Abstract
The transcriptional regulator CcpN of Bacillus subtilis has been recently characterized as a repressor of two gluconeogenic genes, gapB and pckA, and of a small noncoding regulatory RNA, sr1, involved in arginine catabolism. Deletion of ccpN impairs growth on glucose and strongly alters the distribution of intracellular fluxes, rerouting the main glucose catabolism from glycolysis to the pentose phosphate (PP) pathway. Using transcriptome analysis, we show that during growth on glucose, gapB and pckA are the only protein-coding genes directly repressed by CcpN. By quantifying intracellular fluxes in deletion mutants, we demonstrate that derepression of pckA under glycolytic condition causes the growth defect observed in the ccpN mutant due to extensive futile cycling through the pyruvate carboxylase, phosphoenolpyruvate carboxykinase, and pyruvate kinase. Beyond ATP dissipation via this cycle, PckA activity causes a drain on tricarboxylic acid cycle intermediates, which we show to be the main reason for the reduced growth of a ccpN mutant. The high flux through the PP pathway in the ccpN mutant is modulated by the flux through the alternative glyceraldehyde-3-phosphate dehydrogenases, GapA and GapB. Strongly increased concentrations of intermediates in upper glycolysis indicate that GapB overexpression causes a metabolic jamming of this pathway and, consequently, increases the relative flux through the PP pathway. In contrast, derepression of sr1, the third known target of CcpN, plays only a marginal role in ccpN mutant phenotypes.
Regulation of metabolism enables microbes to grow efficiently on a large variety of carbon sources. Partly, these regulation processes ensure efficient resource allocation by expressing substrate-specific transporters and catabolic enzymes only when the substrate is present (38). However, regulation must also avoid simultaneous activity of incompatible or counteracting reactions. A well-known example is the simultaneous presence of ATP-consuming and ATP-generating enzymes that would lead to ATP-dissipating futile cycles (6, 10, 36). Another relevant case is the expression of isoenzymes with different cofactor specificities (9, 21, 27, 33, 38) that may catalyze opposite fluxes through key reactions (7, 9, 11, 16). A particularly challenging situation is therefore the switch from glycolytic to gluconeogenic substrates, where large fluxes through the metabolism backbone have to be reversed because the two groups of substrates enter metabolism at two opposite ends.
While allosteric regulation of enzyme activities plays an important role in fine-tuning and rapid adaptation to nutritional changes, transcriptional regulation is probably the main mechanism that allows the organism to respond metabolically to a nutrient shift by promoting the operation of a new optimal subset of reactions and adjusting the fluxes through the different central pathways for the establishment of a new steady state (26). One of the most thoroughly studied control mechanisms is carbon catabolite repression, by which the presence of a preferred substrate, usually glucose, represses the uptake and metabolism of alternative carbon sources (5, 18, 39, 41) and globally alters the expression of numerous genes (4, 19, 42, 43). In Bacillus subtilis, catabolite repression is primarily mediated by CcpA, a transcriptional regulator that exerts a broad control on central pathways and numerous distinct aspects of cellular metabolism by directly or indirectly controlling the expression of ca. 10% of the genes (17, 23, 24, 30, 32, 38).
More recently, CcpN has been described as an additional mediator of carbon catabolite repression in B. subtilis, which acts on the genes encoding the gluconeogenic enzymes GapB [NADP(H)-dependent glyceraldehyde-3-phosphate dehydrogenase] and PckA (phosphoenolpyruvate [PEP] carboxykinase) and thereby prevents fluxes through these enzymes in the presence of glycolytic substrates (37). In addition, CcpN represses the transcription of the untranslated regulatory RNA SR1, which is involved in fine-tuning arginine catabolism via the control of the translation of the transcriptional activator AhrC (22, 29). Hence, CcpN is thought to play a metabolic role under both glycolytic and gluconeogenic conditions, but its main function as repressor is clearly exerted during growth on glycolytic substrates (37). Moreover, the ccpN gene is cotranscribed with the downstream yqfL gene, the product of which modulates the absolute level of gapB and pckA (and probably sr1 as well) transcription in a CcpN-dependent manner (37).
By quantifying the intracellular flux distributions in 137 B. subtilis single gene knockout mutants we demonstrated previously that these distributions in central carbon metabolism are very robust against genetic lesions during exponential growth on glucose (12). However, the ccpN mutant was a striking exception, being the sole case with an entirely different flux distribution that included the pentose phosphate (PP) pathway, glycolysis, anaplerosis, and the tricarboxylic acid (TCA) cycle. The global nature of the flux response to a ccpN knockout thus raised the question whether more genes are under direct or indirect control of this repressor or whether derepressed gapB and pckA transcription suffices to elicit the observed drastic flux changes.
To identify all CcpN-responsive genes, we monitored the impact of a ccpN knockout on the B. subtilis transcriptome and fluxome during exponential growth on glucose, where CcpN exhibits its full repressing activity. To elucidate the flux-altering mechanisms, we decoupled the individual effects of the deregulated expression of each CcpN target gene on intracellular metabolism by constructing B. subtilis knockout and overexpressing mutants and measuring their intracellular flux responses by 13C-labeling experiments (35).
MATERIALS AND METHODS
Strains and growth conditions.
Wild-type B. subtilis 168 and mutants thereof were used throughout the present study (Table 1). sr1 mutants were constructed by using plasmid pINT1 (29), which carries a cat gene conferring resistance to chloramphenicol flanked by the upstream and downstream regions of the sr1 wild-type gene (29). The IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible Phyperspank promoter from plasmid pDR111 (David Rudner, unpublished data) was used to overexpress gapB at the ectopic amyE locus. For this, a HindIII-SphI fragment carrying the entire gapB gene and its RBS (obtained by PCR using the primers gapBrbsHind [5′-CCCAAGCTTCATAATTGATAAGGGGTGTCCAAC-3′] and gapBstopSph [5′-ACATGCATGCTTATACAGCAGACGGATGTTTCATTC-3′]) was inserted into pDR111 to give pIC586. For physiological and flux analyses, 5 ml of Luria-Bertani complex medium was inoculated with a single colony from selective plates and grown at 37°C for 7 h. These precultures were used to inoculate 5 ml of M9 minimal medium (20) precultures with 50 mg of tryptophan/liter and 5 g of glucose/liter and grown overnight. Both precultures were grown in selective media containing 0.25 mg of phleomycin/liter, 0.5 mg of erythromycin/liter, or 100 mg spectinomycin/liter, where necessary. To reduce polar effects on downstream genes, 1 mM IPTG was added to the cultures of gapA mutants (11). For 13C-labeling experiments, 500-ml baffled shake flasks containing 30 ml of M9 minimal medium were inoculated with at maximum 1% (vol/vol) of M9 preculture and batch cultures were grown at 37°C on a gyratory shaker at 250 rpm. The medium was supplemented with 50 mg of tryptophan/liter and 3 g of [1-13C]glucose (99%; Cambridge Isotope Laboratories)/liter or a mixture of 0.6 g of [U-13C]glucose (99%; Cambridge Isotope Laboratories)/liter and 2.4 g of unlabeled glucose/liter as the sole carbon source. Overexpression of gapB under the control of the Phyperspank promoter was induced by the addition of 1 mM IPTG. Cofeeding experiments were performed in M9 minimal medium supplemented with tryptophan (50 mg/liter), glucose (5 g/liter), aspartate (0.5 g/liter), malate (2.7 g/liter), glutamate (2 g/liter), or succinate (1 g/liter) or in C minimal medium (3) supplemented with tryptophan (50 mg/liter), glucose (5 g/liter), aspartate (0.2 g/liter), or malate (0.2 g/liter).
TABLE 1.
Strains used in this study
| Strain | Nomenclature | Genotype | Source or reference |
|---|---|---|---|
| GM1596 | Wild type | trpC2 | Lab stock (168CA) |
| GM1560 | ccpN* | trpC2 ΔccpN (nonpolar) amy::PgapB′-lacZ(cat) | PS1722; Servant et al. (37) |
| GM1599 | ccpN | trpC2 Δ(ccpN-yqfL)::phleo | PS1679; Servant et al. (37) |
| GM2607 | ccpN gapB | trpC2 Δ(ccpN-yqfL)::phleo ΔgapB::pMUTIN2(ery) | GM1599 × GM1500 DNA |
| GM2611 | ccpN gapB pckA | trpC2 Δ(ccpN-yqfL)::phleo ΔgapB::pMUTIN2(ery) ΔpckA::spec | GM2607 × GM2664 DNA |
| GM2609 | ccpN pckA | trpC2 Δ(ccpN-yqfL)::phleo ΔpckA::spec | GM1599 × GM2664 DNA |
| GM2664 | pckA | trpC2 ΔpckA::spec | Lab stock |
| GM2677 | sr1 | trpC2 Δsr1::cat | GM1596 × pINT1 |
| GM2678 | ccpN sr1 | trpC2 Δ(ccpN-yqfL)::phleo Δsr1::cat | GM1599 × GM2677 DNA |
| GM2690 | ccpN gapB | trpC2 Δ(ccpN-yqfL)::phleo ΔgapB::pEC23(kan) | GM1599 × GM2648 DNA |
| GM1500 | gapB | trpC2 ΔgapB::pMUTIN2(ery) | Fillinger et al. (11) |
| GM2648 | gapB | trpC2 ΔgapB::pEC23(kan) | GM1500 × pEC23 |
| GM2691 | ccpN gapB pckA | trpC2 Δ(ccpN-yqfL)::phleo ΔgapB::pEC23(kan) ΔpckA::spec | GM2611 × GM2648 DNA |
| GM1501 | gapA | trpC2 ΔgapA::pMUTIN2(ery) | Fillinger et al. (11) |
| GM2676 | ccpN gapA | trpC2 Δ(ccpN-yqfL)::phleo ΔgapA::pMUTIN2(ery) | GM1599 × GM1501 DNA |
| GM2679 | ccpN gapA sr1 | trpC2 Δ(ccpN-yqfL)::phleo ΔgapA::pMUTIN2(ery) Δsr1::cat | GM2678 × GM2676 DNA |
| GM2705 | pckA | trpC2 ΔpckA::neo | Lab stock |
| GM2721 | amyE::Phyp | trpC2 amyE::Phyperspank/lacI (spec) | GM1596 × pDR111 |
| GM2723 | amyE::Phyp-gapB | trpC2 amyE::Phyperspank-gapB/lacI (spec) | GM1596 × pIC586 |
| GM2725 | amyE::PhyppckA | trpC2 amyE::Phyperspank/lacI (spec) ΔpckA::neo | GM2705 × GM2721 DNA |
| GM2727 | amyE::Phyp-gapB pckA | trpC2 amyE::Phyperspank-gapB/lacI (spec) ΔpckA::neo | GM2705 × GM2723 DNA |
| GM2730 | ccpN sr1 gapB pckA | trpC2 Δ(ccpN-yqfL)::phleo ΔgapB::pEC23(kan) ΔpckA::spec Δsr1::cat | GM2691 × GM2677 DNA |
| GM2739 | sr1 pckA | trpC2 ΔpckA::spec Δsr1::cat | GM2664 × GM2677 DNA |
| GM2740 | sr1 gapB pckA | trpC2 ΔpckA::spec Δsr1::cat ΔgapB::pEC23(kan) | GM2739 × GM2648 DNA |
For transcriptome analyses, 5-ml precultures in TSS minimal medium [50 mM Tris-HCl (pH 7.5), 15 mM (NH4)2SO4, 8 mM MgSO4·7H2O, 27 mM KCl, 7 mM trisodium citrate, 0.6 mM KH3PO4, 2 mM CaCl2, trace FeSO4·H2O, trace MnSO4, 5 mM glutamate] (15) plus 10 g of glucose/liter as a carbon source and supplemented with tryptophan (50 mg/liter) were inoculated at different dilutions with a single colony from a Luria-Bertani or NB (nutrient broth [8 g/liter], KCl [1 g/liter], 1 mM MgSO4) selective plate, followed by incubation overnight at 37°C with shaking. Cultures (25 ml) of the same medium in 250-ml baffled flasks were inoculated from one preculture in early transition phase at an optical density of 0.05 and incubated at 37°C on a gyratory shaker at 250 rpm.
Determination of physiological parameters.
Bacterial growth was monitored by reading the optical density at 600 nm (A600). The exponential growth phase and the corresponding maximum growth rate (μmax) were identified from a semilogarithmic plot of A600 versus time. The cellular dry weight (cdw) was determined at the mid-exponential-growth phase (A600 of ∼1.5) for each strain. For this purpose, 10-ml culture aliquots were centrifuged at 3,000 × g in preweighed glass tubes, washed once with 0.9% NaCl, dried at 85°C for 24 h, and cooled to room temperature under vacuum. Glucose and acetate concentrations were determined enzymatically using commercial kits. Specific uptake and secretion rates were calculated by linear regression of the substrate or product versus the biomass concentration during the exponential growth phase, multiplied by the μmax.
Metabolic flux analysis.
To access the 13C patterns in proteinogenic amino acids, cell pellets from 2 ml of culture were harvested at an A600 of 1 to 1.5, hydrolyzed in 6 M HCl, and derivatized with N-(tert-butyldimethylsilyl)-N-methyl-trifluoroacetamide as described previously (13). Derivatized amino acids were analyzed for 13C-labeling patterns with a series 8000GC combined with an MD800 mass spectrometer (Fisons Instruments). Flux ratio analysis and subsequent 13C-constrained net flux analysis (13, 14) were conducted by using the software package FiatFlux (44). Briefly, flux ratios are directly calculated from 13C patterns and then used together with measured rates as constraints to estimate the flux distribution from the stoichiometric matrix.
Assay of glyceraldehyde-3-phosphate dehydrogenase activity.
Cells were harvested in mid-exponential growth phase by centrifugation at 4°C and washed twice with 0.9% NaCl. Biomass pellets were kept at −20°C until further analysis. For enzymatic assays cells were 10-fold concentrated in lysis buffer (100 mM Tris-HCl [pH 7.5], 5 mM MgCl2, 1 mM dithiothreitol, and 4 mM phenylmethylsulfonyl fluoride) and disrupted by passage through a French press cell at 4°C. Cell-free lysates were obtained by centrifugation at 23,000 × g and 10 min at 4°C. Supernatant was used for the enzymatic assay in a reaction buffer (125 mM triethanolamine, 5 mM l-cysteine, 20 mM potassium arsenate, 50 mM K2HPO4 [pH 9.2]) at 25°C with glyceraldehyde-3-phosphate (3 mM) and NAD(P)+ (1 mM). Reduction of NAD(P)+ was monitored spectrophotometrically at 340 nm (11). Protein concentration was determined by using Coomassie Plus protein reagent (Pierce) according to the manufacturer's manual.
Genetic methods.
B. subtilis was transformed as described previously (1), except that glucose was replaced by l-malate in the transformation media to transform strains disrupted for ccpN and/or gapA. Selection of the transformants was carried out on Luria-Bertani medium containing erythromycin (0.4 mg/liter), spectinomycin (100 mg/liter), or chloramphenicol (5 mg/liter) or on NB containing phleomycin (0.25 mg/liter).
Transcriptome analysis.
High-quality RNA preparation, radiolabeled cDNA synthesis, and hybridization were performed as described previously (8). Cell pellets for RNA extraction were obtained from exponentially growing cultures (A600 = 0.5). Panorama gene macroarrays containing full-length PCR products of all 4,107 B. subtilis protein-coding genes in duplicate and a cDNA labeling kit were provided by Sigma-Genosys and used as recommended by the manufacturer. The comparison between the wild type and the nonpolar ccpN mutant was repeated four times with three independent RNA preparations and three pairs of macroarrays. The comparison between the wild type and the ccpN-yqfL mutant was repeated twice with two independent RNA preparations and two pairs of macroarrays. The data analysis was performed by using Array Vision software (Imaging Research) for signals and background quantifications and Genespring software (Silicon Genetics) for further comparisons. A normalization procedure was performed in two steps: the overall spot normalization function of Array Vision was used to calculate normalized intensity values of individual spots per array to allow for comparison between two filters for each gene, and Genespring was used for normalization of each gene to itself to allow comparison of all values obtained from all of the experiments for each gene. A variance analysis was performed, and the contrasts were tested by using a Student t test to obtain P values. Only the genes corresponding to a false discovery rate (FDR) inferior to 10% were considered (meaning that there are fewer than 10% of false positives among the genes considered).
Metabolite measurement.
For metabolite quantification, samples were taken during exponential growth on glucose. A portion (1 ml) of culture broth were transferred into a 1.5-ml tube. Cells were spun down for exactly 20 s at 14,000 rpm at room temperature in a tabletop centrifuge. The supernatant was discarded, and the pellet was quickly frozen in liquid nitrogen. Until further analysis, pellets were kept at −80°C. For metabolite extraction, pellets were transferred to liquid nitrogen. Portions (1 ml) of prewarmed extraction solution (75% [vol/vol] ethanol buffered with 10 mM acetate [pH 7.2]) and 5 μl of 1 mM (wt/vol) sodium glutarate were added to the pellet. The samples were incubated at 85°C and 1,000 rpm in a thermomixer for exactly 3 min and then kept on ice until centrifugation. The samples were then centrifuged at 14,000 rpm at 4°C for 10 min. The supernatants were transferred to a new 1.5-ml tube, and the solvent was evaporated in a SpeedDry vacuum concentrator (Martin Christ Gefriertrocknungsanlagen GmbH, Germany) at room temperature for 6 h. A total of 20 μl of 2% methoxamine-HCl in pyridine was added to the dried samples, followed by incubation for 90 min at 40°C. Then, 15 μl was transferred to a new vial for analysis. Prior to analysis, the samples were automatically derivatized with N-methyl-N-(trimethylsilyl)trifluoroacetamide for 30 min at 60°C by an MPS2 autosampler (Gerstel, Germany). The samples were analyzed on an Agilent 6890N gas chromatograph (column, HP5-MS [Hewlett-Packard]) combined with a Pegasus III (Leco, Germany) time-of-flight mass spectrometer. The data were processed with the Leco ChromaTOF 2.32 software (J. C. Ewald and N. Zamboni, unpublished data).
Microarray data accession number.
The complete set of microarray data is available at the Gene Omnibus Expression database under accession number GSE11937.
RESULTS
Transcriptome analysis.
Three genetic targets of CcpN, namely, gapB, pckA, and sr1, had been previously identified (28, 37). To identify further possible CcpN targets that could contribute to the major flux redistribution observed in a ccpN knockout strain (12), we performed a comparative transcriptome analysis of a ccpN mutant and the wild type in minimal glucose medium. Since the flux alteration was shown to be identical in a nonpolar ccpN mutant and in a double ccpN-yqfL mutant (data not shown), we analyzed these two mutants to identify genes whose transcription was similarly affected in both mutants relative to the wild type. A statistical analysis of the results, when considering an FDR limit of 10%, identified only five genes differentially expressed in the ccpN-yqfL double mutant and three in the nonpolar ccpN mutant compared to the wild-type strain (Table 2). Importantly, gapB and pckA were the only genes common to both lists. The fact that gapB and pckA clearly showed up validated the experiment. The other genes detected only in one mutant (yhcI, yurM, cotE, and ansA) were not considered as probable candidates for direct regulation by CcpN because (i) no other gene of the putative corresponding operons (yhcABCDEFGHI, yurONML, and ansAB) was affected in one or the other mutant, (ii) cotE is a σE-dependent gene expressed late in the sporulation process, and (iii) no putative CcpN binding sequence (28, 37) could be detected upstream of these genes (or the first gene of the putative corresponding operons). sr1 could not be identified because only protein-coding sequences are represented on the macroarrays. The mRNA concentration of the known direct target of sr1, ahrC, is not affected by sr1 concentration; only the elongation of its translation is affected (22). Because neither arginine nor ornithine was present in the medium, it was also expected that the indirect targets of sr1, the rocABC and rocDEF operons (22), would not be expressed differentially. Thus, this transcriptome comparison indicated that GapB and/or PckA are likely to mediate the effect of ccpN knockout on central carbon fluxes. It cannot be excluded, however, that sr1 or CcpN have other regulatory, nontranscriptional effects that could directly modulate protein concentration or activity.
TABLE 2.
Genes differentially expressed in ccpN-yqfL or nonpolar ccpN mutant versus wild-type strains (only genes with an FDR lower than 0.1 are shown)
| Comparison and genea | P | Mutant/wild-type ratio | FDR |
|---|---|---|---|
| WT vs ccpN-yqfL mutant | |||
| yhcI | 1.5 × 10−5 | 0.55 | 0.06 |
| pckA | 3.6 × 10−5 | 37.32 | 0.07 |
| yurM | 3.8 × 10−5 | 4.22 | 0.05 |
| gapB | 7.2 × 10−5 | 28.59 | 0.07 |
| ansA | 1.2 × 10−4 | 3.19 | 0.1 |
| WT vs nonpolar ccpN mutant | |||
| gapB | 1.9 × 10−5 | 23.38 | 0.03 |
| cotE | 2.5 × 10−5 | 7.55 | 0.03 |
| pckA | 5.7 × 10−5 | 12.05 | 0.05 |
WT, wild type.
Metabolic fluxes in CcpN mutants.
The reduced growth rate (37) and the alteration of the entire intracellular flux distribution (12) observed in a ccpN knockout strain indicated that CcpN is important to maintain the global flux distribution that allows efficient catabolism of glucose in wild-type B. subtilis. Thus far, however, only relative fluxes through metabolic pathways were reported with three main findings: (i) the ratio between glycolysis and the PP pathway was shifted toward the latter, (ii) the TCA cycle flux was shifted from a strongly catabolic to a mostly anabolic use, and (iii) the gluconeogenic PckA flux was increased. For a deeper analysis, we determined the absolute molecular fluxes through all pathways of central metabolism using 13C-constrained metabolic flux analysis (14) in ccpN mutants.
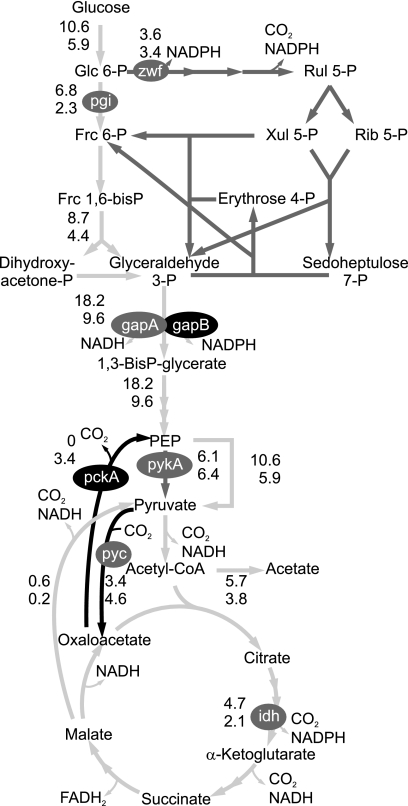
The ccpN mutation roughly halves the growth rate on glucose as sole carbon source. Accordingly, most absolute in vivo fluxes in central metabolism were significantly lower in the mutant (Fig. 1), but the PP pathway and the pyruvate kinase (PykA) fluxes remained at the same absolute level. Hence, the PP pathway becomes the main pathway for glucose catabolism because of a decreased glycolytic flux. As expected from our previous characterization of CcpN functions (37) and the present transcriptome analysis, a significant flux through PckA was found in the ccpN mutant, whereas this flux was below the detection limit in the wild type. Also, the TCA cycle replenishing flux through the pyruvate carboxylase (PycA) was increased in the mutant (Fig. 1). This was somewhat unexpected because pycA transcription is constitutive (2; this study) and because no allosteric regulation of PycA other than activation by acetyl coenzyme A is known. Consequently, a significant fraction of the pyruvate molecules cycle through PycA, PckA, and PykA, thereby dissipating one ATP per cycle.
FIG. 1.
Net fluxes in mmol gcdw−1 h−1 for B. subtilis wild type (top values) and the ccpN mutant (bottom values). Light gray, gray, and black arrows highlight lower, equal, and higher molecular fluxes, respectively, in the mutant. Gray and black ovals indicate equal and higher transcription of key genes, respectively. pgi, phosphoglucose isomerase; zwf: phosphoglucose dehydrogenase; gapA/B, glyceraldehyde-3-P dehydrogenase A/B; pykA, pyruvate kinase; pckA, PEP carboxykinase; pyc, pyruvate carboxylase; idh, isocitrate dehydrogenase.
Obviously, the entire flux distribution is radically affected by the ccpN mutation, but the transcriptome data indicated that CcpN acts exclusively via the gluconeogenic genes gapB and pckA. Since ccpN is cotranscribed with yqfL, a positive modulator of gapB and pckA expression that acts via CcpN (37), the flux effect could be influenced via YqfL. However, we can exclude such an YqfL-dependent effect on the observed phenotypes because identical expression patterns of gapB and pckA have been observed in both nonpolar and polar ccpN mutants (37), as well as identical flux distributions (data not shown).
Contribution of sr1 to the phenotype of the ccpN mutant.
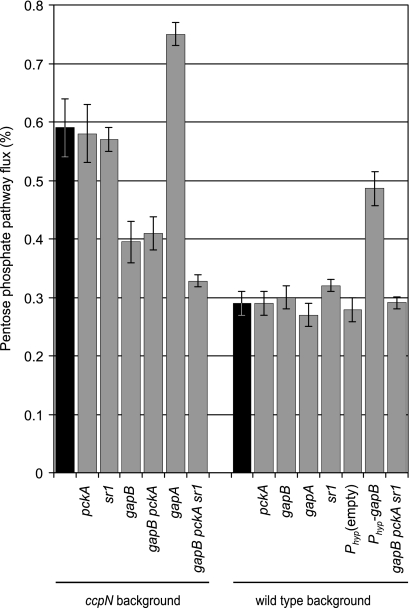
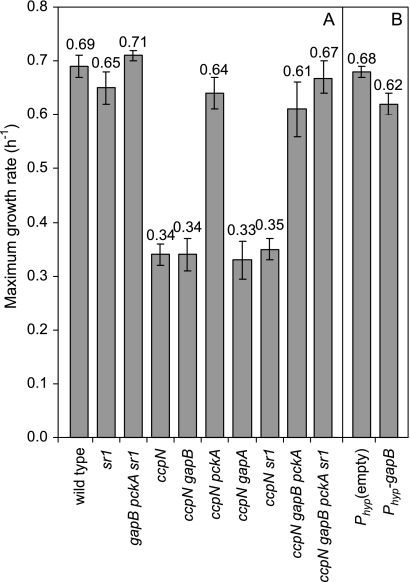
Of the three known targets of CcpN, sr1, a noncoding regulatory RNA, was the least expected to alter the flux distribution and/or growth rate, because none of the known targets of sr1 appears to be required under the chosen conditions. Nevertheless, the mode of action of sr1, modulating translation and thus protein concentrations, could potentially influence fluxes. An sr1 deletion was thus introduced into the ccpN mutant strain. The flux analysis clearly showed that the split between glycolysis and the PP pathway is identical in the ccpN and the ccpN sr1 mutants, while this split ratio in an sr1 mutant is identical to that of the wild type (Fig. 2). Furthermore, the sr1 mutation does not affect the growth rate on glucose when introduced into a wild-type or ccpN background (Fig. 3A). Thus, sr1 does not contribute to either the flux rerouting or the growth defect of the ccpN mutant, and pckA and gapB seem to be the only genes responsible for the observed phenotype of a ccpN mutant.
FIG. 2.
Percentage of glucose catabolized through the PP pathway as opposed to glycolysis. The values are derived through 13C-based flux analysis akin to Fig. 1. Black bars indicate the ccpN and wild-type control values for each panel.
FIG. 3.
Maximum growth rates of wild-type B. subtilis and knockout mutants (A) and wild-type gapB-overexpressing and control strains (B) on glucose minimal medium.
The remaining open question was then how derepression of the two protein encoding targets of CcpN could modulate flux through distant pathways such as the TCA cycle and the PP pathway. To elucidate the individual contributions of gapB and pckA expression, we constructed additional mutant strains by deleting these genes in the ccpN background (Table 1).
Contribution of pckA expression to the phenotype of the ccpN mutant.
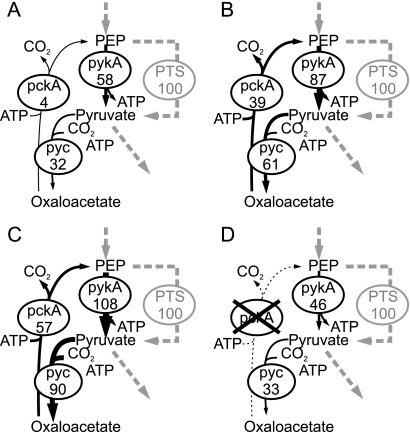
During growth on glucose, derepression of pckA effectively causes a futile cycle (Fig. 4A and B) that has a significant growth-retarding effect because a knockout of pckA in the ccpN background both prevents the futile cycle (Fig. 4D) and restores the wild-type growth rate (Fig. 3A). Similarly, a wild-type-like growth rate was observed when a pckA knockout was introduced into the ccpN gapB double-mutant strain (Fig. 3A). Since the pckA derepression effect on fluxes was mostly local and confined to the futile cycle, i.e., changes in the flux of the anaplerotic, gluconeogenic, and PykA reaction (see Table S1 in the supplemental material, ccpN and ccpN gapB mutants), the growth-reducing effect could be caused by either associated ATP dissipation or a drain on TCA cycle intermediates.
FIG. 4.
Fluxes through the futile cycle in the PEP-pyruvate-oxaloacetate triangle of wild type (A), ccpN mutant (B), ccpN gapB mutant (C), and ccpN pckA mutant (D). Fluxes are given in percentages of the glucose uptake rates that were for 10.6, 5.9, 6.4, and 9.8 mmol g−1 h−1, respectively.
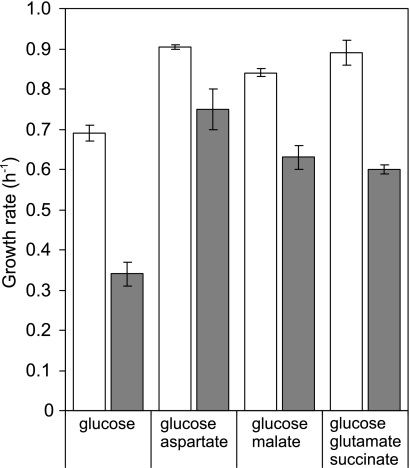
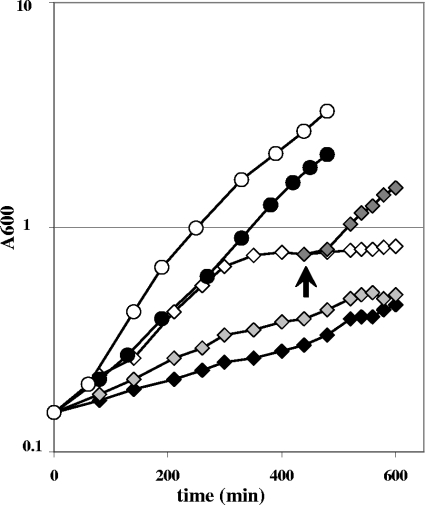
To test the latter possibility, we grew the strains on mixtures of glucose and substrates that feed directly into the TCA cycle. With glucose as the sole carbon source, the ccpN mutant grew at about half the wild-type rate. When cultured on glucose-aspartate, glucose-malate, or glucose-glutamate-succinate mixtures, the ccpN mutant indeed grew faster than on glucose alone. The recovery was most pronounced in a glucose-aspartate mixture with an almost wild-type-like growth rate (Fig. 5). The drain on TCA cycle intermediates therefore appears to be the main cause for the slower growth of the ccpN mutant. In particular, the more efficient metabolic suppression with aspartate and, to a lesser extent, with malate than with succinate-glutamate suggests that the very high PckA flux limits the amount of oxaloacetate available for aspartate synthesis by the aspB-encoded aspartate aminotransferase (AspB) and/or for reaction with acetyl coenzyme A to fuel the TCA cycle. These hypotheses were tested by supplementing the minimal glucose medium with aspartate at a low concentration sufficient to compensate for an aspartate auxotrophy but not as a carbon source. Such a supplementation allowed a wild-type growth rate for the ccpN mutant, although addition of malate at the same low concentration led only to a slightly faster growth of the mutant strain (Fig. 6), indicating that aspartate synthesis is indeed limiting in the ccpN mutant. Growth of the mutant with a low concentration of aspartate was as fast as that of the wild type but stopped earlier (as that of an auxotroph strain deficient in the first step of aspartate synthesis [data not shown]). This early plateau resulted from depletion of the initially added aspartate, and growth could be resumed by adding aspartate in low concentrations again (Fig. 6). Together, these results suggest that the PckA activity reduces the pool size of oxaloacetate, which in turn limits the synthesis of aspartate.
FIG. 5.
Maximum growth rates of wild-type B. subtilis (white bars) and ccpN mutant (gray bars) in minimal medium with mixtures of carbon substrates. Concentrations were as follows: glucose, 5 g/liter; aspartate, 0.5 g/liter; malate, 2.7 g/liter; glutamate, 2 g/liter; and succinate, 1 g/liter.
FIG. 6.
Growth curves of the wild-type (circles) and ccpN mutant (diamonds) strains in glucose minimal medium (solid symbols) supplemented with aspartate (0.2 g/liter; open symbols) or malate (gray symbols). Growth of the ccpN mutant strain resumed when aspartate at a low concentration was added again (dark gray symbols [the arrow indicates the addition of aspartate]) after the early plateau. Similar results were obtained from three independent experiments; the results from one representative experiment are shown here.
Contribution of gapB expression to the phenotype of the ccpN mutant.
In contrast to the clearly negative impact of PckA activity during growth on glucose, the phenotype caused by the presence of GapB was less clear. With respect to growth, disruption of gapB had no effect, either in the ccpN gapB double mutant or in the ccpN gapB pckA triple mutant (Fig. 3A). We therefore conclude that the derepression of gapB is not responsible for the reduced growth of a ccpN mutant.
On the other hand, GapB is clearly the major cause for the higher PP pathway flux in the ccpN mutant because deleting gapB in a ccpN background strongly reduces the flux through the PP pathway, although not quite to wild-type level (Fig. 2). Also the ccpN pckA gapB triple mutant showed the same reduction of PP pathway flux, indicating that the PP pathway flux is modulated independently of pckA. The remaining slight difference in PP pathway flux between these mutants and the wild type was abolished when an sr1 deletion was introduced into the ccpN gapB pckA triple mutant. Thus, SR1 had no contribution to the high PP flux in a ccpN knockout mutant (compare ccpN and ccpN sr1 mutants in Fig. 2) but a slight and rather unexpected one when neither GapB nor PckA are expressed.
If the high PP pathway flux indeed is triggered by derepressed gapB, overexpression of gapB in the wild-type background should result in an increased PP pathway flux as well. We therefore introduced the gapB gene downstream of a strong promoter into the ectopic amyE locus of the wild-type strain (Table 1). The measured GapB activity in this overexpressing strain during growth in glucose minimal medium was similar to that in the ccpN mutant (76 ± 8 U/g versus 80 ± 8 U/g, respectively), while the empty construct showed virtually no activity (4 ± 2 U/g). Along with the increased GapB activity and a wild-type growth rate (Fig. 3B), this strain also showed a higher PP pathway flux (Fig. 2), confirming the role of GapB in modulating the PP pathway flux. Despite the comparable GapB activities, however, the flux increase in the GapB-overproducing strain was not as prominent as in a ccpN mutant (Fig. 2). A similar increase was observed when gapB was overexpressed in a pckA background (data not shown). This demonstrated that GapB overproduction is the main cause of the high PP pathway flux.
Influence of differential gapA and gapB expression on the PP pathway.
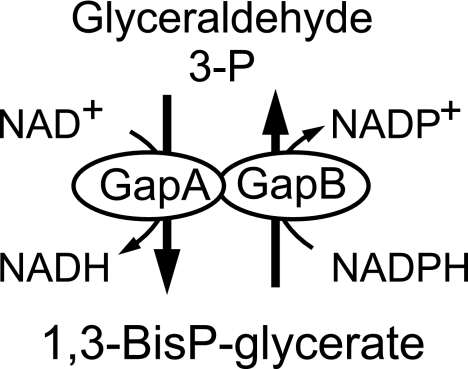
Although the previous results establish that GapB derepression is the main cause of the PP pathway flux increase during growth on glucose, they do not explain why. Two possibilities can be envisaged. First, GapB differs from GapA in being NADP(H)-dependent and in efficiently catalyzing the gluconeogenic reduction of 1,3-bis-phospho-glycerate to glyceraldehyde-3-phosphate (11). Although the actual flux direction of each isoenzyme cannot be resolved using 13C tracer experiments, the simultaneous activity of GapA and GapB may give rise to a so-called redox cycle that would cause a net conversion of NADPH to NADH (Fig. 7). A high PP pathway flux leading to an increased formation of NADPH could thus be implemented to counterbalance such a NADPH-to-NADH conversion. To test this possibility, we constructed a ccpN gapA double mutant, forcing the entire glycolytic flux through the NADPH-forming GapB reaction. The gapA disruption used (11) was obtained by insertion of the pMUTIN plasmid, an element that allows the downstream genes of the gapA operon (31) to be expressed from an IPTG-induced promoter. In the presence of IPTG, the ccpN gapA strain grew at the same rate as the ccpN mutant (Fig. 3A). Thus, GapB is expressed in this strain to a level that compensates for the absence of GapA. A gapA single mutant, in contrast, has a much lower growth rate, presumably because gapB is repressed by CcpN in the presence of glucose (11). In the wild-type background, neither gap gene knockout had an influence on the relative flux through the PP pathway. Unexpectedly, despite the predicted high NADPH production by GapB in the ccpN gapA double mutant, the relative flux through the PP pathway was even higher than in the ccpN mutant (Fig. 2). Thus, NADPH consumption via a putative GapA-GapB redox cycle is not the main cause of the high PP pathway flux in ccpN mutants.
FIG. 7.
Putative redox cycle.
Another possibility is that derepressed GapB catalyzes the gluconeogenic flux during growth on glucose, thereby eliciting, indirectly, a higher flux through the PP pathway. If this was the case, one would expect increased intracellular concentrations of metabolites in the upper part of glycolysis, up to glucose-6-phosphate (G6P), the branch point metabolite of glycolysis versus PP pathway. By determining the intracellular concentrations of ten relevant central carbon intermediates, we indeed observed higher G6P and fructose-6-phosphate (F6P) concentrations in the ccpN mutant compared to the wild type (Fig. 8). Furthermore, also the 6-phosphogluconate (6PG) pool was larger in the mutant, although not as prominent as for G6P and F6P pools. In contrast, the size of the ribose-phosphate pools in the PP pathway were virtually the same in the two strains (Fig. 8). Reduced pool sizes for most of the TCA cycle intermediates in the ccpN mutant are most likely the consequence of the lower TCA cycle flux in the ccpN mutant (Fig. 1). Altogether, these data indicate that the driving force for the GapB-dependent flux change is not cofactor balancing but a metabolic jamming in the upper part of glycolysis.
FIG. 8.
Metabolite concentrations in B. subtilis wild type (white boxes) and ccpN mutant (gray boxes) during exponential growth on glucose minimal medium. Triplicate samples at three different time points during the exponential growth phase were taken for metabolite analysis. The box plots shown in the figure were thus calculated from nine points for each metabolite. The box plots represent the median (horizontal line inside the box), the upper and lower quartiles (upper and lower ends of the box), and the extreme values (shown as whiskers). Circles represent outliers. G6P, glucose-6-phosphate; F6P, fructose-6-phosphate; 6PG, 6-phosphogluconate; glycerol-P, glycerol-3-phosphate; alpha-KG, alpha-ketoglutarate.
DISCUSSION
The recently identified regulator CcpN is a major regulator of central carbon metabolism, primarily by repressing the gluconeogenic genes gapB and pckA (36). Knocking out ccpN causes two major changes: a drastically reduced growth rate on glucose and a greatly altered flux distribution. We demonstrated here that the observed growth defect is caused by the very high PckA activity synthesized in the ccpN mutant. The cause of this PckA-based effect might be the ATP-dissipating futile cycle through PEP, pyruvate, and oxaloacetate triggered by PckA. The ATP expenses of this cycle amount to 8 to 10% of the total ATP generation in the ccpN mutant, as estimated from the metabolic fluxes assuming a P-to-O ratio of 1. Although this may seem substantial, much more energy is lost in a B. subtilis qox mutant with an inefficient respiratory chain (45). Nevertheless, such a mutant still grows as fast as the wild type. Thus, ATP dissipation via this futile cycle seemed not to be the reason for the reduced growth rate of the ccpN mutant.
Instead, we demonstrate that the increased PckA flux causes a shortage of TCA cycle intermediates. Even though the ccpN mutant also has an increased anaplerotic flux through the constitutively expressed PycA (4, 9), this is not sufficient to counteract the gluconeogenic flux through the PckA and thus to feed the TCA cycle. As expected, the growth rate increased when the ccpN mutant was cofed with glucose plus substrates that feed directly into the TCA cycle such as malate or glutamate-succinate (Fig. 5). More precisely, we identified AspB as the most limiting reaction since the addition of aspartate led to wild type-like growth. This finding indicates that aspartate synthesis is limited by the oxaloacetate availability. Although oxaloacetate could not be measured directly, the lower TCA cycle intermediate pools in a ccpN mutant compared to the wild type suggest also a reduced oxaloacetate pool. Furthermore, AspB, the aspartate aminotransferase of the thermophilic Bacillus sp. strain YM-2, has been shown to have a Km value for oxaloacetate that is 2 orders of magnitude higher than that of the B. subtilis CitZ citrate synthase (25, 34, 40). This explains why oxaloacetate availability is primarily insufficient for aspartate synthesis and much less limiting for the synthesis of citrate and further TCA cycle intermediates.
The most prominent change in the flux distribution of the ccpN mutant was the switch from glycolysis to PP pathway as the main catabolic route. By testing a ccpN pckA double mutant, which showed the same relative PP pathway flux as in the ccpN mutant, we could exclude the flux shift to be a growth rate artifact (Fig. 2). Furthermore, overexpression of gapB in the wild type, as well as in the pckA background led to an increased PP pathway flux at the wild-type growth rate, which proved that, in some particular cases, changing the expression of a single gene is sufficient to modulate the flux of a distal pathway.
Two competing hypotheses might explain the change in the PP pathway flux of a ccpN mutant: (i) a cofactor-dependent mode and (ii) a carbon metabolite-dependent mode. An NADPH-consuming redox cycle established by simultaneous GapA and GapB activity (Fig. 7) may contribute to the enhanced PP pathway in the ccpN mutant, but several evidences render this unlikely. First, deletion of gapB in the ccpN mutant background does not fully restore the wild-type flux through PP pathway. Second, in the ccpN gapA double mutant, GapB alone catalyzes the net glycolytic flux, with less, if any, NADPH consumption (in the gluconeogenic direction) compared to the ccpN mutant. In principle, GapB could also exhibit NAD-dependent activity, which could give rise to a futile cycle in combination with its NADP-dependent activity even in the absence of GapA. However, because GapB is much less efficient than GapA for the NAD-dependent conversion of G3P into 1,3-diphosphoglycerate (11), the net conversion of NADPH into NADH is expected to be much lower in the ccpN gapA double mutant than in the ccpN single mutant. Nevertheless, despite this lower consumption of NADPH, the PP pathway is even higher in the ccpN gapA than in the ccpN mutant. Thus, NADPH balancing is not the main cause for the higher PP pathway flux.
The second, cofactor-independent hypothesis is a slowdown of the upper glycolysis elicited by a strong backflux through GapB. According to the mass action law, the resulting high intracellular G6P concentration would then lead to a higher PP pathway flux. Indeed, the two- to threefold-increased pool sizes of G6P and much higher G6P/6PG than G6P/F6P concentration gradients determined in the ccpN mutant (Fig. 8) strongly support this mode of action. This would also explain the further increase in PP pathway flux in a ccpN gapA double mutant (Fig. 2). The absence of the preferred glycolytic glyceraldehyde-3-phosphate dehydrogenase could lead to a further increase of metabolites in the upper glycolysis, which in turn would lead to the very high PP pathway flux of the ccpN gapA double mutant. From our results we thus conclude that metabolic jamming of the upper part of glycolysis is the main cause of the redirection of the flux through the PP pathway. However, our results do not allow us to exclude the possibility that an as-yet-unknown posttranscriptional or allosteric mechanism strengthens the mass action law effect in enhancing the activity of one or several PP pathway enzymes in response to higher concentrations of the upper glycolysis metabolites. A transcriptional mechanism, however, is unlikely since our transcriptional analysis revealed no change in PP pathway mRNAs, in agreement with previously published studies that always found this pathway constitutively transcribed (2). Finally, an as-yet-unknown gapB-independent regulatory effect of CcpN, abolished in the ccpN mutant, could have a role in the increase of the PP pathway flux besides the mass action law in response to higher G6P concentration. Such a hypothetical mechanism could explain why the overexpression of gapB in a wild-type context did not fully mimic a ccpN knockout regarding the PP pathway flux (65% versus 100% increase, respectively), despite an equivalent amount of GapB activity measured in the cells. Nevertheless, we demonstrated that the multifaceted phenotype of a ccpN knockout strain, i.e., low growth rate and high PP pathway flux, can be fully suppressed by the inactivation of the three known targets of CcpN, namely, gapB, pckA, and sr1 (Fig. 2 and 3). Clearly, this does not argue in favor of another, as-yet-unknown, regulatory role of CcpN on one or several other targets.
It was not surprising to find no contribution of SR1 to the high PP pathway flux of a ccpN mutant (Fig. 2, compare the ccpN and ccpN sr1 mutants) because its only known target is AhrC, a transcriptional regulator of the rocABC and rocDEF operons involved in arginine catabolism (21). However, unexpectedly, the PP pathway flux in a ccpN gapB pckA sr1 quadruple mutant was significantly lower, and in fact identical to that of the wild-type strain, than in the triple ccpN gapB pckA mutant. These results indicate that SR1 had no contribution to the high PP flux in a ccpN knockout mutant but a slight one when neither GapB nor PckA is expressed. With no obvious role of the roc operon in the flux through the PP pathway, this suggests that AhrC or more probably SR1 could have other targets that would contribute, under certain conditions, to the repartition of the carbon flow between glycolysis and PP pathway.
In conclusion, our genetic, transcriptomic, fluxomic, and metabolomic data allowed to elucidate the complex phenotype of B. subtilis missing the transcriptional regulator CcpN during growth on glucose. Our analysis reveals (i) that the induced gluconeogenic flux through the PckA cannot be fully compensated by the anaplerotic reaction through the PycA, thus preventing a sufficient aspartate synthesis which leads to reduced growth, and (ii) that the derepressed synthesis of GapB leads to a metabolic jamming, resulting in high hexose phosphate concentrations that cause the increase of the PP pathway flux. The mass action law alone or its combination with a yet-unknown control mechanism of the PP pathway activity orchestrates the response to higher metabolite concentration in the upper part of the glycolysis. Thus, a strong PP pathway flux can be predicted in wild-type B. subtilis under gluconeogenic conditions where CcpN repressor activity is inhibited, as observed in our recent flux measurements (unpublished data). Altogether, the present study provides a detailed understanding of the complex physiological role of CcpN in the control of the central carbon fluxes.
Supplementary Material
Acknowledgments
We are very grateful to Sabine Brantl, David Rudner, and Boris Belitsky for the generous gifts of pINT1, pDR111, and the BB1462 B. subtilis aspB mutant strain, respectively. We thank Stéphane Robin for help in statistical analysis of the transcriptome data, Jennifer Ewald for providing the GC-TOF protocol and assistance during the analysis, and Matthieu Jules for critical reading of the manuscript. We also thank Roelco Kleijn for providing the metabolite extraction protocol and help in data analysis.
Financial support was received from the French Agence Nationale pour la Recherche (grant DynamoCell).
Footnotes
Published ahead of print on 27 June 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Anagnostopoulos, C., and I. P. Crawford. 1961. Transformation studies on the linkage of markers in the tryptophan pathway in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 47378-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aymerich, S., A. Goelzer, and V. Fromion. 2007. Transcriptional controls of the central carbon metabolism in Bacillus subtilis, p. 29-73. In Y. Fujita (ed.), Global regulatory networks in Bacillus subtilis. Transworld Research Network, Trivandrum, India.
- 3.Aymerich, S., G. Gonzy-Treboul, and M. Steinmetz. 1986. 5′-Noncoding region sacR is the target of all identified regulation affecting the levansucrase gene in Bacillus subtilis. J. Bacteriol. 166993-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blencke, H. M., G. Homuth, H. Ludwig, U. Mader, M. Hecker, and J. Stulke. 2003. Transcriptional profiling of gene expression in response to glucose in Bacillus subtilis: regulation of the central metabolic pathways. Metab. Eng. 5133-149. [DOI] [PubMed] [Google Scholar]
- 5.Brückner, R., and F. Titgemeyer. 2002. Carbon catabolite repression in bacteria: choice of the carbon source and autoregulatory limitation of sugar utilization. FEMS Microbiol. Lett. 209141-148. [DOI] [PubMed] [Google Scholar]
- 6.Chao, Y. P., and J. C. Liao. 1994. Metabolic responses to substrate futile cycling in Escherichia coli. J. Biol. Chem. 2695122-5126. [PubMed] [Google Scholar]
- 7.Chao, Y. P., R. Patnaik, W. D. Roof, R. F. Young, and J. C. Liao. 1993. Control of gluconeogenic growth by pps and pck in Escherichia coli. J. Bacteriol. 1756939-6944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doan, T., and S. Aymerich. 2003. Regulation of the central glycolytic genes in Bacillus subtilis: binding of the repressor CggR to its single DNA target sequence is modulated by fructose-1,6-bisphosphate. Mol. Microbiol. 471709-1721. [DOI] [PubMed] [Google Scholar]
- 9.Doan, T., P. Servant, S. Tojo, H. Yamaguchi, G. Lerondel, K. Yoshida, Y. Fujita, and S. Aymerich. 2003. The Bacillus subtilis ywkA gene encodes a malic enzyme and its transcription is activated by the YufL/YufM two-component system in response to malate. Microbiology 1492331-2343. [DOI] [PubMed] [Google Scholar]
- 10.Emmerling, M., M. Dauner, A. Ponti, J. Fiaux, M. Hochuli, T. Szyperski, K. Wüthrich, J. E. Bailey, and U. Sauer. 2002. Metabolic flux responses to pyruvate kinase knockout in Escherichia coli. J. Bacteriol. 184152-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fillinger, S., S. Boschi-Muller, S. Azza, E. Dervyn, G. Branlant, and S. Aymerich. 2000. Two glyceraldehyde-3-phosphate dehydrogenases with opposite physiological roles in a nonphotosynthetic bacterium. J. Biol. Chem. 27514031-14037. [DOI] [PubMed] [Google Scholar]
- 12.Fischer, E., and U. Sauer. 2005. Large-scale in vivo flux analysis shows rigidity and suboptimal performance of Bacillus subtilis metabolism. Nat. Genet. 37636-640. [DOI] [PubMed] [Google Scholar]
- 13.Fischer, E., and U. Sauer. 2003. Metabolic flux profiling of Escherichia coli mutants in central carbon metabolism using GC-MS. Eur. J. Biochem. 270880-891. [DOI] [PubMed] [Google Scholar]
- 14.Fischer, E., N. Zamboni, and U. Sauer. 2004. High-throughput metabolic flux analysis based on GC-MS derived 13C-constraints. Anal. Biochem. 325308-316. [DOI] [PubMed] [Google Scholar]
- 15.Fouet, A., and A. L. Sonenshein. 1990. A target for carbon source-dependent negative regulation of the citB promoter of Bacillus subtilis. J. Bacteriol. 172835-844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Foy, J. J., and J. K. Bhattacharjee. 1978. Biosynthesis and regulation of fructose-1,6-bisphosphatase and phosphofructokinase in Saccharomyces cerevisiae grown in the presence of glucose and gluconeogenic carbon sources. J. Bacteriol. 136647-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fujita, Y., Y. Miwa, S. Tojo, and M. Hirooka. 2007. Carbon catabolite control and metabolic networks are mediated by the CcpA protein in Bacillus subtilis, p. 92-110. In Y. Fujita (ed.), Global regulatory networks in Bacillus subtilis. Transworld Research Network, Trivandrum, India.
- 18.Gancedo, J. M. 1998. Yeast carbon catabolite repression. Microbiol. Mol. Biol. Rev. 62334-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gutierrez-Rios, R. M., J. A. Freyre-Gonzalez, O. Resendis, J. Collado-Vides, M. Saier, and G. Gosset. 2007. Identification of regulatory network topological units coordinating the genome-wide transcriptional response to glucose in Escherichia coli. BMC Microbiol. 753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harwood, C. R., and S. M. Cutting. 1990. Molecular biological methods for Bacillus. John Wiley & Sons, Ltd., Chichester, England.
- 21.Haselbeck, R. J., and L. McAlister-Henn. 1993. Function and expression of yeast mitochondrial NAD- and NADP-specific isocitrate dehydrogenases. J. Biol. Chem. 26812116-12122. [PubMed] [Google Scholar]
- 22.Heidrich, N., A. Chinali, U. Gerth, and S. Brantl. 2006. The small untranslated RNA SR1 from the Bacillus subtilis genome is involved in the regulation of arginine catabolism. Mol. Microbiol. 62520-536. [DOI] [PubMed] [Google Scholar]
- 23.Henkin, T. M. 1996. The role of CcpA transcriptional regulator in carbon metabolism in Bacillus subtilis. FEMS Microbiol. Lett. 1359-15. [DOI] [PubMed] [Google Scholar]
- 24.Henkin, T. M., F. J. Grundy, W. L. Nicholson, and G. H. Chambliss. 1991. Catabolite repression of alpha-amylase gene expression in Bacillus subtilis involves a trans-acting gene product homologous to the Escherichia coli lacl and galR repressors. Mol. Microbiol. 5575-584. [DOI] [PubMed] [Google Scholar]
- 25.Jin, S., and A. L. Sonenshein. 1994. Identification of two distinct Bacillus subtilis citrate synthase genes. J. Bacteriol. 1764669-4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kresnowati, M. T., W. A. van Winden, M. J. Almering, A. ten Pierick, C. Ras, T. A. Knijnenburg, P. Daran-Lapujade, J. T. Pronk, J. J. Heijnen, and J. M. Daran. 2006. When transcriptome meets metabolome: fast cellular responses of yeast to sudden relief of glucose limitation. Mol. Syst. Biol. 249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lerondel, G., T. Doan, N. Zamboni, U. Sauer, and S. Aymerich. 2006. YtsJ has the major physiological role of the four paralogous malic enzyme isoforms in Bacillus subtilis. J. Bacteriol. 1884727-4736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Licht, A., and S. Brantl. 2006. Transcriptional repressor CcpN from Bacillus subtilis compensates asymmetric contact distribution by cooperative binding. J. Mol. Biol. 364434-448. [DOI] [PubMed] [Google Scholar]
- 29.Licht, A., S. Preis, and S. Brantl. 2005. Implication of CcpN in the regulation of a novel untranslated RNA (SR1) in Bacillus subtilis. Mol. Microbiol. 58189-206. [DOI] [PubMed] [Google Scholar]
- 30.Lorca, G. L., Y. J. Chung, R. D. Barabote, W. Weyler, C. H. Schilling, and M. H. Saier, Jr. 2005. Catabolite repression and activation in Bacillus subtilis: dependency on CcpA, HPr, and HprK. J. Bacteriol. 1877826-7839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ludwig, H., G. Homuth, M. Schmalisch, F. M. Dyka, M. Hecker, and J. Stulke. 2001. Transcription of glycolytic genes and operons in Bacillus subtilis: evidence for the presence of multiple levels of control of the gapA operon. Mol. Microbiol. 41409-422. [DOI] [PubMed] [Google Scholar]
- 32.Moreno, M. S., B. L. Schneider, R. R. Maile, W. Weyler, and M. H. Saier, Jr. 2001. Catabolite repression mediated by the CcpA protein in Bacillus subtilis: novel modes of regulation revealed by whole-genome analyses. Mol. Microbiol. 391366-1381. [DOI] [PubMed] [Google Scholar]
- 33.Murai, T., M. Tokushige, J. Nagai, and H. Katsuki. 1971. Physiological functions of NAD- and NADP-linked malic enzymes in Escherichia coli. Biochem. Biophys. Res. Commun. 43875-881. [DOI] [PubMed] [Google Scholar]
- 34.Paulus, H. 1993. Biosynthesis of the aspartate family of amino acids, p. 237-267. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria. American Society for Microbiology, Washington, DC.
- 35.Sauer, U. 2006. Metabolic networks in motion: 13C-based flux analysis. Mol. Syst. Biol. 262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sauer, U., and B. J. Eikmanns. 2005. The PEP-pyruvate-oxaloacetate node as the switch point for carbon flux distribution in bacteria. FEMS Microbiol. Rev. 29765-794. [DOI] [PubMed] [Google Scholar]
- 37.Servant, P., D. Le Coq, and S. Aymerich. 2005. CcpN (YqzB), a novel regulator for CcpA-independent catabolite repression of Bacillus subtilis gluconeogenic genes. Mol. Microbiol. 551435-1451. [DOI] [PubMed] [Google Scholar]
- 38.Sonenshein, A. L. 2007. Control of key metabolic intersections in Bacillus subtilis. Nat. Rev. Microbiol. 5917-927. [DOI] [PubMed] [Google Scholar]
- 39.Stülke, J., and W. Hillen. 1999. Carbon catabolite repression in bacteria. Curr. Opin. Microbiol. 2195-201. [DOI] [PubMed] [Google Scholar]
- 40.Sung, M. H., K. Tanizawa, H. Tanaka, S. Kuramitsu, H. Kagamiyama, K. Hirotsu, A. Okamoto, T. Higuchi, and K. Soda. 1991. Thermostable aspartate aminotransferase from a thermophilic Bacillus species: gene cloning, sequence determination, and preliminary X-ray characterization. J. Biol. Chem. 2662567-2572. [PubMed] [Google Scholar]
- 41.Warner, J. B., and J. S. Lolkema. 2003. CcpA-dependent carbon catabolite repression in bacteria. Microbiol. Mol. Biol. Rev. 67475-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Westergaard, S. L., A. P. Oliveira, C. Bro, L. Olsson, and J. Nielsen. 2007. A systems biology approach to study glucose repression in the yeast Saccharomyces cerevisiae. Biotechnol. Bioeng. 96134-145. [DOI] [PubMed] [Google Scholar]
- 43.Yoshida, K., K. Kobayashi, Y. Miwa, C. M. Kang, M. Matsunaga, H. Yamaguchi, S. Tojo, M. Yamamoto, R. Nishi, N. Ogasawara, T. Nakayama, and Y. Fujita. 2001. Combined transcriptome and proteome analysis as a powerful approach to study genes under glucose repression in Bacillus subtilis. Nucleic Acids Res. 29683-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zamboni, N., E. Fischer, and U. Sauer. 2005. FiatFlux-a software for metabolic flux analysis from 13C-glucose experiments. BMC Bioinformatics 6209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zamboni, N., and U. Sauer. 2003. Knockout of the high-coupling cytochrome aa3 oxidase reduces TCA cycle fluxes in Bacillus subtilis. FEMS Microbiol. Lett. 226121-126. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.