Abstract
Legionella pneumophila is an intracellular pathogen that has been shown to utilize the Icm/Dot type IV secretion system for pathogenesis. This system was shown to be composed of Icm/Dot complex components, accessory proteins, and a large number of translocated substrates. In this study, comparison of the icmQ regulatory regions from many Legionella species revealed a conserved regulatory sequence that includes the icmQ −10 promoter element. Mutagenesis of this conserved regulatory element indicated that each of the nucleotides in it affects the level of expression of the icmQ gene but not in a uniform fashion. A genomic analysis discovered that four additional genes in L. pneumophila contain this conserved regulatory sequence, which was found to function similarly in these genes as well. Examination of these four genes indicated that they are dispensable for intracellular growth, but two of them were found to encode new Icm/Dot translocated substrates (IDTS). Comparison of the genomic regions encoding these two IDTS among the four available L. pneumophila genomic sequences indicated that one of these genes is located in a hypervariable genomic region, which was shown before to contain an IDTS-encoding gene. Translocation analysis that was performed for nine proteins encoded from this hypervariable genomic region indicated that six of them are new IDTS which are translocated into host cells in an Icm/Dot-dependent manner. Furthermore, a bioinformatic analysis indicated that additional L. pneumophila genomic regions that contain several neighboring IDTS-encoding genes are hypervariable in gene content.
Legionella pneumophila, the causative agent of Legionnaires' disease, is an intracellular pathogen that multiplies in nature inside amoebae and in human macrophages during disease (29, 48). The icm/dot type IV secretion system is the major virulence system known in L. pneumophila. This system consists of 26 Icm/Dot proteins, 18 of which are homologous to conjugation-related proteins and probably constitute the secretion complex itself (reviewed in reference 52). Two of the Icm/Dot proteins that do not contain homologous proteins in conjugative systems are IcmR and IcmQ, and they were shown previously to be essential components of the Icm/Dot system (53). IcmQ was shown to self-polymerize and to possess pore-forming activity, and IcmR was shown to function as a chaperon of IcmQ and to regulate its activity (16, 17); in addition, both proteins were found not to be translocated into host cells (14). Previously, we analyzed the genomic region expected to contain these two genes from many Legionella species and our analysis revealed the presence of a conserved icmQ gene and a large hypervariable gene family named fir genes (functional homologues of icmR), located at the icmR genomic position (19, 20). Although hypervariable in sequence, the fir genes from all of these Legionella species were found, together with their corresponding icmQ genes, to function similarly during infection and most of them were found to be regulated by the CpxR and PmrA response regulators, while other fir genes were found to be regulated by one of these regulators (such as the L. pneumophila icmR gene, which was found to be regulated by CpxR) (18).
The CpxR response regulator is part of a two-component system which includes the CpxA cognate inner membrane sensor histidine kinase (47). It has been found that this two-component system is activated in Escherichia coli by periplasmic stress, such as accumulation of misfolded proteins in the bacterial periplasm (46). The PmrA response regulator is also part of a two-component system, which includes the PmrB cognate inner membrane sensor histidine kinase. The PmrAB system has also been found to be present in different pathogenic bacteria, such as Salmonella enterica serovar Typhimurium (25), Pseudomonas aeruginosa (39), and Erwinia carotovora (30). This system in S. enterica was shown to be responsible for the induction of genes that encode enzymes that are involved in the modification of bacterial lipopolysaccharide as a response to specific stimuli from the environment, such as extracytoplasmic Fe3+ and low pH, thus gaining resistance to host antimicrobial peptides (55). In L. pneumophila, the CpxRA and PmrAB two-component systems were shown to directly regulate the expression of substrates of the Icm/Dot secretion system (1, 56).
During recent years, a large number of proteins (RalF, LidA, Lep, Sid, Vip, Wip, Ylf, Leg, Vpd, Drr, Ceg, and LubX) were shown to translocate into host cells via the Icm/Dot secretion system (reviewed in reference 44); in most cases, the function of these proteins during infection is not known, but a few of them already have an associated function (3, 12, 31-34, 37, 38, 42, 43). Even though a large number of proteins were shown to be translocated into host cells by the Icm/Dot secretion system (more then 70 proteins), a similar number of proteins was also suggested to be putative translocated substrates based on different bioinformatic analyses (5, 15, 56). The validated Icm/Dot translocated substrates (IDTS) as well as the putative ones were found to contain several common features, including the following: (i) most of the IDTS contain no sequence homology to any known proteins, but a few of them contain homology to eukaryotic proteins; (ii) many of the IDTS contain one or more paralogs in the L. pneumophila genome; and (iii) some of the IDTS-encoding genes are located close to one another.
One of the approaches used before to identify new IDTS in L. pneumophila was to identify conserved regulatory elements of known IDTS, use these conserved regulatory elements to perform genomic searches to identify additional genes that contain these sequences, and examine the genes identified to determine whether they encode new IDTS (1, 56). In this report we identified and characterized the icmQ regulatory signature and used this sequence to identify seven new IDTS, six of which are located one next to the other in a hypervariable genomic region. We suggest that L. pneumophila hypervariable genomic regions tend to encode IDTS.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The L. pneumophila strains used in this study were L. pneumophila JR32, a streptomycin-resistant, restriction-negative mutant of L. pneumophila Philadelphia-1, which is a wild-type strain in terms of intracellular growth (49); GS3011, an icmT mutant (57); HK-PQ1, a pmrA mutant (56); OG3002, a cpxR mutant (21); LM1367, an rpoS mutant (26); OG3001, a letA mutant (22); ZT-001, an lpg0541 mutant; ZT-003, an lpg1836 mutant; ZT-004, an lpg1962 mutant; and ZT-005, an lpg1983 mutant. The E. coli strains used were MC1022, containing the M15 deletion of the lacZ gene (9), and MC1061, containing a deletion in the lacZ gene (10). Bacterial media, plates, and antibiotic concentrations were used as described previously (53). For the plasmids and primers used in this study, see Tables S1 and S2 in the supplemental material, respectively.
Construction of lacZ translational fusions.
To generate lacZ translational fusions, the regulatory regions of the genes of interest (about 300 bp) were amplified by PCR with the primers described in Table S2 in the supplemental material. The PCR products were then digested with BamHI and EcoRI, cloned into pGS-lac-02, and sequenced to generate the plasmids listed in Table S1 in the supplemental material. The levels of expression from these plasmids were measured by a β-galactosidase assay as previously described (40, 56).
Construction of site-directed mutations.
To generate substitutions in the regulatory regions of the genes examined, site-directed mutagenesis was performed on the consensus sequences by a PCR overlap extension approach (28) in a manner similar to that described before (56). In all of the mutations constructed, the changes were always A to C, C to A, G to T, and T to G (except in one case where a G-to-A change was performed). The primers used for the mutagenesis are described in Table S2 in the supplemental material, and the plasmids resulting from the site-directed mutagenesis are listed in Table S1 in the supplemental material.
Construction of plasmids for allelic exchange.
To generate a deletion substitution in the L. pneumophila lpg0541 and lpg1983 genes, a 1-kb DNA fragment located on each side of the deletion planned was amplified by PCR using the primers listed in Table S2 in the supplemental material. The two fragments that were amplified for each gene were cloned into pUC-18 digested with HincII to generate the plasmids listed in Table S1 in the supplemental material and sequenced to confirm that no mutations were incorporated during the PCR. The resulting plasmids were digested to take out the inserts and used for a four-way ligation that contains the kanamycin (Km) resistance cassette (Pharmacia) and the pUC-18 vector to generate the plasmids listed in Table S1 in the supplemental material. To generate insertion mutations in lpg1836 and lpg1962, a 2-kb fragment was amplified by PCR using the primers listed in Table S2 in the supplemental material and cloned into pUC-18 to generate the plasmids listed in Table S1 in the supplemental material. The two resulting plasmids were digested with SmaI (for lpg1836) and HpaI (for lpg1962), and the Km resistance cassette was cloned into the genes. The four resulting plasmids containing the Km resistance cassette were digested with PvuII, and the resulting fragments were cloned into the pLAW344 allelic exchange vector digested with EcoRV to generate the plasmids listed in Table S1 in the supplemental material, which were used for allelic exchange of lpg0541, lpg1836, lpg1962, and lpg1983, carried out as described before (53).
Construction of cyaA fusions.
The plasmid pMMB-cyaA-C (56) was used for the cloning of all cyaA fusions constructed. All genes examined were amplified by PCR using a pair of primers (see Table S2 in the supplemental material) containing suitable restriction sites at the 5′ end. The PCR products were subsequently digested with the relevant enzymes and cloned into the pMMB-cyaA-C vector to generate the plasmids listed in Table S1 in the supplemental material. All of the inserts were sequenced to verify that no mutations were incorporated during the PCR.
Analysis of mRNA transcription start site.
RNA preparation was performed as described elsewhere (21). To determine the transcription start site of the gene of interest, 5′ rapid amplification of cDNA ends (RACE) (Invitrogen) was performed as described by the manufacturer. The RACE-GSP primer (see Table S2 in the supplemental material) was used for generating the cDNA; the RACE-1 primer (see Table S2 in the supplemental material), together with the AA (abridged-anchor) primer supplied with the kit, were used for the first PCR; and the AUA (abridged universal amplification) primer supplied with the kit and the RACE-2 primer (see Table S2 in the supplemental material) were used for the nested PCR. The resulting fragment from the second PCR was subsequently digested with SalI and BamHI and cloned into pUC-18 digested with the same enzymes, and several different clones were sequenced to determine the transcription start site of the mRNA.
Quantitative RT-PCR analysis.
RNA preparation was performed as described elsewhere (21). Total RNA was digested with an RNase-free DNase reagent (Promega). The reverse transcription-PCR (RT-PCR) contained the following components: 10 μg of total RNA, 20 pmol of all of the reverse primers (see Table S2 in the supplemental material), 40 U of RNase inhibitor (RNasin; Promega), and 10 U of avian myeloblastosis virus reverse transcriptase (Promega). The reaction mix was incubated for 5 min at 45°C, and cDNA synthesis was conducted at 37°C for 1 h. A 1/50 dilution of the RT reaction mix was used for PCR using the following cycle conditions: 30 s at 95°C, 30 s at 50°C, and 30 s at 72°C for extension. The number of cycles varied according to the level of expression of the various mRNAs, and no-RT controls were performed for each of the genes examined.
Intracellular-growth and translocation analyses.
Intracellular-growth assays with Acanthamoeba castellanii and with HL-60-derived macrophages were performed similarly to those previously described (54). A CyaA translocation assay was performed as described before (1), and the presence of the CyaA fusion proteins was detected by Western blot analysis using monoclonal anti-CyaA 3D1 (Santa Cruz Biotechnology, Inc.) diluted 1:500 and goat anti-mouse immunoglobulin G conjugated to horseradish peroxidase (Jackson Immunoresearch Laboratories, Inc.) diluted 1:10,000.
Bioinformatic analyses.
Genomic analysis for finding the genes containing the icmQ regulatory element was performed using the RSAT web site (http://rsat.ulb.ac.be/rsat/). Homology searches were performed using NCBI BLAST (2).
RESULTS
icmQ genes from different Legionella species contain a conserved regulatory element.
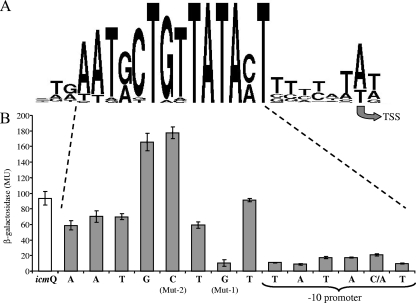
The fir-icmQ region was previously cloned from many Legionella species, and the fir genes were found to contain conserved regulatory elements in their upstream regulatory region which serve as binding sites for the CpxR and PmrA regulators (18). An analysis of the L. pneumophila icmQ gene that was performed before indicated that the gene contains a regular −10 promoter element, most likely recognized by the RpoD sigma factor (23). Analysis of the upstream regulatory regions of the icmQ genes from all of the Legionella species indicated above revealed a conserved sequence located immediately upstream from the −10 promoter element of the icmQ genes (Fig. 1), but no PmrA or CpxR sites were identified. The −10 promoter element of each of the different icmQ genes was found to consist of the sequence TATACT or TATAAT (positions −12 to −7) (the positions indicated are related to the transcription start site), and immediately upstream of this sequence (positions −20 to −13) the sequence AATRCTGT (R is A or G) was found (Fig. 1). It was shown before for several bacterial promoters that a 5′-TGN-3′ motif (positions −15 to −13), located immediately upstream from the −10 promoter element, functions as part of the promoter −10 regulatory element necessary for transcription initiation, and this motif was named “extended −10” (4, 7, 50). The icmQ regulatory element described here was found to contain this motif at the same location that was described before for extended −10 promoter elements, but it was found to be part of a larger conserved element (Fig. 2A).
FIG. 1.
The regulatory regions of the icmQ genes from different Legionella species contain a conserved regulatory element. The regulatory sequences of the icmQ genes from 27 Legionella species were aligned. The regulatory elements are in bold, the L. pneumophila icmQ transcription start site is marked in bold and underlined, and the distances to the first ATG are indicated. The conserved regulatory element is indicated at the bottom (R is A or G, and M is A or C). The Legionella species from which the icmQ regulatory regions were aligned (and their accession numbers) are L. longbeachae (Lng, AY512558), L. gratiana (Gra, AY860642), L. worsleiensis (Wor, AY860646), L. shakespearei (Sha, AY860647), L. moravica (Mor, AY860644), L. quateirensis (Qua, AY860645), L. waltersii (Wal, AY860648), L. pneumophila (Lpn, Y12705), L. hackeliae (Hac, AY753534), L. jamestowniensis (Jam, AY860649), L. brunensis (Bru, AY860650), L. feeleii (Fee, AY753535), L. jordanis (Jor, AY860651), L. fairfieldensis (Fai, AY860653), L. lansingensis (Lan, AY860652), L. micdadei (Mic, AY512559), L. maceachernii (Mac, AY860654), L. nautarum (Nau, AY860655), L. drozanskii (Dro, AY860662), L. birminghamensis (Bir, AY860641), L. quinlivanii (Qui, AY860656), L. erythra (Ery, AY860658), L. rubrilucens (Rub, AY860659), L. spiritensis (Spi, AY860657), L. israelensis (Isr, AY860663), L. londiniensis (Lon, AY860660), and L. adelaidensis (Ade, AY860661).
FIG. 2.
Mutation analysis of the conserved icmQ regulatory signature. (A) Sequence logo of the conserved icmQ signature, representing the most conserved nucleotides in the sequence. The transcription start site (TSS) that was determined for the L. pneumophila icmQ gene is indicated. (B) Each of the nucleotides in the conserved icmQ regulatory signature was mutated (as described in Materials and Methods), and the effects of the different mutations are shown in comparison to the wild-type levels of expression (white bar). The levels of expression were measured at early stationary phase by using a β-galactosidase assay as described in Materials and Methods. The results (Miller units [MU]) are the averages ± standard deviations of at least three independent experiments.
The icmQ conserved motif affects the level of expression of the icmQ gene.
To determine the importance of the consensus sequence identified for the expression of the icmQ gene, we used the L. pneumophila icmQ::lacZ fusion and mutated each of the conserved nucleotides in the motif (see Materials and Methods); the results of this analysis are presented in Fig. 2B. As was shown before, all of the mutations in the −10 promoter element itself resulted in a dramatic reduction in the level of expression of the icmQ::lacZ fusion (23). A similar result was obtained with the G residue which is part of the extended −10 promoter, and a less pronounced reduction in the level of expression was observed with the upstream T residue, which is also part of the extended −10 sequence (Fig. 2B). However, the mutations constructed in the G and C residues located immediately upstream from the TGN extended −10 motif resulted in an increase (twofold) in the level of expression of the icmQ::lacZ fusion (Fig. 2B). The mutations constructed in the residues located further upstream in the consensus (AAT) (Fig. 2A) resulted in a minor effect on the level of expression of the icmQ::lacZ fusion (Fig. 2B). These results clearly indicate that each of the residues in the RCTGTATAMT motif (R is G or A, and M is A or C) affects the expression of the icmQ gene and can be considered a conserved icmQ regulatory signature.
Identification of new genes containing the icmQ regulatory signature.
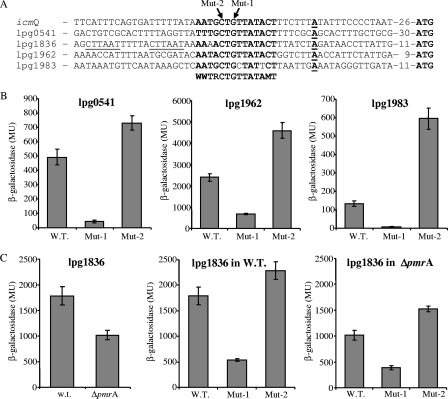
The relative conservation of the icmQ regulatory signature found in the different Legionella species, together with the fact that IcmQ is a component of the major L. pneumophila pathogenesis system, led us to search for additional genes that contain this conserved regulatory element in the L. pneumophila genome. A genomic search was performed using a whole-genome DNA pattern analysis, allowing one nucleotide mismatch in nucleotides that had a dramatic effect on the level of expression of the icmQ gene and up to two changes in the nucleotides that had a minor effect on the level of expression, as was deduced from the results presented in Fig. 2B. The analysis performed resulted in the identification of four L. pneumophila genes that contain the icmQ regulatory signature (Fig. 3A): (i) lpg0541, a predicted transmembrane protein located next to milX (lpg0540) (27) (MilX [PhtA] belongs to a family of phagosomal transporters which were shown before to be required for intracellular growth [51]); (ii) lpg1836 (ceg25), which was suggested before as a putative IDTS-encoding gene, since it contains a PmrA regulatory element (several other Ceg proteins were shown before to be translocated into host cells [1, 56]); (iii) lpg1962, a predicted peptidyl-prolyl cis-trans isomerase which was found to be located in a hypervariable genomic region (see below); and (iv) lpg1983, whose product contains no significant homology to other proteins.
FIG. 3.
The icmQ regulatory signature is found in four additional genes in the L. pneumophila genome. (A) Sequence alignment of the regulatory regions of icmQ, lpg0541, lpg1836 (ceg25), lpg1962, and lpg1983. The putative consensus sequence is in boldface, the transcription start sites were determined for all of the genes and are marked in bold and underlined, the putative PmrA binding site is underlined for lpg1836 (ceg25), and the distances to the first ATG are indicated. (B) The regulatory element identified is functional also in the four new genes found. The expression levels of lacZ fusions containing the wild-type (W.T.) regulatory region as well as two mutations (Mut-1 and Mut-2) (Fig. 2) in the regulatory signatures of lpg0541, lpg1962, and lpg1983 were determined at early stationary phase. (C) Analysis of the expression of lpg1836 (ceg25) in the wild-type strain and the pmrA deletion mutant (ΔpmrA) (left). The effects of the two mutations, indicated in panel B, on the level of expression of lpg1836 in the wild-type strain (middle) and in the pmrA deletion mutant (right) are shown. The levels of expression were determined using the β-galactosidase assay as described in Materials and Methods. Statistical analyses of the expression of the wild-type fusion and each of the mutants (Mut-1 and Mut-2) were performed using a standard t test, and the results were found to be significantly different (P < 0.0001) in all of them. The results (Miller units [MU]) are the averages ± standard deviations of at least three independent experiments.
The icmQ regulatory signature is involved in the regulation of the new genes identified.
To examine the involvement of the icmQ regulatory signature in the regulation of the four new genes identified, the transcription start site of each gene was determined using the RACE system, and it was found to be located at the same position as in the icmQ gene, indicating that the −10 promoter element of these genes constitutes the downstream 6 nucleotides of the consensus sequence identified (Fig. 3A). In addition, to determine the involvement of the upstream part of the consensus, which is in part unique for the icmQ regulatory signature, two mutations were constructed. The mutations were constructed in the 2 nucleotides that resulted in the most dramatic effect on the level of expression of the icmQ gene—the first (Mut-1) decreased and the second (Mut-2) increased the level of expression of the icmQ gene (Fig. 2B). As can be seen in Fig. 3B and C (middle), a result similar to that obtained with the icmQ gene was also obtained with the four new genes examined, i.e., mutation Mut-1 caused a reduction in the level of expression and mutation Mut-2 caused an increase in the level of expression.
The position of the transcription start sites together with the effect of the two mutations constructed on the level of expression of the four new genes clearly indicates that the regulatory sequence identified functions as the regulatory element of these four genes as well.
Lpg1836 is regulated by PmrA and by the new regulatory element.
As indicated above, lpg1836 (ceg25) was suggested before as a potential IDTS since it was found to contain a PmrA regulatory element (Fig. 3A). To determine if this gene is regulated by PmrA, we examined its level of expression in a pmrA deletion mutant, and a level of expression lower than that for the wild-type strain was obtained (Fig. 3C, left). In order to examine whether the icmQ regulatory signature found in this gene and the PmrA regulatory element affect one another, we examined the level of expression of the lpg1836::lacZ fusion containing the two mutations in the icmQ regulatory signature also in the pmrA deletion mutant (Fig. 3C, right). The results obtained clearly indicate that the two mutations examined had the same effect in the wild-type strain and in the pmrA deletion mutant (even though the overall level of expression was lower in the pmrA mutant), indicating that both the PmrA regulator and the regulatory element identified are involved in the regulation of ceg25.
The four new genes identified are dispensable for intracellular growth.
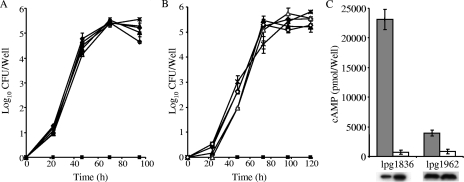
Since the icmQ gene belongs to the main pathogenesis system of Legionella and it was found to be required for intracellular multiplication, we examined whether the four new genes identified are required for intracellular growth. For this purpose, null mutations were introduced in each of the four new genes and the resulting mutants were examined for intracellular growth in the protozoan host A. castellanii and in HL-60-derived human macrophages. As can be seen in Fig. 4A and B, the four new genes identified were found to be dispensable for intracellular growth in the two hosts examined.
FIG. 4.
Involvement of the four new genes in L. pneumophila pathogenesis. Null mutants constructed with mutations in the four new genes (lpg0541, filled triangle; lpg1836, open triangle; lpg1962, ×; and lpg1983, open diamond) were examined for intracellular growth in A. castellanii (A) and HL-60-derived human macrophages (B) in comparison to growth in the wild-type strain (filled diamond) and an icmT deletion mutant (filled square). (C) Translocation of CyaA fusions into HL-60-derived human macrophages was examined for the four new genes from the wild-type strain JR32 (gray) and the icmT mutant (white), and the lpg1836 (ceg25) and lpg1962 CyaA fusions were found to be translocated into host cells in an Icm/Dot-dependent manner. cAMP, cyclic AMP.
lpg1836 and lpg1962 are substrates of the Icm/Dot system.
Since it was shown before that construction of deletion mutants for many of the known IDTS produced no intracellular-growth phenotype, we also examined whether the genes identified encode new IDTS by constructing fusions to the N-terminal domain of the Bordetella pertussis adenylate cyclase (cyaA fusions). Analysis of these fusions for translocation into HL-60-derived human macrophages indicated very clearly that lpg1836 (ceg25) and lpg1962 translocated into host cells in an icm/dot-dependent manner (Fig. 4C). The two other genes examined (lpg0541 and lpg1983) did not show any translocation when examined in the wild-type strain or the icmT mutant (data not shown). All of the CyaA fusions had similar levels in the wild type and the icmT mutant, as deduced by Western blot analysis using an anti-CyaA antibody (Fig. 4C).
The ceg25 gene, which was found here to be translocated into host cells, is currently the seventh ceg gene (together with ceg7, ceg10, ceg18, ceg23, ceg29, and ceg33) that has been found to be translocated into host cells in an Icm/Dot-dependent manner (1, 56).
lpg1962 is located in a hypervariable genomic region.
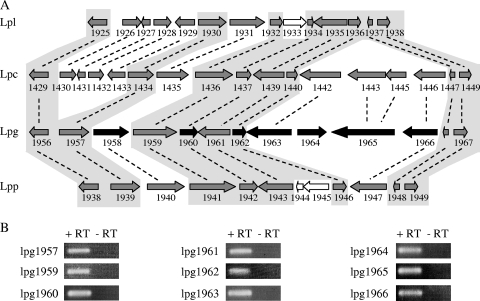
Previously, it was shown that L. pneumophila IDTS-encoding genes are sometimes located in close proximity to one another (34, 35). This observation led us to compare the genomic regions of the two genes that were found to encode new IDTS (lpg1836 and lpg1962) in the four L. pneumophila genomes available (Philadelphia-1, Lens, Paris, and Corby) (11, 13, 24). lpg1836 was found to be located in a conserved genomic region, but the analysis of the region containing lpg1962 revealed that this IDTS is encoded from a hypervariable genomic region (Fig. 5A). The boundaries of this region can be set between two conserved genes: lpg1956 (l-Ala-dl-Glu-epimerase) and lpg1967 (a transcriptional regulator from the TetR family). The numbers of genes found between these two conserved genes and the gene contents differ dramatically among the four L. pneumophila genomes sequenced, and in L. pneumophila Philadelphia-1, one of these genes (lpg1958 [legL5]) was shown before to be translocated into host cells (15). In two sites in this region, a group of three to four genes was found to be present in two out of the four genomes (lpg1958, lpg1963, lpg1965, lpg1966, lpc1430, lpc1431, lpc1432, lpc1433, and lpc1435), one gene was found to be present in three out of the four genomes (lpg1966), and another gene was found only in one of the genomes (lpg1964) (Fig. 5A). In addition, a transposase-encoding gene was found to be present in two of the genomes, but not in the same location and not the same transposase in both genomes (Fig. 5A).
FIG. 5.
lpg1962 is located in a hypervariable genomic region. (A) Schematic presentation of the genes located between lpg1956 and lpg1967 of L. pneumophila Lens (Lpl), L. pneumophila Corby (Lpc), L. pneumophila Philadelphia-1 (Lpg), and L. pneumophila Paris (Lpp). Homologous genes are indicated by dashed lines, genes that encode IDTS are colored in black (lpg1958 [legL5], lpg1960 [lirA], lpg1962 [lirB], lpg1963 [lirC], lpg1964 [lirD], lpg1965 [lirE], and lpg1966 [lirF]), and genes encoding transposases/integrases are marked in white. A gray background indicates genes that are present in all four strains. (B) All of the genes located in this hypervariable region are expressed. The expression of the genes was determined using RT-PCR; the PCR was carried out for 25 cycles for all of the genes (+RT). Equivalent amounts of RNA were used for RT reactions without reverse transcriptase and then examined by PCR for the same number of cycles (−RT), and no product was obtained.
The genes found in the hypervariable genomic region are expressed.
The high variability of the genes in this region (Fig. 5A) and the fact that many of the genes were not present in all of the genomes might suggest that the region contains pseudogenes or genes that are not expressed. To examine whether the genes located in the hypervariable region of L. pneumophila Philadelphia-1 genome are expressed, we used quantitative RT-PCR and found that all of them are expressed (Fig. 5B).
Six of the genes located in the hypervariable region encode new IDTS.
Since two genes located in this region (lpg1958 [legL5] and lpg1962) were found to be translocated into host cells (14) (Fig. 4C), we examined the rest of the genes in this hypervariable region for translocation. CyaA fusions were constructed for all of the genes located between lpg1956 and lpg1967 (besides lpg1958 [legL5]), and they were examined for translocation into host cells from the wild-type strain as well as from an icmT deletion mutant. As can be seen clearly in Fig. 6, six of the genes examined (lpg1960, lpg1962, lpg1963, lpg1964, lpg1965, and lpg1966) were found to be translocated into host cells in an icm/dot-dependent manner. In all of the fusions examined, the levels of the CyaA fusion proteins were found to be similar in the wild type and the icmT mutant, as deduced by Western blot analysis using an anti-CyaA antibody (Fig. 6). The new translocated genes were designated lir for Legionella IDTS region (Table 1).
FIG. 6.
Icm/Dot-dependent translocation of the proteins encoded by genes located at the variable region. Wild-type strain JR32 (gray bars) and icmT mutant GS3011 (white bars) harboring the CyaA fusion proteins (indicated below the bars) were used to infect HL-60-derived human macrophages, and the cyclic AMP (cAMP) levels of the infected cells were determined (as described in Materials and Methods). The data are the means for the amount of cAMP per well obtained, and the error bars indicate standard deviations. A Western blot analysis using anti-CyaA antibody is presented below each bar, indicating that similar levels of the CyaA fusions were present in wild-type JR32 (left) and the icmT mutant (right) used for infection.
TABLE 1.
Genes located in the lpg1962 hypervariable genomic region
| Locus tag | Gene designationa | Size (aa) | Strain(s) in which gene was presentb | Paralog | Domainc |
|---|---|---|---|---|---|
| lpg1957 | 439 | L, P, C | |||
| lpg1958 | legL5 | 534 | P | LRR | |
| lpg1959 | 664 | L*, P, C | |||
| lpg1960 | lirA | 257 | L*, P, C | ||
| lpg1961 | 504 | L, P, C | |||
| lpg1962 | lirB | 188 | L, P, C | PPIASE | |
| lpg1963 | lirC | 699 | C | ||
| lpg1964 | lirD | 436 | |||
| lpg1965 | lirE | 977 | C | lpg1966 | |
| lpg1966 | lirF | 521 | P, C | lpg1965 |
lir, Legionella Icm/Dot translocated substrate region.
L, L. pneumophila strain Lens; P, L. pneumophila strain Paris; C, L. pneumophila strain Corby; *, part of the gene was present.
LRR, leucine-rich repeat; PPIASE, cyclophilin-type peptidyl-prolyl cis-trans isomerase.
Our results indicate that of the seven IDTS that were found to be located in this hypervariable genomic region, only one (lpg1962 [lirB]) was found to be present in all four sequenced L. pneumophila genomes (Table 1). This was also the only gene that was found to contain the conserved icmQ regulatory signature in the regulatory region of the homologous genes in all four genomes.
The level of expression of several new IDTS is affected by PmrA, CpxR, RpoS, and/or LetA.
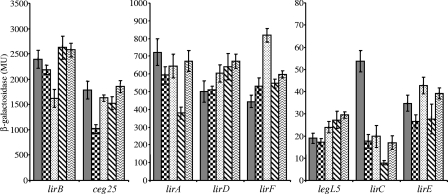
In previous studies, many IDTS were shown to be regulated by the PmrA and/or the CpxR response regulator (1, 56); other IDTS were shown to be upregulated at stationary phase, and those are probably regulated by the RpoS and/or LetA regulator, which was shown to be required for intracellular growth (22, 26, 27, 36). To examine whether the new IDTS identified are also regulated by one or more of these four regulators, lacZ fusions were constructed for all IDTS found in the variable region and their levels of expression were examined in the wild-type strain as well as in deletion mutants of the four regulators indicated (Fig. 7). The analysis performed revealed that most of the genes located in the hypervariable region are expressed at low levels. In addition, the levels of expression of several of the new IDTS were affected by the deletions in the regulators examined. The deletion in cpxR was found to affect the levels of expression of lirB and lirF but in opposite directions (it was shown before that CpxR functions as a repressor or an activator of IDTS [1]). The deletion in rpoS was found to reduce the level of expression of lirA and lirC, as was shown before for sidG and ralF (56). Moreover, the level of expression of lirC was reduced in comparison to that in the wild-type strain in all of the regulators examined, and the most pronounced effect was found with the rpoS deletion mutant. This result might indicate that the four regulators examined are part of a regulatory network. Examination of the lir gene regulatory regions revealed a putative, regular −10 promoter element (data not shown), but no PmrA or CpxR regulatory elements were identified. The ways in which the regulators examined affect the levels of expression of several of the lir genes are currently not known, but the effect most likely occurs in an indirect manner by a putative regulator which might be regulated by one or more of the regulators examined.
FIG. 7.
Examination of the effects of different regulators on the IDTS identified. The levels of expression of the eight IDTS lacZ fusions (lirA, lirB, lirC, lirD, lirE, lirF, legL5, and ceg25) in wild-type strain JR32 (gray), a pmrA deletion mutant (checkers), a cpxR deletion mutant (dots), an rpoS deletion mutant (stripes), and a letA deletion mutant (waves) were examined. The levels of expression were determined using a β-galactosidase assay, as described in Materials and Methods. The results (Miller units [MU]) are the averages ± standard deviations (error bars) of at least three different experiments.
DISCUSSION
Analysis of the icmQ regulatory region from many Legionella species resulted in the identification of a conserved regulatory element (nucleotides −17 to −7, RCTGNTATAMT) which includes the −10 promoter element (nucleotides −12 to −7, TATAMT), an extended −10 promoter element (nucleotides −15 to −13, TGN), and two additional highly conserved nucleotides (positions −17 and −16, RC) (Fig. 2). Mutations constructed in each of these nucleotides affected the level of expression of an icmQ::lacZ fusion. The role of the TGN motif located at positions −15 to −13 is well known, and these nucleotides constitute a common extended −10 motif (4, 7, 50). However, conserved nucleotides at positions −16 and −17 are not common and mutations in these sites resulted in an increase in the level of expression (Fig. 2). The effect of the nucleotides located at positions −16 and −17 on the level of expression of the downstream gene was examined before for three E. coli promoters (6, 41), and the analysis indicated that these nucleotides affect promoter strength in the case of promoters containing an extended −10 motif or regular promoters containing a −35 element (6, 41). The analysis performed with E. coli indicated that the sequence RCTG (R is A or G) at positions −17 to −14 reduced the level of expression two- to fivefold in comparison to that of the TCTG and GATG sequences (6, 41). In the case of the L. pneumophila icmQ promoter as well as the four other genes that were found to contain the sequence RCTG (R is A or G) at positions −17 to −14, a 1.5- to fourfold increase in the level of expression was obtained when the sequence was changed to TCTG or GATG, results which are identical to those obtained with E. coli (the E. coli wild-type promoters examined were identical to the mutated icmQ promoter). In addition, we examined the effect of changing the GCTG sequence present in the L. pneumophila icmQ gene (in all four L. pneumophila genomes available) to the sequence present in other Legionella icmQ genes and in the two IDTS identified (ACTG), and no increase in the level of expression in comparison to that of the wild-type icmQ sequence was observed (78.8 ± 5.1 Miller units), indicating that the G and A nucleotides at position −17 have the same effect on the level of expression of the downstream gene (the same result was also obtained with the E. coli promoter analysis described above). This comparison indicates that the icmQ gene and the four other genes examined contain a promoter element in which the nucleotides at positions −17 and −16 result in a promoter less active than promoters that contain most of the other nucleotides in these positions, since promoters containing the nucleotides RTTG and RCTG were found to be the least active of all promoters (6, 41). This analysis predicts that the five genes containing this regulatory element that were examined in our study will have low levels of expression, and indeed this was found to be the case for three of these genes (icmQ, lpg0541, and lpg1983); however, the two other genes (lpg1836 [ceg25] and lpg1962 [lirB]) were found to have high levels of expression (Fig. 3). In the case of ceg25, this can be explained by the observation that this gene was found to be regulated also by the PmrA activator (Fig. 7); in the case of lirB, it is currently not known which activator regulates its expression, but its expression was reduced in the cpxR deletion mutant (Fig. 7), a result that might indicate that this gene is indeed regulated by a yet-unknown activator.
The examination of the genes containing the icmQ regulatory signature resulted in the identification of lpg1962, or lirB, which was found to be located in a hypervariable genomic region (Fig. 5A). Analysis of the genes located in this region indicated that they are all expressed, and most of them (7 genes out of 10) were found to be translocated into host cells in an Icm/Dot-dependent manner (Table 1 and Fig. 6). In addition, the levels of expression of a few of the genes located in this region were found to be affected by known regulators of IDTS (Fig. 7). Moreover, this genomic region was found to be hypervariable compared to the corresponding regions from the three other L. pneumophila strains for which a complete genomic sequence is available (Fig. 5A). To examine whether this feature is also found in other genomic regions that contain several IDTS-encoding genes, we looked for additional such regions, and five regions were identified (besides the genomic region that was presented in Fig. 5A [lpg1957 to lpg1966]). The regions identified were (i) lpg1144 to lpg1158 {this region contains 15 genes, 3 of which were shown to encode IDTS [lpg1144 (cegC3), lpg1148, and lpg1158] and 1 of which was suggested to encode a putative IDTS [lpg1155]}, (ii) lpg2144 to lpg2157 {this region contains 14 genes, 4 of which were shown to encode IDTS [lpg2144 (legAU13), lpg2155 (sidJ), lpg2156 (sdeB), and lpg2157 (sdeA)] and 2 of which were suggested to encode putative IDTS [lpg2153 and lpg2154]}, (iii) lpg2398 to lpg2411 (Fig. 8A) {this region contains 14 genes, 4 of which were shown to encode IDTS [lpg2400 (legL7), lpg2407, lpg2409 (ceg29), and lpg2410 (vpdA)]}, (iv) lpg2452 to lpg2465 {this region contains 14 genes, 4 of which were shown to encode IDTS [lpg2452 (legA14), lpg2456 (legA15), lpg2464 (sidM or drrA), and lpg2465 (sidD)]}, and (v) lpg2824 to lpg2832 (Fig. 8B) {this region contains nine genes, three of which were shown to encode IDTS [lpg2829 (sidH), lpg2830 (lubX), and lpg2831 (vipD)] and one of which was suggested to encode a putative IDTS [lpg2826 (ceg31)]}. Not even one of these regions was found to be identical among the four sequenced L. pneumophila genomes. Five of these regions were found to be hypervariable (Fig. 5A and 8A and B and data not shown), and the genomic region between lpg1144 and lpg1158 was found to be relatively conserved in the four genomes. The variability of the five regions described includes the IDTS-encoding genes themselves, and the most striking situation is presented in Fig. 8B, in which the sidH gene (lpg2829) is functional and encodes an IDTS in L. pneumophila Philadelphia-1 (27), while it is absent in strains Lens and Corby and an insertion of transposase- and integrase-encoding genes (lpp2884 and lpp2885) was found to be located within this gene in strain Paris. Genes encoding transposases were also found in the region, as presented in Fig. 5A. We think that the variability of the genomic regions described and the high density of IDTS-encoding genes within these regions might indicate that there are hot spots in the L. pneumophila genome that are used for the acquisition of new genes, most likely by horizontal gene transfer, and that some of these genes later adapt and participate in the manipulation of the host cell during infection.
FIG. 8.
Additional regions encoding known IDTS are hypervariable. Schematic presentation of the genomic regions between lpg2398 and lpg2411 (A) and between lpg2824 and lpg2832 (B) in L. pneumophila Lens (Lpl), L. pneumophila Corby (Lpc), L. pneumophila Philadelphia-1 (Lpg), and L. pneumophila Paris (Lpp). Homologous genes are indicated by dashed lines, genes that encode known IDTS are colored in black (lpg2400 [legL7], lpg2407, lpg2409 [ceg29], lpg2410 [vpdA], lpg2829 [sidH], lpg2830 [lubX], and lpg2831 [vipD]), the predicted IDTS-encoding gene lpg2826 (ceg31) is hatched, and genes encoding transposases/integrases are marked in white. A gray background indicates genes that are present in all four strains.
Until now, several features were found to be common for IDTS. (i) Deletion in most of the IDTS-encoding genes examined was found to have no intracellular-growth phenotype (8, 15, 35, 43, 45). (ii) Most of the IDTS are L. pneumophila-unique genes not present in other bacteria, and some of them are homologous to eukaryotic proteins (5, 15, 35, 43, 56). (iii) Even though unique for L. pneumophila, many of the IDTS contain one or more paralogs in the L. pneumophila genome (8, 35, 45). (iv) IDTS-encoding genes are sometimes located next to one another (34, 35), which is also evident from our study. (v) Some of the genomic regions containing IDTS-encoding genes are hypervariable (Fig. 5A and 8A and B). These features and the fact that the number of IDTS found in L. pneumophila is very large (44) might indicate that these bacteria encounter an ongoing challenge when growing intracellularly in a variety of protozoan hosts. It might be that during the course of evolution these bacteria come upon the challenge of the enormous diversity of their protozoan hosts by an ongoing incorporation of changes, which occur randomly, into genomic hot spots that encode IDTS and that in this way they change and increase the repertoire of their IDTS. The ways in which these changes occur and are maintained in the genome are questions for future research.
Supplementary Material
Acknowledgments
We thank Ohad Gal-Mor, Michal Rasis, and Hani Kotzer for plasmid construction.
This work was supported by a grant (882/07) from the Israeli Science Foundation (to G.S.).
Editor: J. B. Bliska
Footnotes
Published ahead of print on 11 August 2008.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Altman, E., and G. Segal. 2008. The response regulator CpxR directly regulates the expression of several Legionella pneumophila icm/dot components as well as new translocated substrates. J. Bacteriol. 1901985-1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 253389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banga, S., P. Gao, X. Shen, V. Fiscus, W. X. Zong, L. Chen, and Z. Q. Luo. 2007. Legionella pneumophila inhibits macrophage apoptosis by targeting pro-death members of the Bcl2 protein family. Proc. Natl. Acad. Sci. USA 1045121-5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barne, K. A., J. A. Bown, S. J. Busby, and S. D. Minchin. 1997. Region 2.5 of the Escherichia coli RNA polymerase sigma70 subunit is responsible for the recognition of the ′extended −10′ motif at promoters. EMBO J. 164034-4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bruggemann, H., C. Cazalet, and C. Buchrieser. 2006. Adaptation of Legionella pneumophila to the host environment: role of protein secretion, effectors and eukaryotic-like proteins. Curr. Opin. Microbiol. 986-94. [DOI] [PubMed] [Google Scholar]
- 6.Burr, T., J. Mitchell, A. Kolb, S. Minchin, and S. Busby. 2000. DNA sequence elements located immediately upstream of the −10 hexamer in Escherichia coli promoters: a systematic study. Nucleic Acids Res. 281864-1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Camacho, A., and M. Salas. 1999. Effect of mutations in the “extended −10” motif of three Bacillus subtilis sigmaA-RNA polymerase-dependent promoters. J. Mol. Biol. 286683-693. [DOI] [PubMed] [Google Scholar]
- 8.Campodonico, E. M., L. Chesnel, and C. R. Roy. 2005. A yeast genetic system for the identification and characterization of substrate proteins transferred into host cells by the Legionella pneumophila Dot/Icm system. Mol. Microbiol. 56918-933. [DOI] [PubMed] [Google Scholar]
- 9.Casadaban, M. J., J. Chou, and S. N. Cohen. 1980. In vitro gene fusions that join an enzymatically active β-galactosidase segment to amino-terminal fragments of exogenous proteins: Escherichia coli plasmid vectors for the detection and cloning of translational initiation signals. J. Bacteriol. 143971-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Casadaban, M. J., and S. N. Cohen. 1980. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J. Mol. Biol. 138179-207. [DOI] [PubMed] [Google Scholar]
- 11.Cazalet, C., C. Rusniok, H. Bruggemann, N. Zidane, A. Magnier, L. Ma, M. Tichit, S. Jarraud, C. Bouchier, F. Vandenesch, F. Kunst, J. Etienne, P. Glaser, and C. Buchrieser. 2004. Evidence in the Legionella pneumophila genome for exploitation of host cell functions and high genome plasticity. Nat. Genet. 361165-1173. [DOI] [PubMed] [Google Scholar]
- 12.Chen, J., K. S. de Felipe, M. Clarke, H. Lu, O. R. Anderson, G. Segal, and H. A. Shuman. 2004. Legionella effectors that promote nonlytic release from protozoa. Science 3031358-1361. [DOI] [PubMed] [Google Scholar]
- 13.Chien, M., I. Morozova, S. Shi, H. Sheng, J. Chen, S. M. Gomez, G. Asamani, K. Hill, J. Nuara, M. Feder, J. Rineer, J. J. Greenberg, V. Steshenko, S. H. Park, B. Zhao, E. Teplitskaya, J. R. Edwards, S. Pampou, A. Georghiou, I. C. Chou, W. Iannuccilli, M. E. Ulz, D. H. Kim, A. Geringer-Sameth, C. Goldsberry, P. Morozov, S. G. Fischer, G. Segal, X. Qu, A. Rzhetsky, P. Zhang, E. Cayanis, P. J. De Jong, J. Ju, S. Kalachikov, H. A. Shuman, and J. J. Russo. 2004. The genomic sequence of the accidental pathogen Legionella pneumophila. Science 3051966-1968. [DOI] [PubMed] [Google Scholar]
- 14.Coers, J., J. C. Kagan, M. Matthews, H. Nagai, D. M. Zuckman, and C. R. Roy. 2000. Identification of Icm protein complexes that play distinct roles in the biogenesis of an organelle permissive for Legionella pneumophila intracellular growth. Mol. Microbiol. 38719-736. [DOI] [PubMed] [Google Scholar]
- 15.de Felipe, K. S., S. Pampou, O. S. Jovanovic, C. D. Pericone, S. F. Ye, S. Kalachikov, and H. A. Shuman. 2005. Evidence for acquisition of Legionella type IV secretion substrates via interdomain horizontal gene transfer. J. Bacteriol. 1877716-7726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dumenil, G., and R. R. Isberg. 2001. The Legionella pneumophila IcmR protein exhibits chaperone activity for IcmQ by preventing its participation in high-molecular-weight complexes. Mol. Microbiol. 401113-1127. [DOI] [PubMed] [Google Scholar]
- 17.Dumenil, G., T. P. Montminy, M. Tang, and R. R. Isberg. 2004. IcmR-regulated membrane insertion and efflux by the Legionella pneumophila IcmQ protein. J. Biol. Chem. 2794686-4695. [DOI] [PubMed] [Google Scholar]
- 18.Feldman, M., and G. Segal. 2007. A pair of highly conserved two-component systems participates in the regulation of the hypervariable FIR proteins in different Legionella species. J. Bacteriol. 1893382-3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feldman, M., and G. Segal. 2004. A specific genomic location within the icm/dot pathogenesis region of different Legionella species encodes functionally similar but nonhomologous virulence proteins. Infect. Immun. 724503-4511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feldman, M., T. Zusman, S. Hagag, and G. Segal. 2005. Coevolution between nonhomologous but functionally similar proteins and their conserved partners in the Legionella pathogenesis system. Proc. Natl. Acad. Sci. USA 10212206-12211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gal-Mor, O., and G. Segal. 2003. Identification of CpxR as a positive regulator of icm and dot virulence genes of Legionella pneumophila. J. Bacteriol. 1854908-4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gal-Mor, O., and G. Segal. 2003. The Legionella pneumophila GacA homolog (LetA) is involved in the regulation of icm virulence genes and is required for intracellular multiplication in Acanthamoeba castellanii. Microb. Pathog. 34187-194. [DOI] [PubMed] [Google Scholar]
- 23.Gal-Mor, O., T. Zusman, and G. Segal. 2002. Analysis of DNA regulatory elements required for expression of the Legionella pneumophila icm and dot virulence genes. J. Bacteriol. 1843823-3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Glockner, G., C. Albert-Weissenberger, E. Weinmann, S. Jacobi, E. Schunder, M. Steinert, J. Hacker, and K. Heuner. 2008. Identification and characterization of a new conjugation/type IVA secretion system (trb/tra) of Legionella pneumophila Corby localized on two mobile genomic islands. Int. J. Med. Microbiol. 298411-428. [DOI] [PubMed] [Google Scholar]
- 25.Groisman, E. A. 2001. The pleiotropic two-component regulatory system PhoP-PhoQ. J. Bacteriol. 1831835-1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hales, L. M., and H. A. Shuman. 1999. The Legionella pneumophila rpoS gene is required for growth within Acanthamoeba castellanii. J. Bacteriol. 1814879-4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hammer, B. K., E. S. Tateda, and M. S. Swanson. 2002. A two-component regulator induces the transmission phenotype of stationary-phase Legionella pneumophila. Mol. Microbiol. 44107-118. [DOI] [PubMed] [Google Scholar]
- 28.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 7751-59. [DOI] [PubMed] [Google Scholar]
- 29.Horwitz, M. A., and S. C. Silverstein. 1980. Legionnaires' disease bacterium (Legionella pneumophila) multiplies intracellularly in human monocytes. J. Clin. Investig. 60441-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hyytiainen, H., S. Sjoblom, T. Palomaki, A. Tuikkala, and E. Tapio Palva. 2003. The PmrA-PmrB two-component system responding to acidic pH and iron controls virulence in the plant pathogen Erwinia carotovora ssp. carotovora. Mol. Microbiol. 50795-807. [DOI] [PubMed] [Google Scholar]
- 31.Ingmundson, A., A. Delprato, D. G. Lambright, and C. R. Roy. 2007. Legionella pneumophila proteins that regulate Rab1 membrane cycling. Nature 450365-369. [DOI] [PubMed] [Google Scholar]
- 32.Kubori, T., A. Hyakutake, and H. Nagai. 2008. Legionella translocates an E3 ubiquitin ligase that has multiple U-boxes with distinct functions. Mol. Microbiol. 671307-1319. [DOI] [PubMed] [Google Scholar]
- 33.Laguna, R. K., E. A. Creasey, Z. Li, N. Valtz, and R. R. Isberg. 2006. A Legionella pneumophila-translocated substrate that is required for growth within macrophages and protection from host cell death. Proc. Natl. Acad. Sci. USA 10318745-18750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu, Y., and Z. Q. Luo. 2007. The Legionella pneumophila effector SidJ is required for efficient recruitment of endoplasmic reticulum proteins to the bacterial phagosome. Infect. Immun. 75592-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luo, Z. Q., and R. R. Isberg. 2004. Multiple substrates of the Legionella pneumophila Dot/Icm system identified by interbacterial protein transfer. Proc. Natl. Acad. Sci. USA 101841-846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lynch, D., N. Fieser, K. Gloggler, V. Forsbach-Birk, and R. Marre. 2003. The response regulator LetA regulates the stationary-phase stress response in Legionella pneumophila and is required for efficient infection of Acanthamoeba castellanii. FEMS Microbiol. Lett. 219241-248. [DOI] [PubMed] [Google Scholar]
- 37.Machner, M. P., and R. R. Isberg. 2007. A bifunctional bacterial protein links GDI displacement to Rab1 activation. Science 318974-977. [DOI] [PubMed] [Google Scholar]
- 38.Machner, M. P., and R. R. Isberg. 2006. Targeting of host rab GTPase function by the intravacuolar pathogen Legionella pneumophila. Dev. Cell 1147-56. [DOI] [PubMed] [Google Scholar]
- 39.McPhee, J. B., S. Lewenza, and R. E. Hancock. 2003. Cationic antimicrobial peptides activate a two-component regulatory system, PmrA-PmrB, that regulates resistance to polymyxin B and cationic antimicrobial peptides in Pseudomonas aeruginosa. Mol. Microbiol. 50205-217. [DOI] [PubMed] [Google Scholar]
- 40.Miller, J. H. 1972. Experiments in molecular biology. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 41.Mitchell, J. E., D. Zheng, S. J. Busby, and S. D. Minchin. 2003. Identification and analysis of ′extended −10′ promoters in Escherichia coli. Nucleic Acids Res. 314689-4695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Murata, T., A. Delprato, A. Ingmundson, D. K. Toomre, D. G. Lambright, and C. R. Roy. 2006. The Legionella pneumophila effector protein DrrA is a Rab1 guanine nucleotide-exchange factor. Nat. Cell Biol. 8971-977. [DOI] [PubMed] [Google Scholar]
- 43.Nagai, H., J. C. Kagan, X. Zhu, R. A. Kahn, and C. R. Roy. 2002. A bacterial guanine nucleotide exchange factor activates ARF on Legionella phagosomes. Science 295679-682. [DOI] [PubMed] [Google Scholar]
- 44.Ninio, S., and C. R. Roy. 2007. Effector proteins translocated by Legionella pneumophila: strength in numbers. Trends Microbiol. 15372-380. [DOI] [PubMed] [Google Scholar]
- 45.Ninio, S., D. M. Zuckman-Cholon, E. D. Cambronne, and C. R. Roy. 2005. The Legionella IcmS-IcmW protein complex is important for Dot/Icm-mediated protein translocation. Mol. Microbiol. 55912-926. [DOI] [PubMed] [Google Scholar]
- 46.Raivio, T. L., and T. J. Silhavy. 2001. Periplasmic stress and ECF sigma factors. Annu. Rev. Microbiol. 55591-624. [DOI] [PubMed] [Google Scholar]
- 47.Raivio, T. L., and T. J. Silhavy. 1997. Transduction of envelope stress in Escherichia coli by the Cpx two-component system. J. Bacteriol. 1797724-7733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rowbotham, T. J. 1980. Preliminary report on the pathogenicity of Legionella pneumophila for freshwater and soil amoebae. J. Clin. Pathol. 331179-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sadosky, A. B., L. A. Wiater, and H. A. Shuman. 1993. Identification of Legionella pneumophila genes required for growth within and killing of human macrophages. Infect. Immun. 615361-5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sanderson, A., J. E. Mitchell, S. D. Minchin, and S. J. Busby. 2003. Substitutions in the Escherichia coli RNA polymerase sigma70 factor that affect recognition of extended −10 elements at promoters. FEBS Lett. 544199-205. [DOI] [PubMed] [Google Scholar]
- 51.Sauer, J. D., M. A. Bachman, and M. S. Swanson. 2005. The phagosomal transporter A couples threonine acquisition to differentiation and replication of Legionella pneumophila in macrophages. Proc. Natl. Acad. Sci. USA 1029924-9929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Segal, G., M. Feldman, and T. Zusman. 2005. The Icm/Dot type-IV secretion systems of Legionella pneumophila and Coxiella burnetii. FEMS Microbiol. Rev. 2965-81. [DOI] [PubMed] [Google Scholar]
- 53.Segal, G., and H. A. Shuman. 1997. Characterization of a new region required for macrophage killing by Legionella pneumophila. Infect. Immun. 655057-5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Segal, G., and H. A. Shuman. 1999. Legionella pneumophila utilize the same genes to multiply within Acanthamoeba castellanii and human macrophages. Infect. Immun. 672117-2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tamayo, R., A. M. Prouty, and J. S. Gunn. 2005. Identification and functional analysis of Salmonella enterica serovar Typhimurium PmrA-regulated genes. FEMS Immunol. Med. Microbiol. 43249-258. [DOI] [PubMed] [Google Scholar]
- 56.Zusman, T., G. Aloni, E. Halperin, H. Kotzer, E. Degtyar, M. Feldman, and G. Segal. 2007. The response regulator PmrA is a major regulator of the icm/dot type IV secretion system in Legionella pneumophila and Coxiella burnetii. Mol. Microbiol. 631508-1523. [DOI] [PubMed] [Google Scholar]
- 57.Zusman, T., G. Yerushalmi, and G. Segal. 2003. Functional similarities between the icm/dot pathogenesis systems of Coxiella burnetii and Legionella pneumophila. Infect. Immun. 713714-3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.