Abstract
The colonization of the gastrointestinal tract of patients by the opportunistic gram-negative bacillus Klebsiella pneumoniae generally occurs prior to the development of nosocomial infections. Mutant strain C-81 was isolated owing to its reduced capacity to colonize the digestive tract in a murine model following transposon mutagenesis (N. Maroncle, D. Balestrino, C. Rich, and C. Forestier, Infect. Immun. 70:4729-4734, 2002). Nucleotide sequence analysis showed that the transposon had inserted into the first open reading frame, eefA, of a three-gene locus (eefABC) whose homologue encodes a tripartite efflux pump in Enterobacter aerogenes (M. Masi, J. M. Pages, C. Villard, and E. Pradel, J. Bacteriol. 187:3894-3897, 2005), and this operon includes an additional short (183-bp) potential open reading frame, eefX, upstream of eefA. In vivo assays showed that a ΔeefA isogenic mutant strain normally colonized the gastrointestinal tract in single-strain tests but was significantly impaired in competition against wild-type strain LM21. Although the cecum was the compartment with the highest number of CFU, the ΔeefA mutant also was detected in the stomach in numbers smaller than those of the wild-type strain. The expression of this potential efflux pump could not be linked to any antimicrobial drug resistance phenotype, but it conferred on the bacteria an acid tolerance response to inorganic acid. The expression of the eef promoter region, measured via a lacZ reporter construction, was slightly induced by an acidic environment and also by hyperosmolarity but not by the presence of bile salts. These results suggest that an efflux pump can confer measurable ecological benefits on K. pneumoniae in an environment with high competition potential.
Klebsiella pneumoniae is an opportunistic pathogen responsible for many nosocomial infections, and most clinical strains exhibit high levels of resistance to a wide variety of structurally unrelated antibiotics. Epidemiological studies have shown that, whatever the infection site, the first stage in nosocomial infections due to K. pneumoniae consists of the colonization of the patient's gastrointestinal (GI) tract (6, 16). This pathogen therefore has to sense and respond to numerous different environments in order to survive and, consequently, to persist in the GI tract of the host. The first major barrier encountered following oral consumption is stomach acidity. The bacteria then enter the small intestine, where they encounter stresses associated with volatile fatty acids, variations in pH and osmolarity, and competition with endogenous flora. Using signature-tagged mutagenesis (STM) in a murine model, we previously identified 13 genes encoding factors required for the in vivo colonization of the GI tract by K. pneumoniae LM21 (14). All of these mutants were selected for their inability to colonize the murine intestinal tract under competitive conditions. One of the identified genes potentially encoded a protein involved in a cryptic efflux pump, EefABC, previously described in Enterobacter aerogenes (18). This pump is thought to belong to the resistance-nodulation-division (RND) family and to be composed of an inner membrane protein, EefB, a periplasmic intermediate, EefA, and an outer membrane protein, EefC (18). No specific function has been attributed to this structure so far, whose expression is repressed by the H-NS repressor in the heterologous host Escherichia coli (18). In addition to the export of antimicrobials, several recent publications have highlighted the natural physiological role of efflux pumps in exporting noxious substances out of the bacterial cell, thereby allowing the bacteria to survive in hostile environments and facilitating intestinal colonization (11, 13, 24, 30). In this report, we characterized the eef-like operon of K. pneumoniae and investigated the role of the potentially encoded efflux pump in the colonization of the GI tract by this bacterial species, using both in vitro and in vivo models.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains are listed in Table 1. Clinical isolate K. pneumoniae LM21 has been described previously (8). Unless otherwise stated, Escherichia coli and Klebsiella pneumoniae strains were stored at −20°C and −80°C, respectively, in Luria-Bertani (LB) medium (Difco, Paris) containing 20% glycerol and grown either in LB broth or on LB agar plates at 37°C. When needed, media were supplemented with relevant antibiotics: amoxicillin (50 μg/ml), kanamycin (50 μg/ml), tetracyclin (30 μg/ml), and spectinomycin (50 μg/ml). Bacterial growth was monitored by measuring the optical density at 620 nm, and the numbers of CFU were quantified by plating serial dilutions of the suspension on LB agar plates.
TABLE 1.
Bacterial strains and plasmids used in this study
| Plasmid or strain | Description | Source or reference |
|---|---|---|
| Klebsiella pneumoniae | ||
| LM21 | Wild-type strain; Apr | 9 |
| C-81 | LM21 with mini-Tn5 inserted in eefA; Apr, Kmr | 17 |
| LM21Spt | LM21 ΔSHV1::aadA7; Spr | This study |
| LM21ΔeefA | LM21 ΔSHV1::aadA7 ΔeefA::aph3; Spr, Kmr | This study |
| LM21ΔeefAApr | LM21 ΔeefA::aph3; Kmr, Apr, Spr (spontaneous mutant) | This study |
| LM21ciseefXA | LM21 ΔSHV1::Φ (eef-eefX-eefA-aadA7) ΔeefA::aph3; Spr, Kmr | This study |
| Φ (eef-lacZ) | LM21 ΔeefA::lacZ-aph3; Kmr | This study |
| LM21CH199 | LM21 containing the pKOBEG199 plasmid; Apr, Tcr | |
| Escherichia coli K-12 | ||
| JM109 | F′ traD36 proA+B+lacIq Δ(lacZ)M15/Δ(lac-proAB) glnV44 e14−gyrA96 recA1 relA1 endA1 thi hsdR17 | |
| C600 | F−thr-1 leuB6 thi-1 lacY1 glnV44 rfbD1 fhuA21 | 27 |
| CH158 | Pop3ybeW::aph3′ | 5 |
| Plasmid | ||
| pKOBEG199 | Plasmid with λ red genes under the control of the arabinose-inducible promoter | 2 |
DNA manipulations.
Primers eefseq1-5′/3′, eefseq2-5′/3′, and eefseq3-5′/3′ (Table 2) were designed from the complete sequence of the eef operon of E. aerogenes (AJ508047) and the K. pneumoniae MGH78578 genomic sequence (CP000647) and were used to amplify and sequence a 3,204-bp fragment of the eef-like operon from K. pneumoniae LM21. Chromosomal DNA was prepared from wild-type K. pneumoniae LM21 using the Nucleospin tissue kit (Macherey-Nagel, Hoerdt, France). Amplifications were performed using platinum Taq DNA polymerase high fidelity (Invitrogen, Cergy Pontoise, France) according to the manufacturer's instructions. The PCR fragments were purified with the Nucleospin extract II kit (Macherey-Nagel). Sequences were obtained commercially from Genome Express (Meylan, France) and assembled with the previously determined sequence of the fragment upstream from the mini-Tn5 insertion site in the C-81 mutant genome. Sequence homologies were determined using the BLAST 2.2.17 algorithm from the National Center for Biotechnology Information.
TABLE 2.
Oligonucleotides used in this study
| Primer | Primer sequence (5′-3′) |
|---|---|
| eefX5′ | CTCACCACAAAACCAGAGG |
| eefX3′ | AAACGAGGGAATAGGTTGGG |
| eefA3′ | TTCGGTAAACAGGCGTTTCT |
| eefAB13′ | GCCTTCACTGTCTGTCCGG |
| eefAfin5′ | GACACCTACGGCGATAAATG |
| eefBdeb3′ | AGCTGAAGTAGAGCAGATGG |
| eefBfin5′ | TGACCTCGCTGGCGTTTATC |
| eefCdeb3′ | TCAGTTCGGCATCAACGGTG |
| eefseq1-5′ | AAATACATCATAGCTCCCATCG |
| eefseq1-3′ | CTCAGCACCAGCCATTTATC |
| eefseq2-5′ | CGATGCCGCCAACAAAGTTG |
| eefseq2-3′ | AAGTTCTGCAGGAACAGATAC |
| eefseq3-5′ | CGAAGGACAGCACCGAGTT |
| eefseq3-3′ | GCACGTTCATCAGGCTGTCA |
| RNA16S-1 | ATGACCAGCCACACTGGAAC |
| RNA16S-2 | CTTCCTCCCCGCTGAAAGTA |
| BamHIPeef5′ | CGGGATCCCGTATCGCGCTGCCGAATGGT |
| EcoRIPeef3′ | GGAATTCCATCTCCTCTGGTTTTGTGGT |
| GB5′ | AAAGCCACGTTGTGTCTCAA |
| GBnp3′ | TTAGAAAAACTCATCGAGCA |
| eefA.GB5′ | CCTATTCCCTCGTTTTTAAATAAACGGGGCGGACGTCTTATATCTTGAGAAAATATAATGAAAGCCACGTTGTGTCTCAA |
| eefA.GBnp3′ | TCAGAATGGCGATCACCCAGGCAAAGACCGGGCGACGCACGAAGAAACGGGAGAACATCATTAGAAAAACTCATCGAGCA |
| lacZ5′ | ATGACCATGATTACGGATTCAC |
| Km.lacZ3′ | ATCAGAGATTTTGAGACACAACGTGGCTTTATGGCCTGCCCGGTTATTAT |
| eefA.lacZ5′ | CAACCTATTCCCTCGTTTTTAAATAAACGGGGCGGACGTCTTATATCTTGAGAAAATATAATGACCATGATTACGGATTCAC |
| deblacZ3′ | TTCTCCGGCGCGTAAAAATG |
| aadA7ampli5′ | CACTGGCATTTAATAACGCGTC |
| aadA7ampli3′ | TTAATCACTTTACTTTTATC |
| SHV1spect5′ | TGTATTGTGGTTATGCGTTATATTCGCCTGTGTATTATCTCCCTGTTAGCCACCCTGCCGCACTGGCATTTAATAACGCGTC |
| SHV1spect3′ | TATCGCCCGGCACGCCTGCGAGGTGCTGCGGGCCGGATAACGCGCGCGGCCACCGCCGGGTTAATCACTTTACTTTTATC |
| SHV1eefA3′ | TGTATTGTGGTTATGCGTTATATTCGCCTGTGTATTATCTCCCTGTTAGCCACCCTGCCGAGCTGAAGTAGAGCAGATGG |
| spect-eefA5′ | TTCGGCGCAATTGACGCGTTATTAAATGCCAGTGTATCGCGCTGCCGAATGGT |
To create an eefA-defective mutant of K. pneumoniae LM21, an eefA gene deletion mutant strain was made by allelic exchange, replacing the wild-type chromosomal eefA sequence with a selectable kanamycin resistance cassette. This mutated copy was generated by a PCR procedure. The kanamycin cassette was amplified using primers eefA.GB5′ and eefA.GBnp3′ and template DNA from E. coli strain CH158 harboring the kanamycin resistance gene in its chromosome (the aph3 gene from pUC-4K; a gift from J. M. Ghigo). The resulting 1,097-bp aph3-eefA cassette, flanked by short portions (60 bp) of the eefA 3′ and 5′ regions, was electroporated in the K. pneumoniae LM21 pKOBEG199-containing strain CH199 (Table 1). Mutants were selected on LB agar containing kanamycin and checked by PCR, which was performed with the specific primers eefX5′-GBnp3′ and GB5′-eefBdeb3′ (Table 2). One selected clone was cultured in LB broth containing kanamycin, and the loss of the pKOBEG199 plasmid then was screened by subculture on LB medium containing tetracycline. This mutant (designated LM21ΔeefAApr) was used in an in vivo cocolonization experiment with the cis-complemented mutant (designated LM21ciseefXA) described below. In a second step using the same procedure, the spectinomycin resistance cassette (aadA7) was introduced into this mutant by replacing the chromosomal SHV1bla gene, which gave rise to mutant LM21ΔeefA.
A similar strategy was used to create the eefA cis-complemented mutant. The chromosomal SHV1bla gene of mutant LM21ΔeefA was replaced with a DNA fragment containing the spectinomycin resistant cassette (aadA7) fused with a 1,731-bp region of the K. pneumoniae eef operon encompassing the potential promoter region and the two open reading frames (ORFs) eefX and eefA, thereby giving rise to LM21ciseefXA. Each mutant then was used to perform murine colonization assays and in vitro stress response assays.
Real-time reverse transcription-PCR (RT-PCR) was performed using a LightCycler (Roche Applied Science) with RNA extracted as described by Gosink et al. (10) and treated twice with RNase free DNase I (Roche Applied Science) to remove any contaminating genomic DNA. Total RNA was reverse transcribed and amplified by using primers specific to eefXAB mRNA (eefX5′/eefX3′, eefseq1-3′/eefA3′, and eefseq3-5′/eefseq2-3′) and 16S rRNA (RNA 16S-1/RNA 16S-2) (Table 2). The amplification of a single expected product was confirmed by melting curve analysis and electrophoresis on 2% agarose gels. eef mRNA levels and 16S rRNA (endogenous control) were quantified using RNA master SYBR green I (Roche Diagnostics) with 100 ng of total RNA.
To generate lacZ fusions with the potential eef promoter region, a lacZ fragment amplified from E. coli C600 genomic DNA using eef-lacZ5′ and Km.lacZ3′ primers was fused with the kanamycin-resistant aph3 cassette and incorporated in the chromosomal DNA of strain LM21 by double allelic exchange with the eefA gene as described above. The influence of stresses on the expression of eef was measured on bacteria grown overnight in LB medium supplemented with the appropriate compounds, i.e., HCl (pH 3.0), organic acid (pH 5.5), 1% bile salts, 0.125 M NaCl, and 0.5 M NaCl. Enzymatic activity was assayed in triplicate for each condition as described by Miller and was expressed in arbitrary Miller units (22).
Stress response assays and susceptibility tests. (i) Acid tolerance assays.
The behavior of Klebsiella strains under both inorganic and organic acid conditions was assessed according to the method previously described by Merrell and Camili (21). Briefly, each test strain was grown overnight in LB broth and then diluted to reach an optical density at 620 nm of 0.2. The bacterial cells from 1 ml of the suspensions were harvested by centrifugation (16,000 × g for 1 min at room temperature) and resuspended in 100 μl of LB broth (pH 7.0). Samples were divided into two aliquots: half of the cells were mixed with 950 μl of LB broth, pH 7.0 (unadapted cells), and the other half with either 950 μl of LB broth, pH 4.0 (HCl, inorganic acid response), or 950 μl of LB broth supplemented with 0.05× organic acid cocktail, pH 5.5 (organic acid response and 1× cocktail comprising 87 mM acetic acid, 25 mM butyric acid, and 37 mM propionic acid). After 1 h of incubation at 37°C with aeration, the adapted (pH 4.0 and 5.5 assays) and the unadapted (pH 7.0) cultures were pelleted at 16,000 × g for 1 min, and the cells were resuspended in either 1 ml of LB broth, pH 3.0 (inorganic acid shock with HCl), or 1 ml of LB broth supplemented with 0.3× organic acid cocktail (pH 4.3; organic acid shock). Viable cell counts were determined at t = 0 and after 60 min of incubation at 37°C by making serial dilutions in saline and plating aliquots onto LB agar plates that were incubated overnight at 37°C. The number of surviving microorganisms was determined by dividing the total number of viable cells after the hour-long acid shock by the initial number of viable cells at t = 0, and the percentage of adaptation was determined by dividing the results of adapted assays by those of the unadapted assays.
(ii) Osmotic and bile challenge assays.
The bile used in this work was purchased from Sigma (Sigma-Aldrich Chimie, Saint Quentin Fallavier, France). It is a crude ox bile extract that contains sodium salts of taurocholic, glycocholic, deoxycholic, and cholic acids. For the growth assay, 5 ml of LB broth with or without the addition of various concentrations of bile (1 and 5%, wt/vol) or NaCl (0.25 and 0.5 M) was inoculated with cells from an overnight culture to reach 4 × 107 CFU/ml. The cultures were incubated aerobically with shaking at 37°C for 5 and 3 h, respectively, and the number of viable bacteria was determined by plating serial dilutions onto LB agar. Results are expressed as the ratio of the number of CFU obtained from LB cultures containing different concentrations of bile or NaCl to the number of CFU obtained from control cultures (LB alone).
(iii) Susceptibility tests.
The MICs of bile salts, detergents (sodium dodecyl sulfate and N-lauroyl sarkosine), and antibiotics (rifampin, gentamicin, amoxicillin, ceftazidime, tetracycline, and erythromycin) for K. pneumoniae LM21, its ΔeefA mutant, and the cis-complemented mutant were determined using the standard microtiter broth dilution method in Mueller-Hinton broth with an inoculum of 5 × 105 CFU/ml. Microtiters were incubated for 24 h at 37°C. The compounds used in these assays were purchased from Sigma (Sigma-Aldrich Chimie, Saint Quentin Fallavier, France).
Intestinal colonization assays.
Female specific-pathogen-free mice (OF1; 8 to 18 weeks old; 20 g; Iffa Credo L'Arbresle France) were used. They were individually caged and given sterile water containing 1 g of spectinomycin per liter throughout the experiment and had ad libitum access to feed. After 24 h, 200-μl bacterial suspensions containing about 108 CFU/ml were given intragastrically.
For the 10-day colonization experiment, cages were cleaned daily. On day 1 and subsequently every day after inoculation, feces were collected and homogenized in saline, and serial dilutions were plated onto selective media. For individual colonization, six mice were infected with the strain and another six mice with each mutant strain. For competition assays, the LM21ΔeefA or LM21ciseefXA strain was mixed with the strain LM21Spt in equal amounts in 200 μl sterile water and infected intragastrically into six mice. The removed feces were plated onto spectinomycin-containing LB plates to measure the total number of CFU and onto Sp-Km-containing LB plates to measure the number of mutant CFU. From these numbers, the exact input ratio of mutant to wild type was calculated for inoculum and feces contents. In addition, we tested the behavior of the cis-complemented strain (LM2ciseefXA) together with an eefA-deficient mutant strain in a competition assay. Owing to the similarity of the antibiotic resistance phenotype of the cis-complemented strain (LM21ciseefXA) and the LM21ΔeefA mutant (both Spr and Kmr), we used in this assay the mutant LM21ΔeefAApr, which was obtained during the course of the LM21ΔeefA mutant synthesis and which still harbored the SHV1-encoding gene (Apr). Therefore, the number of CFU of each mutant strain in this assay was determined by plating samples of feces onto Ap- and Sp-containing plates, respectively.
To determine which parts of the GI tract were colonized by Klebsiella pneumoniae and either its LM21ΔeefA or LM21ciseefXA mutant, three mice in each group of cocolonization assays were euthanized by cervical dislocation on day 12 after feeding, and the stomach, jejunum, ileum, cecum, and colon were removed. Their contents were weighed and homogenized in saline. Bacterial concentrations of each part were determined by plating serial dilutions of the intestinal contents onto LB agar plates supplemented with antibiotic. Results are expressed as the number of CFU per g of intestinal content.
Nucleotide sequence accession number.
The sequence of K. pneumoniae LM21 has been deposited in GenBank under accession number EU126024.
RESULTS
Identification and features of the eefXABC operon in K. pneumoniae.
The 3,768-bp DNA fragment sequenced from Klebsiella pneumoniae LM21 (GenBank accession number EU126024) shared 99.15 and 90.96% identity with the K. pneumoniae MGH78578 genomic sequence and Enterobacter aerogenes BW16627 eefABC tripartite efflux pump operon, respectively. This 3,768-bp sequence revealed two complete ORFs, designated eefX and eefA, and a partial sequence of a third ORF, called eefB, located in the same coding strand and positioned tandemly on the chromosome. Part of a putative acetyltransferase-encoding gene also emerged upstream of the eef genes and was located in the opposite direction on the complementary strand. eefX (nucleotide positions 399 to 584) and eefA (nucleotide positions 619 to 1743) encoded 61 and 374 amino acids (aa), respectively. No significant similarity was found in the databases for eefX or its deduced amino acid sequence. A consultation of databases using blastx showed that the amino acid sequence of EefA was 96.42% identical and 98.35% similar to that of the EefA lipoprotein of E. aerogenes. The complete K. pneumoniae MGH78578 genome sequence was used to determine the putative sequence following the partial K. pneumoniae LM21 eefB gene. The analysis of the sequence thus obtained revealed the complete 3,108-bp eefB gene and, downstream, another 1,365-bp ORF designated eefC and located tandemly on the same coding strand as eefB. The amino acid sequences of the corresponding 1,035- and 454-aa products were 98.94 and 98.24% similar to E. aerogenes EefB inner membrane protein and EefC outer membrane protein, respectively (Fig. 1).
FIG. 1.
Schematic representation of the eef gene clusters from E. aerogenes (lower) and their homologs in K. pneumoniae (upper) and their environments. Open arrows indicate the direction of transcription. The 3,768-bp K. pneumoniae sequence determined in this study includes the two complete ORFs eefX and eefA and the 5′ region of eefB; the 3′ regions of eefB and eefC correspond to the sequence from the K. pneumoniae strain whose genome has been entirely sequenced (MGH75758). The levels of homology between the two deduced amino acid sequences are indicated in percentages. Black arrows indicate predicted promoters, and mini-Tn5 corresponds to the transposon insertion in the original STM mutant C-81. The arrowheads denote primers used to amplify by RT-PCR the junctions between the ORFs: (a) primers eefX5′ and eefA3′, 433 bp; (b) eefAfin5′ and eefBdeb3′, 378 bp; and (c) eefBfin5′ and eefCdeb3′, 501 bp. The lower part of the figure shows the RT-PCR products by an ethidium bromide-stained agarose gel obtained using the K. pneumoniae wild-type genomic DNA as the template. Only products b and c were detected when the STM mutant C-81 genomic extract was used as the template DNA (data not shown).
The transposon insertion in mutant C-81 occurred within codon 28 of eefA, encoding Gln. The stop codon (TGA) of eefA overlays with the start codon (ATG) of eefB, while the stop codon of eefB and the start codon of eefC seemingly are separated by 3 bp. The overlap between the genes and the lack of predicted stem-loop structures between the ORFs suggested that the genes are organized into an operon. A promoter consensus sequence was detected upstream of the eefX start codon (−35, CTTTCC; −10, TCGTATAAT; SoftBerry BPROM [http://linux1.softberry.com]). The analysis of the transcriptional units was performed by RT-PCR using primers hybridizing within two consecutive ORFs, namely eefX5′-eefA3′ for junction eefX-A (amplicon a), eefAfin5′-eefBdeb3′ for junction eefA-B (b), and eefBfin5′-eefCdeb3′ for junction eefB-C (c) ( Fig. 1). Using RNA extracted from the K. pneumoniae LM21 wild-type strain as the template, the reactions were positive for amplicons a, b, and c (Fig. 1), whereas only the amplicons b and c were detected using the total RNA of the template transposon mutant C-81. The results suggested that the four ORFs belong to a single transcriptional unit.
We then looked for the expression of the eef genes in the mutants by real-time RT-PCR. In mutants LM21ΔeefA and LM21ΔeefAciseefXA, the specific eefB transcripts were present in much larger quantities (about 100-fold more) than in the wild-type strain (Fig. 2). This overexpression probably was due to the presence of the strong promoter of the kanamycin resistance gene within the cassette. A fourfold increase in the expression of the specific eefXA messenger was observed in the cis-complemented mutant LM21ciseefXA, whereas the LM21ΔeefA mutant exhibited an amount of transcript of eefX similar to that of the wild-type strain (Fig. 2).
FIG. 2.
Quantitative real-time expression of eefX, eefA, and eefB in the ΔeefA and the cis-complemented (cisΔeefXA) mutants compared to their expression in wild-type strain LM21. The expression of eefX, eefA, and eefB was measured by the ΔΔCT method, in which the 16S rRNA was used as an internal reference and wild-type cells were used as the calibrator. Levels of change (n-fold) in eefX, eefA, and eefB with respect to the wild type are shown for the ΔeefA and the cis-complemented mutants.
Intestinal murine colonization assays.
To investigate the influence of this Eef potential efflux pump in K. pneumoniae intestinal tract colonization, the ΔeefA mutant, the cis-complemented mutant, and the parent strain were fed to mice individually or mixed at a ratio of 1:1 (competition), and the bacterial counts of K. pneumoniae in feces were recorded for 10 days. The analysis of the in vitro growth of each of these mutants, both individually and in competition, did not reveal any impairment or inhibition (data not shown). In addition, we checked that the presence of a spectinomycin-encoding cassette did not affect the in vivo colonization abilities of K. pneumoniae LM21 in a competition assay with the unmarked wild-type strain (data not shown). In an individual colonization assay (Fig. 3A), the ΔeefA and the cis-complemented LM21ciseefXA mutants were as effective colonizers as the parent strain and colonized the intestine at levels of about 5 × 109 CFU per g of feces. As no difference was observed in the colonization ability when they were fed individually to mice, each was given in combination with the parent strain to investigate the role of the eef products during competition. In competition assays, the ΔeefA mutant colonized at levels of about 108 CFU per g of feces (Fig. 3B), a highly statistically significant decrease compared to the levels of the parent strain (P < 0.01 by the Student's test). Similar results were obtained when the transposon insertion mutant (C-81) was tested, even 21 days postinoculation (data not shown). In a competition colonization assay involving the wild-type strain and the cis-complemented mutant (LM21ciseefXA), a statistically significant difference (P < 0.05) also was observed, but only at days 5, 6, 7, and 9 postinoculation (Fig. 3C). In addition, competition colonization assays involving the cis-complemented strain together with the ΔeefA mutant (LM21ΔeefAApr) clearly showed that the ΔeefA mutant was outcompeted (Fig. 3D).
FIG. 3.
Colonization properties in the murine model of wild-type K. pneumoniae LM21 (filled squares), the isogenic mutant strains LM21ΔeefA (filled triangles) and LM21ΔeefAApr (open triangles), and the cis-complemented mutant LM21ciseefXA (filled circles). The colonization properties of the strains are shown individually (A) and as the competition between the wild type and ΔeefA (B), between the wild type and ciseefXA (C), and between ciseefXA and ΔeefAApr (D). *, P < 0.05; **, P < 0.01.
To determine which parts of the digestive tract were colonized mostly by K. pneumoniae LM21 and its mutants, two sets of mice fed with the wild-type strain and either the LM21ΔeefA or LM21ciseefXA mutant were sacrificed on day 12 after being fed. The analysis of the intestinal samples indicated that, whatever the strain tested, K. pneumoniae organisms were recovered mainly from the stomach, cecum, and colon samples (Fig. 4A and B). The greatest difference in the level of colonization (2 log) was observed in samples from the stomach between the LM21ΔeefA mutant and the wild-type strain.
FIG. 4.
Bacterial loads determined by plating and counting colonies (in CFU) of the wild-type strain (filled squares), the ΔeefA mutant (filled triangles), and the cis-complemented ciseefXA mutant (open diamonds) in different parts of the mouse GI tract 12 days after the oral administration of the strains. The wild-type strain was fed in competition with the ΔeefA mutant (A) or with the cis-complemented ciseefXA mutant (B) to groups of three mice. Each symbol represents the number of CFU per gram of intestinal content from one mouse, and the means of the values are represented by a bar.
Resistance to GI stresses and susceptibility tests.
To determine whether the potential Eef efflux pump of K. pneumoniae LM21 played a role when the bacteria encountered the specific GI stress, we measured the level of expression of a transcriptional Φ(eef-lacZ) fusion inserted into the K. pneumoniae LM21 chromosome. The expression of the K. pneumoniae wild-type endogenous β-galactosidase was quite low (5.46 ± 2.8 Miller units), and none of the stress conditions tested had any significant influence on its expression (Table 3). Statistically significant increases, up to a 64-fold order of magnitude, were observed when the bacteria harboring the Φ(eef-lacZ) fusion were grown in the presence of acids, both organic and inorganic, and in high-osmolarity conditions, whereas no activation of transcription was observed in the presence of 1% bile salts (Table 3).
TABLE 3.
Effects of several stresses on the expression of β-galactosidase by wild-type K. pneumoniae LM21 and a chromosomal Φ (eef-lacZ) fusion
| Stress | Activity for straina:
|
|
|---|---|---|
| K. pneumoniae LM21::Φ (eef-lacZ) | K. pneumoniae LM21 | |
| Control | 0.7 ± 0.6 | 5.46 ± 2.8 |
| HCl (pH 3) | 48.3* ± 4.7 | 3.35 ± 1.2 |
| Inorganic acid (pH 5.5) | 48.4* ± 2.6 | 6.68 ± 3.4 |
| Bile salts (1%) | 0.2 ± 0.3 | 1.57 ± 0.7 |
| NaCl (0.125 M) | 44.3* ± 0.6 | 8.74 ± 3.9 |
| NaCl (0.5 M) | 39.5* ± 4.9 | 0.00 ± 0.0 |
β-Galactosidase activities are expressed in Miller units as the means of a minimum of three experiments ± standard deviations. Strains initially were grown in LB broth and then diluted in LB broth alone (control) or supplemented with inorganic acid (HCl; pH 3), organic acids (pH 5.5), bile salts (1%), and NaCl (0.125 and 0.5 M). An asterisk indicates that the difference is statistically significant (P < 0.05 by the Student's test).
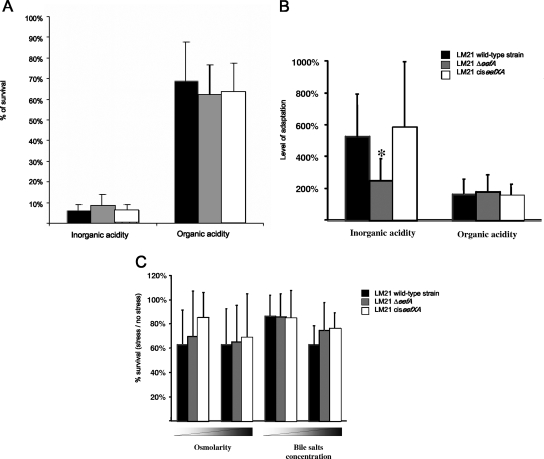
We then determined the behavior of the LM21ΔeefA mutant and its cis-complemented counterpart in acid tolerance experiments, with both inorganic (HCl) and organic acid, i.e., a cocktail of butyric, acetic, and propionic acids. When bacteria were directly exposed to an acidic environment (HCl at pH 3.0 or inorganic acids at pH 4.3), the percentages of survival ranged between 5.78 and 8.75% for the inorganic challenge and between 62.49 and 68.63% for the organic challenge, with no statistically significant difference among the three strains (Fig. 5A). The same acid shocks then were performed with a previous 1-h adaptation in sublethal concentrations of either HCl (pH 4.0) or inorganic acids (pH 5.5). In these assays, the LM21ΔeefA mutant showed a reduction in its ability to adapt to the acidic environment compared to that of the wild-type strain but only in the testing with the inorganic (HCl) substrate, and this phenotype was restored with the cis-complemented strain (Fig. 5B).
FIG. 5.
Stress sensitivities of the LM21 wild-type strain (black bars), the ΔeefA mutant (gray bars), and the cis-complemented mutant (white bars). (A) The percent acid survival of unadapted cells to both inorganic and organic acids was measured by dividing the number of viable cells after a 1-h-long acid shock (HCl, pH 3.0, or organic acids, pH 4.3) by the initial number of viable cells at t = 0. (B) The level of adaptation corresponds to the ratio of the survival of adapted cells, i.e., acid shocked after a 1-h adaptation in sublethal conditions, to the survival of unadapted cells. (C) The percentage of resistance to NaCl (0.25 and 0.5 M) and crude ox bile salt extract (1 and 5%) was calculated by comparison with the numbers of viable cells in LB medium alone. The data represent mean values from three independent determinations.
We also compared the abilities of the wild-type strain and the mutants to grow in LB medium alone and supplemented with different concentrations of either bile salts or NaCl. No difference was observed in the growth capacities of the three strains (Fig. 5C).
In addition, we determined the MICs of bile salts and other detergents and antibiotics for the wild-type strain and the LM21ΔeefA mutant. No difference was observed between the two strains (data not shown), and there was no evidence of putative substrates.
DISCUSSION
Efflux pumps have been detected in a wide variety of bacterial species, and their most commonly observed function is to counter the toxicity of antimicrobial agents, antibiotics, disinfectants, or dyes (24). Most of these multidrug transporters belong to the RND ATP-independent group, which mediates the efflux of the toxic compounds from the cell in a coupled exchange with protons (26). The model of RND efflux pumps of E. coli is composed of two specific proteins, an inner membrane protein (AcrA) and a periplasmic intermediate (AcrB), that allow the transport of drugs to the outer membrane protein (TolC) (31, 34). A few AcrAB multidrug efflux systems have been characterized in Klebsiella spp. (7, 20, 23, 28), but the sequencing data of K. pneumoniae strain MGH78578 show that many others potentially are present, as in closely related E. coli species (3). In this study, we characterized a gene cluster in K. pneumoniae LM21 that exhibits strong homologies with the EefABC RND efflux system-encoding operon, initially described in E. aerogenes (18). Although the regulation of the expression of this RND pump has been thoroughly investigated and requires the H-NS regulatory protein (18), no specific substrate has been assigned to this efflux system. A mutant of K. pneumoniae LM21 with disruption in the Eef-like genes initially was selected during the course of STM screening by using a murine intestine colonization model (14). By testing an isogenic eefA-deficient mutant in this animal model, we were able to reproduce the impairment of colonization ability. However, this defect occurred only when the mutant was administered together with the wild-type strain, i.e., in a situation of competition. The initial STM screening per se included a fierce competition struggle, since all transposon insertion mutants to be tested first were administered in pools of 24, and then the attenuated mutants were tested in equal ratios with the wild-type strain (14). When administered alone, mutant C-81, with the mini-Tn5 derivative inserted inside the eefA-like gene, behaved like the ΔeefA isogenic mutant and colonized the intestine at levels identical to those obtained with the wild-type strain (data not shown). cis complementation of the mutation restored the in vivo colonization phenotype, but only partially, probably owing to an impaired expression of eefB, eefC, and eefX in the construct. Overall, our findings indicate that this potential efflux system plays a role during competitive events involving related strains within the bacterial intestinal ecosystem.
In order to determine the substrates specifically associated with this efflux pump in K. pneumoniae, we first determined the behavior of both the wild-type strain and its ΔeefA isogenic mutant when exposed to several antimicrobial drugs. No difference in the MICs was observed with any of the antibiotic or antiseptic molecules tested (data not shown). We then performed tests reproducing some of the stresses encountered in vivo by the bacteria during the GI colonization process, i.e., contact with inorganic and organic acids and bile salts and in conditions of high osmolarity. A statistically significant difference was observed between the ΔeefA isogenic mutant and its parental strain in the number of bacteria remaining viable after the challenge of adapted cells with the inorganic acid compound HCl. Since this molecule is predominant in the stomach, and because the greatest differences in colonization levels between the two strains were observed in this organ, it seems likely that the Eef-like pump of K. pneumoniae helps the bacteria cross this highly hostile environment during the colonization process. It is unlikely that the acid tolerance response shown by K. pneumoniae, and previously described by Aghi and Chhiber (1), is due directly to the potential Eef efflux pump. One possible explanation is the presence of a shared regulator mechanism that would control the expression of both acid tolerance-related genes and efflux pump genes (19). Were this the case, then the export of a specific substrate by the Eef pump concomitant with an intracellular accumulation of protons would be linked to an acid tolerance response mediated by a global/common regulator, which could prepare the bacterial cells for the environmental stresses subsequently confronted in the intestine. The global regulator protein H-NS has been shown to repress the expression of the EefABC efflux pump of E. aerogenes (18), and several genes whose expression is modulated by H-NS are responsive to changes in environmental conditions in E. coli (11). However, no difference in the expression of the eef genes was detected between K. pneumoniae LM21 expressing a dominant-negative E. coli hns allele (32) and the wild-type strain (data not shown). These results suggest that there is a more complex or specific system that regulates eef expression in Klebsiella. Some efflux pumps have a relatively high basal level of expression, whereas others are inducible by the transported substrate. The RND efflux pump genes often are regulated by the gene product of an adjacent and divergently transcribed regulatory gene (25). No such gene has been detected in the K. pneumoniae eef operon, but the role of the eefX gene encoding a putative 61-aa-long protein, absent in the E. aerogenes eef operon, currently is being investigated.
Although no biological function has been attributed to the Eef efflux pump, we demonstrated that it conferred an advantage to the bacteria during the colonization of the digestive tract in a murine model. The GI tract of humans contains bile salts and fatty acids, and Ma et al. showed that the AcrAB efflux pump of E. coli was involved in bacterial survival in the presence of these compounds (15). More recently, studies performed with Helicobacter pylori and Campylobacter jejuni have shown that efflux pumps are required for the in vivo colonization of the gastric tract by these pathogens (12, 14, 30). The maintenance of a metal homeostasis that modulates urease activity could be responsible for the survival of H. pylori in the gastric environment, and the multidrug CmeABC of C. jejuni mediates bile salt resistance, which is required for the successful colonization of the intestinal tract in chickens (13, 30). We showed in the present study that the bile resistance phenotype was not impaired in the ΔeefA K. pneumoniae mutant compared to that of the wild-type strain. Alternative explanations include an intricate interplay between acidic and other undetected stress responses that might regulate the levels of expression of these transporters. Nevertheless, our findings add to the growing body of evidence that tripartite efflux pumps allow the survival of the bacterium within hostile environments and, hence, contribute to the pathogenicity of the organism.
Editor: V. J. DiRita
Footnotes
Published ahead of print on 21 July 2008.
REFERENCES
- 1.Aghi, P., and S. Chhiber. 1999. Adaptative acid tolerance in Klebsiella pneumoniae. Ind. J. Med. Microbiol. 1781-84. [Google Scholar]
- 2.Balestrino, D., J. A. Haagensen, C. Rich, and C. Forestier. 2005. Characterization of type 2 quorum sensing in Klebsiella pneumoniae and relationship with biofilm formation. J. Bacteriol. 1872870-2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 2771453-1474. [DOI] [PubMed] [Google Scholar]
- 4.Reference deleted.
- 5.Chaveroche, M. K., J. M. Ghigo, and C. d'Enfert. 2000. A rapid method for efficient gene replacement in the filamentous fungus Aspergillus nidulans. Nucleic Acids Res. 28E97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Champs, C., M. P. Sauvant, C. Chanal, D. Sirot, N. Gazuy, R. Malhuret, J. C. Baguet, and J. Sirot. 1989. Prospective survey of colonization and infection caused by expanded-spectrum-beta-lactamase-producing members of the family Enterobacteriaceae in an intensive care unit. J. Clin. Microbiol. 272887-2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fang, C. T., H. C. Chen, Y. P. Chuang, S. C. Chang, and J. T. Wang. 2002. Cloning of a cation efflux pump gene associated with chlorhexidine resistance in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 462024-2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Favre-Bonte, S., A. Darfeuille-Michaud, and C. Forestier. 1995. Aggregative adherence of Klebsiella pneumoniae to human intestine-407 cells. Infect. Immun. 631318-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Favre-Bonte, S., B. Joly, and C. Forestier. 1999. Consequences of reduction of Klebsiella pneumoniae capsule expression on interactions of this bacterium with epithelial cells. Infect. Immun. 67554-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gosink, M. M., N. M. Franklin, and G. P. Roberts. 1990. The product of the Klebsiella pneumoniae nifX gene is a negative regulator of the nitrogen fixation (nif) regulon. J. Bacteriol. 1721441-1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hommais, F., E. Krin, C. Laurent-Winter, O. Soutourina, A. Malpertuy, J.-P. Le Caer, A. Danchin, and P. Bertin. 2001. Large-scale monitoring of pleiotropic regulation of gene expression by the prokaryotic nucleoid-associated protein, HN-S. Mol. Microbiol. 4020-3612. [DOI] [PubMed] [Google Scholar]
- 12.Lin, J., M. Akiba, O. Sahin, and Q. Zhang. 2005. CmeR functions as a transcriptional repressor for the multidrug efflux pump CmeABC in Campylobacter jejuni. Antimicrob. Agents Chemother. 491067-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin, J., and A. Martinez. 2006. Effect of efflux pump inhibitors on bile resistance and in vivo colonization of Campylobacter jejuni. J. Antimicrob. Chemother. 58966-972. [DOI] [PubMed] [Google Scholar]
- 14.Lin, J., O. Sahin, L. O. Michel, and Q. Zhang. 2003. Critical role of multidrug efflux pump CmeABC in bile resistance and in vivo colonization of Campylobacter jejuni. Infect. Immun. 714250-4259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma, D., D. N. Cook, M. Alberti, N. G. Pon, H. Nikaido, and J. E. Hearst. 1995. Genes acrA and acrB encode a stress-induced efflux system of Escherichia coli. Mol. Microbiol. 1645-55. [DOI] [PubMed] [Google Scholar]
- 16.Markowitz, S. M., J. M. Veazey, Jr., F. L. Macrina, C. G. Mayhall, and V. A. Lamb. 1980. Sequential outbreaks of infection due to Klebsiella pneumoniae in a neonatal intensive care unit: implication of a conjugative R plasmid. J. Infect. Dis. 142106-112. [DOI] [PubMed] [Google Scholar]
- 17.Maroncle, N., D. Balestrino, C. Rich, and C. Forestier. 2002. Identification of Klebsiella pneumoniae genes involved in intestinal colonization and adhesion using signature-tagged mutagenesis. Infect. Immun. 704729-4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Masi, M., J. M. Pages, C. Villard, and E. Pradel. 2005. The eefABC multidrug efflux pump operon is repressed by H-NS in Enterobacter aerogenes. J. Bacteriol. 1873894-3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Masuda, N., and G. M. Church. 2003. Regulatory network of acid resistance genes in Escherichia coli. Mol. Microbiol. 48699-712. [DOI] [PubMed] [Google Scholar]
- 20.Mazzariol, A., J. Zuliani, G. Cornaglia, G. M. Rossolini, and R. Fontana. 2002. AcrAB efflux system: expression and contribution to fluoroquinolone resistance in Klebsiella spp. Antimicrob. Agents Chemother. 463984-3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Merrell, D. S., and A. Camilli. 1999. The cadA gene of Vibrio cholerae is induced during infection and plays a role in acid tolerance. Mol. Microbiol. 34836-849. [DOI] [PubMed] [Google Scholar]
- 22.Miller, J. H. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratories, Cold Spring Harbor, NY.
- 23.Ogawa, W., M. Koterasawa, T. Kuroda, and T. Tsuchiya. 2006. KmrA multidrug efflux pump from Klebsiella pneumoniae. Biol. Pharm. Bull. 29550-553. [DOI] [PubMed] [Google Scholar]
- 24.Piddock, L. J. 2006. Clinically relevant chromosomally encoded multidrug resistance efflux pumps in bacteria. Clin. Microbiol. Rev. 19382-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poole, K., and R. Srikumar. 2001. Multidrug efflux in Pseudomonas aeruginosa: components, mechanisms and clinical significance. Curr. Top. Med. Chem. 159-71. [DOI] [PubMed] [Google Scholar]
- 26.Putman, M., H. W. van Veen, and W. N. Konings. 2000. Molecular properties of bacterial multidrug transporters. Microbiol. Mol. Biol. Rev. 64672-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raleigh, E. A., N. E. Murray, H. Revel, R. M. Blumenthal, D. Westaway, A. D. Reith, P. W. Rigby, J. Elhai, and D. Hanahan. 1988. McrA and McrB restriction phenotypes of some E. coli strains and implications for gene cloning. Nucleic Acids Res. 161563-1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ruzin, A., M. A. Visalli, D. Keeney, and P. A. Bradford. 2005. Influence of transcriptional activator RamA on expression of multidrug efflux pump AcrAB and tigecycline susceptibility in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 491017-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reference deleted.
- 30.Stähler, F. N., S. Odenbreit, R. Haas, J. Wilrich, A. H. Van Vliet, J. G. Kusters, M. Kist, and S. Bereswill. 2006. The novel Helicobacter pylori CznABC metal efflux pump is required for cadmium, zinc, and nickel resistance, urease modulation, and gastric colonization. Infect. Immun. 743845-3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tseng, T. T., K. S. Gratwick, J. Kollman, D. Park, D. H. Nies, A. Goffeau, and M. H. Saier, Jr. 1999. The RND permease superfamily: an ancient, ubiquitous and diverse family that includes human disease and development proteins. J. Mol. Microbiol. Biotechnol. 1107-125. [PubMed] [Google Scholar]
- 32.Williams, R. M., S. Rimsky, and H. Buc. 1996. Probing the structure, function, and interactions of the Escherichia coli H-NS and StpA proteins by using dominant negative derivatives. J. Bacteriol. 1784335-4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reference deleted.
- 34.Zgurskaya, H. I., and H. Nikaido. 2000. Multidrug resistance mechanisms: drug efflux across two membranes. Mol. Microbiol. 37219-225. [DOI] [PubMed] [Google Scholar]