Abstract
Live attenuated bacterial strains expressing heterologous antigens represent an attractive vaccine development strategy. However, the use of drug resistance genes for the selection of expression plasmids introduced into live vectors poses theoretical health risks. Therefore, we developed a novel approach for plasmid selection based on immunity to the antimicrobial peptide microcin H47 (MccH47). Two expression plasmids encoding the reporter green fluorescent protein (GFPuv) were constructed; selection markers comprised either mchI, conferring immunity to MccH47 (pGEN222I), or bla (encoding β-lactamase), conferring conventional resistance to ampicillin (pGEN222). GFPuv-specific serum immunoglobulin G (IgG) antibody responses were analyzed in mice immunized intranasally either with Salmonella enterica serovar Typhi CVD 908-htrA or Shigella flexneri 2a CVD 1208S live vector and were boosted parenterally with purified GFPuv. Similar IgG antibody responses were observed for both pGEN222 and pGEN222I when either CVD 1208S or CVD 908-htrA(pGEN222I) was used as the carrier. Interestingly, CVD 908-htrA(pGEN222I) elicited a significantly higher IgG response than CVD 908-htrA(pGEN222). We also compared the priming potential of homologous priming either with CVD 908-htrA(pGEN222I) or CVD 1208S(pGEN222I) to heterologous priming first with CVD 908-htrA(pGEN222I) and then with CVD 1208S(pGEN222I) and vice versa. Immunization with two unrelated live vectors significantly enhanced the IgG responses compared to responses engendered by homologous CVD 908-htrA(pGEN222I) but not to those of CVD 1208S(pGEN222I). MccH47 offers an alternate system for plasmid selection in bacterial live vectors that greatly improves their clinical acceptability. Furthermore, the success of the heterologous priming strategy supports the feasibility of the future development of multivalent live vector-based immunization strategies against multiple human pathogens.
With the advent of recombinant bioengineering techniques, it is possible to genetically attenuate pathogenic bacteria to create safe and immunogenic oral vaccines. Such vaccines then can be further engineered to express protective antigens from unrelated human pathogens, creating multivalent live vector vaccine strains. Typically, these foreign proteins are expressed within live vectors from multicopy expression plasmids that do not encode transfer functions and are not considered to be self transmissible. Antibiotic resistance markers usually are inserted into expression plasmids to facilitate the selection and detection of live vectors carrying expression plasmids after their introduction. Until recently, these resistance markers were not considered to pose a risk of interfering with clinical antimicrobial therapy for three important reasons: (i) the expression plasmids (and accompanying resistance genes) could not be efficiently mobilized from live vector donors to a recipient (22), (ii) the plasmid markers used encoded resistance to antibiotics not in widespread medical use, and (iii) with no relevant antibiotic selective pressure, even rare plasmid transfers would not lead to de novo resistances becoming established within a new bacterial population (22).
However, a growing body of evidence now clearly points to an inherent plasticity in the bacterial genome of intestinal microbes that allows for rapid adaptation to environmental pressures using a striking variety of genetic mechanisms (6, 16, 40). Indeed, intestinal bacteria have been proposed to act as a reservoir for mobile resistance cassettes and associated genes of metabolic importance, which can be exchanged and maintained between resident flora of intestinal biofilms (23) and might also be acquired or horizontally transferred to various genera of bacteria passing through the colon (41). Given the inherent unpredictability of plasmid mobilization between enteric strains and the possibility of stable propagation in the absence of selection, the prospect of unintended and unforeseen genetic events compromising critical antimicrobial therapies cannot be formally excluded. Such risk is unacceptable if alternatives to antibiotic selection can be developed.
Several antibiotic-free plasmid selection systems have been described. Auxotrophy complementation represents a selection system that draws much attention. If a strain is auxotrophic to an essential metabolite due to a chromosomal gene mutation, this otherwise lethal mutation can be complemented with a functional, plasmid-borne copy of the gene. This approach has been applied in numerous bacterial species, such as Lactococcus lactis (15), Salmonella spp. (25, 45), Vibrio cholerae (39), and Escherichia coli (8). In a variation of this theme, a novel operator-repressor titration system controlling the chromosomal synthesis of an essential metabolite uses plasmids to titrate off the repressor; the loss of such plasmids no longer titrates the repressor, and the synthesis of the essential gene ceases, leading to the death of the bacterium (14). It also has been proposed that the selection of plasmids could be accomplished if such plasmids encoded antisense RNA to block the transcription of a lethal gene within the recipient chromosome (31). However, all of these strategies for antibiotic-free plasmid selection involve the reengineering or modification of the bacterial strains. This may lead to the overattenuation of the vaccine strain itself, which may result in the poor immunogenicity of the live vector vaccine (5). Here, we report a novel alternative approach for plasmid selection that avoids the genetic modification of the vaccine strain and involves immunity against an antimicrobial peptide called microcin H47 (MccH47).
MccH47 is produced by a natural E. coli isolate (21). Microcins have a broad spectrum of bactericidal activity against Enterobacteriaceae, including Escherichia, Salmonella, Shigella, Citrobacter, Klebsiella, and Enterobacter (28, 30). The synthesis of MccH47 is encoded by mchB within an ∼10.5-kb mch47 operon (GenBank accession number AJ009631) (33). The mch47 peptide is synthesized as a 75-residue precursor that is processed during secretion to a 60-residue mature extracellular peptide of 4.9 kDa (36). The mch47 operon also encodes an ATP binding cassette (ABC) export system specific for H47 (2), a catecholate siderophore production system that is proposed to enhance MccH47 uptake by target bacteria (1, 33), and an immunity protein that is required for bacterial self immunity (10, 37). MccH47 targets the proton channel of the F0 portion of ATP synthase, which abolishes the regulated entry of protons, leading to the depolarization of target cell membranes (38, 48). Bacterial self immunity is conferred by a 69-amino-acid highly hydrophobic protein encoded by mchI. MchI is anchored within the cytoplasmic membrane by two transmembrane regions and binds to MccH47 to prevent cell death (37). Since there are currently no existing clinical applications for MccH47, we chose to develop an MchI-based plasmid selection method for use in clinically proven strains of attenuated Salmonella and Shigella live vectors.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
All plasmid constructions were maintained and recovered either in E. coli DH5α (Invitrogen Life Technologies, Carlsbad, CA) or E. coli XL1-Blue (Stratagene, La Jolla, CA). Selection with ampicillin was used, where appropriate, at a concentration of 50 μg/ml. Plates were incubated at 30°C for 24 to 36 h to obtain isolated colonies ∼2 mm in diameter to minimize any toxicity of reporter green fluorescent protein (GFPuv) expression in live vectors.
Unless otherwise indicated, E. coli and Salmonella strains used in this study were grown in Luria-Bertani (LB) medium. Since the S. enterica serovar Typhi strain CVD 908-htrA is an auxotrophic derivative of wild-type strain Ty2 with deletions in aroC, aroD, and htrA (46), LB medium for this strain was supplemented with 2,3-dihydroxybenzoic acid (Sigma, St. Louis, MO) as previously described (11, 17). When grown on solid medium, plasmid-bearing derivatives of CVD 908-htrA were streaked from frozen (−70°C) master stocks onto 2× LB agar containing 2% (wt/vol) Bacto tryptone, 1% (wt/vol) Bacto yeast extract, and 50 mM NaCl (2× LB50 agar).
Shigella flexneri 2a strain CVD 1208S, which harbors attenuating mutations in guaBA, set, and sen (4), was grown on Hy-Soy medium (1% [wt/vol] Hy-Soy, 0.5% [wt/vol] Hi-Yeast 444 [Quest Sheffield, Chicago, IL], 150 mM NaCl) supplemented with Congo red and 0.005% (wt/vol) guanine. This animal-free medium has been chosen in accordance with regulatory guidelines aimed at reducing the theoretical and remote risk of transmissible spongiform encephalopathies by vaccines intended for human use (19). The optimization of growth conditions for CVD 908-htrA in an animal-free formulation is currently under way.
Molecular genetic techniques.
Standard techniques were used for plasmid constructions (42). Unless otherwise noted, Taq DNA polymerase (Invitrogen, San Diego, CA) or Vent DNA polymerase (New England BioLabs, Beverly, MA) was used in PCRs. CVD 908-htrA and CVD 1208S strains were electroporated with recombinant plasmids as previously described (12). Isolated transformants were swabbed onto supplemented 2× LB50 or Hy-Soy agar, respectively, and incubated at 30°C for 20 h. Frozen master stocks were prepared by harvesting bacteria into LB or Hy-Soy medium with 20% glycerol without further supplementation, followed by freezing them at −70°C.
Cloning of the E. coli mchI gene.
An mchI genetic cassette was constructed that consisted of a variant synthetic lacUV5 promoter controlling the transcription of mchI. This construction was accomplished in a three-step PCR. Primers 1 and 2 (Table 1), with 30 bp of complementary sequence, were used in an initial PCR 1 to generate a 90-bp promoter cassette encoding a transcriptionally weakened 33C variant of the lacUV5 promoter (18) and optimized ribosome binding site (35). This initial promoter cassette was specifically engineered such that the modified lacUV5 promoter (51 bp) was flanked by NheI and XbaI restriction sites to allow easy replacement with a stronger promoter cassette if the transcription of mchI was insufficient to allow selection with MccH47. A promoterless 244-bp mchI genetic cassette was synthesized in PCR 2 using primer 3 (33 bp complementary to primer 2 for final assembly in final overlapping PCR) and primer 4 with crude total template DNA from the MccH47-producing strain E. coli RYC1000(pEX4) (21) (a kind gift from Klaus Hantke, Institut für Organische Chemie, Universität Tübingen, Tübingen, Germany). The final 301-bp mchI gene was assembled as a genetic cassette flanked by NheI sites using primers 1 and 4 with the products of PCRs 1 and 2. The desired product was inserted into pSMART-LCKan (Lucigen Corporation, Middleton, WI) to create pSMARTmchI, which was recovered in E. coil DH5α.
TABLE 1.
Primers used in the construction and testing of mchI expression plasmids
| Primer | Sequencea | Cassette created | Templateb | Reference or source |
|---|---|---|---|---|
| 1 | 5′-GCTAGCGGGCACCCCAGGCTTTCCACTTTATGCTTCCGGCTCGTATAATGTGTGGAATCT-3′ | lacUV5 promoter variant 33C | NA | 18 |
| 2 | 5′-TAACTCATTTTTTTTTCCTCCTTATTTTCTAGATTCCACACATTATACGAGCCGGAAGCA-3′ | lacUV5 promoter variant 33C | NA | 18 |
| 3 | 5′-TCTAGAAAATAAGGAGGAAAAAAAAATGAGTTATAAAAAACTGTACCAATTGACGGCTATAT-3′ | mchI | RYC1000(pEX4) | 37 |
| 4 | 5′-GCTAGCCTATTAACTTCCGTTTTTCCAGCCTGACCAGACCAAAATCTTTCGAATCCCATGA-3′ | mchI | RYC1000(pEX4) | 37 |
| 5 | 5′-GTAAACTTGGTCTGACAGGTACCAATGCTTAATCAGTG-3′ | pGENMch | This study | |
| 6 | 5′-CACTGATTAAGCATTGGTACCTGTCAGACCAAGTTTAC-3′ | pGENMch | This study | |
| 7 | 5′-CAAATAGGGGTACCGCGCACATTTCCCCG-3′ | pGENMch | This study | |
| 8 | 5′-CGGGGAAATGTGCGCGGTACCCCTATTTG-3′ | pGENMch | This study |
Relevant restriction sites are designated in boldface (see the text for details); ribosome binding sites and start codons are in italics.
NA, not applicable.
Construction of expression plasmids.
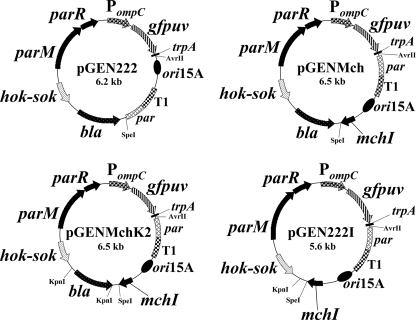
The mchI cassette was inserted subsequently as an NheI fragment into the previously described pGEN222 expression plasmid that contains the plasmid maintenance functions hok-sok and parA (comprised of parM and parR) (12). We originally intended to insert this NheI cassette into the unique AvrII site of pGEN222, behind the trpA transcriptional terminator, such that the expression of MchI would be controlled solely by the weakened synthetic variant lacUV5 promoter. However, we were never able to obtain this construction. After trial and error, we were able to obtain the insertion of mchI into pGEN222 only after genetically reversing the orientation of the origin of replication in pGEN222 (containing ori15A, the T1 terminator, and par cassettes) and inserting mchI into the original AvrII target site adjacent to the ori15A origin of replication, creating pGENMch (Fig. 1). To facilitate the removal of the bla gene, two KpnI sites were introduced upstream and downstream of bla in pGENMch by the QuikChange II-E site-directed mutagenesis kit (Stratagene, La Jolla, CA). Two pairs of complementary primers (primers 5 and 6 as well as 7 and 8) (Table 1) were designed to accomplish this modification of pGENMch. The resulting clone was designated pGENMchK2. The bla gene was removed by KpnI digestion, and the plasmid was religated to create pGEN222I (Fig. 1). The expected DNA sequences of the resulting constructs were confirmed by DNA sequence analysis.
FIG. 1.
Genetic maps of isogenic expression plasmids encoding the prokaryotic codon-optimized GFPuv. Key restriction sites used for constructions are shown. Abbreviations: PompC, modified osmotically controlled ompC promoter from E. coli; gfpuv, gene encoding prokaryotic codon-optimized GFPuv; T1, transcriptional terminator from the rrnB rRNA operon of E. coli; par, passive partitioning function from pSC101; ori15A, origin of replication from p15A providing an expected copy number of ∼15 per chromosomal equivalent; bla, β-lactamase gene conferring resistance to ampicillin; mchI, gene coding for the immunity protein MchI conferring resistance to MccH47; hok-sok, postsegregational killing locus from the multiple antibiotic resistance R plasmid pR1; and parM and parR, two loci comprising the parA active partitioning system from pR1.
Selection of E. coli and Shigella transformants containing mchI-carrying plasmids on solid medium.
All constructions of expression plasmids carrying both the GFPuv operon and mchI were carried out using selection for resistance to ampicillin. Final non-drug-resistant constructs were selected for on solid medium impregnated with MccH47 from crude culture supernatants following the final deletion of the bla allele.
A master stock of crude extract containing secreted MccH47 was prepared from overnight cultures of the MccH47-producing strain RYC1000(pEX4), which was grown in a modified M63 broth medium consisting of KH2PO4 (13.6 g/liter), (NH4)2SO4 (2 g/liter), FeSO4·7H2O (0.5 mg/liter), MgSO4·7H2O (0.25 g/liter) and supplemented with 0.2% (wt/vol) glucose, 1 μg/ml vitamin B1 (thiamine hydrochloride), 0.1% (wt/vol) Casamino Acids, and ampicillin (50 μg/ml), and then the solution was adjusted to pH 7.0 with KOH. Aliquots (25 ml) of M63 medium in 250-ml baffle flasks were inoculated with 1 ml RYC1000(pEX4) overnight culture and incubated at 37°C for 24 h with vigorous aeration at 250 rpm. After centrifugation, each supernatant was sterilized using a 0.2 μM Nalgene syringe filter (Nalge Company, Rochester, NY) and subsequently concentrated down to 1 ml using a Centricon Plus-20 centrifugal filter (NMWL 10000; Millipore, Bedford, MA). The final master stock was created by pooling 1-ml concentrates from 20 M63 cultures. The MIC of this master stock then was determined for E. coli DH5α and S. flexneri 2a CVD 1208S, and clear zones of growth inhibition were observed at a 1/40 dilution for DH5α and a 1/5 dilution for CVD 1208S. Frozen aliquots of this stock were maintained at −70°C until needed. After determining crude extract MICs, MccH47-supplemented solid medium was prepared using 2× LB50 and was used to select for MccH47-resistant clones following the transformation of DH5α, XL1-Blue, or CVD 1208S with MchI-coding plasmids.
Selection of CVD 908-htrA transformants containing mchI-carrying plasmids on solid medium.
We observed that S. enterica serovar Typhi strains were not as sensitive to the bactericidal effects of MccH47 as either E. coli or Shigella strains, and they required much higher concentrations of MccH47 than we could achieve using concentrated culture filtrates for the selection of transformants (data not shown). Therefore, a cross-streaking method (20, 48) was employed using 2× LB50 plates for the selection of CVD 908-htrA transformants receiving plasmids in which bla had been deleted.
Briefly, a single colony of the MccH47-producing strain RYC1000(pEX4) was inoculated into 2.5 ml of 1× LB50 supplemented with ampicillin (50 μg/ml). The tube was incubated for 6 h with shaking at 37°C. Subsequently, a sterile cotton swab was dipped into the culture suspension, and excess fluid was squeezed out by pressing the swab against the tube. The swab then was used to inoculate a 1-cm streak across a 2× LB50 agar plate, which was incubated for 24 h at 37°C. Bacterial growth was removed by scraping sterile glass slides across the surface of the plate. The remaining bacteria were killed by chloroform vapor. Cultures of CVD 908-htrA transformants were streaked at right angles to the original band of growth now impregnated with MccH47. Sensitive strains were unable to grow in the immediate vicinity of the MccH47 gradient, while the growth of transformants was not inhibited.
After restreaking bacteria recovered from the immediate vicinity of the MccH47 gradient for isolated colonies, transformants were conclusively identified using a patch test (48). Briefly, this test was performed by seeding a lawn of each candidate transformant onto 2× LB50 plates and then stabbing the lawn with the MccH47-producing strain, RYC1000(pEX4). After overnight incubation at 37°C, zones of growth inhibition were observed for CVD 908-htrA parent strains, while the growth of transformants was not affected.
Plasmid stability test.
To determine the persistence in vitro of pGEN222 or pGEN222I in a growing population of Salmonella and Shigella live vectors, bacterial cultures were passaged for 4 days (96 h) without antibiotic selection. Frozen stocks were streaked onto appropriately supplemented solid medium without selection and incubated at 30°C for 48 h to obtain isolated colonies. Two to three fluorescing colonies then were inoculated into 20 ml liquid medium without selection and incubated with shaking at 225 rpm overnight at 30°C (0-h starting cultures for serial passages). Overnight starter cultures then were diluted 1:50 into fresh nonselective medium, incubated for 24 h at 37°C, and then serially passaged every 24 h in the same way for 96 h total. Viable counts on nonselective solid medium were determined for hours 0, 2, 4, 6, 8, 10, 24, 48, 72, and 96. Plasmid stability was reported as the percentage of fluorescing colonies compared to the total number of CFU.
Western immunoblotting.
Western immunoblot analysis was carried out as previously described (13). Samples were prepared from cultures grown at 30°C to an optical density at 600 nm (OD600) of approximately 0.8. Briefly, an equal volume of the culture was mixed with 2× Laemmli sample buffer (Bio-Rad, Hercules, CA) containing 5% (vol/vol) 2-mercaptoethanol and was resolved using sodium dodecyl sulfate-12% (vol/vol) polyacrylamide gels. Proteins then were electroblotted onto a polyvinyl difluoride membrane (Bio-Rad, Hercules, CA). The detection of GFPuv was carried out using a murine GFPuv primary antibody at a dilution of 1:15,000 (BD Biosciences/Clontech, Palo Alto, CA) and a peroxidase-labeled affinity-purified goat anti-mouse secondary antibody at a dilution of 1:10,000 (Kirkegaard & Perry Laboratories, Inc., Gaithersburg, MD). Immunoblots were developed using an ECL plus detection system (GE Healthcare, Piscataway, NJ), and blots were exposed to Kodak X-OMAT XAR-2 film.
Vaccination of mice.
Groups of 10 to 15 female BALB/c mice (Charles River Laboratories, Wilmington, MA), aged 6 to 8 weeks, were inoculated via the intranasal (i.n.) route (7, 13, 29) by placing 10 μl of a vaccine suspension containing 1 × 109 to 3 × 109 CFU into the right and left nares on days 0 and 14. Mice were boosted intramuscularly (i.m.) with 0.5 μg of recombinant GFPuv protein (BD Biosciences/Clontech, Palo Alto, CA) absorbed to 0.5 mg of alhydrogel on day 42. Serum samples from mice were collected by retroorbital bleeding on days 0, 14, 28, 42, 49, 56, and 70. Sera were stored at −70°C until analyzed. The methods used in these animal studies were approved by the Institutional Animal Care and Use Committee of the University of Maryland, Baltimore.
Measurement of antibodies in mouse sera.
Total serum immunoglobulin G (IgG) antibodies against GFPuv protein were measured by an enzyme-linked immunosorbent assay (ELISA). Briefly, 96-well plates were coated with 100 μl of GFPuv antigen in carbonate buffer (pH 6.9) at 2 μg/ml. Plates were blocked overnight with 10% (wt/vol) milk (Nestle USA, Inc., Glendale, CA) in phosphate-buffered saline (PBS) and washed six times with PBS containing 0.05% (vol/vol) Tween 20 (PBST) after being coated and blocked. To determine endpoint titers, sera were tested in serial twofold dilutions using 10% (wt/vol) milk in PBST (PBSTM), starting at a 1:50 dilution. Specific antibodies were detected with goat anti-mouse IgG-horseradish peroxidase conjugate (Roche Diagnostics Corporation, Indianapolis, IN) diluted 1:1,000 in PBSTM. The substrate solution used was TMB microwell peroxidase (Kirkegaard & Perry Laboratories, Inc., Gaithersburg, MD). After 15 min of incubation, the reaction was stopped with the addition of 100 μl of 1 M H3PO4, and the OD450 was measured with an ELISA reader (Multiskan Ascent; Thermo Labsystem, Helsinki, Finland). Sera were run in duplicate; negative and positive controls were included in each assay. IgG endpoint titers were calculated from linear regression curves as the inverse of the serum dilution that produced an OD of 0.2 U above that of the blank and were expressed in ELISA units per milliliter. Seroconversion was defined as a fourfold increase in the antibody titer after immunization.
Statistical analysis.
Logarithms (base 10) of antibody titers measured at different time points (days 28, 42, 49, 56, and 70) were used in analyses, since the logarithms were more nearly normally distributed than untransformed titers. Groups of mice were compared over all time points simultaneously using multivariate analysis of variance with the Wilks lambda criterion; when only two groups were compared, this is equivalent to Hotelling's T square. Comparisons for a particular day were done using analysis of variance, which for the comparison of two groups is equivalent to the Student's t test. All P values reported are two-sided; P < 0.05 was considered statistically significant. Analyses were done using NCSS 2007 (Number Cruncher Statistical Systems, Kaysville, UT).
RESULTS
Construction of expression plasmids encoding MchI.
Based on the published data for the mch47 operon carried by plasmid pEX4 in E. coli strain RYC1000 (37), primers were designed to amplify a promoterless 244-bp mchI genetic cassette encoding the highly hydrophobic 69-residue MccH47 immunity protein. To avoid the potentially lethal high-level synthesis of the immunity protein, the expression of mchI was controlled by engineering a weakened constitutive variant of the lacUV5 promoter to regulate the transcription of the mchI open reading frame. After repeated unsuccessful attempts to insert the final 301-bp mchI NheI cassette into the previously described medium-copy-number pGEN222 expression plasmid (∼15 copies per chromosomal equivalent [12]), we reengineered the region of this plasmid encoding the origin of replication to finally allow the insertion of mchI adjacent to ori15A, creating pGENMch (Fig. 1). It is likely that the transcription of mchI within pGENMch is increased by the RNA II promoter within ori15A (47), suggesting that the lacUV5 variant promoter engineered into the mchI cassette was too weak.
Selection of MccH47-resistant transformants on solid medium.
A preliminary analysis of E. coli strains DH5α and XL1-Blue and S. flexneri 2a CVD 1208S showed comparable sensitivity to MccH47. However, CVD 908-htrA displayed only a moderate sensitivity to MccH47 (data not shown). Based on these observations, separate selection procedures to select for MccH47-resistant transformants of E. coli/S. flexneri or for those of S. enterica serovar Typhi were developed. After the deletion of the bla cassette encoding resistance to ampicillin, the final pGEN222I plasmid encoding resistance only to MccH47 was readily recovered in DH5α, XL1-Blue, or CVD 1208S cells after simply plating them on solid medium impregnated with MccH47 and identifying isolated colonies.
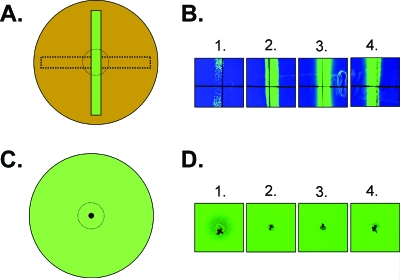
In contrast, the selection of MccH47-resistant CVD 908-htrA required the use of a cross-streaking procedure to enrich for transformants in the immediate vicinity of the MccH47 deposition (Fig. 2A), followed by patch testing to confirm resistance to MccH47 (Fig. 2C). Growth in the MccH47 zone of the cross-streak was restreaked onto solid medium without MccH47 to recover single colonies. Isolated colonies recovered using this enrichment procedure were screened for the expression of GFPuv, and approximately 55% of these isolates expressed GFPuv.
FIG. 2.
MccH47 sensitivity assays for CVD 908-htrA carrying plasmids pGEN222 (lanes 1), pGENMch (lanes 2), pGENMchK2 (lanes 3), and pGEN222I (lanes 4). (A and B) The cross-streaking method for resistance to MccH47. An overnight culture of RYC1000(pEX4) was swabbed from left to right across 2× LB50 plates and allowed to grow overnight at 37°C to impregnate the medium with MccH47 (A, dotted horizontal rectangle). Excess bacterial growth then was removed from the surface, and remaining bacteria were lysed with chloroform. CVD 908-htrA carrying plasmids expressing GFPuv with or without MchI then were streaked orthogonally to the MccH47 zone (A, vertical green rectangle), and plates were again incubated overnight at 37°C. The presence or absence of zones of clearing near the MccH47 region are represented as dotted circles in the graphic; panel B documents these zones for plasmid-bearing CVD 908-htrA live vectors. (C and D) Patch tests for resistance to MccH47. 2× LB50 plates were seeded with a lawn of CVD 908-htrA carrying plasmids expressing GFPuv with or without MchI (C, green circle). The MccH47-expressing strain RYC1000(pEX4) was spotted in the middle of each plate, and plates were incubated at 37°C overnight. The presence or absence of zones of clearing are represented by dotted circles; panel D documents these zones for plasmid-bearing CVD 908-htrA live vectors.
Final resistance profiles for CVD 908-htrA strains carrying plasmids expressing GFPuv either with or without MchI are shown in Fig. 2 using the methods of cross-streaking (Fig. 2B) and patch testing (Fig. 2D). The clearing in the region of MccH47 deposition for the MccH47-sensitive CVD 908-htrA(pGEN222) strain is observed in lane 1 of Fig. 2B using the cross-streak technique, although some breakthrough of GFPuv-expressing colonies is evident. In contrast, all strains expressing the MchI immunity protein grow as luxuriant streaks through the area impregnated with MccH47 (Fig. 2B, lanes 2 to 4). As expected, zones of clearing in the patch test are again obvious for the MccH47-sensitive CVD 908-htrA(pGEN222) strain (Fig. 2D, lane 1), whereas all strains carrying plasmids expressing the MchI immunity protein (lanes 2 to 4) are confluent up to the patch of RYC1000(pEX4).
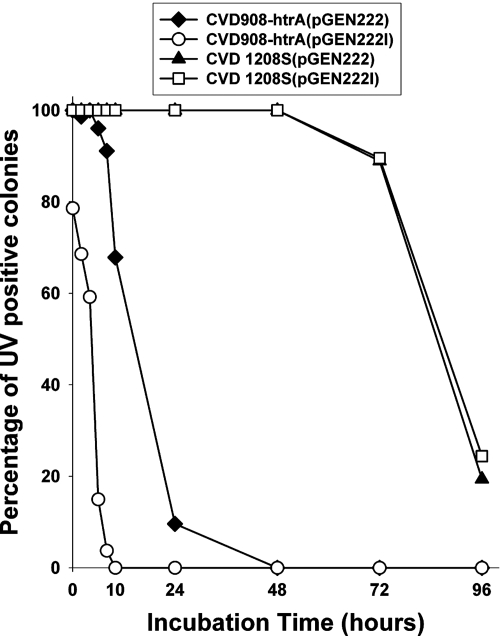
Plasmid stability test.
The retention of pGEN222I compared to that of pGEN222 during the growth of CVD 908-htrA and CVD 1208S transformants in vitro was assessed by serial passage at 37°C in liquid medium without antibiotic selection. As shown in Fig. 3, pGEN222I proved unstable in S. enterica serovar Typhi CVD 908-htrA and was cured within 10 h without selection; the retention of pGEN222 was slightly better, with complete loss occurring at 48 h. However, S. flexneri 2a CVD 1208S exhibited superior plasmid retention, with 100% of the population retaining either pGEN222 or pGEN222I after 48 h of passage in selection-free medium. Greater than 90% plasmid retention for both pGEN222 and pGEN222I still was observed after 72 h, but it dropped to approximately 20% at 96 h.
FIG. 3.
In vitro stability of pGEN222 and pGEN222I plasmids in CVD 908-htrA and CVD 1208S live vectors. Live vector strains were passaged at 37°C from an overnight starter culture (grown without selection) every 24 h for 96 h in antibiotic-free liquid medium. Plasmid stability is reported as the percentage of fluorescing colonies from the total number of CFU plated on nonselective medium.
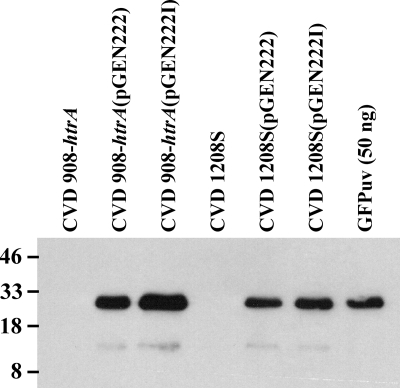
Expression of GFPuv.
We then compared GFPuv expression levels of conventional pGEN222 to those of nonantibiotic pGEN222I plasmids in both S. flexneri and S. enterica serovar Typhi vaccine strains. As shown in Fig. 4, comparable levels of GFPuv expression were detected in immunoblots of whole-cell lysates from both strains carrying either expression plasmid; analogous bands with a relative molecular mass of 27 kDa also could be seen in polyacrylamide gels stained with Coomassie brilliant blue (data not shown). As expected, no protein was recognized by anti-GFPuv antibody in whole-cell lysates from CVD 908-htrA and CVD 1208S.
FIG. 4.
Western immunoblot analysis of whole bacterial lysates of CVD 908-htrA (lane 1), CVD 908-htrA(pGEN222) (lane 2), CVD 908-htrA(pGEN222I) (lane 3), CVD 1208S (lane 4), CVD 1208S(pGEN222) (lane 5), CVD 1208S(pGEN222I) (lane 6), and GFPuv protein (50 ng) (lane 7). Numbers at the left denote relative molecular masses in kilodaltons. The detection of GFPuv was carried out using murine polyclonal primary antibody.
Antibody responses to GFPuv elicited by microcin- and ampicillin-selected plasmids carried in S. enterica serovar Typhi CVD 908-htrA and S. flexneri 2a CVD 1208S live vectors.
We analyzed the total GFPuv-specific IgG levels evoked by CVD 908-htrA and CVD 1208S live vectors carrying conventional expression plasmids selected using ampicillin (pGEN222) and plasmids selected by conferring resistance to MccH47 (pGEN222I). Sixty mice were allocated into six groups (10 mice per group) and vaccinated according to the regimens shown in Table 2. Antibody levels were clearly higher in mice immunized with plasmid-containing live vectors than in those immunized with empty live vectors (P < 0.0001, both overall [multivariate analysis of variance] and for each day [analysis of variance]).
TABLE 2.
Percentage of seroconverted mice and GMTs after vaccination with CVD 908-htrA or CVD 1208S live vectors carrying plasmids selected either by resistance to ampicillin (pGEN222) or MccH47 (pGEN222I)
| Group | Immunization regimena
|
% Seroconverted miceb (GMT)c
|
||||||
|---|---|---|---|---|---|---|---|---|
| Prime 1 | Prime 2 | Boost | Day 28 | Day 42 | Day 49 | Day 56 | Day 70 | |
| 1 | CVD 908-htrA | CVD 908-htrA | GFPuv | 0 (10) | 0 (22) | 0 (39) | 90 (4,140) | 90 (9,257) |
| 2 | CVD 908-htrA(pGEN222) | CVD 908-htrA(pGEN222) | GFPuv | 20 (43) | 20 (74) | 100 (6,305) | 100 (113,341) | 100 (118,622) |
| 3 | CVD 908-htrA(pGEN222I) | CVD 908-htrA(pGEN222I) | GFPuv | 100 (14,092) | 100 (39,082) | 100 (869,134) | 100 (1,174,129) | 100 (1,096,698) |
| 4 | CVD 1208S | CVD 1208S | GFPuv | 0 (30) | 0 (31) | 0 (41) | 80 (4,088) | 100 (19,810) |
| 5 | CVD 1208S(pGEN222) | CVD 1208S(pGEN222) | GFPuv | 100 (46,051) | 100 (50,720) | 100 (914,116) | 100 (1,189,972) | 100 (1,390,420) |
| 6 | CVD 1208S(pGEN222I) | CVD 1208S(pGEN222I) | GFPuv | 100 (85,244) | 100 (74,695) | 100 (995,557) | 100 (1,125,476) | 100 (1,207,469) |
Animals received primary immunizations on days 0 and 14 and booster immunizations on day 42, as described in Materials and Methods.
The percentage of mice that developed reciprocal serum IgG anti-GFPuv titers after vaccination. Sera were tested from twofold serial dilutions starting at 1:50.
GMTs of serum anti-GFPuv are shown in ELISA units.
We hypothesized that equivalent GFPuv-specific IgG serum antibody responses would be induced by bacterial hosts carrying either pGEN222 or pGEN222I, since copy numbers, stabilization functions, and antigen expression cassettes were identical for both expression plasmids. However, as reported in Table 2, while 100% of mice immunized with two doses of CVD 908-htrA(pGEN222I), CVD 1208S(pGEN222), or CVD 1208S(pGEN222I) exhibited seroconversion on day 28, only 20% of mice immunized with CVD 908-htrA(pGEN222) seroconverted. A week after being boosted with GFPuv protein (day 49), all of these mice showed seroconversion. The induction of GFPuv-specific IgG antibody was two to three orders of magnitude lower in mice primed with CVD 908-htrA(pGEN222) than in mice primed with the other plasmid-containing vectors after the administration of the second priming dose (P < 0.0001, both overall and at each day). This difference in titers persisted 7 days after boosting but narrowed to approximately a 10-fold difference 2 weeks after boosting. There was no overall difference in anti-GFPuv IgG titers among the groups of mice immunized with CVD 908-htrA(pGEN222I), CVD 1208S(pGEN222), or CVD 1208S(pGEN222I) (P = 0.18). In pairwise comparisons of live attenuated Salmonella vectors containing pGEN222 and pGEN222I, GFPuv-specific IgG antibody titers were significantly higher in mice immunized with CVD 908-htrA(pGEN222I) than in those immunized with CVD 908-htrA(pGEN222) (overall P = 0.0002; P < 0.0001 for each specific day).
Responses were similar between the two groups of mice in which attenuated Shigella was used as the carrier for antigen delivery (overall P = 0.61; P = 0.20 to 0.86 for comparisons at specific days). Ratios of geometric mean titers (GMTs) in these two groups [CVD 1208S(pGEN222)/CVD 1208S(pGEN222I)] ranged from 0.54 at day 28 to 1.15 at day 70, with upper limits of 95% confidence intervals for the ratios ranging from 1.44 at day 28 to 2.17 at day 70. Although antibody responses generally were higher in mice immunized with CVD 1208S(pGEN222I) than in mice immunized with CVD 908-htrA(pGEN222I) (Table 2), the difference was significant only at day 28 (P = 0.007; in the overall comparison, P = 0.15).
As expected, all mice immunized with CVD 908-htrA, CVD 908-htrA(pGEN222), or CVD 908-htrA(pGEN222I) mounted strong antibody responses against Salmonella lipopolysaccharide (LPS) (data not shown). Similarly, all mice immunized with CVD 1208S, CVD 1208S(pGEN222), or CVD 1208S(pGEN222I) mounted strong antibody responses against Shigella LPS (data not shown).
Antibody responses to GFPuv elicited in mice primed i.n. with Salmonella followed by Shigella live vector (or vice versa) and those primed against GFPuv using two doses of a single live vector vaccine.
In an attempt to improve the priming strategy and efficiency of our live vectors, we investigated the homologous priming potential of both attenuated CVD 908-htrA and CVD 1208S strains carrying pGEN222I and compared them to the potential of heterologous priming first with CVD 908-htrA(pGEN222I) and then with CVD 1208S(pGEN222I), or vice versa. BALB/c mice were randomly assorted into five groups of 15 mice and immunized i.n. on days 0 and 14, with subsequent boosting with GFPuv protein on day 42. As expected, all mice immunized with CVD 908-htrA mounted IgG serum antibody responses against Salmonella LPS, and all mice immunized with CVD 1208S displayed IgG antibody responses against Shigella LPS (data not shown).
As shown in Table 3, only 80% of mice seroconverted to GFPuv by day 42 after receiving two doses of CVD 908-htrA(pGEN222I), while 100% of mice in all other groups vaccinated with live vectors had seroconverted by day 28. The highest responses after priming with live vectors were observed on day 28 in mice mucosally immunized with two doses of CVD 1208S(pGEN222I) (GMT = 46,344), and they were more than 20-fold higher than the titers seen in mice primed with two doses of CVD 908-htrA(pGEN222I); these relative differences narrowed after boosting with GFPuv. The higher responses with S. flexneri homologous priming than with S. enterica serovar Typhi homologous priming were statistically significant overall (P = 0.005) and at days 28 (P < 0.0001), 42 (P = 0.0007), and 49 (P = 0.046). Applying Fisher's procedure for combining P values to the overall results for this experiment (P = 0.005) and the P value of 0.15 for the same comparison in the previous experiment, we have a significant overall difference (P = 0.006), with relative antibody levels being much higher after CVD 1208S priming than those after priming with CVD 908-htrA live vectors, but the levels become much closer after boosting with GFPuv. As expected, mice primed with only two doses of PBS did not seroconvert until 2 weeks after being boosted with GFPuv, but titers remained at least 100-fold lower than those for mice primed with any live vector combination carrying pGEN222I.
TABLE 3.
Percentage of seroconverted mice and GMTs after immunization with CVD 908-htrA and CVD 1208S live vectors carrying pGEN222I using a homologous or heterologous vaccination approach
| Group | Immunization regimena
|
% Seroconverted miceb (GMT)c
|
||||||
|---|---|---|---|---|---|---|---|---|
| Prime 1 | Prime 2 | Boost | Day 28 | Day 42 | Day 49 | Day 56 | Day 70 | |
| 1 | CVD 908-htrA(pGEN222I) | CVD 908-htrA(pGEN222I) | GFPuv | 73.3 (1,807) | 80 (2,579) | 100 (203,368) | 100 (461,503) | 100 (593,347) |
| 2 | CVD 1208S(pGEN222I) | CVD 1208S(pGEN222I) | GFPuv | 100 (46,344) | 100 (33,848) | 100 (637,600) | 100 (795,562) | 100 (891,711) |
| 3 | CVD 908-htrA(pGEN222I) | CVD 1208S(pGEN222I) | GFPuv | 100 (14,658) | 100 (13,097) | 100 (911,902) | 100 (1,147,824) | 100 (1,243,812) |
| 4 | CVD 1208S(pGEN222I) | CVD 908-htrA(pGEN222I) | GFPuv | 100 (20,458) | 100 (20,298) | 100 (600,911) | 100 (876,346) | 100 (1,066,344) |
| 5 | PBS | PBS | GFPuv | 0 (16) | 0 (25) | 0 (15) | 100 (4,883) | 100 (17,250) |
Animals received primary immunizations on days 0 and 14 and booster immunizations on day 42, as described in Materials and Methods.
The percentage of mice that developed reciprocal serum IgG anti-GFPuv titers after vaccination. Sera were tested from twofold serial dilutions starting at 1:50.
GMTs of serum anti-GFPuv are shown in ELISA units.
Priming with the CVD 908-htrA/CVD 1208S combination produced the highest GFPuv-specific GMT in mice on day 49 (Table 3) (GMT = 911,902), 7 days after being boosted with purified antigen, and titers for this combination remained somewhat higher through day 70. However, differences between the two heterologous priming combinations were not statistically significant, either overall (P = 0.40) or for any specific day (P = 0.072 for day 49 and 0.26 to 0.52 for other days). Although the patterns of antibody titers in mice with heterologous priming appear somewhat different from those of priming with two doses of CVD 1208S, there were no significant differences in antibody levels among the three groups, either overall (P = 0.10) or at specific days (P = 0.087 to 0.22).
DISCUSSION
We have developed a nonantibiotic plasmid selection system for use with attenuated Salmonella and Shigella live vector vaccine strains that is based on resistance to MccH47. Microcins are relatively small ribosomally synthesized antimicrobial peptides (typically less than 10 kDa) that may be bactericidal or bacteriostatic (27, 34). There are no existing clinical uses for microcins in general, or MccH47 specifically, possibly due to the difficulty in achieving biologically inhibitory concentrations of microcin in vivo (9). Therefore, the use of microcins in place of antibiotics for plasmid selection would improve the acceptability of such plasmid-based live vector vaccines by ensuring that the unintended transfer of antibiotic resistance genes to pathogenic bacteria could not compromise therapeutic treatments in a clinical setting. Another significant advantage to using this approach is that microcin-selected plasmids can be readily introduced into a variety of currently available live vectors without further genetic manipulation of the live vector chromosome.
To demonstrate the feasibility of using MccH47 for plasmid selection, we constructed a pair of expression plasmids (pGEN222 and pGEN222I) in which selection was accomplished solely through conventional resistance to ampicillin or through resistance to MccH47 that was conferred by the MccH47 immunity protein MchI. To easily confirm and monitor the presence of these plasmids within live vectors, both constructs encoded the fluorescent reporter protein GFPuv and were derived from the medium-copy-number expression plasmid pGEN222 (12). This original expression plasmid was chosen for study because it possesses a plasmid maintenance system comprised of a parA plasmid-partitioning operon (to enhance plasmid inheritance after the division of a bacterium) and a hok-sok postsegregational killing function (to remove plasmidless bacteria). Together, these maintenance functions are intended to promote uniform plasmid inheritance and prevent faster-growing plasmidless bacteria from overtaking a growing population of live vectors.
The inhibitory characteristics of MccH47 against the live vector strains used in this study varied. While E. coli and S. flexneri 2a strains displayed similar levels of sensitivity to MccH47, CVD 908-htrA displayed only moderate sensitivity. Since MccH47 is not commercially available, it was necessary to prepare concentrated crude supernatants from cultures secreting MccH47 and to carry out MIC assays for each strain. Assay results confirmed that much higher concentrations of MccH47 crude extract were necessary to inhibit the background growth of CVD 908-htrA during selection (data not shown). Therefore, while transformants of E. coli and Shigella could be directly selected on solid medium containing inhibitory concentrations of MccH47, MccH47-resistant CVD 908-htrA strains could be enriched for using only the cross-streak procedure (Fig. 2A and B), with subsequent restreaking for the isolation and confirmation of plasmid acquisition using a patch test (Fig. 2C and D). The exact mechanism of MccH47 uptake by susceptible strains remains unclear. Azpiroz and Lavina (1) have proposed that for susceptible E. coli strains, mature MccH47 has to be joined with glycosylated dihydroxybenzoylserine, a breakdown product of enterobactin (or a related iron-complexing siderophore), prior to entering target cells through the catecholate receptor proteins Cir, Fiu, and FepA. It is possible that differences in the amino acid sequences of these or related receptors in S. enterica serovar Typhi account at least in part for the reduced susceptibility of S. enterica serovar Typhi to MccH47.
Our strategy for immunization using these improved live vectors was based on a two-phase heterologous prime/boost approach involving sequential administration (in a live vector priming phase and a purified protein boosting phase) of the same antigen (GFPuv) in two different vaccine formulations by different routes (i.n. and i.m.). Heterologous prime/boost strategies recently have been shown to significantly improve the immunogenicity of both eukaryotic and prokaryotic foreign antigens delivered by attenuated S. enterica serovar Typhi live vectors (7, 24, 49). The results of our study further support the efficacy of the heterologous prime/boost strategy, as we observed a drastic elevation of GFPuv-specific IgG responses after GFPuv protein boosting (Tables 2 and 3).
We expected that an equivalent GFPuv-specific IgG antibody response would be observed in mice immunized with live vector strains bearing expression plasmids selected either with ampicillin (pGEN222) or MccH47 (pGEN222I). This assumption was based on the relative genetic identity of both expression plasmids, as well as the comparable levels of GFPuv expression in attenuated Salmonella and Shigella live vectors demonstrated in vitro (Fig. 4). While this expectation turned out to be correct when using attenuated Shigella flexneri 2a strains as live vectors, significantly higher GFPuv-specific IgG responses were seen in mice primed with S. enterica serovar Typhi CVD 908-htrA(pGEN222I) than for those primed with the conventional CVD 908-htrA(pGEN222) live vector (Table 2).
Results from the in vitro plasmid stability study (Fig. 3) may partially explain these results. The relative stability of our expression plasmids in CVD 1208S live vectors correlates nicely with the observed immunogenicity of GFPuv delivered by CVD 1208S carrying these plasmids in vivo. However, both expression plasmids were unstable in the attenuated S. enterica serovar Typhi CVD 908-htrA live vector, with pGEN222I being completely lost after 10 h and pGEN222 completely cured after 48 h. Since the instability of these expression plasmids in vitro also is likely to be the case in vivo, the superior immunogenicity of GFPuv delivered by S. enterica serovar Typhi live vectors carrying pGEN222I plasmids compared to that of live vectors carrying pGEN222 seems surprising. However, Pickett et al. (32) have shown that antibody responses to a foreign antigen occur despite the fact that viable counts of plasmid-bearing strains in an i.n. inoculum drop from 109 to 102 CFU in mice within 18 h after immunization. We hypothesize that the priming of the murine immune system by S. enterica serovar Typhi live vectors in some cases occurs quite early after i.n. immunization, before plasmid instability exerts a significant effect. However, we cannot rule out the possibility that the lack of strict isogenicity between pGEN222I and pGEN222 (Fig. 2) has influenced differences in immunogenicity between CVD 908-htrA(pGEN222I) and CVD 908-htrA(pGEN222), although whatever effect influences S. enterica serovar Typhi apparently has no effect on S. flexneri 2a live vectors.
Several reports have suggested that enhanced immunogenicity with heterologous prime/boost strategies are observed because such strategies avoid the induction of anti-vector immunity (both T and B cell mediated) that occurs with repeated immunizations using the same vaccine (26); the anti-vector immunity induced following the first dose blunts any further increases in immunogenicity following additional vaccinations (3, 43). We therefore attempted to demonstrate the improved priming capacity of our live vectors by comparing the immunogenicity of pGEN222I when delivered by two different live vectors in the priming phase to that delivered by two doses of the same live vector. In both groups of mice receiving heterologous priming, we observed an excellent induction of GFPuv-specific IgG responses. In addition, there was no clear difference in immune responses according to the order of the two strains used in heterologous priming. Although heterologous priming apparently was not superior to priming with two doses of S. flexneri-based CVD 1208S (pGEN222I), our data clearly show that heterologous priming improves foreign antigen-specific humoral immune responses in mice primed using S. enterica serovar Typhi-based live vectors. A similar approach to improving cellular immunity against a foreign antigen by immunization with two different serovars of Salmonella enterica (Typhimurium and Dublin) recently was reported by Sevil Domènech et al. (44). However, to our knowledge, the current work represents the first attempt to elicit an antigen-specific humoral response by presenting a common foreign antigen to the immune system using three distinct vaccination formulations (two unrelated live vector primes and a subunit protein boost).
In summary, we have developed a novel nonantibiotic plasmid selection system for use in attenuated bacterial live vectors that is based on the susceptibility of strains to MccH47. The implementation of this selection method is straightforward, requiring no additional manipulation of the live vector chromosome, and it has been successfully tested in the clinically proven vaccine strains S. enterica serovar Typhi CVD 908-htrA and S. flexneri 2a CVD 1208S. The success of our heterologous prime/boost strategy involving priming with two unrelated live vector strains followed by boosting with individual antigens supports the feasibility of the future development of multivalent live vector-based immunization strategies against multiple human pathogens.
Acknowledgments
This research was supported by grant 5 RO1 AI29471 (M.M.L.), the Mid-Atlantic Regional Center for Excellence for Biodefense and Emerging Infectious Diseases Research grant U54 AI57168 (M.M.L.), and research contract NO1 AI45251 (M.M.L.).
We thank Klaus Hantke from the Institut für Organische Chemie, Universität Tübingen, Tübingen, Germany, for kindly providing the MccH47-producing E. coli strain RYC1000(pEX4) that was used in these studies and Sharon Tennant for helpful reviews of the manuscript.
This work is dedicated to the memory of Ellen Galen, who always loved to hear stories from the laboratory.
Editor: A. J. Bäumler
Footnotes
Published ahead of print on 28 July 2008.
REFERENCES
- 1.Azpiroz, M. F., and M. Lavina. 2004. Involvement of enterobactin synthesis pathway in production of microcin H47. Antimicrob. Agents Chemother. 481235-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Azpiroz, M. F., E. Rodriguez, and M. Lavina. 2001. The structure, function, and origin of the microcin H47 ATP-binding cassette exporter indicate its relatedness to that of colicin V. Antimicrob. Agents Chemother. 45969-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barouch, D. H., M. G. Pau, J. H. Custers, W. Koudstaal, S. Kostense, M. J. Havenga, D. M. Truitt, S. M. Sumida, M. G. Kishko, J. C. Arthur, B. Korioth-Schmitz, M. H. Newberg, D. A. Gorgone, M. A. Lifton, D. L. Panicali, G. J. Nabel, N. L. Letvin, and J. Goudsmit. 2004. Immunogenicity of recombinant adenovirus serotype 35 vaccine in the presence of pre-existing anti-Ad5 immunity. J. Immunol. 1726290-6297. [DOI] [PubMed] [Google Scholar]
- 4.Barry, E. M., J. Wang, T. Wu, T. Davis, and M. M. Levine. 2006. Immunogenicity of multivalent Shigella-ETEC candidate vaccine strains in a guinea pig model. Vaccine 243727-3734. [DOI] [PubMed] [Google Scholar]
- 5.Baud, D., J. Benyacoub, V. Revaz, M. Kok, F. Ponci, M. Bobst, R. Curtiss III, P. De Grandi, and D. Nardelli-Haefliger. 2004. Immunogenicity against human papillomavirus type 16 virus-like particles is strongly enhanced by the PhoPc phenotype in Salmonella enterica serovar Typhimurium. Infect. Immun. 72750-756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burrus, V., and M. K. Waldor. 2004. Shaping bacterial genomes with integrative and conjugative elements. Res. Microbiol. 155376-386. [DOI] [PubMed] [Google Scholar]
- 7.Chinchilla, M., M. F. Pasetti, S. Medina-Moreno, J. Y. Wang, O. G. Gomez-Duarte, R. Stout, M. M. Levine, and J. E. Galen. 2007. Enhanced immunity to Plasmodium falciparum circumsporozoite protein (PfCSP) by using Salmonella enterica serovar Typhi expressing PfCSP and a PfCSP-encoding DNA vaccine in a heterologous prime-boost strategy. Infect. Immun. 753769-3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fiedler, M., and A. Skerra. 2001. proBA complementation of an auxotrophic E. coli strain improves plasmid stability and expression yield during fermenter production of a recombinant antibody fragment. Gene 274111-118. [DOI] [PubMed] [Google Scholar]
- 9.Frana, T. S., S. A. Carlson, D. C. Rauser, B. D. Jones, B. J. Fergen, and R. W. Griffith. 2004. Effects of microcin 24-producing Escherichia coli on shedding and multiple-antimicrobial resistance of Salmonella enterica serotype Typhimurium in pigs. Am. J. Vet. Res. 651616-1620. [DOI] [PubMed] [Google Scholar]
- 10.Gaggero, C., F. Moreno, and M. Lavina. 1993. Genetic analysis of microcin H47 antibiotic system. J. Bacteriol. 1755420-5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galen, J. E., O. G. Gomez-Duarte, G. A. Losonsky, J. L. Halpern, C. S. Lauderbaugh, S. Kaintuck, M. K. Reymann, and M. M. Levine. 1997. A murine model of intranasal immunization to assess the immunogenicity of attenuated Salmonella Typhi live vector vaccines in stimulating serum antibody responses to expressed foreign antigens. Vaccine 15700-708. [DOI] [PubMed] [Google Scholar]
- 12.Galen, J. E., J. Nair, J. Y. Wang, S. S. Wasserman, M. K. Tanner, M. B. Sztein, and M. M. Levine. 1999. Optimization of plasmid maintenance in the attenuated live vector vaccine strain Salmonella Typhi CVD 908-htrA. Infect. Immun. 676424-6433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galen, J. E., L. Zhao, M. Chinchilla, J. Y. Wang, M. F. Pasetti, J. Green, and M. M. Levine. 2004. Adaptation of the endogenous Salmonella enterica serovar Typhi clyA-encoded hemolysin for antigen export enhances the immunogenicity of anthrax protective antigen domain 4 expressed by the attenuated live-vector vaccine strain CVD 908-htrA. Infect. Immun. 727096-7106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garmory, H. S., M. W. Leckenby, K. F. Griffin, S. J. Elvin, R. R. Taylor, M. G. Hartley, J. A. Hanak, E. D. Williamson, and R. M. Cranenburgh. 2005. Antibiotic-free plasmid stabilization by operator-repressor titration for vaccine delivery by using live Salmonella enterica serovar Typhimurium. Infect. Immun. 732005-2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glenting, J., S. M. Madsen, A. Vrang, A. Fomsgaard, and H. Israelsen. 2002. A plasmid selection system in Lactococcus lactis and its use for gene expression in L. lactis and human kidney fibroblasts. Appl. Environ. Microbiol. 685051-5056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gootz, T. D. 2006. The forgotten gram-negative bacilli: what genetic determinants are telling us about the spread of antibiotic resistance. Biochem. Pharmacol. 711073-1084. [DOI] [PubMed] [Google Scholar]
- 17.Hone, D. M., A. M. Harris, S. Chatfield, G. Dougan, and M. M. Levine. 1991. Construction of genetically defined double aro mutants of Salmonella Typhi. Vaccine 9810-816. [DOI] [PubMed] [Google Scholar]
- 18.Kobayashi, M., K. Nagata, and A. Ishihama. 1990. Promoter selectivity of Escherichia coli RNA polymerase: effect of base substitutions in the promoter −35 region on promoter strength. Nucleic Acids Res. 187367-7372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kotloff, K. L., J. K. Simon, M. F. Pasetti, M. B. Sztein, S. L. Wooden, S. Livio, J. P. Nataro, W. C. Blackwelder, E. M. Barry, W. Picking, and M. M. Levine. 2007. Safety and immunogenicity of CVD 1208S, a live, oral ΔguaBA Δsen Δset Shigella flexneri 2a vaccine grown on animal-free media. Hum. Vaccin. 3268-275. [DOI] [PubMed] [Google Scholar]
- 20.Lai, P. S., Y. F. Ngeow, S. D. Puthucheary, and C. W. Wang. 1983. Comparison of two methods for bacteriocin typing of Serratia marcescens. J. Clin. Microbiol. 171-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laviña, M., C. Gaggero, and F. Moreno. 1990. Microcin H47, a chromosome-encoded microcin antibiotic of Escherichia coli. J. Bacteriol. 1726585-6588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Levine, M. M., J. B. Kaper, H. Lockman, R. E. Black, M. L. Clements, and S. Falkow. 1983. Recombinant DNA risk assessment studies in humans: efficacy of poorly mobilizable plasmids in biologic containment. J. Infect. Dis. 148699-709. [DOI] [PubMed] [Google Scholar]
- 23.Licht, T. R., B. B. Christensen, K. A. Krogfelt, and S. Molin. 1999. Plasmid transfer in the animal intestine and other dynamic bacterial populations: the role of community structure and environment. Microbiology 1452615-2622. [DOI] [PubMed] [Google Scholar]
- 24.Londoño-Arcila, P., D. Freeman, H. Kleanthous, A. M. O'Dowd, S. Lewis, A. K. Turner, E. L. Rees, T. J. Tibbitts, J. Greenwood, T. P. Monath, and M. J. Darsley. 2002. Attenuated Salmonella enterica serovar Typhi expressing urease effectively immunizes mice against Helicobacter pylori challenge as part of a heterologous mucosal priming-parenteral boosting vaccination regimen. Infect. Immun. 705096-5106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McNeill, H. V., K. A. Sinha, C. E. Hormaeche, J. J. Lee, and C. M. Khan. 2000. Development of a nonantibiotic dominant marker for positively selecting expression plasmids in multivalent Salmonella vaccines. Appl. Environ. Microbiol. 661216-1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McShane, H., and A. Hill. 2005. Prime-boost immunisation strategies for tuberculosis. Microbes Infect. 7962-967. [DOI] [PubMed] [Google Scholar]
- 27.Moreno, F., J. L. San Millan, C. Hernandez-Chico, and R. Kolter. 1995. Microcins. Biotechnology 28307-321. [DOI] [PubMed] [Google Scholar]
- 28.Papagianni, M. 2003. Ribosomally synthesized peptides with antimicrobial properties: biosynthesis, structure, function, and applications. Biotechnol. Adv. 21465-499. [DOI] [PubMed] [Google Scholar]
- 29.Pasetti, M. F., R. Salerno-Goncalves, and M. B. Sztein. 2002. Salmonella enterica serovar Typhi live vector vaccines delivered intranasally elicit regional and systemic specific CD8+ major histocompatibility class I-restricted cytotoxic T lymphocytes. Infect. Immun. 704009-4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Patzer, S. I., M. R. Baquero, D. Bravo, F. Moreno, and K. Hantke. 2003. The colicin G, H and X determinants encode microcins M and H47, which might utilize the catecholate siderophore receptors FepA, cir, fiu and IroN. Microbiology 1492557-2570. [DOI] [PubMed] [Google Scholar]
- 31.Pfaffenzeller, I., J. Mairhofer, G. Striedner, K. Bayer, and R. Grabherr. 2006. Using ColE1-derived RNA I for suppression of a bacterially encoded gene: implication for a novel plasmid addiction system. Biotechnol. J. 1675-681. [DOI] [PubMed] [Google Scholar]
- 32.Pickett, T. E., M. F. Pasetti, J. E. Galen, M. B. Sztein, and M. M. Levine. 2000. In vivo characterization of the murine intranasal model for assessing the immunogenicity of attenuated Salmonella enterica serovar Typhi strains as live mucosal vaccines and as live vectors. Infect. Immun. 68205-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poey, M. E., M. F. Azpiroz, and M. Lavina. 2006. Comparative analysis of chromosome-encoded microcins. Antimicrob. Agents Chemother. 501411-1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pons, A. M., I. Lanneluc, G. Cottenceau, and S. Sable. 2002. New developments in non-post translationally modified microcins. Biochimie 84531-537. [DOI] [PubMed] [Google Scholar]
- 35.Ringquist, S., S. Shinedling, D. Barrick, L. Green, J. Binkley, G. D. Stormo, and L. Gold. 1992. Translation initiation in Escherichia coli: sequences within the ribosome-binding site. Mol. Microbiol. 61219-1229. [DOI] [PubMed] [Google Scholar]
- 36.Rodríguez, E., C. Gaggero, and M. Lavina. 1999. The structural gene for microcin H47 encodes a peptide precursor with antibiotic activity. Antimicrob. Agents Chemother. 432176-2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rodríguez, E., and M. Lavina. 1998. Genetic analysis of microcin H47 immunity. Can. J. Microbiol. 44692-697. [DOI] [PubMed] [Google Scholar]
- 38.Rodríguez, E., and M. Lavina. 2003. The proton channel is the minimal structure of ATP synthase necessary and sufficient for microcin h47 antibiotic action. Antimicrob. Agents Chemother. 47181-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ryan, E. T., T. I. Crean, S. K. Kochi, M. John, A. A. Luciano, K. P. Killeen, K. E. Klose, and S. B. Calderwood. 2000. Development of a ΔglnA balanced lethal plasmid system for expression of heterologous antigens by attenuated vaccine vector strains of Vibrio cholerae. Infect. Immun. 68221-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Salyers, A. A., and C. F. Amabile-Cuevas. 1997. Why are antibiotic resistance genes so resistant to elimination? Antimicrob. Agents Chemother. 412321-2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Salyers, A. A., A. Gupta, and Y. Wang. 2004. Human intestinal bacteria as reservoirs for antibiotic resistance genes. Trends Microbiol. 12412-416. [DOI] [PubMed] [Google Scholar]
- 42.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 43.Santra, S., Y. Sun, J. G. Parvani, V. Philippon, M. S. Wyand, K. Manson, A. Gomez-Yafal, G. Mazzara, D. Panicali, P. D. Markham, D. C. Montefiori, and N. L. Letvin. 2007. Heterologous prime/boost immunization of rhesus monkeys by using diverse poxvirus vectors. J. Virol. 818563-8570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sevil Domènech, V. E., K. Panthel, S. E. Winter, and H. Russmann. 2008. Heterologous prime-boost immunizations with different Salmonella serovars for enhanced antigen-specific CD8 T-cell induction. Vaccine 261879-1886. [DOI] [PubMed] [Google Scholar]
- 45.Tacket, C. O., S. M. Kelly, F. Schodel, G. Losonsky, J. P. Nataro, R. Edelman, M. M. Levine, and R. Curtiss III. 1997. Safety and immunogenicity in humans of an attenuated Salmonella Typhi vaccine vector strain expressing plasmid-encoded hepatitis B antigens stabilized by the asd-balanced lethal vector system. Infect. Immun. 653381-3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tacket, C. O., M. B. Sztein, G. A. Losonsky, S. S. Wasserman, J. P. Nataro, R. Edelman, D. Pickard, G. Dougan, S. N. Chatfield, and M. M. Levine. 1997. Safety of live oral Salmonella Typhi vaccine strains with deletions in htrA and aroC aroD and immune response in humans. Infect. Immun. 65452-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tomizawa, J. 1990. Control of ColE1 plasmid replication intermediates in the binding of RNA I and RNA II. J. Mol. Biol. 212683-694. [DOI] [PubMed] [Google Scholar]
- 48.Trujillo, M., E. Rodriguez, and M. Lavina. 2001. ATP synthase is necessary for microcin H47 antibiotic action. Antimicrob. Agents Chemother. 453128-3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vindurampulle, C. J., L. F. Cuberos, E. M. Barry, M. F. Pasetti, and M. M. Levine. 2004. Recombinant Salmonella enterica serovar Typhi in a prime-boost strategy. Vaccine 223744-3750. [DOI] [PubMed] [Google Scholar]