Abstract
The transformation of the enteropathogenic bacterium Yersinia pseudotuberculosis into the plague bacillus, Yersinia pestis, has been accompanied by extensive genetic loss. This study focused on chromosomal regions conserved in Y. pseudotuberculosis and lost during its transformation into Y. pestis. An extensive PCR screening of 78 strains of the two species identified five regions (R1 to R5) and four open reading frames (ORFs; orf1 to orf4) that were conserved in Y. pseudotuberculosis and absent from Y. pestis. Their conservation in Y. pseudotuberculosis suggests a positive selective pressure and a role during the life cycle of this species. Attempts to delete two ORFs (orf3 and orf4) from the chromosome of strain IP32953 were unsuccessful, indicating that they are essential for its viability. The seven remaining loci were individually deleted from the IP32953 chromosome, and the ability of each mutant to grow in vitro and to kill mice upon intragastric infection was evaluated. Four loci (orf1, R2, R4, and R5) were not required for optimal growth or virulence of Y. pseudotuberculosis. In contrast, orf2, encoding a putative pseudouridylate synthase involved in RNA stability, was necessary for the optimal growth of IP32953 at 37°C in a chemically defined medium (M63S). Deletion of R1, a region predicted to encode the methionine salvage pathway, altered the mutant pathogenicity, suggesting that the availability of free methionine is severely restricted in vivo. R3, a region composed mostly of genes of unknown functions, was necessary for both optimal growth of Y. pseudotuberculosis at 37°C in M63S and for virulence. Therefore, despite their loss in Y. pestis, five of the nine Y. pseudotuberculosis-specific chromosomal loci studied play a role in the survival, growth, or virulence of this species.
Yersinia pseudotuberculosis, an enteropathogenic bacterium, has given rise within the last 1,500 to 20,000 years to a clonal variant, Yersinia pestis, the causative agent of plague (2, 4, 21). From an enteropathogen transmitted by the oral route and present in the environment, Y. pseudotuberculosis rapidly evolved into a flea-transmitted pathogen with a marked increase in virulence.
One of the major genetic differences between these two species was the acquisition by Y. pestis of two specific plasmids, pFra and pPla. The 101-kb pFra plasmid encodes functions necessary for survival in and colonization of the flea midgut (22), while pPla (9.6 kb) is thought to facilitate Y. pestis dissemination from the intradermal site of flea inoculation (49). Therefore, acquisition of these plasmids has certainly been a key step in the emergence of an arthropod-transmitted bacterium.
Whole-genome comparisons of Y. pseudotuberculosis IP32953 with several Y. pestis isolates showed that, as expected, most of the chromosomal backbone is highly conserved, with 75% of predicted proteins sharing greater than 97% identity in the two species (4). A notable difference between the Y. pseudotuberculosis and Y. pestis chromosomes is a burst of insertion sequences (IS) in the latter. Although a total of 17 IS elements belonging to the insertion sequences IS100, IS185, and IS1541 are present in the Y. pseudotuberculosis genome, 108 to 176 copies of these elements are found in Y. pestis (4, 5, 11, 38, 50). A preliminary analysis of gene differences across a panel of nine strains of Y. pseudotuberculosis and 13 strains of Y. pestis revealed that the transformation of an enteropathogen into the hypervirulent plague bacillus was accompanied by the acquisition of only a few new genetic materials (4), one of them being a functional filamentous phage genome (12). However, most remarkably, this transformation was characterized by the loss of genetic material. The Y. pseudotuberculosis IP32953 genome contains 317 genes that are absent from those of Y. pestis CO92 and KIM. Furthermore, 208 genes present in the two species have been inactivated during the transformation process (4), suggesting that genetic loss has been more important than the acquisition of new genes in this process.
In a recent work, a Y. pseudotuberculosis-specific chromosomal region (i.e., a region present in all Y. pseudotuberculosis strains and absent from all Y. pestis isolates tested) was shown to encode a DNA methyltransferase which plays a role in Y. pseudotuberculosis virulence (41). In the present study, we sought to identify the other Y. pseudotuberculosis-specific regions and investigate their role in the survival, growth, and virulence of this species.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Bacterial strains and plasmids used in the present study for mutant constructions are listed in Table 1. A set of 31 Y. pseudotuberculosis strains serotypes I to V and of various geographical origins (see Table S1 in the supplemental material) and of 47 Y. pestis strains isolated from different plague foci in the world and belonging to the three classical biovars (data not shown) was used to test the specificity of the chromosomal regions. Bacteria were grown in Luria-Bertani (LB) medium or M63S broth [0.1 M KH2PO4, 0.2% (NH4)2SO4, 0.02% MgSO4, 0.00005% FeSO4, 0.2% glucose, 0.2% Casamino Acids] or on LB or LB-Sac (without NaCl and supplemented with 10% sucrose) agar plates. Yersinia strains were grown at 28 or 37°C and Escherichia coli strains at 37°C. Chloramphenicol (25 μg/ml), kanamycin (30 μg/ml), or spectinomycin (50 μg/ml) was added to the medium when necessary.
TABLE 1.
Bacterial strains and plasmids used for mutant construction
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| Y. pseudotuberculosis | ||
| IP32953 | Serotype O:1b (France) | Institut Pasteur |
| IP32953(pKOBEG-sacB) | pKOBEG-sacB introduced into IP32953 | 40 |
| IP32953Δorf1 | Allelic exchange between YPTB2793 and a kan cassette | This study |
| IP32953Δorf2 | Allelic exchange between YPTB1058 and a kan cassette | This study |
| IP32953ΔR1 | Allelic exchange between the YPTB0872-YPTB0878 region and a kan cassette | This study |
| IP32953ΔR2 | Allelic exchange between the YPTB2180-YPTB2183 region and a kan cassette | This study |
| IP32953ΔR3 | Allelic exchange between the YPTB2193-YPTB2201 region and a kan cassette | This study |
| IP32953ΔR4 | Allelic exchange between the YPTB2205-YPTB2207 region and a kan cassette | This study |
| IP32953ΔR5 | Allelic exchange between the YPTB2490-YPTB2497 region and a kan cassette | This study |
| IP32953(pBAD-orf4) | pBAD43-orf4 introduced into IP32953 by electroporation | This study |
| IP32953(pBAD-orf3) | pBAD43-orf3 introduced into IP32953 by electroporation | This study |
| IP32953Δorf4(pBAD-orf4) | Allelic exchange between the chromosomal copy of YPTB3368 and a kan cassette | This study |
| IP32953Δorf3(pBAD-orf3) | Allelic exchange between the chromosomal copy of YPTB1495 and a kan cassette | This study |
| E. coli DH5α | F−endA1 hsdR17supE44 thi-1 recA1 gyrA relA1 Δ(lacIZA-argF)U169 deoR [φ80dlac(lacZ)M15] | Biolabs |
| Plasmids | ||
| pKOBEG-sacB | repA cat araC pBAD exo bet gam sacB; Cmr | 13 |
| pUC4K | bla oriR6K oriT cos rpsL kan; Ampr Kanr | Amersham |
| pBAD43 | Low copy number, paraB; Cmr | 19 |
| pBAD43-orf4 | ORF4 with its promoter region cloned into pBAD43 | This study |
| pBAD43-orf3 | ORF3 with its promoter region cloned into pBAD43 | This study |
aCmr, chloramphenicol resistance; Ampr, ampicillin resistance; Kanr, kanamycin resistance.
DNA and gene manipulation.
Genomic and plasmid DNA were extracted with an Isoquick nucleic acid extraction kit (Isoquick) and a Qiagen plasmid miniprep kit (Qiagen), respectively. Recombinant plasmid DNA and PCR fragments were introduced into Yersinia or E. coli by electroporation as described previously (6). PCRs were performed as previously described (41), using the primers listed in Table S2 in the supplemental material for the screening of Y. pseudotuberculosis-specific loci and in Table S3 in the supplemental material for mutant constructions and verification.
Construction of Y. pseudotuberculosis mutants.
Each target chromosomal region was deleted in Y. pseudotuberculosis strains by allelic exchange with a kan cassette, following the long flanking homologous region PCR procedure (13). Briefly, a three-step PCR allowed the amplification of a linear PCR fragment composed of the kan gene with its promoter region and flanked by ∼500-bp homologs to the sequences flanking each Y. pseudotuberculosis-specific region. This fragment was introduced by electroporation into strain IP32953(pKOBEG-sacB), and correct allelic exchange between the PCR fragment and the chromosomal target gene was checked by PCR with different primer pairs (see Table S3 in the supplemental material). Y. pseudotuberculosis IP32953 clones with the appropriate mutation were then streaked onto LB-Sac agar plates to select for derivatives cured of pKOBEG-sacB.
Cloning procedures.
orf3 and orf4 with their promoter regions were amplified with the primer pairs 431Abis/431B and 430Abis/430B, respectively, and cloned into the HindIII and KpnI sites of plasmid pBAD43. Recombinant plasmids (pBAD-orf3 and pBAD43-orf4) were introduced by electroporation into E. coli DH5α, digested with KpnI to check their size, and then introduced into Y. pseudotuberculosis IP32953(pKOBEG-sacB).
In silico analyses.
The presence and conservation of the Y. pseudotuberculosis-specific regions identified in IP32953 (4) were searched in the sequenced genomes of Y. pseudotuberculosis IP31758 (15) and YPIII (GenBank no. CP000950; U.S. DOE Joint Genome Institute) and of Y. pestis CO92 (38), KIM (11), Nepal516 (5), Antiqua (5), Microtus 91001 (50), Angola (CP000901; The Institute for Genomic Research), and pestoides F (CP000668; U.S. DOE Joint Genome Institute), using MaGe software (https://www.genoscope.cns.fr/agc/mage).
Animal infections.
Prior to infection, the presence of the pYV virulence plasmid and high-pathogenicity island was systematically verified in each strain by PCR with the primer pairs 160A/160B (yopM) and 18/19 (irp2) (see Table S2 in the supplemental material). Groups of ten 4-week-old C57BL/6 female mice (Janvier) were infected intragastrically (i.g.) with 0.2 ml of bacterial suspensions. Lethality was recorded daily for 3 weeks.
RESULTS AND DISCUSSION
Identification of Y. pseudotuberculosis-specific chromosomal loci.
A comparison of the genome of Y. pseudotuberculosis IP32953 to those of Y. pestis CO92 and KIM and a subsequent PCR screening of a panel of 19 strains of Y. pestis and Y. pseudotuberculosis previously identified two solitary open reading frame (ORFs) and five regions (chromosomal segment composed of two or more adjacent ORFs) that were specific to Y. pseudotuberculosis (4). In the present study, to further delineate Y. pseudotuberculosis-specific regions, a larger panel of 78 strains (47 strains of Y. pestis belonging to the three classical biovars and 31 strains of Y. pseudotuberculosis of serotypes I to V and of various geographical origins [see Table S1 in the supplemental material]) were used. The chromosomal loci tested were selected in order to (i) delineate each Y. pseudotuberculosis-specific region, (ii) reconsider ORFs that were previously found to be present in at least eight of the nine Y. pseudotuberculosis strains previously analyzed, and (iii) reexamine ORFs previously considered as non-Y. pseudotuberculosis-specific because a weak amplification product was detected in one or two Y. pestis isolates (4). This screening did not include R6, a Y. pseudotuberculosis-specific region encoding a type I restriction-modification system, because this region has been recently characterized (41).
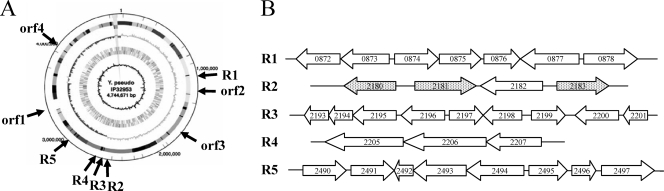
Screening by PCR of the panel of 78 isolates with 30 primer sets (see Table S2 in the supplemental material) allowed to retain 22 Y. pseudotuberculosis-specific genes corresponding to three solitary ORFs (orf1 to orf3) and four regions (R1, R3, R4, and R5) (see Table S1 in the supplemental material). Furthermore, region R2 contained three genes that were unequally distributed in various strains and one ORF that was systematically present in all Y. pseudotuberculosis isolates studied (see Table S1 in the supplemental material). Since this feature was similar to that observed for the DNA methyltransferase locus, which was shown to encode a virulence-associated factor in Y. pseudotuberculosis (41), R2 was kept for further analysis. Finally, although orf4 did not fully meet the criteria required for specificity since it was detected in all but two Y. pseudotuberculosis isolates (see Table S1 in the supplemental material), this ORF was retained because of its frequent distribution (94%) in the species. Altogether, four ORFs (orf1 to orf4) and five regions (R1 to R5) were thus selected for further analysis (Table 2). The positions of these nine loci on the IP32953 chromosome are indicated in Fig. 1A, and the genetic organizations of the five regions are presented in Fig. 1B.
TABLE 2.
Y. pseudotuberculosis-specific regions and ORF
| Specific ORF or region | YPTB no.a | Previous region no. | Presence (%) in:
|
|
|---|---|---|---|---|
| Y. pestis | Y. pseudotuberculosis | |||
| orf1 | 2793 | 25 | 0 | 100 |
| orf2 | 1058 | 8 | 0 | 100 |
| orf3 | 1495 | 12 | 0 | 100 |
| orf4 | 3368 | 29 | 0 | 94 |
| R1 | 0872 to 0878 | 7 | 0 | 100 |
| R2 | 2180 to 2183 | 17 | 0 | 74-100b |
| R3 | 2193 to 2201 | 18+19 | 0 | 100 |
| R4 | 2205 to 2207 | 20 | 0 | 100 |
| R5 | 2490 to 2497 | 22 | 0 | 100 |
As defined previously (4).
Only one ORF was present in all Y. pseudotuberculosis strains tested.
FIG. 1.
(A) Location of the nine Y. pseudotuberculosis-specific loci on the IP32953 chromosomal map. (B) Genetic organization of the five Y. pseudotuberculosis-specific regions. Numbers inside arrows indicate the YPTB designation assigned to the IP32953 genes. Gray arrows correspond to genes that are not found in all Y. pseudotuberculosis strains.
Analysis of the Y. pseudotuberculosis-specific chromosomal regions.
As a first approach to the understanding of the role of these nine chromosomal loci on Y. pseudotuberculosis physiology, a search for putative functions derived from the identification of homologous genes in other bacterial genomes was carried out. Furthermore, the presence, conservation, and synteny of each specific region of IP32953 were examined in the sequenced genomes of Y. pseudotuberculosis strains IP31758 and YPIII and of Y. pestis strains CO92, KIM, Microtus 91001, Nepal516, Antiqua, Angola, and Pestoides F. The nine specific regions or ORFs identified were individually deleted from the chromosome of strain IP32953, and the ability of the mutants to grow in vitro in a complex (LB) or chemically defined (M63S) medium at 28°C (optimal temperature for Y. pseudotuberculosis growth in vitro) or 37°C (temperature of the mammalian host) was determined twice independently and was compared to that of the wild-type strain IP32953. The importance of each specific locus on Y. pseudotuberculosis virulence was also evaluated by monitoring the lethality of groups of 10 C57BL/6 mice infected i.g. with 104 CFU of either the wild-type strain IP32953 or the various deletion derivatives.
Four loci are not required for Y. pseudotuberculosis growth and virulence.
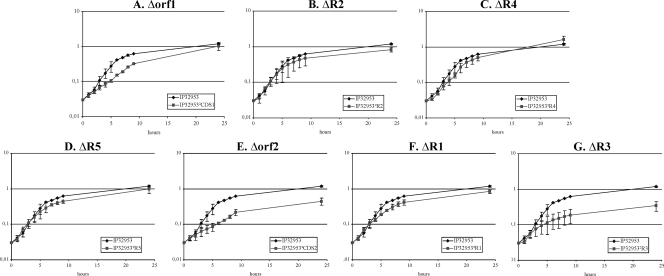
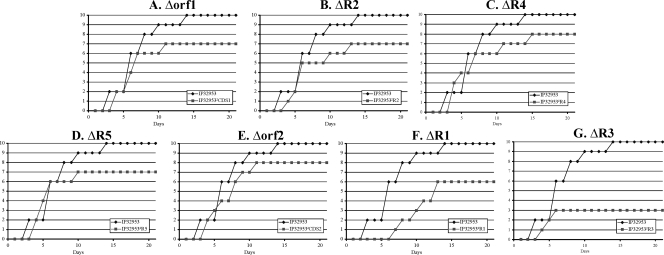
Among the nine chromosomal regions or ORFs analyzed, four loci (orf1, R2, R4, and R5) were found to be dispensable for Y. pseudotuberculosis growth in vitro (Fig. 2). IP32953Δorf1 exhibited a slight delay during the exponential growth phase in M63S at 37°C, but this delay disappeared at the stationary phase (Fig. 2A). Furthermore, although the number of dead animals was slightly lower for each of the four mutants (7 or 8 dead mice out of 10) than for the wild-type strain (10 of 10 dead animals) (Fig. 3A to D), this difference was not statistically significant (P > 0.05 with the χ2 test). This suggests that these four chromosomal loci do not play a major role in Y. pseudotuberculosis growth and pathogenicity, at least under the conditions used in the present study.
FIG. 2.
Growth curves in M63S at 37°C of the seven IP32953 derivatives, each with a Y. pseudotuberculosis-specific locus deleted. Standard deviations of two independent experiments are represented by vertical bars.
FIG. 3.
Kinetics of mouse lethality upon i.g. injection of 104 CFU of IP32953 and the various deletion mutants. Ten mice were infected with each strain and mortality was recorded daily.
orf1 (YPTB2793) is a 1,287-bp sequence which potentially encodes a member of the NCS2 family of uracil/xanthine transporter (see Table S4 in the supplemental material). The NCS2 family consists of over 50 currently sequenced proteins from gram-negative and gram-positive bacteria, archaea, fungi, plants, and animals (10, 14, 46). Uracil/xanthine permeases include permeases for diverse substrates such as xanthine, uracil, and vitamin C, but many members of this family are functionally uncharacterized and may transport other substrates. Several genes that putatively have the same function are present on the Y. pseudotuberculosis IP32953 genome. They include YPTB0031, an NCS2 family xanthine/uracil:H+ symporter; YPTB3600, a putative xanthine/uracil permease; and YPTB3953, a member of the xanthine/uracil permease family. Therefore, it may be hypothesized that deletion of csd1 had no impact on Y. pseudotuberculosis growth and virulence because of the presence of other genes with similar functions.
R2 is a 4,875-bp long region composed of four genes: YPTB2180 to YPTB2183 (Fig. 1B and see Table S4 in the supplemental material). The only Y. pseudotuberculosis-specific gene, YPTB2182, is of unknown function. The three other genes were missing in a few strains and one of them (YPTB2181) was present but truncated in YPIII (see Table S1 in the supplemental material). YPTB2181 is also of unknown function. Based on sequence homologies, YPTB2180 is predicted to encode an adenosine deaminase which plays an important role in the purine pathway (25, 42). When preformed purine samples are present in the growth medium, they are readily taken up by specific transport systems and used for nucleotide synthesis, while de novo purine synthesis is shut down (34, 35). In E. coli, Salmonella enterica serovar Typhimurium (34), and Bacillus cereus (16), adenine is converted to hypoxanthine, with the intermediate formation of adenosine and inosine catalyzed by a purine nucleoside phosphorylase and an adenosine deaminase. The fact that the absence of this pathway did not affect Y. pseudotuberculosis growth and virulence suggests that the de novo purine synthesis pathway might be sufficient to provide the necessary amount of these molecules, both in vitro and in vivo, and could explain why only 80% of the Y. pseudotuberculosis strains analyzed retained this gene. YPTB2183 is homologous to members of the GntR family of transcription factors, one of the most prevalent superfamilies of transcription factors, which regulates a very diverse set of operons and regulons (20, 44).
R4 has a size of 3,608 bp and is composed of three genes: YPTB2205 to YPTB2207 (Fig. 1B and see Table S4 in the supplemental material). YPTB2207 encodes a hypothetical protein. Both YPTB2205 and YPTB2206 are homologous to genes coding for the permease subunit of ABC sugar/ribose transporters. Since another ABC sugar transporter operon (YPTB3229 to YPTB3231) is present on the IP32953 genome, it may fulfill the same function as R4, thus explaining the absence of effect of the deletion of this region on Y. pseudotuberculosis growth and pathogenicity.
R5 is a 11,701-bp long region formed of eight genes: YPTB2490 to YPTB2497 (Fig. 1B and see Table S4 in the supplemental material). Two genes (YPTB2492 and YPTB2496) are of unknown function. YPTB2491 is a putative proton-dependent di-tripeptide transporter belonging to the POT family (proton-dependent oligopeptide transport) (39), while YPTB2497 is predicted to be a cation/proton antiporter. R5 carries four ORFs homologous to genes involved in periplasmic glucan biosynthesis: YPTB2490 (htrB), a lipid A biosynthesis lauroyl acyltransferase gene; YPTB2493 (mdoH) and YPTB2494 (mdoG), two genes required for the synthesis of periplasmic branched glucans; and YPTB2495, a pseudogene homologous to genes participating in glucan biosynthesis. The periplasm of gram-negative bacteria is formed of a concentrated gel-like matrix containing the murein sacculus, a variety of specialized proteins found exclusively in this compartment, and the osmoregulated periplasmic glucans (OPGs). OPGs are important intrinsic components of gram-negative bacteria envelope, which can be essential under extreme conditions. Mutants defective in OPG synthesis have a highly pleiotropic phenotype, indicative of an overall alteration of their envelope properties. Strains of the plant pathogens Agrobacterium tumefaciens, Pseudomonas syringae, Erwinia chrysanthemi, and Sinorhizobium meliloti defective in OPG synthesis have been shown to be attenuated or avirulent for plants (3, 32, 37). Similarly, mammalian pathogens such as Salmonella enterica serovar Typhimurium and Pseudomonas aeruginosa with mutations altering OPG biosynthesis are severely impaired in their virulence (29, 54). Since one of the genes involved in glucan biosynthesis carried by the R5 region is a pseudogene, this suggests that this pathway is not functional in Y. pseudotuberculosis and may thus explain the absence of effect of R5 deletion on growth and virulence of this species. However, glucan biosynthesis is a crucial process for many bacteria, and it may be surprising that this pathway is dispensable in Y. pseudotuberculosis, especially since no other locus with a similar function was identified on the IP32953 chromosome.
Therefore, four specific chromosomal loci (orf1, R2, R4, and R5) are dispensable for in vitro growth and virulence of Y. pseudotuberculosis IP32953. Nevertheless, the fact that they are present in all Y. pseudotuberculosis strains tested suggests that they have been subjected to a positive selective pressure. For two of them (orf1 and R4), the absence of effect observed upon their inactivation could be explained by the presence of other chromosomal genes having potentially similar functions. For the last two specific loci (R2 and R5), it may be hypothesized that their functions are dispensable for in vivo and in vitro growth, under the conditions used in the present study, but that they are important under other circumstances, such as during Y. pseudotuberculosis survival and multiplication in the environment.
orf2 is necessary for optimal growth of Y. pseudotuberculosis at 37°C in a chemically defined medium.
Deletion of orf2 (YPTB1058) had no impact on the growth of the mutant strain when the bacteria were grown in LB medium or in M63S medium at 28°C (data not shown) but resulted in a reproducible longer doubling time and a lower bacterial load when the bacteria were grown at 37°C in M63S (Fig. 2E). This suggests that orf2 participates in the acquisition of molecules that are missing in a chemically defined medium such as M63S and that are important, in a temperature-dependent manner, for bacterial metabolism.
orf2 (290 bp) shares the highest homology (80% amino acid identity, 88% amino acid similarity) with YjbC (NP290656), a pseudouridylate synthase of E. coli O157:H7 (see Table S4 in the supplemental material). Deletion of some synthase genes (rluA, rulD, and truB) was shown to impair E. coli growth (17, 18, 43). This enzyme is involved in converting uracil bases to pseudouridine. Cellular RNAs contain a number of posttranscriptionally modified nucleosides, the most common of which is pseudouridine (28). Pseudouridine is found in RNAs (such as rRNA, tRNA, and snRNA) whose functions depend on a stable tertiary structure (36). In E. coli, 11 synthases belonging to five families account for all of the known rRNA and tRNA pseudouridine residues. Similarly, the genome of Y. pseudotuberculosis IP32953 carries 11 putative pseudouridylate synthase genes: YPTB0482 (truB), YPTB0638 (rluA), YPTB0772 (ygbO), YPTB0846 (rulD), YPTB1301 (rsuA), YPTB2135, YPTB2428 (ymfC), YPTB2478 (rluC), YPTB2618 (truA), YPTB3009, and YPTB1058 (orf2). Each of the synthase families has a specific activity which does not overlap with the function of the other families. This may explain the observed in vitro growth defect of the IP32953Δorf2 mutant despite the high number of synthase genes on the IP32953 chromosome.
The IP32953Δorf2 mutant had a virulence comparable to that of the wild-type strain (Fig. 3E). This may indicate either that the function of orf2 is not essential for Y. pseudotuberculosis pathogenicity in our model of murine infection or that during in vivo multiplication, some other synthase families may compensate the absence of orf2.
R1 participates to Y. pseudotuberculosis pathogenicity.
R1 is a 6,545-bp region that carries seven genes (YPTB0872 to YPTB0878) (Table 2 and Fig. 1B). Deletion of R1 did not impair the in vitro growth of IP32953ΔR1, even in M63S at 37°C (Fig. 2F), but affected the virulence of the mutant strain upon i.g. infection. Although all 10 animals infected with wild-type IP32953 died, only six mice infected with IP32953ΔR1 did so (Fig. 3F). The difference was statistically significant (P < 0.05 with the χ2 test). Furthermore, the mean time to death was delayed for the IP32953ΔR1 derivative (10 days) compared to the wild-type strain (5 days) (Fig. 3F).
The in silico analysis suggests that R1 is involved in the methionine salvage pathway (see Table S4 in the supplemental material). Methionine is an essential amino acid and is required for a number of cellular functions, including the initiation of protein synthesis; the methylation of DNA, rRNA, and xenobiotics; and the biosynthesis of cysteine, phospholipids, and polyamines. Polyamines, organic molecules with multiple amino groups, are synthesized in large amounts in rapidly growing cells. During polyamine synthesis of spermidine from putrescine and spermine from spermidine, methionine is consumed in a one-to-one stoichiometry with the by-product formation of methylthioadenosine. Since the amount of methionine is typically limiting in cells and de novo synthesis of methionine is energetically expensive, it is important for microorganisms to be able to recycle this amino acid.
Based on the similarities with the characterized genes of Klebsiella pneumoniae (7-9, 33, 56), it can be predicted that in Y. pseudotuberculosis the YPTB0874 product makes the initial step of the methionine salvage pathway in which methylthioadenosine is degraded into methylthioribose. YPTB0878, a methylthioribose kinase, phosphorylates methylthioribose in methylthioribose-1-phosphate. YPTB0877 transforms methylthioribose-1-phosphate to 2,3-diketo-5-methylthio-1-phosphopentene. YPTB0875 is an E-1 enzyme that transforms 2,3-diketo-5-methylthio-1-phosphopentene to 1,2-dihydroxy-3-keto-5-methylthiopentene. YPTB0876 is an ARD/ARD′ enzyme that oxidizes the 1,2-dihydroxy-3-keto-5-methylthiopentene to 2-oxo-4-methylthiobutanoic acid. Finally, YPTB0873 is an aminotransferase that transforms 2-oxo-4-methylthiobutanoic acid to l-methionine.
It was previously shown that compounds interfering with methionine synthesis inhibit the growth of K. pneumoniae (53). This was not the case here for Y. pseudotuberculosis, which grew equally well in the presence or absence of the R1 region. However, since the media used in the present study contained exogenous methionine, the methionine salvage pathway was most likely not induced, thus explaining the absence of in vitro growth defect of the IP32953ΔR1 mutant. However, our results indicate that this salvage pathway is important during growth of Y. pseudotuberculosis in an animal host, suggesting that the availability of methionine is severely restricted in the tissues of infected animals.
The analysis of nine sequenced genomes of Y. pestis and Y. pseudotuberculosis showed a high conservation (perfect synteny and >99% amino acid identity for each gene product) of R1 in the two Y. pseudotuberculosis strains (see Table S1 in the supplemental material) and confirmed its absence in the classical Y. pestis biovars. Interestingly, R1 was present and conserved (>98.5% amino acid identity for each gene product) in two Y. pestis strains: Pestoides (YPDSF2878-2884) and Angola (A3327-A3333). These two strains belong to branch 0 of the Y. pestis evolutionary tree (1) and therefore are considered to be an early step in Y. pestis differentiation. These results thus indicate that R1 was present in the ancestral Y. pestis strain and was eliminated at a later stage of its evolution, before its divergence into branches 1 and 2.
R3 is necessary for optimal growth at 37°C in M63S and is essential for Y. pseudotuberculosis virulence.
R3 is a 11,408-bp region composed of nine genes (YPTB2193 to YPTB2201) (Table 2 and Fig. 1B). Similarly to the IP32953Δorf2 mutant, IP32953ΔR3 displayed, at 37°C in M63S only, an impaired ability to multiply (Fig. 2G). Furthermore, the IP32953ΔR3 mutant was severely affected in its pathogenicity for mice since only 3 of 10 animals (P < 0.05 with the χ2 test) died of the infection (Fig. 3G).
Among the nine genes carried by R3, five (YPTB2193, YPTB2194, YPTB2198, YPTB2199, and YPTB2201) encode hypothetical proteins of unknown function (see Table S4 in the supplemental material), one of which (YPTB2198) is truncated in IP31758 (see Table S1 in the supplemental material). Of the four remaining genes, three are predicted to encode proteins involved in diverse pathways: an oxidoreductase (YPTB2195), an aldehyde dehydrogenase (YPTB2197), and an aminotransferase (YPTB2200). The last gene (YPTB2196) is homologous to genes encoding sigma-54 transcriptional regulators. Based on the high number of hypothetical genes and on the absence of clearly identified function conferred by R3, it is hardly possible to predict which of the genes carried by R3 are important for Y. pseudotuberculosis growth and pathogenicity.
Of note, the first ORF of the R3 region (YPTB2196) is present and conserved (100% amino acid identity) in the genome of strain Pestoides F (YPDSF0862), while it is absent from the other Y. pestis sequenced genomes. This argues for a sequential loss of the R3 region during the early steps of Y. pestis differentiation rather than for a loss en block of the entire region.
orf3 and orf4 are vital for Y. pseudotuberculosis IP32953.
Despite several attempts, inactivation of orf3 and orf4 by allelic exchange following the classical long flanking homologous region PCR procedure systematically failed, suggesting that these two sequences are crucial for Y. pseudotuberculosis viability. To confirm this hypothesis, each ORF (with its promoter) was cloned into pBAD43 and the recombinant plasmids were introduced individually into IP32953 (Table 1). In this background, replacement of the chromosomal orf3 or orf4 copy by a kan cassette occurred, indicating that the presence in trans of a functional gene was required to inactivate the chromosomal copy. Therefore, both orf3 and orf4 appear to be essential for Y. pseudotuberculosis IP32953 viability.
orf3 (1,749 bp) has no known function (see Table S4 in the supplemental material) but possesses a LysM (lysin motif) domain which is found in a variety of enzymes involved in bacterial cell wall degradation (23) and which promotes binding to peptidoglycan. It may thus be hypothesized that orf3 plays a role in the maintenance of the cell wall structure of Y. pseudotuberculosis. However, the analysis of the YPIII and IP31758 genomes indicates that this ORF is absent from the former and is truncated due to a frameshift in the latter.
orf4 (588 bp) has also no known function (see Table S4 in the supplemental material), but its C-terminal domain is similar to that found in two-component transcriptional regulators. This domain contains DNA and RNA polymerase binding sites (30, 55).
Therefore, the Y. pseudotuberculosis genome contains two genes of unknown functions that are required for Y. pseudotuberculosis IP32953 viability. However, since orf3 is absent from strain YPIII and mutated in strain IP31758 and since orf4 was not detected in two of the 31 Y. pseudotuberculosis strains screened by PCR, the presence of these two ORFs might not be required in all isolates of this species. A similar strain-to-strain variation was observed with the Y. pseudotuberculosis dam gene encoding a DNA adenine methyl transferase. This gene could be deleted from the chromosome of strain IP32953 (51) but was essential for the viability of strain YPIII (24). This also points to the fact that, due to the genetic polymorphism of Y. pseudotuberculosis, the results of the functional studies performed with the various derivatives of strain IP32953 might not be systematically extended to the entire species.
Conclusions.
We were able to identify and characterize here nine chromosomal loci that were lost by Y. pestis but that were subjected to a positive selective pressure in Y. pseudotuberculosis, probably because they are important at some stages of its life cycle. It is surprising, at least for the five loci that were shown to play a role in Y. pseudotuberculosis survival, growth, or virulence, that these regions were eliminated from the Y. pestis genome. This could be due to the presence of newly acquired regions fulfilling similar functions in Y. pestis. For instance, it is known that acquisition of the plasmid pPla had conferred adhesion and invasion properties to Y. pestis (26, 27), possibly explaining the inactivation of genes encoding another adhesin (YadA) and invasin (45, 47, 48). Since several loci, acquired or lost by Y. pestis, are of unknown function, it is not possible to check this hypothesis. However, this was not the case for the few Y. pseudotuberculosis-specific genes with a predicted function (orf2 and R1). No genes with a similar activity were identified on the Y. pestis chromosome. Alternatively, these gene losses may reflect a process of reductive evolution. These loci may have been lost because Y. pestis has a much more restricted epidemiological cycle (rodent-flea-rodent) than its Y. pseudotuberculosis progenitor, with no long-term persistence in the environment. The specific regions may also be required during the enteric life of Y. pseudotuberculosis and may thus be dispensable for Y. pestis. Although the separation of the Y. enterocolitica and Y. pseudotuberculosis branches occurred much earlier (0.4 to 1.9 million years) than the emergence of Y. pestis (1,500 to 20,000 years) (2), it is remarkable to note that six of these loci are partially (R2 and R3) or entirely (orf1, orf2, R1, and R5) present in the sequenced genome of Y. enterocolitica strain 8081 (52). Since Y. pseudotuberculosis and Y. enterocolitica are two enteropathogens with similar epidemiological cycles, this further reinforces the hypothesis of a role of these loci during their enteric and/or environmental life. Finally, it is also possible that some of these regions have been eliminated because they were deleterious for Y. pestis pathogenicity or flea transmission (black hole theory) (31).
These eliminations occurred at different steps during the transformation of Y. pseudotuberculosis into Y. pestis. Since all nine specific regions were absent from the classical Y. pestis biovars, they were most likely lost along branch 0, before the split of Y. pestis into branches 1 and 2. Based on their presence or absence in pestoides strains, it can also be hypothesized that seven of the specific loci were lost soon after the divergence of Y. pestis from Y. pseudotuberculosis, that R3 was progressively eliminated at a later step, and that R1 was the last specific region to be lost.
A role in the survival, growth, or virulence of Y. pseudotuberculosis IP32953 was thus identified for five of the nine Y. pseudotuberculosis-specific loci identified. A function was predicted for two of these six loci, based on in silico analyses. However, the four other regions are mainly composed of genes coding for hypothetical proteins. The identification of these new and not-yet-characterized genetic determinants will be useful for a better understanding of Y. pseudotuberculosis physiology and/or pathogenesis.
Supplementary Material
Acknowledgments
F.P. was the recipient of a grant from the Ministère de la Recherche et de la Technologie (France).
Editor: J. B. Bliska
Footnotes
Published ahead of print on 4 August 2008.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Achtman, M., G. Morelli, P. Zhu, T. Wirth, I. Diehl, B. Kusecek, A. J. Vogler, D. M. Wagner, C. J. Allender, W. R. Easterday, V. Chenal-Francisque, P. Worsham, N. R. Thomson, J. Parkhill, L. E. Lindler, E. Carniel, and P. Keim. 2004. Microevolution and history of the plague bacillus, Yersinia pestis. Proc. Natl. Acad. Sci. USA 10117837-17842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Achtman, M., K. Zurth, C. Morelli, G. Torrea, A. Guiyoule, and E. Carniel. 1999. Yersinia pestis, the cause of plague, is a recently emerged clone of Yersinia pseudotuberculosis. Proc. Natl. Acad. Sci. USA 9614043-14048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Breedveld, M. W., and K. J. Miller. 1994. Cyclic beta-glucans of members of the family Rhizobiaceae. Microbiol. Rev. 58145-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chain, P. S., E. Carniel, F. W. Larimer, J. Lamerdin, P. O. Stoutland, W. M. Regala, A. M. Georgescu, L. M. Vergez, M. L. Land, V. L. Motin, R. R. Brubaker, J. Fowler, J. Hinnebusch, M. Marceau, C. Medigue, M. Simonet, V. Chenal-Francisque, B. Souza, D. Dacheux, J. M. Elliott, A. Derbise, L. J. Hauser, and E. Garcia. 2004. Insights into the evolution of Yersinia pestis through whole-genome comparison with Yersinia pseudotuberculosis. Proc. Natl. Acad. Sci. USA 10113826-13831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chain, P. S., P. Hu, S. A. Malfatti, L. Radnedge, F. Larimer, L. M. Vergez, P. Worsham, M. C. Chu, and G. L. Andersen. 2006. Complete genome sequence of Yersinia pestis strains Antiqua and Nepal516: evidence of gene reduction in an emerging pathogen. J. Bacteriol. 1884453-4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Conchas, R. F., and E. Carniel. 1990. A highly efficient electroporation system for transformation of Yersinia. Gene 87133-137. [DOI] [PubMed] [Google Scholar]
- 7.Cornell, K. A., R. W. Winter, P. A. Tower, and M. K. Riscoe. 1996. Affinity purification of 5-methylthioribose kinase and 5-methylthioadenosine/S-adenosylhomocysteine nucleosidase from Klebsiella pneumoniae. Biochem. J. 317(Pt. 1)285-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dai, Y., T. C. Pochapsky, and R. H. Abeles. 2001. Mechanistic studies of two dioxygenases in the methionine salvage pathway of Klebsiella pneumoniae. Biochemistry 406379-6387. [DOI] [PubMed] [Google Scholar]
- 9.Dai, Y., P. C. Wensink, and R. H. Abeles. 1999. One protein, two enzymes. J. Biol. Chem. 2741193-1195. [DOI] [PubMed] [Google Scholar]
- 10.de Koning, H., and G. Diallinas. 2000. Nucleobase transporters. Mol. Membr. Biol. 1775-94. [DOI] [PubMed] [Google Scholar]
- 11.Deng, W., V. Burland, G. Plunkett III, A. Boutin, G. F. Mayhew, P. Liss, N. T. Perna, D. J. Rose, B. Mau, S. Zhou, D. C. Schwartz, J. D. Fetherston, L. E. Lindler, R. R. Brubaker, G. V. Plano, S. C. Straley, K. A. McDonough, M. L. Nilles, J. S. Matson, F. R. Blattner, and R. D. Perry. 2002. Genome sequence of Yersinia pestis KIM. J. Bacteriol. 1844601-4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Derbise, A., V. Chenal-Francisque, F. Pouillot, C. Fayolle, M. C. Prevost, C. Medigue, B. J. Hinnebusch, and E. Carniel. 2007. A horizontally acquired filamentous phage contributes to the pathogenicity of the plague bacillus. Mol. Microbiol. 631145-1157. [DOI] [PubMed] [Google Scholar]
- 13.Derbise, A., B. Lesic, D. Dacheux, J. M. Ghigo, and E. Carniel. 2003. A rapid and simple method for inactivating chromosomal genes in Yersinia. FEMS Immunol. Med. Microbiol. 38113-116. [DOI] [PubMed] [Google Scholar]
- 14.Diallinas, G., L. Gorfinkiel, H. N. Arst, Jr., G. Cecchetto, and C. Scazzocchio. 1995. Genetic and molecular characterization of a gene encoding a wide specificity purine permease of Aspergillus nidulans reveals a novel family of transporters conserved in prokaryotes and eukaryotes. J. Biol. Chem. 2708610-8622. [DOI] [PubMed] [Google Scholar]
- 15.Eppinger, M., M. J. Rosovitz, W. F. Fricke, D. A. Rasko, G. Kokorina, C. Fayolle, L. E. Lindler, E. Carniel, and J. Ravel. 2007. The complete genome sequence of Yersinia pseudotuberculosis IP31758, the causative agent of Far East scarlet-like fever. PLoS Genet. 3e142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gabellieri, E., S. Bernini, L. Piras, P. Cioni, E. Balestreri, G. Cercignani, and R. Felicioli. 1986. Purification, stability, and kinetic properties of highly purified adenosine deaminase from Bacillus cereus NCIB 8122. Biochim. Biophys. Acta 884490-496. [DOI] [PubMed] [Google Scholar]
- 17.Gutgsell, N., N. Englund, L. Niu, Y. Kaya, B. G. Lane, and J. Ofengand. 2000. Deletion of the Escherichia coli pseudouridine synthase gene truB blocks formation of pseudouridine 55 in tRNA in vivo, does not affect exponential growth, but confers a strong selective disadvantage in competition with wild-type cells. RNA 61870-1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gutgsell, N. S., M. P. Deutscher, and J. Ofengand. 2005. The pseudouridine synthase RluD is required for normal ribosome assembly and function in Escherichia coli. RNA 111141-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 1774121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haydon, D. J., and J. R. Guest. 1991. A new family of bacterial regulatory proteins. FEMS Microbiol. Lett. 63291-295. [DOI] [PubMed] [Google Scholar]
- 21.Hinchliffe, S. J., K. E. Isherwood, R. A. Stabler, M. B. Prentice, A. Rakin, R. A. Nichols, P. C. F. Oyston, J. Hinds, R. W. Titball, and B. W. Wren. 2003. Application of DNA microarrays to study the evolutionary genomics of Yersinia pestis and Yersinia pseudotuberculosis. Genome Res. 132018-2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hinnebusch, B. J., A. E. Rudolph, P. Cherepanov, J. E. Dixon, T. G. Schwan, and A. Forsberg. 2002. Role of Yersinia murine toxin in survival of Yersinia pestis in the midgut of the flea vector. Science 296733-735. [DOI] [PubMed] [Google Scholar]
- 23.Joris, B., S. Englebert, C. P. Chu, R. Kariyama, L. Daneo-Moore, G. D. Shockman, and J. M. Ghuysen. 1992. Modular design of the Enterococcus hirae muramidase-2 and Streptococcus faecalis autolysin. FEMS Microbiol. Lett. 70257-264. [DOI] [PubMed] [Google Scholar]
- 24.Julio, S. M., D. M. Heithoff, D. Provenzano, K. E. Klose, R. L. Sinsheimer, D. A. Low, and M. J. Mahan. 2001. DNA adenine methylase is essential for viability and plays a role in the pathogenesis of Yersinia pseudotuberculosis and Vibrio cholerae. Infect. Immun. 697610-7615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaplan, N. O., S. P. Colowick, and M. M. Ciotti. 1952. Enzymatic deamination of adenosine derivatives. J. Biol. Chem. 194579-591. [PubMed] [Google Scholar]
- 26.Kienle, Z., L. Emody, C. Svanborg, and P. W. O'Toole. 1992. Adhesive properties conferred by the plasminogen activator of Yersinia pestis. J. Gen. Microbiol. 1381679-1687. [DOI] [PubMed] [Google Scholar]
- 27.Lahteenmaki, K., R. Virkola, A. Saren, L. Emody, and T. K. Korhonen. 1998. Expression of plasminogen activator Pla of Yersinia pestis enhances bacterial attachment to the mammalian extracellular matrix. Infect. Immun. 665755-5762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Limbach, P. A., P. F. Crain, and J. A. McCloskey. 1994. Summary: the modified nucleosides of RNA. Nucleic Acids Res. 222183-2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mahajan-Miklos, S., M. W. Tan, L. G. Rahme, and F. M. Ausubel. 1999. Molecular mechanisms of bacterial virulence elucidated using a Pseudomonas aeruginosa-Caenorhabditis elegans pathogenesis model. Cell 9647-56. [DOI] [PubMed] [Google Scholar]
- 30.Martinez-Hackert, E., and A. M. Stock. 1997. The DNA-binding domain of OmpR: crystal structures of a winged helix transcription factor. Structure 5109-124. [DOI] [PubMed] [Google Scholar]
- 31.Maurelli, A. T., R. E. Fernandez, C. A. Bloch, C. K. Rode, and A. Fasano. 1998. “Black holes” and bacterial pathogenicity: a large genomic deletion that enhances the virulence of Shigella spp. and enteroinvasive Escherichia coli. Proc. Natl. Acad. Sci. USA 953943-3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mukhopadhyay, P., J. Williams, and D. Mills. 1988. Molecular analysis of a pathogenicity locus in Pseudomonas syringae pv. syringae. J. Bacteriol. 1705479-5488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Myers, R. W., J. W. Wray, S. Fish, and R. H. Abeles. 1993. Purification and characterization of an enzyme involved in oxidative carbon-carbon bond cleavage reactions in the methionine salvage pathway of Klebsiella pneumoniae. J. Biol. Chem. 26824785-24791. [PubMed] [Google Scholar]
- 34.Neuhard, J., and P. Nygaard. 1987. Purine samples and pyrimidines, p. 445-473. In F. Neidhardt, J. Ingraham, K. Lowe, B. Magasanik, M. Schaecher, and H. Umbarger (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology, vol. 1. American Society for Microbiology, Washington, DC. [Google Scholar]
- 35.Nygaard, P. 1993. Purine and pyrimidine salvage pathways, p. 359-378. In A. Sonenshein, J. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria. American Society for Microbiology, Washington, DC.
- 36.Ofengand, J., A. Malhotra, J. Remme, N. S. Gutgsell, M. Del Campo, S. Jean-Charles, L. Peil, and Y. Kaya. 2001. Pseudouridines and pseudouridine synthases of the ribosome. Cold Spring Harbor Symp. Quant. Biol. 66147-159. [DOI] [PubMed] [Google Scholar]
- 37.Page, F., S. Altabe, N. Hugouvieux-Cotte-Pattat, J. M. Lacroix, J. Robert-Baudouy, and J. P. Bohin. 2001. Osmoregulated periplasmic glucan synthesis is required for Erwinia chrysanthemi pathogenicity. J. Bacteriol. 1833134-3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parkhill, J., B. W. Wren, N. R. Thomson, R. W. Titball, M. T. Holden, M. B. Prentice, M. Sebaihia, K. D. James, C. Churcher, K. L. Mungall, S. Baker, D. Basham, S. D. Bentley, K. Brooks, A. M. Cerdeno-Tarraga, T. Chillingworth, A. Cronin, R. M. Davies, P. Davis, G. Dougan, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Leather, S. Moule, P. C. Oyston, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Genome sequence of Yersinia pestis, the causative agent of plague. Nature 413523-527. [DOI] [PubMed] [Google Scholar]
- 39.Paulsen, I. T., and R. A. Skurray. 1994. The POT family of transport proteins. Trends Biochem. Sci. 19404. [DOI] [PubMed] [Google Scholar]
- 40.Pouillot, F., A. Derbise, M. Kukkonen, J. Foulon, T. K. Korhonen, and E. Carniel. 2005. Evaluation of O-antigen inactivation on Pla activity and virulence of Yersinia pseudotuberculosis harbouring the pPla plasmid. Microbiology 1513759-3768. [DOI] [PubMed] [Google Scholar]
- 41.Pouillot, F., C. Fayolle, and E. Carniel. 2007. A putative DNA adenine methyltransferase is involved in Yersinia pseudotuberculosis pathogenicity. Microbiology 1532426-2434. [DOI] [PubMed] [Google Scholar]
- 42.Powell, J. F., and J. R. Hunter. 1956. Adenosine deaminase and ribosidase in spores of Bacillus cereus. Biochem. J. 62381-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Raychaudhuri, S., L. Niu, J. Conrad, B. G. Lane, and J. Ofengand. 1999. Functional effect of deletion and mutation of the Escherichia coli rRNA and tRNA pseudouridine synthase RluA. J. Biol. Chem. 27418880-18886. [DOI] [PubMed] [Google Scholar]
- 44.Rigali, S., A. Derouaux, F. Giannotta, and J. Dusart. 2002. Subdivision of the helix-turn-helix GntR family of bacterial regulators in the FadR, HutC, MocR, and YtrA subfamilies. J. Biol. Chem. 27712507-12515. [DOI] [PubMed] [Google Scholar]
- 45.Rosqvist, R., M. Skurnik, and H. Wolf-Watz. 1988. Increased virulence of Yersinia pseudotuberculosis by two independent mutations. Nature 334522-524. [DOI] [PubMed] [Google Scholar]
- 46.Saier, M. H., Jr., B. H. Eng, S. Fard, J. Garg, D. A. Haggerty, W. J. Hutchinson, D. L. Jack, E. C. Lai, H. J. Liu, D. P. Nusinew, A. M. Omar, S. S. Pao, I. T. Paulsen, J. A. Quan, M. Sliwinski, T. T. Tseng, S. Wachi, and G. B. Young. 1999. Phylogenetic characterization of novel transport protein families revealed by genome analyses. Biochim. Biophys. Acta 14221-56. [DOI] [PubMed] [Google Scholar]
- 47.Simonet, M., B. Riot, N. Fortineau, and P. Berche. 1996. Invasin production by Yersinia pestis is abolished by insertion of an IS200-like element within the inv gene. Infect. Immun. 64375-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Skurnik, M., and H. Wolf-Watz. 1989. Analysis of the yopA gene encoding the Yop1 virulence determinants of Yersinia spp. Mol. Microbiol. 3517-529. [DOI] [PubMed] [Google Scholar]
- 49.Sodeinde, O. A., Y. V. B. K. Subrahmanyam, K. Stark, T. Quan, Y. D. Bao, and J. D. Goguen. 1992. A surface protease and the invasive character of plague. Science 2581004-1007. [DOI] [PubMed] [Google Scholar]
- 50.Song, Y., Z. Tong, J. Wang, L. Wang, Z. Guo, Y. Han, J. Zhang, D. Pei, D. Zhou, H. Qin, X. Pang, J. Zhai, M. Li, B. Cui, Z. Qi, L. Jin, R. Dai, F. Chen, S. Li, C. Ye, Z. Du, W. Lin, J. Yu, H. Yang, P. Huang, and R. Yang. 2004. Complete genome sequence of Yersinia pestis strain 91001, an isolate avirulent to humans. DNA Res. 11179-197. [DOI] [PubMed] [Google Scholar]
- 51.Taylor, V. L., R. W. Titball, and P. C. Oyston. 2005. Oral immunization with a dam mutant of Yersinia pseudotuberculosis protects against plague. Microbiology 1511919-1926. [DOI] [PubMed] [Google Scholar]
- 52.Thomson, N. R., S. Howard, B. W. Wren, M. T. Holden, L. Crossman, G. L. Challis, C. Churcher, K. Mungall, K. Brooks, T. Chillingworth, T. Feltwell, Z. Abdellah, H. Hauser, K. Jagels, M. Maddison, S. Moule, M. Sanders, S. Whitehead, M. A. Quail, G. Dougan, J. Parkhill, and M. B. Prentice. 2006. The complete genome sequence and comparative genome analysis of the high pathogenicity Yersinia enterocolitica strain 8081. PLoS Genet. 2e206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tower, P. A., L. L. Johnson, A. J. Ferro, J. H. Fitchen, and M. K. Riscoe. 1991. Synergistic activity of 5-trifluoromethylthioribose and inhibitors of methionine synthesis against Klebsiella pneumoniae. Antimicrob. Agents Chemother. 351557-1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Valentine, P. J., B. P. Devore, and F. Heffron. 1998. Identification of three highly attenuated Salmonella typhimurium mutants that are more immunogenic and protective in mice than a prototypical aroA mutant. Infect. Immun. 663378-3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wietzorrek, A., and M. Bibb. 1997. A novel family of proteins that regulates antibiotic production in streptomycetes appears to contain an OmpR-like DNA-binding fold. Mol. Microbiol. 251181-1184. [DOI] [PubMed] [Google Scholar]
- 56.Wray, J. W., and R. H. Abeles. 1995. The methionine salvage pathway in Klebsiella pneumoniae and rat liver. Identification and characterization of two novel dioxygenases. J. Biol. Chem. 2703147-3153. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.