec translocon for other gram-negative pathogens, we hypothesized that the Sec system represents a mechanism by which R. typhi proteins, including potential virulence factors, are secreted into the extracytoplasmic environment. The objective of this study was to identify and characterize such Sec-dependent R. typhi extracytoplasmic proteins in the context of a mammalian in vitro model of infection.
Because the N-terminal signal sequences of Sec-dependent preproteins have a distinct tripartite amino acid composition (31), various computer programs (e.g., LipoP, SignalP, and Phobius) have been designed to identify proteins containing these signal peptides. In silico prediction of signal peptides combined with the laboratory-based E. coli alkaline phosphatase (PhoA) assay have been used to identify bacterial signal-peptide-containing extracytoplasmic proteins (20). In particular, this assay has been used to successfully identify extracytoplasmic proteins in several bacteria, including Pseudomonas aeruginosa (20), Helicobacter pylori (5), Bacillus subtilis (28), Actinobacillus actinomycetemcomitans (46), and the intracellular bacterium Mycobacterium tuberculosis (49). In this communication, we report the use of web-based algorithms to screen the entire R. typhi strain Wilmington genome for open reading frames (ORFs) predicted to encode proteins containing signal peptides. The putative signal peptides identified were then tested using the E. coli-based PhoA assay. Quantitative, real-time reverse transcription PCR (RT-qPCR) was then used to measure the gene expression profiles of those R. typhi extracytoplasmic proteins (recognized through the PhoA fusion assay) during an in vitro infection in HeLa cells to identify proteins with potential roles in infection and virulence in the mammalian host.
MATERIALS AND METHODS
In silico analysis of R. typhi signal peptides.
Amino acid sequences for all predicted ORFs of the Rickettsia typhi strain Wilmington genome (GenBank accession no. AE017197) (23) were analyzed for the presence of an N-terminal signal sequence by using the following programs: SignalP version 3.0 with hidden Markov model (HMM) and neural network (NN) algorithms (4), LipoP version 1.0 (17), and Phobius (18). All R. typhi protein annotations referred to in this work are based on those designated by McLeod et al. (23).
Growth, maintenance, and isolation of R. typhi.
All work with live rickettsiae was performed in a biosafety level 3 facility. R. typhi strain Wilmington organisms were propagated in low-subculture (<25 passages) HeLa cells (ATCC number CCL-2), maintained in Dulbecco's modification of Eagle's medium supplemented with 5% heat-inactivated fetal bovine serum and kept at 34°C with 5% CO2. When the desired level of R. typhi infection was reached, the rickettsiae were partially purified from the host cells. The infected HeLa cells were lysed by sonication, using two 30-s pulses at setting 6.5 with a Fisher Scientific model 100 sonic dismembrator. After sonication, the suspension was spun at 1,000 × g at 4°C for 5 min to pellet the host cell debris. The rickettsiae-containing supernatant was then spun at 17,000 × g at 4°C for 10 min to pellet the R. typhi. The rickettsial pellet was washed one to three times in 1 ml ice-cold sucrose-phosphate-glutamate buffer (SPG: 218 mM sucrose, 3.76 mM potassium phosphate monobasic, 7.1 mM potassium phosphate dibasic, 4.9 mM potassium glutamate). The viable R. typhi organisms were then quantified using a LIVE/DEAD BacLight bacterial viability kit (Molecular Probes), per the manufacturer's instructions. The purified rickettsiae were then either used immediately for infections or frozen at −80°C in SPG.
R. typhi genomic DNA extraction.
Partially purified R. typhi pellets were incubated at 90°C for 15 min, after which the heat-killed bacteria were taken into a biosafety level 2 laboratory. R. typhi genomic DNA was extracted using a Wizard genomic DNA purification kit (Promega), following the manufacturer's instructions for isolation of genomic DNA from gram-negative bacteria. Each DNA preparation was tested for the presence of R. typhi DNA by PCR, using degenerate primers for the rickettsial 17-kDa gene (forward primer, 5′-GCT CTT GCA ACT TCT ATG TT-3′; reverse primer, 5′-CAT TGT TCG TCA GGT TGG CG-3′). PCR amplicons were sequenced to confirm the presence of R. typhi genomic DNA.
Site-directed mutagenesis of pJDT vectors.
The BamHI restriction site in the alkaline phosphatase gene fusion vectors pJDT1, pJDT2, and pJDT3 (kindly provided by Robert Hancock from the University of British Columbia) (24) was transformed into a MfeI site using a QuikChange site-directed mutagenesis kit (Stratagene). For the pJDT2 vector, the extra nucleotide following the original BamHI site was also altered to prevent the introduction of a stop codon prior to the ′phoA gene. Mutagenesis was achieved using the primers listed in Fig. 1B and following the manufacturer's instructions, with the resulting plasmids named pJDT1-SDM, pJDT2-SDM, and pJDT3-SDM.
FIG. 1.
The alkaline phosphatase (′phoA) gene fusion vectors. (A) The composition of the pJDT and pJDT-SDM ′phoA gene fusion vectors. Restriction sites are underlined, and nucleotides highlighted in gray boxes represent the extra nucleotides added to generate three different reading frames. The first six amino acids of the ′PhoA protein are listed below the DNA sequence, along with the two amino acids introduced by the BamHI or MfeI sites and the frame-altering nucleotides. (B) The primers for site-directed mutagenesis of the pJDT vectors to convert the BamHI restriction sites into MfeI sites. Altered nucleotides are in bold and underlined, and the MfeI sites are in green.
Alkaline phosphatase gene fusion assays.
For each putative R. typhi signal peptide to be tested, primers were designed to amplify the genomic DNA sequence encoding the signal peptide, with flanking SacI, BamHI, or MfeI restriction sites incorporated into the primers. The primer pairs for each signal peptide are listed in Table S2 in the supplemental material, with the size of most signal peptide amplicons being <200 bp. The resulting amplicons were sequentially digested with the appropriate restriction enzymes. For some signal peptide amplicons, it was necessary to first clone the PCR products into the pCR4-TOPO vector using a TOPO TA cloning kit (Invitrogen).
Digested signal peptide amplicons were then ligated to the appropriate pJDT vector to result in an in-frame gene fusion between the signal peptide and the ′phoA gene (see schematic of vectors in Fig. 1A) and transformed into chemically competent DH5α E. coli (Invitrogen). Transformants were recovered and screened for PhoA activity on indicator LB agar containing 90 μg/ml 5-bromo-4-chloro-3-indolyl phosphate (BCIP, the chromogenic PhoA substrate), 75 mM Na2HPO4 (to repress endogenous phosphatase activity), and 100 μg/ml ampicillin. Plasmids from blue (indicating PhoA activity) and white (indicating lack of PhoA activity) clones were purified and sequenced using primers that flank the insertion site (forward primer, 5′-CAG GAA ACA GCT ATG AC-3′; reverse primer, 5′-CGC TAA GAG AAT CAC GCA GAG C-3′) to confirm that in-frame gene fusions were generated with no other sequence mutations.
HeLa cell infections and R. typhi RNA extraction.
R. typhi organisms were added to HeLa cells at a multiplicity of infection (MOI) of approximately 10; infected cells were incubated at 37°C with 5% CO2. At four days postinfection, the rickettsiae were isolated by partial purification for RNA extraction. The resulting bacterial pellet was resuspended in the lysis buffer RLT Plus (supplemented with β-mecaptoethanol) from the AllPrep DNA/RNA mini kit (Qiagen). RNA extraction was performed using the AllPrep kit, including the on-column DNase treatment, per the manufacturer's instructions. All RNA preparations were checked for the presence of DNA contamination by PCR using primers targeting the R. typhi rpsL (RT0119) gene (see primer sequences in Table S3 in the supplemental material) and also the human β-actin gene (forward primer, 5′-ATG GGT CAG AAG GAT TCC-3′; reverse primer, 5′-GTG TGG TGC CAG ATT TTC-3′). RNA samples verified to be free of DNA were then used for real-time qRT-PCR (see below).
Real-time RT-PCR procedure and data analysis.
A two-step, real-time RT-qPCR technique was used. The RNA was first converted to cDNA using the SuperScript III First-Strand Synthesis SuperMix for RT-qPCR (Invitrogen), following the manufacturer's instructions. The resulting cDNA was used directly as a template in the real-time qPCR samples, which were prepared using the SYBR GreenER qPCR SuperMix Universal (Invitrogen) per the manufacturer's instructions, but with adjustments of the samples for a total reaction volume of 10 μl. PCR cycles were performed in a Stratagene Mx3000P qPCR machine with the following cycle program: 50°C for 2 min; 95°C for 10 min; 40 amplification cycles of 95°C for 15 s, 55°C or 60°C (annealing temperature) for 60 s, and 72°C for 30 s (fluorescence reading point); and a melting analysis of 95°C for 60 s, followed by 55°C for 30 s, with a fluorescence reading taken at every degree temperature increase back up to 95°C. Primer pairs used are detailed in Table S3 in the supplemental material. Amplicons from each qPCR primer pair were sequenced to confirm specific amplification of the target cDNA. All primers used for qPCR were purified using Illustra MicroSpin G-25 columns (GE Healthcare), per the manufacturer's directions. For every run, each sample was tested in triplicate with a corresponding no-template (i.e., nuclease-free water as template) control sample.
The data were corrected for the reference dye and background fluorescence using the Stratagene MxPro analysis program. The efficiency of the primers in each reaction tube was calculated as described by Ramakers et al. (35), whereby a linear regression on the log(adjusted fluorescence) per cycle number was used to estimate the efficiency of the primers for cDNA amplification. Any sample with a primer efficiency value of less than 1.8 was not used in the analysis, as primer pairs yielding an amplication efficiency below this level are not considered accurate for quantification (22). In addition, any samples with product detected in their corresponding no-template control or with nonspecific products and/or primer dimers detected in the dissociation curve analysis were also not included the final analyses. Threshold values (and thus the corresponding CT values) were determined by the MxPro program, although all threshold values were manually confirmed.
Because real-time RT-qPCR was used to answer a yes/no question regarding gene expression and was not used to compare gene transcript levels between different experimental conditions, the R. typhi genes of interest were normalized to the R. typhi rpsL gene only. This gene encodes the 30S ribosomal protein S12 and was chosen as the reference gene because of its known expression during HeLa cell infections (our unpublished results). For each reaction, the quantity of the initial template was calculated by dividing the threshold fluorescence value by the efficiency raised to the CT cycle value (6). Samples with adjusted fluorescence levels that were not able to cross the threshold value were considered not expressed within this particular experimental system. For every run, each sample was tested in triplicate; the three quantity values calculated for each sample were averaged, and this average quantity was then divided by the average quantity for the reference sample rpsL. The final expression value is therefore the relative quantity of the gene of interest compared to that of the rpsL gene, with the expression of rpsL thus set at one.
RESULTS
In silico analysis of R. typhi signal peptides.
All proteins predicted to be encoded by the R. typhi strain Wilmington genome (23) were analyzed for the presence of an N-terminal signal peptide using the programs LipoP version 1.0 (17), Phobius (18), and SignalP version 3.0 with HMM and NN algorithms (4). Between these four analyses, a total of 191 individual R. typhi proteins were predicted to contain signal peptides (Table 1; also see Table S4 in the supplemental material), comprising 23% of the total estimated protein repertoire of this organism. However, of these 191 putative extracytoplasmic proteins, only 50 were predicted by all four programs, indicating considerable variability in the results generated by the different algorithms. LipoP, the only program to distinguish between lipoprotein and nonlipoprotein signal sequences, designated 16 R. typhi ORFs to encode lipoproteins with signal peptides cleaved by a type II signal peptidase; 12 of these putative lipoproteins were predicted by all other algorithms to be extracytoplasmic, although with almost no consensus regarding the signal peptide cleavage site. LipoP seemed to be the most stringent of the prediction algorithms, identifying only 67 extracytoplasmic proteins in the R. typhi proteome, with only one signal peptide not predicted by any of the other programs (see Table S4 in the supplemental material). In contrast, the most relaxed algorithm appeared to be the SignalP NN model, which predicted 136 proteins to contain signal peptides, 49 of which were not predicted by any other program. The discrepant nature of these in silico results highlighted the need for a biological system for experimental validation of these signal peptide predictions.
TABLE 1.
R. typhi signal peptide predictions and PhoA assay resultsa
| Signal Peptide Analysis | Signal peptide prediction by the following algorithms:
|
Total unique proteins with predicted signal peptides | |||
|---|---|---|---|---|---|
| Phobius | LipoP 1.0 | SignalP 3.0 (NN) | SignalP 3.0 (HMM) | ||
| Proteins with predicted signal peptides | 106 | 67 | 136 | 106 | 191 |
| Signal peptides tested (′PhoA assay) | 74 | 58 | 77 | 73 | 102 |
| Positive signal peptides | 66 | 55 | 65 | 66 | 84 |
| Percent positive signal peptides of those tested | 89% | 95% | 84% | 90% | 82% |
For each algorithm used to identify putative R. typhi signal peptides, the predicted number of signal peptides, the number tested by PhoA fusions, and the number found to function in E. coli are listed. The consolidated results for the total number of signal peptides identified across all the programs are presented in the final column. Results stratified by all the different combinations of the algorithms are presented in Table S4 in the supplemental material.
Alkaline phosphatase gene fusion assays.
To assess whether or not the putative R. typhi signal peptides identified by the LipoP, Phobius, and SignalP prediction algorithms were directing the export of their cognate proteins via the Sec translocon, we utilized the E. coli phoA gene fusion assay (20, 24). This system exploits two characteristics of PhoA: the enzyme is only active after translocation through the Sec system into the periplasm, and the phosphatase activity can be easily detected with the addition of a chromogenic substrate (BCIP) to the bacterial medium (20, 24). In this assay, DNA encoding signal peptides of interest is inserted just upstream of and in frame with the E. coli phoA gene lacking its native signal peptide sequence (annotated as ′phoA). The resulting fusion protein thus contains the N-terminal signal peptide of interest fused to the ′PhoA moiety, and the function of the putative signal peptide can be assessed by monitoring its ability to direct the ′PhoA moiety to the Sec apparatus for translocation into the periplasm, allowing for phosphatase activity, resulting in dark-blue bacterial colonies.
We tested 102 of the 191 putative signal peptides predicted by the different computer algorithms. This subset comprised the 50 signal peptides predicted by all four programs and an additional 52 signal peptides from proteins annotated as hypothetical that were predicted by any program (see Table S1 in the supplemental material). Because 30% of the R. typhi proteome is annotated as hypothetical (250/838 of the total predicted proteins) (23), we decided to follow up on any hypothetical protein identified in our in silico screen, in an effort to identify novel proteins with potential roles in rickettsial infection and virulence. We cloned the selected 102 putative signal peptides to create in-frame fusions with ′phoA using the pJDT vector series first described by Mdluli et al. (24), and a second series of vectors we created by using site-directed mutagenesis to replace the BamHI site in the pJDT vectors with a MfeI site (Fig. 1). After transformation into DH5α E. coli, we found that 84 (82%) of the putative R. typhi signal peptides tested were able to successfully direct secretion of ′PhoA into the bacterial periplasm, suggesting that their cognate proteins are likewise secreted via the rickettsial Sec system (Tables 1 and 2).
TABLE 2.
PhoA assay resultsa
| Locus tag | Predicted protein product | Locus tag | Predicted protein product |
|---|---|---|---|
| PhoA-positive samples | PhoA-positive samples | ||
| RT0013 | Probable zinc/manganese ABC transporter binding protein ZnuA | RT0382 | Hypothetical protein |
| RT0015 | 190-kDa antigen Sca1 | RT0383 | Rickettsial conserved hypothetical protein |
| RT0028 | VirB6-like protein | RT0388 | Soluble lytic transglycosylase domain-containing protein |
| RT0029 | VirB6-like protein | RT0390 | Carboxypeptidase IIW (LcdA) |
| RT0030 | VirB6-like protein | RT0393 | Hypothetical protein |
| RT0043 | Hypothetical proteinb | RT0395 | Rickettsial conserved hypothetical protein |
| RT0044 | VacJ lipoprotein | RT0399 | Rickettsial conserved hypothetical protein |
| RT0052 | 190-kDa antigen Sca2 | RT0406 | Hypothetical protein |
| RT0056 | Hypothetical protein | RT0417 | Hypothetical protein |
| RT0057 | OmpW-like outer membrane protein | RT0438 | Cell surface antigen Sca3 |
| RT0064 | Rickettsial conserved hypothetical proteinc | RT0444 | Hypothetical protein |
| RT0080 | Rickettsial conserved hypothetical protein | RT0445 | Rickettsial conserved hypothetical protein |
| RT0103 | Hypothetical protein | RT0465 | Rickettsial conserved hypothetical protein |
| RT0136 | Conserved hypothetical proteind | RT0475 | Rickettsial conserved hypothetical protein |
| RT0150 | Outer membrane protein Omp1 | RT0491 | Rickettsial conserved hypothetical protein |
| RT0174 | Probable lipoprotein | RT0492 | Rickettsial conserved hypothetical protein |
| RT0178 | Rickettsial conserved hypothetical protein | RT0546 | Hypothetical protein |
| RT0187 | Hypothetical protein | RT0548 | 2-Octaprenyl-6-methoxyphenyl hydroxylase |
| RT0188 | Rickettsial conserved hypothetical protein | RT0551 | Rickettsial conserved hypothetical protein |
| RT0216 | Outer membrane protein TolC | RT0561 | Hypothetical protein |
| RT0218 | Rickettsial conserved hypothetical protein | RT0565 | Protein export protein PrsA |
| RT0222 | Hypothetical protein | RT0601 | Hypothetical protein |
| RT0224 | Hypothetical protein | RT0667 | Rickettsial conserved hypothetical protein |
| RT0247 | Rickettsial conserved hypothetical protein | RT0692 | Rickettsial conserved hypothetical protein |
| RT0260 | Rickettsial conserved hypothetical protein | RT0694 | Hypothetical protein |
| RT0263 | Cytochrome c1 heme protein FbcH | RT0697 | Hypothetical protein |
| RT0276 | Hypothetical protein | RT0699 | Rickettsial outer membrane protein B (rOmpB/Sca5) |
| RT0279 | Rickettsial conserved hypothetical protein | RT0711 | Conserved hypothetical protein |
| RT0281 | VirB9 protein of the type IV secretion system | RT0733 | Rickettsial conserved hypothetical protein |
| RT0286 | Rickettsial conserved hypothetical protein | RT0758 | Peptidoglycan-associated lipoprotein Pal |
| RT0287 | Rickettsial conserved hypothetical protein | RT0761 | Hypothetical protein |
| RT0291 | Outer membrane antigenic lipoprotein B (NlpD) | RT0767 | Rickettsial conserved hypothetical protein |
| RT0301 | Rickettsial conserved hypothetical protein | RT0773 | Rickettsial conserved hypothetical protein |
| RT0312 | Hypothetical protein | RT0775 | Rickettsial conserved hypothetical protein |
| RT0331 | Probable efflux transporter | RT0797 | Hypothetical protein |
| RT0338 | Rickettsial conserved hypothetical protein | RT0807 | Rickettsial conserved hypothetical protein (phospholipase D; Pld) |
| RT0341 | Hypothetical protein | RT0810 | Rickettsial conserved hypothetical protein |
| RT0348 | Hypothetical protein | RT0815 | Possible outer surface protein |
| RT0355 | Hypothetical protein | RT0816 | Possible outer surface protein |
| RT0358 | Rickettsial conserved hypothetical protein | RT0821 | Rickettsial 17-kDa surface antigen |
| RT0377 | d-Alanyl-d-alanine carboxypeptidase DacF | RT0825 | Hypothetical protein |
| RT0379 | Possible periplasmic protein | RT0858 | Hypothetical protein |
| PhoA-negative samples | PhoA-negative samples | ||
| RT0011 | Hypothetical protein | RT0432 | NADP-thioredoxin reductase TrxB1 |
| RT0039 | Rickettsial conserved hypothetical protein | RT0459 | Rickettsial conserved hypothetical protein |
| RT0047 | Hypothetical protein | RT0554 | Rickettsial conserved hypothetical protein |
| RT0083 | Hypothetical protein | RT0563 | Rickettsial conserved hypothetical protein |
| RT0182 | Rickettsial conserved hypothetical protein | RT0621 | Possible permease |
| RT0199 | Hypothetical protein | RT0756 | Hypothetical protein |
| RT0258 | Hypothetical protein | RT0853 | Hypothetical protein |
| RT0319 | Hypothetical protein | RT0862 | Conserved hypothetical protein |
| RT0347 | Hypothetical protein | RT0866 | Hypothetical protein |
The R. typhi proteins with predicted N-terminal signal peptides that functioned in the PhoA system (84 PhoA-positive samples), as well as those putative signal peptides that did not function in E. coli (18 PhoA-negative samples).
“Hypothetical protein” refers to a predicted ORF with no database matches in any other organism.
“Rickettsial conserved hypothetical protein” refers to a predicted ORF with matches only in other rickettsial sequences.
“Conserved hypothetical protein” refers to an ORF that has matches to proteins of unknown function in organisms other than rickettsiae.
Eighteen of the tested R. typhi signal peptides were not able to direct Sec-dependent protein secretion; the resulting lack of PhoA enzymatic activity thus yielded white E. coli colonies, and all but two of the nonfunctional signal peptides were from hypothetical proteins. Based on the results from proteins with known or proposed functions (i.e., those proteins not annotated as hypothetical), the E. coli-based ′phoA gene fusion system seemed to recognize R. typhi cytoplasmic and extracytoplasmic proteins appropriately, indicating that this assay can serve as a useful resource for the identification of Sec-secreted rickettsial proteins.
In vitro gene expression of R. typhi extracytoplasmic proteins.
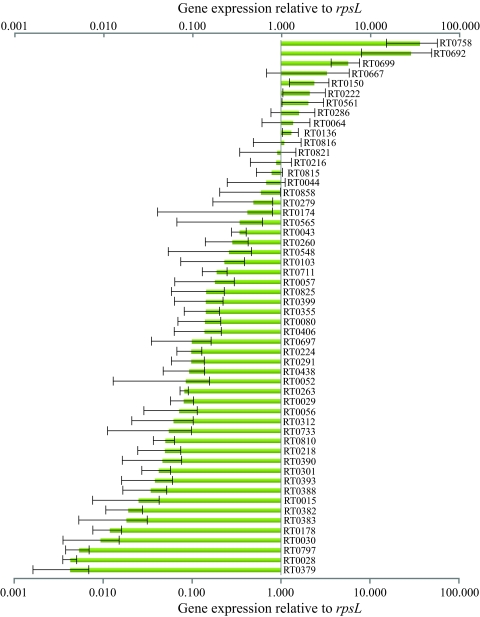
To identify those Sec-dependent R. typhi proteins potentially involved during mammalian infections, we analyzed the corresponding gene expression during infections in HeLa cells. The cells were infected with an MOI of approximately 10, and the rickettsiae were isolated 4 days postinfection; following RNA extraction, gene expression was analyzed using two-step, real-time RT-qPCR. Due to either low efficiency or amplification of nonspecific products, suitable qPCR primer pairs were not obtained for 16 genes, which were therefore not analyzed (see Table S5 in the supplemental material). However, of the remaining 68 putative extracytoplasmic proteins identified by the PhoA fusion assay, 54 corresponding genes were expressed during the HeLa cell infections, while gene expression was not detected for 14 of the Sec-dependent proteins (Fig. 2; also see Table S5 in the supplemental material). When expression levels of all analyzed genes were normalized to rpsL gene expression, the pal (RT0758) gene was the most highly expressed in HeLa cells. Among the 10 genes with the highest relative expression, 7 encoded hypothetical proteins, as well as rompB (RT0699) and omp1 (RT0150). Overall, a majority of the extracytoplasmic proteins exhibited gene expression during the in vitro infections in HeLa cells, indicating that many of these proteins, including many hypothetical proteins, could be playing important roles in the context of mammalian infections.
FIG. 2.
Gene expression of R. typhi extracytoplasmic proteins in HeLa cells. HeLa cells were infected with R. typhi at an MOI of ∼10, and the bacteria were isolated for RNA extraction at 4 days postinfection. Two-step, real-time RT-qPCR was used to assess gene expression of the putative R. typhi extracytoplasmic proteins relative to the rpsL gene. The mean relative expression and the standard errors are presented for each gene; results are from three independent infections. Individual data points are listed in Table S5 in the supplemental material.
DISCUSSION
The fundamental processes responsible for rickettsial infection, growth, and pathogenesis in humans are inadequately understood at the molecular level. Here, we report the combined use of a bioinformatics screen, an E. coli-based functional assay, and analysis of cognate rickettsial gene expression to define Sec-dependent R. typhi extracytoplasmic proteins that could be involved in mammalian infections and virulence.
Our initial step was to perform an in silico screen with the annotated R. typhi Wilmington genome (23) to identify proteins predicted to contain an N-terminal signal peptide to direct them to the Sec translocon. Of all the 191 R. typhi proteins predicted to have signal peptides, we chose to further examine 102 signal peptides that were either predicted by all four programs or predicted by any program and annotated as hypothetical. To assess the functionality of these putative signal peptides, we utilized an E. coli-based alkaline phosphatase gene fusion system. By using this system, we found that 84 of the 102 signal peptides tested were able to direct the Sec-dependent secretion of ′PhoA into the periplasm, suggesting that the cognate proteins of these signal peptides are also extracytoplasmic in R. typhi (Table 2). Sixty of these proteins with functional signal peptides were annotated as hypothetical, and the remaining 24 were proteins with an assigned homolog and/or function. Among these 24 proteins were the R. typhi surface cell antigen (Sca) proteins Sca1-3 and Sca5. These proteins are thought to be autotransporters, which are Sec-dependent proteins that direct their own translocation across the outer membrane (25). Sca5 (rOmpB) has been shown to be expressed on the surface of both spotted fever group and typhus group rickettsiae (2, 14). The signal peptide from the R. typhi 17-kDa surface antigen was positively identified by our screen, and this lipoprotein has also been recognized on the surface of spotted fever group and typhus group rickettsiae (2, 50). The R. typhi phopholipase D (Pld) signal peptide was also positive; this protein has been identified as a putative cell surface virulence factor with a potential role in rickettsial escape from the phagocytic vacuole (36, 48).
R. typhi signal peptides from a VirB9 homolog (RT0281) and three VirB6-like proteins (RT0028-0030) were also found to function in E. coli. These proteins are part of the type IV secretion system; although this system is not yet well characterized in rickettsiae, in the prototypical Agrobacterium Vir type IV secretion system apparatus, translocation of both VirB6 and VirB9 across the inner membrane has been characterized as Sec dependent, with VirB6 localizing to the inner membrane and VirB9 to the outer membrane (3, 47).
Our screen also identified functional signal peptides from proteins with characterized homologs in other bacteria, such as the d-alanyl-d-alanine carboxypeptidase DacF (RT0377), which is known to be involved in Bacillus subtilis cell wall synthesis and is thought to be extracytoplasmic, although this has not yetbeen demonstrated experimentally (39). An FbcH homolog (RT0263) was also identified; this protein is part of the cytochrome bc1 complex and has been found to contain an N-terminal signal peptide and localize in the inner membrane of the alphaproteobacterium Bradyrhizobium japonicum (44). An R. typhi homolog of the carboxypeptidase IIW (LcdA; RT0390) had a functional signal peptide. In Bacillus megaterium, LcdA is active in the membrane and has been shown to both hydrolyze d-alanine from peptidoglycan and also incorporate diaminopimelate into the cell wall of Bacillus megaterium (8). Our screen also identified an R. typhi homolog of the lipoprotein NlpD (RT0291). Lipidation has been confirmed for this protein in both E. coli and Bartonella bacilliformis, and NlpD was shown to be immunogenic during human infections with B. bacilliformis (16, 26), suggesting that it is surface exposed.
The R. typhi outer membrane protein Omp1 (RT0150) is homologous to E. coli YaeT, which is part of an outer membrane complex involved in outer membrane biogenesis (51). The signal peptide from the R. typhi protein annotated as the outer membrane porin OmpW (RT0057) was functional; in Salmonella enterica serovar Typhimurium, this protein has been shown to function as the outer membrane component of an efflux pump system (11). Our screen also identified the R. typhi homolog of the peptidoglycan-associated lipoprotein (Pal; RT0758), which is anchored to the outer membrane and recognized as a potential vaccine candidate from Haemophilus influenzae (27). In addition, Pal is also known to be released into serum by E. coli during sepsis (21).
Additional R. typhi proteins with functional signal peptides were annotated as PrsA (RT0565), TolC (RT0216), VacJ (RT0044), and ZnuA (RT0013). PrsA has been characterized as a membrane-bound lipoprotein essential for growth and protein secretion in Bacillus subtilis (45). TolC is a highly studied protein in E. coli and other bacteria; this protein is located in the outer membrane and forms a homotrimeric pore spanning the periplasm for the direct export of substrates from the bacterial cytoplasm (19). VacJ has been found to be a surface-exposed lipoprotein in Shigella flexneri, with a potential role in intracellular spreading (43), and ZnuA is a known periplasmic zinc transporter in E. coli (52).
There were three R. typhi proteins with positive signal peptides that had been annotated with putative functions that have yet to be demonstrated experimentally: RT0381, RT0388, and RT0548. RT0331 is annotated as a probable efflux transporter; bacterial efflux proteins have been extensively studied and are localized to the outer membrane (29). RT0388 is annotated as a soluble lytic transgylcosylase; in gram-negative bacteria, lytic transglycosylases are localized to the periplasm where they are involved in the breakdown of peptidoglycan (40). RT0548 is annotated as a 2-octaprenyl-6-methoxyphenyl hydroxylase; one protein in E. coli characterized with this enzymatic function is the UbiH protein involved in the generation of ubiquinone, which is a membrane-associated biosynthetic pathway (53).
Among the 18 R. typhi putative signal peptides that were not capable of directing the Sec-dependent export of ′PhoA in E. coli, 16 of these proteins are annotated as hypothetical. The signal peptide from the R. typhi homolog of a NADP-thioredoxin reductase, TrxB, was also negative in our screen. This enzyme is known to be cytosolic in E. coli (30); therefore, we believe that this enzyme is also likely cytosolic in rickettsiae. The remaining negative signal peptide from our screen was from RT0621, annotated as a possible permease. Bacterial permeases are characterized as being extracytoplasmic (7); therefore, if this protein does in fact function as a permease, it is possible that the permease exits the bacterial cytoplasm through a non-Sec-dependent mechanism or that this sample represents a false negative in our PhoA screen.
The Phobius, LipoP, and SignalP algorithms predict the presence of N-terminal signal peptides based on the tripartite structure of known signal peptides, which includes a positively charged amino terminus (N region), a hydrophobic H region that forms a membrane-spanning helix, and a polar carboxyl terminus (C region) (31). Each prediction algorithm identified this structure in the putative R. typhi extracytoplasmic proteins, with the N region being dominated by lysine residues, the H region mainly containing leucine residues, and the C region rich in serine residues, with alanine being the most common residue just prior to the predicted cleavage site (see Table S1 in the supplemental material for the sequences of the predicted cleavage sites). For the 102 R. typhi signal peptides that we functionally tested, we found that the overall sequence organization of those signal peptides that functioned in E. coli was not different from those signal peptides that did not function. However, among the signal peptides that did not function in the heterologous system, those identified by the Phobius and SignalP HMM algorithms were identified as having glycine and tyrosine residues, respectively, in the position just prior to the predicted cleavage site.
Overall, the heterologous PhoA assay appeared to correctly distinguish R. typhi signal peptides from cytoplasmic and Sec-dependent proteins, indicating that this system can be a useful tool in the study and identification of extracytoplasmic proteins in other rickettsiae. Furthermore, based on the results of signal peptides from the proteins with known or hypothesized functions, we believe that our PhoA assay results from the 76 proteins annotated as hypothetical are highly accurate and that this work has provided the first laboratory-based suggestion of their subcellular localization.
In order to hone in on which of the Sec-dependent proteins identified in the PhoA screen could be important during a mammalian infection, we assessed their cognate gene transcription during R. typhi infections in HeLa cells (Fig. 2; also see Table S5 in the supplemental material). During these in vitro infections, expression was detected for the corresponding transcripts of the R. typhi extracytoplasmic proteins OmpW (RT0057), Omp1 (RT0150), TolC (RT0216), FbcH (RT0263), LcdA (RT0390), NlpD (RT0291), Pal (RT0758), and the rickettsial 17-kDa surface antigen (RT0821). The genes encoding all of the tested Sca proteins (Sca1-3 and Sca5) were expressed, as were all of the virB6-like genes. Thirty-eight of the expressed genes corresponded to R. typhi extracytoplasmic proteins annotated as hypothetical; of the 10 most highly expressed R. typhi genes in our HeLa cell infections, 7 were hypothetical. These 10 highly transcribed genes encoded the following proteins: Pal (RT0758; the most highly expressed in our experimental system), RT0692 (hypothetical), RT0699 (Sca5), RT0667 (hypothetical), RT0150 (Omp1), RT0222 (hypothetical), RT0561 (hypothetical), RT0286 (hypothetical), RT0064 (hypothetical), and RT0136 (hypothetical).
Overall, this work has identified a number of R. typhi extracytoplasmic proteins with potential roles in mammalian infection and virulence. Because techniques that involve genetic manipulation cannot readily be used to screen for essential, accessory, and/or virulence-associated rickettsial proteins, other avenues of research must be used for molecular characterization of the rickettsiae and their interactions with host cells. To identify proteins that could be involved in rickettsial infection of mammalian cells, we focused on the identification of the Sec-translocated extracytoplasmic proteins of R. typhi. Starting with a bioinformatics screen to predict which R. typhi proteins contained N-terminal signal peptides, we then assessed the functionality of select R. typhi putative signal peptides in an E. coli-based gene fusion system. Finally, to determine the relevance of the proteins identified from in silico and heterologous work, we examined their cognate gene transcription by R. typhi during infections in HeLa cells. The data generated from this combination of techniques form a significant contribution to the field of rickettsial molecular biology and serve as a platform from which to launch future, in-depth studies of specific proteins.
Supplementary Material
Acknowledgments
This project was supported by award numbers AI017828 and AI059118 from the National Institute of Allergy and Infectious Diseases. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases or the National Institutes of Health.
We thank Robert Hancock from the University of British Columbia for providing us with the pJDT ′phoA gene fusion vectors. We thank Joseph Gillespie for his computational assistance, and we also thank Rebecca Maag and Magda Beier-Sexton for their comments on the manuscript.
Footnotes
Published ahead of print on 18 July 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Alto, N. M. 2008. Mimicking small G-proteins: an emerging theme from the bacterial virulence arsenal. Cell Microbiol. 10566-575. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, B. E. 1990. The 17-kilodalton protein antigens of spotted fever and typhus group rickettsiae. Ann. N. Y. Acad. Sci. 590326-333. [DOI] [PubMed] [Google Scholar]
- 3.Backert, S., and T. F. Meyer. 2006. Type IV secretion systems and their effectors in bacterial pathogenesis. Curr. Opin. Microbiol. 9207-217. [DOI] [PubMed] [Google Scholar]
- 4.Bendtsen, J. D., H. Nielsen, G. Von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340783-795. [DOI] [PubMed] [Google Scholar]
- 5.Bina, J. E., F. Nano, and R. E. Hancock. 1997. Utilization of alkaline phosphatase fusions to identify secreted proteins, including potential efflux proteins and virulence factors from Helicobacter pylori. FEMS Microbiol. Lett. 14863-68. [DOI] [PubMed] [Google Scholar]
- 6.Čikoš, S., A. Bukovská, and J. Koppel. 2007. Relative quantification of mRNA: comparison of methods currently used for real-time PCR data analysis. BMC Mol. Biol. 8113. doi: 10.1186/1471-2199-8-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen, G. N., and J. Monod. 1957. Bacterial permeases. Bacteriol. Rev. 21169-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DasGupta, H., and D. P. Fan. 1979. Purification and characterization of a carboxypeptidase-transpeptidase of Bacillus megaterium acting on the tetrapeptide moiety of the peptidoglycan. J. Biol. Chem. 2545672-5683. [PubMed] [Google Scholar]
- 9.De Buck, E., J. Anné, and E. Lammertyn. 2007. The role of protein secretion systems in the virulence of the intracellular pathogen Legionella pneumophila. Microbiology 1533948-3953. [DOI] [PubMed] [Google Scholar]
- 10.Dreher-Lesnick, S. M., S. M. Ceraul, M. S. Rahman, and A. F. Azad. 2008. Genome-wide screen for temperature-regulated genes of the obligate intracellular bacterium, Rickettsia typhi. BMC Microbiol. 861. doi: 10.1186/1471-2180/8/61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gil, F., F. Ipinza, J. Fuentes, R. Fumeron, J. M. Villarreal, A. Aspée, G. C. Mora, C. C. Vásquez, and C. Saavedra. 2007. The ompW (porin) gene mediates methyl viologen (paraquat) efflux in Salmonella enterica serovar Typhimurium. Res. Microbiol. 158529-536. [DOI] [PubMed] [Google Scholar]
- 12.Gillespie, J. J., K. Williams, M. Shukla, E. E. Snyder, E. K. Nordberg, S. M. Ceraul, C. Dharmanolla, D. Rainey, J. Soneja, J. M. Shallom, N. D. Vishnubhat, R. Wattam, A. Purkayastha, M. Czar, O. Crasta, J. C. Setubal, A. F. Azad, and B. S. Sobral. 2008. Rickettsia phylogenomics: unwinding the intricacies of obligate intracellular life. PLoS ONE 3e2018. doi: 10.1371/journal.pone.0002018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gold, V. A., F. Duong, and I. Collinson. 2007. Structure and function of the bacterial Sec translocon. Mol. Membr. Biol. 24387-394. [DOI] [PubMed] [Google Scholar]
- 14.Hackstadt, T., R. Messer, W. Cieplak, and M. G. Peacock. 1992. Evidence for proteolytic cleavage of the 120-kilodalton outer membrane protein of rickettsiae: identification of an avirulent mutant deficient in processing. Infect. Immun. 60159-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hybiske, K., and R. S. Stephens. 2008. Exit strategies of intracellular pathogens. Nat. Rev. Microbiol. 699-110. [DOI] [PubMed] [Google Scholar]
- 16.Ichikawa, J. K., C. Li, J. Fu, and S. Clarke. 1994. A gene at 59 minutes on the Escherichia coli chromosome encodes a lipoprotein with unusual amino acid repeat sequences. J. Bacteriol. 1761630-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Juncker, A. S., H. Willenbrock, G. Von Heijne, S. Brunak, H. Nielsen, and A. Krogh. 2003. Prediction of lipoprotein signal peptides in gram-negative bacteria. Protein Sci. 121652-1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Käll, L., A. Krogh, and E. L. Sonnhammer. 2004. A combined transmembrane topology and signal peptide prediction method. J. Mol. Biol. 3381027-1036. [DOI] [PubMed] [Google Scholar]
- 19.Koronakis, V., A. Sharff, E. Koronakis, B. Luisi, and C. Hughes. 2000. Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export. Nature 405914-919. [DOI] [PubMed] [Google Scholar]
- 20.Lewenza, S., J. L. Gardy, F. S. Brinkman, and R. E. Hancock. 2005. Genome-wide identification of Pseudomonas aeruginosa exported proteins using a consensus computational strategy combined with a laboratory-based PhoA fusion screen. Genome Res. 15321-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liang, M. D., A. Bagchi, H. S. Warren, M. M. Tehan, J. A. Trigilio, L. K. Beasley-Topliffe, B. L. Tesini, J. C. Lazzaroni, M. J. Fenton, and J. Hellman. 2005. Bacterial peptidoglycan-associated lipoprotein: a naturally occurring toll-like receptor 2 agonist that is shed into serum and has synergy with lipopolysaccharide. J. Infect. Dis. 191939-948. [DOI] [PubMed] [Google Scholar]
- 22.Lutfalla, G., and G. Uze. 2006. Performing quantitative reverse-transcribed polymerase chain reaction experiments. Methods Enzymol. 410386-400. [DOI] [PubMed] [Google Scholar]
- 23.McLeod, M. P., X. Qin, S. E. Karpathy, J. Gioia, S. K. Highlander, G. E. Fox, T. Z. McNeill, H. Jiang, D. Muzny, L. S. Jacob, A. C. Hawes, E. Sodergren, R. Gill, J. Hume, M. Morgan, G. Fan, A. G. Amin, R. A. Gibbs, C. Hong, X. J. Yu, D. H. Walker, and G. M. Weinstock. 2004. Complete genome sequence of Rickettsia typhi and comparison with sequences of other rickettsiae. J. Bacteriol. 1865842-5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mdluli, K. E., J. D. Treit, V. J. Kerr, and F. E. Nano. 1995. New vectors for the in vitro generation of alkaline phosphatase fusions to proteins encoded by G+C-rich DNA. Gene 155133-134. [DOI] [PubMed] [Google Scholar]
- 25.Ngwamidiba, M., G. Blanc, D. Raoult, and P. E. Fournier. 2006. Sca1, a previously undescribed paralog from autotransporter protein-encoding genes in Rickettsia species. BMC Microbiol. 612. doi: 10.1186/1471-2180-6-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Padmalayam, I., T. Kelly, B. Baumstark, and R. Massung. 2000. Molecular cloning, sequencing, expression, and characterization of an immunogenic 43-kilodalton lipoprotein of Bartonella bacilliformis that has homology to NlpD/LppB. Infect. Immun. 684972-4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parsons, L. M., F. Lin, and J. Orban. 2006. Peptidoglycan recognition by Pal, an outer membrane lipoprotein. Biochemistry 452122-2128. [DOI] [PubMed] [Google Scholar]
- 28.Payne, M. S., and E. N. Jackson. 1991. Use of alkaline phosphatase fusions to study protein secretion in Bacillus subtilis. J. Bacteriol. 1732278-2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Piddock, L. J. 2006. Multidrug-resistance efflux pumps—not just for resistance. Nat. Rev. Microbiol. 4629-636. [DOI] [PubMed] [Google Scholar]
- 30.Prinz, W. A., F. Åslund, A. Holmgren, and J. Beckwith. 1997. The role of the thioredoxin and glutaredoxin pathways in reducing protein disulfide bonds in the Escherichia coli cytoplasm. J. Biol. Chem. 27215661-15667. [DOI] [PubMed] [Google Scholar]
- 31.Pugsley, A. P. 1993. The complete general secretory pathway in gram-negative bacteria. Microbiol. Rev. 5750-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rahman, M. S., S. M. Ceraul, S. M. Dreher-Lesnick, M. S. Beier, and A. F. Azad. 2007. The lspA gene, encoding the type II signal peptidase of Rickettsia typhi: transcriptional and functional analysis. J. Bacteriol. 189336-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rahman, M. S., J. A. Simser, K. R. Macaluso, and A. F. Azad. 2003. Molecular and functional analysis of the lepB gene, encoding a type I signal peptidase from Rickettsia rickettsii and Rickettsia typhi. J. Bacteriol. 1854578-4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rahman, M. S., J. A. Simser, K. R. Macaluso, and A. F. Azad. 2005. Functional analysis of secA homologues from rickettsiae. Microbiology 151589-596. [DOI] [PubMed] [Google Scholar]
- 35.Ramakers, C., J. M. Ruijter, R. H. Deprez, and A. F. Moorman. 2003. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 33962-66. [DOI] [PubMed] [Google Scholar]
- 36.Renesto, P., P. Dehoux, E. Gouin, L. Touqui, P. Cossart, and D. Raoult. 2003. Identification and characterization of a phospholipase D-superfamily gene in rickettsiae. J. Infect. Dis. 1881276-1283. [DOI] [PubMed] [Google Scholar]
- 37.Rusch, S. L., and D. A. Kendall. 2007. Interactions that drive Sec-dependent bacterial protein transport. Biochemistry 469665-9673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saier, M. H., Jr. 2006. Protein secretion and membrane insertion systems in gram-negative bacteria. J. Membr. Biol. 21475-90. [DOI] [PubMed] [Google Scholar]
- 39.Scheffers, D. J., and M. G. Pinho. 2005. Bacterial cell wall synthesis: new insights from localization studies. Microbiol. Mol. Biol. Rev. 69585-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scheurwater, E., C. W. Reid, and A. J. Clarke. 2008. Lytic transglycosylases: bacterial space-making autolysins. Int. J. Biochem. Cell Biol. 40586-591. [DOI] [PubMed] [Google Scholar]
- 41.Schroeder, G. N., and H. Hilbi. 2008. Molecular pathogenesis of Shigella spp.: controlling host cell signaling, invasion, and death by type III secretion. Clin. Microbiol. Rev. 21134-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spears, K. J., A. J. Roe, and D. L. Gally. 2006. A comparison of enteropathogenic and enterohaemorrhagic Escherichia coli pathogenesis. FEMS Microbiol. Lett. 255187-202. [DOI] [PubMed] [Google Scholar]
- 43.Suzuki, T., T. Murai, I. Fukuda, T. Tobe, M. Yoshikawa, and C. Sasakawa. 1994. Identification and characterization of a chromosomal virulence gene, vacJ, required for intercellular spreading of Shigella flexneri. Mol. Microbiol. 1131-41. [DOI] [PubMed] [Google Scholar]
- 44.Thöny-Meyer, L., P. James, and H. Hennecke. 1991. From one gene to two proteins: the biogenesis of cytochromes b and c1 in Bradyrhizobium japonicum. Proc. Natl. Acad. Sci. USA 885001-5005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vitikainen, M., I. Lappalainen, R. Seppala, H. Antelmann, H. Boer, S. Taira, H. Savilahti, M. Hecker, M. Vihinen, M. Sarvas, and V. P. Kontinen. 2004. Structure-function analysis of PrsA reveals roles for the parvulin-like and flanking N- and C-terminal domains in protein folding and secretion in Bacillus subtilis. J. Biol. Chem. 27919302-19314. [DOI] [PubMed] [Google Scholar]
- 46.Ward, J., J. Fletcher, S. P. Nair, M. Wilson, R. J. Williams, S. Poole, and B. Henderson. 2001. Identification of the exported proteins of the oral opportunistic pathogen Actinobacillus actinomycetemcomitans by using alkaline phosphatase fusions. Infect. Immun. 692748-2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ward, J. E., D. E. Akiyoshi, D. Regier, A. Datta, M. P. Gordon, and E. W. Nester. 1988. Characterization of the virB operon from an Agrobacterium tumefaciens Ti plasmid. J. Biol. Chem. 2635804-5814. [PubMed] [Google Scholar]
- 48.Whitworth, T., V. L. Popov, X. J. Yu, D. H. Walker, and D. H. Bouyer. 2005. Expression of the Rickettsia prowazekii pld or tlyC gene in Salmonella enterica serovar Typhimurium mediates phagosomal escape. Infect. Immun. 736668-6673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wiker, H. G., M. A. Wilson, and G. K. Schoolnik. 2000. Extracytoplasmic proteins of Mycobacterium tuberculosis—mature secreted proteins often start with aspartic acid and proline. Microbiology 1461525-1533. [DOI] [PubMed] [Google Scholar]
- 50.Williams, J. C., D. H. Walker, M. G. Peacock, and S. T. Stewart. 1986. Humoral immune response to Rocky Mountain spotted fever in experimentally infected guinea pigs: immunoprecipitation of lactoperoxidase 125I-labeled proteins and detection of soluble antigens of Rickettsia rickettsii. Infect. Immun. 52120-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu, T., J. Malinverni, N. Ruiz, S. Kim, T. J. Silhavy, and D. Kahne. 2005. Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell 121235-245. [DOI] [PubMed] [Google Scholar]
- 52.Yatsunyk, L. A., J. A. Easton, L. R. Kim, S. A. Sugarbaker, B. Bennett, R. M. Breece, I. I. Vorontsov, D. L. Tierney, M. W. Crowder, and A. C. Rosenzweig. 2008. Structure and metal binding properties of ZnuA, a periplasmic zinc transporter from Escherichia coli. J. Biol. Inorg. Chem. 13271-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Young, I. G., P. Stroobant, C. G. Macdonald, and F. Gibson. 1973. Pathway for ubiquinone biosynthesis in Escherichia coli K-12: gene-enzyme relationships and intermediates. J. Bacteriol. 11442-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.