Abstract
Cerebral malaria (CM) is a primary cause of malaria-associated deaths among young African children. Yet no diagnostic tools are available that could be used to predict which of the children infected with Plasmodium falciparum malaria will progress to CM. We used the Plasmodium berghei ANKA murine model of experimental cerebral malaria (ECM) and high-density oligonucleotide microarray analyses to identify host molecules that are strongly associated with the clinical symptoms of ECM. Comparative expression analyses were performed with C57BL/6 mice, which have an ECM-susceptible phenotype, and with mice that have ECM-resistant phenotypes: CD8 knockout and perforin knockout mice on the C57BL/6 background and BALB/c mice. These analyses allowed the identification of more than 200 host molecules (a majority of which had not been identified previously) with altered expression patterns in the brain that are strongly associated with the manifestation of ECM. Among these host molecules, brain samples from mice with ECM expressed significantly higher levels of p21, metallothionein, and hemoglobin α1 proteins by Western blot analysis than mice unaffected by ECM, suggesting the possible utility of these molecules as prognostic biomarkers of CM in humans. We suggest that the higher expression of hemoglobin α1 in the brain may be associated with ECM and could be a source of excess heme, a molecule that is considered to trigger the pathogenesis of CM. Our studies greatly enhance the repertoire of host molecules for use as diagnostics and novel therapeutics in CM.
Among the four species of Plasmodium that infect humans, Plasmodium falciparum is the most lethal; it is responsible for more than 90% of deaths, mostly in children under the age of 5 years. One characteristic feature of falciparum malaria is the clinical manifestation termed cerebral malaria (CM), a serious complication that often results in death. Of the approximately 1 million deaths due to malaria per year, mostly in children living in sub-Saharan Africa, the majority are a consequence of CM or severe anemia. A major cause of deaths from CM is thought to be the nonarousable coma, a classical symptom of CM. However, CM, if diagnosed and correctly treated, is not always fatal. A prognostic test that could allow clinicians to predict which of the P. falciparum-infected patients brought to a hospital due to clinical malaria would progress to CM might save the lives of thousands of young children each year.
Susceptibility to CM is believed to result from a combination of parasite and host factors. The ability of the parasite to sequester itself within the deep vascular beds of brain tissue is thought to be the dominant parasite characteristic required for CM (34). In previous studies, associations between a number of host molecules and susceptibility to CM have been established (65). The majority of these genes, including the tumor necrosis factor alpha (TNF-α) and gamma interferon (IFN-γ) genes, encode proteins of the immune system. The development of a prognostic test that could be used to predict the outcome of P. falciparum infection in a susceptible host will require a complete knowledge of the expression pattern of the pathogenesis factors that are specifically triggered during a clinical episode of CM. To obtain a more accurate and complete picture of the pathogenesis of CM, we have identified the repertoire of genes whose levels are altered by measuring their expression in whole-brain tissue in real time and determined the biological associations at the protein level of a few selected molecules in the Plasmodium berghei ANKA murine model of experimental cerebral malaria (ECM).
Like P. falciparum, P. berghei ANKA, a strain causing murine malaria, possesses the ability to sequester itself within the microvasculature; it is the parasite of choice for in vivo studies of CM in mice (17). Infection with P. berghei ANKA parasites results in death for 100% of mice. However, the cause of death differs greatly based on the genetic background of the mouse, and different strains of mice are broadly categorized as susceptible or resistant to CM (17). In susceptible strains (e.g., C57BL/6 and CBA/J), approximately 60 to 100% of mice develop CM 6 to 10 days postinfection, and mice die with relatively low levels of parasitemia (below 10%). The features of ECM in these mice resemble those of P. falciparum clinical disease in humans: hemi- or paraplegia, deviation of the head, ataxia, distressed breathing, and convulsions. In contrast, resistant mice (e.g., BALB/c mice) do not develop any clinical or pathological features resembling those of CM (17) but instead succumb to anemia and hyperparasitemia approximately 15 to 21 days after infection with P. berghei ANKA. Susceptible mice that do not develop ECM follow a clinical course of disease similar to that of resistant mice and eventually die of hyperparasitemia and severe anemia. Furthermore, previous studies have shown that loss of function of CD8 or perforin molecules converts susceptible C57BL/6 mice to resistant mice whose resistance to ECM is comparable to that observed for the BALB/c strain (38, 47, 69). These studies suggest a role for the cell-cell interaction pathways dependent on these molecules in augmenting CM.
The goal of this study was to identify novel host molecules in the brain that are affected in the course of the pathological manifestation of CM. In particular, we wanted to identify novel host molecules that could serve as prognostic biomarkers of CM. Using high-density microarray chips, we compared the whole-brain tissue expression profiles of susceptible C57BL/6 mice with ECM (moribund mice) to those of (i) infected C57BL/6 mice without ECM (nonmoribund mice) and (ii) infected BALB/c mice (resistant mice) during the effector phase of ECM, the phase most pertinent to the management and therapeutic treatment of CM. Furthermore, to determine the mechanisms of CD8- and perforin-mediated pathogenesis of ECM, we also compared the expression profiles of infected C57BL/6 wild-type (WT) mice with ECM to those of (i) infected CD8 gene knockout (CD8-KO) mice and (ii) infected perforin gene knockout (PFP-KO) mice on the C57BL/6 background, both of which are resistant to ECM. Based on differential expression in brain tissue samples, we identified a wide range of host genes that were strongly associated with the manifestation of ECM. Bioinformatic analyses were performed using several computational tools for sequence analysis and contextual functional inference, and genes with significantly altered expression were assigned to several distinct biological functional categories. Several of these molecules belong to families of known immunological relevance that could either serve as novel cytoadherence molecules or contribute directly to the pathogenesis of CM.
MATERIALS AND METHODS
Mice and parasite infections.
Six- to 8-week-old female C57BL/6 WT mice, CD8-KO and PFP-KO mice on a C57BL/6 background, and BALB/c WT mice were purchased from The Jackson Laboratory (Bar Harbor, ME). Parasite infection was induced by intraperitoneal injection of 106 P. berghei ANKA parasites, and mice were monitored for clinical symptoms of ECM (hemi- or paraplegia, deviation of the head, tendency to roll over upon stimulation, ataxia, and convulsions) beginning on day 3 postinfection. In most instances, percentages of parasitemia (calculated as [number of parasitized erythrocytes/total erythrocytes] × 100) were determined by examining Giemsa-stained thin blood films prepared every day beginning on day 3 postinfection and at the time of sample collection. All mice used in these studies were maintained at a National Institute of Allergy and Infectious Diseases (NIAID) animal care facility and were treated in accordance with the guidelines of the Animal Care and Use Committee of the National Institutes of Health (NIH).
Preparation of whole-brain RNA.
Brain tissue samples were collected simultaneously from each group and stored at −80°C until use. For RNA preparation, tissue samples were pulse-homogenized in Tri-Reagent (Molecular Research Center, Cincinnati, OH), and RNA was isolated following two chloroform extractions, isopropanol precipitation, and resuspension in diethyl pyrocarbonate-treated water. The quantity of RNA was determined by optical densitometry, and its quality was evaluated by agarose gel electrophoresis.
Microarray and gene expression analysis.
The global gene expression profiles of moribund C57BL/6 WT mice were compared with those of both nonmoribund (C57BL/6 WT) and resistant (BALB/c, C57BL/6 CD8-KO, and C57BL/6 PFP-KO) mice to determine the array of specific host genes induced and repressed during the expression of ECM. Microarray expression profiles were determined from RNA samples isolated from five to six individual mice per group. cDNA was synthesized from 30 μg of RNA extracted from host brain tissue and then labeled with Cy3 or Cy5 as described previously (40). Briefly, cDNA was labeled with 50 nmol of dUTP-Cy3 or dUTP-Cy5 during a reverse transcription reaction at 42°C for 90 min in a sample mixture containing 300 mM dithiothreitol; 15 mM each dATP, dCTP, dGTP, and dTTP; a mixture of poly(T) and random hexamer primers; and 300 U Superscript II. After incorporation of the label, RNA was degraded by the addition of NaOH, and labeled cDNA was purified and concentrated by ultrafiltration through a Vivaspin 500 column (Sartorius, Goettingen, Germany). Labeled cDNA was then hybridized to a murine oligonucleotide chip containing 16,600 oligonucleotide probes (Qiagen, Valencia, CA), and the slide was scanned by a GenePix microarray scanner. Microarray data were analyzed with GenePix Pro software (version 6.0; Axon Instruments, Inc., Union City, CA) and filtered using the NIAID microarray database tools (http://madb-niaid.cit.nih.gov), and extracted spots were normalized to the precalculated 50th percentile (median).
Western blot analysis.
Protein samples from brain tissue were prepared as a 10% brain homogenate, and tissue-specific p21, metallothionein, and hemoglobin α1 proteins were detected using antibodies specific for recombinant human p21WAF1 protein (Biosource, Camarillo, CA), metallothionein (Abcam Inc.), or hemoglobin α1 (Novus Biologicals Inc.) and a commercially obtained chemiluminescence-linked Western blotting kit (Western Light Tropix, Bedford, MA). Protein bands were visualized following incubation with enhanced chemiluminescence detection reagents, and the integrated optical densities (IOD) for each lane were measured using MetaMorph software, version 6.1.
Gene annotation, ontology, and identification of biological pathways.
Protein sequences were obtained from the nonredundant database of protein sequences (National Center for Biotechnology Information, NIH, Bethesda, MD). Profile searches were conducted using the PSI-BLAST program (3) with either a single sequence or an alignment used as the query, and with a default profile inclusion expectation (E) value threshold of 0.01 (unless specified otherwise), and the searches were iterated until convergence. The multiple alignments used were constructed using the T_Coffee program (39), followed by manual correction based on the PSI-BLAST results. For all searches involving membrane-spanning domains or low-complexity sequences, we used a statistical correction for compositional bias to reduce false-positive results due to the general hydrophobicity of these proteins. A library of a large set of alignments of conserved protein domains, including those from the PFAM database (http://www.sanger.ac.uk/Software/Pfam/index.shtml) as well as an additional set of unpublished conserved domains, was used for domain searches with the hidden Markov models (HMMER package) or with PSI-BLAST (position-specific score matrices). Transmembrane regions were predicted in individual proteins using the TMHMM program, version 2.0, with default parameters. Signal peptides were predicted using the SIGNALP program (www.cbs.dtu.dk/services/SignalP-2.0/). More than 300 profiles of known domains were used to search the mouse proteins by the method described above. The domains collected from these searches were assembled using the scripts from the in-house TASS package. Functions were assigned using the contextual information from the domain architectures and published experimental results. Contextual analysis was performed using the protein-protein and genetic interaction data stored in the BioGrid and VisANT databases. All large-scale sequence analysis procedures were carried out using the TASS package (V. Anantharaman, S. Balaji, and L. Aravind, unpublished data).
RESULTS
P. berghei ANKA infection in genetically susceptible and resistant strains of mice.
In two well-controlled experiments where mice were followed twice every day for clinical symptoms of ECM and blood films were prepared daily, we found that 16 of 20 (80%) mice infected with P. berghei ANKA developed the symptoms of ECM and became moribund between days 5 and 8 postinfection. No symptoms of ECM were observed for 20 CD8-KO and 20 PFP-KO mice up to day 12 postinfection. Similarly, none of the P. berghei ANKA-infected BALB/c mice developed ECM; instead, they developed >80% parasitemia before death. Our results are consistent with the clinical and parasitological outcomes of P. berghei ANKA infections reported previously for WT, CD8-KO, and PFP-KO C57BL/6 mice and for BALB/c mice (5, 16, 17, 30, 38).
Alterations in gene expression profiles related to susceptibility or resistance to ECM.
We compared the global expression profiles of host genes in the brains of individual mice with ECM (C57BL/6 WT moribund mice) to those for mice without ECM (C57BL/6 WT nonmoribund, C57BL/6 CD8-KO resistant, C57BL/6 PFP-KO resistant, and BALB/c resistant mice), in order to identify novel host molecules/biomarkers associated with the pathogenesis of ECM, by performing microarray hybridizations with individual RNAs from brain samples isolated from five to six mice per group. For the comparison of moribund C57BL/6 mice with nonmoribund C57BL/6 mice and resistant BALB/c mice, altered expression of an individual gene was defined as significant only if two criteria were met: (i) moribund mice exhibited an average ≥2-fold increase (upregulation) or decrease (downregulation) in gene expression relative to expression for both nonmoribund and resistant mice (P < 0.05 by a two-tailed Student t test) (criterion 1) and (ii) moribund mice exhibited a ≥2-fold increase (upregulation) or decrease (downregulation) in three of the five arrays per group (criterion 2).
When moribund and nonmoribund C57BL/6 mice were compared, the input data were five arrays and 34,928 arrayed sequences corresponding mainly to protein-coding genes. A total of 34,144 sequences were excluded by criterion 1, and 172 additional genes were excluded by criterion 2. When moribund C57BL/6 mice were compared to resistant BALB/c mice, the input data were five arrays and 34,826 arrayed sequences. In this comparison, 33,543 sequences were excluded by criterion 1 and 169 additional sequences were excluded by criterion 2. A total of 612 genes were considered to show significantly altered expression when moribund C57BL/6 mice were compared to nonmoribund C57BL/6 mice, and 1,114 genes were considered to show significantly altered expression when moribund C57BL/6 mice were compared to resistant BALB/c mice. Of these 1,726 genes, 210 genes (66% upregulated, 34% downregulated) showed significantly altered expression in both the nonmoribund and resistant groups relative to expression in the moribund group. All microarray gene expression data are available in Table S1 in the supplemental material.
For microarray analysis comparing moribund WT to resistant CD8-KO and PFP-KO mice (six mice per group), data were analyzed using the significant analysis of microarrays (SAM) program, and a false-discovery rate (FDR) was chosen that would yield a set of genes that were both statistically significant and also of a size manageable for in-depth functional analysis. For comparison with CD8-KO mice, genes with an FDR of <2.2% and an average twofold increase (upregulation) or decrease (downregulation) in expression were included, yielding a data set of 166 genes (92% upregulated, 8% downregulated). For PFP-KO mice, genes with an FDR of <4.2% and an average twofold increase (upregulation) or decrease (downregulation) in expression were included, yielding a data set of 9 genes (78% upregulated, 22% downregulated).
In order to understand the significance of the altered gene expression patterns, we carried out a number of comparisons of these patterns between different sets of mice. We first compared the expression patterns seen in the nonmoribund C57BL/6 and resistant BALB/c mice to determine if the similar phenotypes of avoidance of ECM in mice with disparate major histocompatibility complex alleles have similar molecular profiles. Second, we compared the expression profiles of these nonmoribund or resistant mice with those of the resistance phenotypes associated with CD8-KO and PFP-KO mice in order to identify potential shared and different pathways leading to resistance in these mice. Comparison of the nonmoribund C57BL/6 and resistant BALB/c mice showed that the two groups have similar patterns of expression changes with respect to the moribund mice (Pearson correlation, ∼0.83). At least 60 genes whose expression patterns in the resistant CD8-KO group were altered relative to those in WT mice with ECM were also present in the list of genes with altered expression in the C57BL/6 nonmoribund and resistant BALB/c mice. This commonality in gene expression patterns suggests that the distinct pathways utilized by these mice to avoid ECM share certain common mechanisms. The resistance phenotype of PFP-KO mice was associated with significant alterations in only nine genes. At face value, this would suggest that the resistance phenotype in PFP-KO mice occurs via a relatively restricted downstream mechanism compared to the other resistance phenotypes. These results should be considered hardly surprising, since the resistance to ECM resulted from the loss of a single genetic locus in the mouse genome.
To glean the functional significance of the genes with altered expression, we performed a comprehensive computational sequence analysis of their protein products, identified all conserved domains present in those products, and predicted subcellular targeting signals such as signal peptides and transmembrane helices. We then combined this with a survey of relevant literature on all genes for which there were published references and with contextual information from high-throughput interaction mapping studies using the Biogrid and VisANT databases (22). As a result, we were able to confidently classify the majority of genes into various functional guilds (Table 1). Using this functional classification and the inferred interactions of the gene products, we pieced together several biological processes that are affected by ECM. We discuss some key examples below.
TABLE 1.
Biomarkers of ECM categorized by function
| Functional category | Fold downregulation or upregulation of genesa in the following mice:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| CD8-KO
|
PFP-KO
|
Resistant BALB/c
|
Nonmoribund C57BL/6
|
|||||
| Down | Up | Down | Up | Down | Up | Down | Up | |
| Apoptosis | 7 | 6 | 2 | 6 | 2 | |||
| Cell cycle | 1 | 1 | ||||||
| Cellular metabolism | 9 | 2 | 4 | 2 | 4 | 2 | ||
| Chaperones | 1 | 2 | 2 | 2 | 2 | 2 | ||
| Chromatin | 7 | 5 | 1 | 5 | 1 | |||
| Cytoskeleton | 7 | 1 | 1 | 11 | 2 | 11 | 2 | |
| DNA repair | 1 | 3 | 3 | |||||
| General stress response/detoxification | 2 | 1 | 1 | |||||
| Hematopoiesis | 1 | 1 | 3 | 1 | 3 | |||
| Immunity | ||||||||
| Cell adhesion and surface molecules | 41 | 1 | 2 | 2 | 26 | 10 | 31 (26 + 5) | 5 |
| Chemokines | 4 | 1 | ||||||
| Other | 4 | 2 | 0 | 1 | 1 | |||
| Signaling: kinases | 5 | 6 | 1 | 6 | 1 | |||
| Signaling | 11 | 8 | 1 | 8 | 1 | |||
| First-messenger (including neurotransmitter) receptors | 4 | 2 | 5 | 1 | 5 | 1 | ||
| Metal carriers | 2 | 2 | 2 | |||||
| Protein trafficking | 3 | 1 | 5 | 5 | 5 | 5 | ||
| RNA metabolism | 4 | 2 | 5 | 7 | 5 | 7 | ||
| Transcription regulators | 11 | 1 | 1 | 1 | 8 | 10 | 8 | 10 |
| Transporters | 3 | 1 | 5 | 3 | 5 | 3 | ||
| Ubiquitin system and proteasome | 4 | 2 | 2 | |||||
| Protein degradation (ubiquitin independent) | 2 | 2 | ||||||
| Miscellaneous | 5 | 6 | 5 | 6 | 5 | |||
Relative to expression by susceptible C57BL/6 mice with ECM. Data for the overrepresented, overexpressed immunity-related class are boldface. Down, downregulation; Up, upregulation.
The puzzle of hemoglobin gene expression and heme oxidation in ECM.
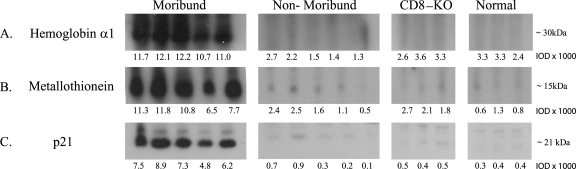
We observed that transcription of the hemoglobin α- and β-chain genes was considerably downregulated in the brain during ECM (Table 2) in comparison to that for nonmoribund and resistant BALB/c mice. Because reticulocytes are the sole source of hemoglobin mRNA, this finding suggested a possible reduction in the circulating-reticulocyte count. This possibility is consistent with previous reports of impaired erythropoiesis in mice infected with lethal murine malaria strains (7, 54) and in clinical cases of severe malaria (27). To further investigate the possible implications of the reduction in hemoglobin chain expression in ECM, we measured the expression of hemoglobin α1 chain protein in the brain tissue. In contrast to transcriptional levels, we found that hemoglobin α1 protein was overexpressed 6.3-fold in moribund mice relative to expression in nonmoribund mice (Fig. 1A). These counterintuitive results suggest the distinct possibility of an imbalance in the number of reticulocytes with respect to mature erythrocytes (which contain hemoglobin protein but not RNA) during ECM. This imbalance, in turn, might be related to the elevated expression of heme oxygenase 1 (Hmox1) that we observed for moribund mice with ECM. Recent studies have shown that deletion of the Hmox1 gene or inhibition of the enzyme results in an elevated incidence of ECM (44), suggesting a role for free heme liberated from lysed erythrocytes in pathogenesis. Taken together, our observations might imply that the increase in the number of mature infected erythrocytes in the brain during ECM might result in an excessive release of heme, which, in turn, could trigger elevated Hmox1 levels as a defensive response. Given the identification of a human microsatellite polymorphism in the Hmox1 promoter that is associated with susceptibility to CM (61), such a mechanism might also be relevant in human CM. In this context, it is of interest that lipocalin 2, an iron-sequestering protein that is present at the blood-brain barrier and prevents bacterial entry into the brain (33), is overexpressed in ECM. It remains to be seen if lipocalin 2 might play a role in sequestering the iron released from ruptured erythrocytes.
TABLE 2.
Biological functions of a subset of ECM-related genes in our data set
| Function and gene | Descriptiona | Expressionb |
|---|---|---|
| Apoptosis | ||
| Bcl2a1a | B-cell leukemia/lymphoma 2-related protein A1a | ↓ CD8-KO |
| Bcl2a1b | B-cell leukemia/lymphoma 2-related protein A1b | ↓ CD8-KO |
| Fas | Fas (TNF receptor superfamily member); extracellular TNF repeats and intracellular death domain | ↓ CD8-KO |
| Cyr61 | Cysteine-rich protein 61; protein with insulin growth factor binding domain, vWC, TSP1, and C-terminal cysteine knot | ↓ Res-BALB/c, nonmoribund |
| Tnfsf10 | TNF (ligand) superfamily, member 10 | ↑ Res-BALB/c, nonmoribund |
| Alox12b | Arachidonate 12-lipoxygenase, 12R type | ↓ Res-BALB/c, nonmoribund |
| Aloxe3 | Arachidonate lipoxygenase 3 | ↓ Res-BALB/c, nonmoribund |
| Cdkn1a | Cyclin-dependent kinase inhibitor 1A (p21) | ↓ CD8-KO, Res-BALB/c, nonmoribund |
| Cellular metabolism | ||
| Sult1a1 | Sulfotransferase family 1A, phenol preferring, member 1 | ↓ CD8-KO, Res-BALB/c, nonmoribund |
| Ugt8a | UDP galactosyltransferase 8A | ↑ CD8-KO, Res-BALB/c, nonmoribund |
| Chaperones | ||
| Fkbp5 | FKBP-type peptidyl-prolyl cis-trans-isomerase with tetratricopeptide repeats | ↓ CD8-KO, Res-BALB/c, nonmoribund |
| Mkks | McKusick-Kaufman syndrome protein; TCP-1/cpn60 chaperonin family | ↑ CD8-KO, Res-BALB/c, nonmoribund |
| Chromatin | ||
| Cenpa | Centromeric histone H3 | ↓ CD8-KO |
| Axud1 | AXIN1 upregulated 1; tesmin/TSO1-like CXC domain | ↓ CD8-KO, Res-BALB/c, nonmoribund |
| Nasp | Nuclear autoantigenic sperm protein isoform 2 | ↑ Res-BALB/c, nonmoribund |
| Hist1h1c | Histone 1, H1c | ↓ Res-BALB/c, nonmoribund |
| Cytoskeleton | ||
| Actb | β-Actin | ↓ CD8-KO, Res-BALB/c, nonmoribund |
| Gfap | Glial fibrillary acidic protein | ↓ CD8-KO |
| Tagln | Transgelin | ↓ CD8-KO, Res-BALB/c, nonmoribund |
| Utrn | Utrophin | ↑ CD8-KO |
| Cfl1 | Cofilin 1, nonmuscle | ↓ CD8-KO, Res-BALB/c, nonmoribund |
| Tubd1 | Tubulin, delta 1 | ↑ Res-BALB/c, nonmoribund |
| DNA repair | ||
| Gadd45b | Growth arrest and DNA damage-inducible 45β | ↓ CD8-KO |
| Ddit4 | DNA damage-inducible transcript 4 | ↓ Res-BALB/c, nonmoribund |
| General stress response/ detoxification | ||
| Cybb | Cytochrome b245 | ↓ CD8-KO |
| Cyp4f18 | Cytochrome P450 | ↓ Res-BALB/c, nonmoribund |
| Hematopoiesis | ||
| Hmox1 | Heme oxygenase (decycling) 1 | ↓ CD8-KO, Res-BALB/c, nonmoribund |
| Hbb-b2 | Hemoglobin β, adult minor chain | ↑ Res-BALB/c, nonmoribund |
| Hba-a1 | Hemoglobin α, adult chain 1 | ↑ Res-BALB/c, nonmoribund |
| Immunity: cell adhesion and surface molecules | ||
| Ctla2b | Cytotoxic T-lymphocyte-associated protein 2β; cathepsin propeptide inhibitor domain | ↓ CD8-KO, Res-BALB/c, nonmoribund |
| Fcgr3a | FcγRIV, immunoglobin receptor | ↓ CD8-KO |
| Lgals3 | Galectin 3 | ↓ CD8-KO |
| Gzmb | Granzyme B; secreted trypsin-like serine protease | ↓ CD8-KO |
| Gzma | Granzyme A; secreted trypsin-like serine protease | ↓ CD8-KO |
| Ctla2a | Cytotoxic T-lymphocyte-associated protein 2α | ↓ CD8-KO, Res-BALB/c, nonmoribund |
| Itm2a | Integral membrane protein 2A, T-cell-specific signaling | ↑ CD8-KO |
| 1700058C13Rik | LBP/BPI/CETP family, N-terminal domain. paralogs involved in inflammation (binds lipopolysaccharides) | ↓ Res-BALB/c, nonmoribund |
| Angptl4 | Fibrinogen/angiopoietin-related protein | ↓ Res-BALB/c, nonmoribund |
| Tlr11 | Toll-like receptor 11 | ↑ Res-BALB/c, nonmoribund |
| 4933433K01Rik | Bacterial phospholipase D LOC194908 | ↑ Res-BALB/c, nonmoribund |
| Cd14 | CD14 antigen | ↓ Res-BALB/c, nonmoribund |
| Fabp7 | Fatty acid binding protein 7, brain; lipocalin domain | ↑ CD8-KO, Res-BALB/c, nonmoribund |
| Immunity: chemokines | ||
| Ccl5 | Chemokine (C-C motif) ligand 5 | ↓ CD8-KO |
| Ccl25 | Chemokine (C-C motif) ligand 25 | ↓ CD8-KO |
| Ccl3 | Chemokine (C-C motif) ligand 3 | ↓ CD8-KO |
| Other immunity-related proteins | ||
| Myd116 | Myeloid differentiation primary response gene 116 | ↓ CD8-KO |
| Glipr2 | GLI pathogenesis-related 2 | ↓ CD8-KO |
| Mllt3 | Myeloid/lymphoid or mixed-lineage-leukemia translocation to 3 | ↑ Res-BALB/c, nonmoribund |
| Signaling | ||
| Arrdc2 | Arrestin domain-containing 2 | ↓ CD8-KO, Res-BALB/c, nonmoribund |
| S100a4 | EF-hand calcium binding protein | ↓ CD8-KO |
| Gbp2 | Guanylate nucleotide binding protein 2 | ↓ Res-BALB/c, nonmoribund |
| Sash1 | SAM and SH3 domain containing 1, adaptor protein | ↓ Res-BALB/c, nonmoribund |
| Ppp1r12c | Protein phosphatase 1, regulatory subunit 12C | ↓ Res-BALB/c, nonmoribund |
| Ppm1f | Protein phosphatase 1F (PP2C domain containing) | ↑ Res-BALB/c, nonmoribund |
| Signaling: kinases | ||
| Map4k1 | Mitogen-activated protein kinase 1 | ↓ CD8-KO |
| Map3k6 | Mitogen-activated protein kinase 6 | ↓ CD8-KO, Res-BALB/c, nonmoribund |
| Phka1 | Phosphorylase kinase α1 isoform 2 | ↑ Res-BALB/c, nonmoribund |
| First-messenger (including neurotransmitter) receptors | ||
| Il2rg | Interleukin 2 receptor, gamma chain | ↓ CD8-KO |
| Fpr-rs2 | Formyl peptide receptor, related sequence 2 | ↓ CD8-KO |
| Gpr34 | G protein-coupled receptor 34 | ↑ CD8-KO |
| Gpr17 | G protein-coupled receptor 17 | ↑ CD8-KO |
| Gprc5a | G protein-coupled receptor, family C, group 5, member A | ↓ Res-BALB/c, nonmoribund |
| Olfr705 | Olfactory receptor 705 | ↓ Res-BALB/c, nonmoribund |
| Bdkrb1 | Bradykinin receptor, β1 | ↓ Res-BALB/c, nonmoribund |
| Npy6r | Neuropeptide Y receptor Y6 | ↓ Res-BALB/c, nonmoribund |
| V1rf3 | Vomeronasal 1 receptor, F3 | ↓ Res-BALB/c, nonmoribund |
| Metal carriers | ||
| Mt2 | Metallothionein 2 | ↓ CD8-KO, Res-BALB/c, nonmoribund |
| Mt1 | Metallothionein 1 | ↓ CD8-KO, Res-BALB/c, nonmoribund |
| Protein trafficking | ||
| Pnpla2 | Transport-secretion protein; patatin-like phospholipase | ↓ CD8-KO, Res-BALB/c, nonmoribund, |
| Rac2 | RAS-related C3 botulinum substrate 2; Rho (Ras homology) subfamily of Ras-like small GTPases | ↓ CD8-KO |
| Tbc1d4 | TBC1 domain family, member 4 | ↑ Res-BALB/c, nonmoribund |
| Arfl4 | ADP-ribosylation factor 4-like | ↓ CD8-KO, Res-BALB/c, nonmoribund |
| RNA metabolism | ||
| Nanos2 | Nanos homolog 2 | ↓ CD8-KO |
| Cirbp | Cold-inducible RNA binding protein | ↓ CD8-KO, Res-BALB/c, nonmoribund |
| Sfpq | Splicing factor proline/glutamine rich | ↑ CD8-KO, Res-BALB/c, nonmoribund |
| Pcf11 | Pre-mRNA cleavage complex II protein | ↑ Res-BALB/c, nonmoribund |
| 4930535B03Rik | CID/RPR (C-terminal domain-interacting) domain protein involved in RNA metabolism | ↓ CD8-KO, Res-BALB/c, nonmoribund |
| Transcription regulators | ||
| Myc | Myc proto-oncogene protein, bHLH DNA-binding domain | ↓ CD8-KO |
| Hmgb2 | High-mobility-group 2 protein; HMG box DNA-binding domain | ↓ CD8-KO |
| Zfp36 | Zinc finger protein 36, C2H2 zinc finger domain protein | ↓ CD8-KO |
| Gata2 | GATA binding protein 2, GATA-type Zn finger DNA-binding domain | ↓ CD8-KO |
| Runx1 | Runt-related transcription factor 1 | ↓ CD8-KO |
| Maff | bZip transcription factor | ↓ CD8-KO |
| Stat1 | STAT (p53 fold) transcription factor | ↓ CD8-KO |
| Tcerg1 | Transcription elongation regulator 1 | ↑ CD8-KO, Res-BALB/c, nonmoribund |
| Rbpsuh | TIG + p53 fold T domain transcription factor, downstream of Notch signaling pathway | ↑ Res-BALB/c, nonmoribund |
| Nfkbia | Inhibitor of NF-κB, ankyrin repeat protein | ↓ CD8-KO, Res-BALB/c, nonmoribund |
| Fosb | FBJ osteosarcoma oncogene B; domain transcription factor | ↓ Res-BALB/c, nonmoribund |
| Jund1 | Jun proto-oncogene-related gene d1 b-Zip; domain transcription factor | ↓ Res-BALB/c, nonmoribund |
| Mafk | bZip transcription factor | ↓ Res-BALB/c, nonmoribund |
| Sox21 | HMG box transcription factor | ↑ Res-BALB/c, nonmoribund |
| Transporters | ||
| Slc1a3 | Solute carrier family 1 (glial high-affinity glutamate) | ↓ CD8-KO |
| Ucp2 | Uncoupling protein 2 (mitochondrial, proton carrier) | ↓ CD8-KO, Res-BALB/c, nonmoribund |
| Slc10a6 | Sodium-dependent organic anion transporter | ↓ Res-BALB/c, nonmoribund |
| Abca8a | ATP-binding cassette, subfamily A | ↑ Res-BALB/c, nonmoribund |
| Ubiquitin system and proteasome | ||
| Herc5 | HECT domain E3 ubiquitin ligase | ↓ CD8-KO |
| Psme4 | Proteasome (prosome, macropain) activator subunit 4 | ↓ Res-BALB/c, nonmoribund |
| Protein degradation (ubiquitin independent) | ||
| Htra4 | HtrA serine peptidase 4 | ↓ Res-BALB/c, nonmoribund |
| Htra1 | HtrA serine peptidase 1 | ↓ Res-BALB/c, nonmoribund |
bHLH, basic helix-loop-helix; HMG, high mobility group; TIG, transcription factor immunoglobulin domain.
Arrows indicate upregulation (↑) or downregulation (↓) in gene expression in resistant BALB/c (Res-BALB/c), nonmoribund C57BL/6 (nonmoribund), and/or CD8-KO mice relative to that in susceptible C57BL/6 mice with ECM. For a complete table showing all microarray gene expression data, see Table S1 in the supplemental material.
FIG. 1.
Biomarkers of ECM as detected by Western blotting. The expression of murine biomarkers of ECM was measured in brain tissue samples from individual moribund (n = 5), nonmoribund (n = 5), CD8-KO (n = 3), and healthy (uninfected) (n = 3) C57BL/6 mice by enhanced chemiluminescence-based Western blot analysis. Expression levels were measured based on the intensities of protein bands for each lane by using MetaMorph software (version 6.1) and are represented as average IOD units. The actual IOD units are the values shown multiplied by 1,000. Details of the specific secondary antibodies and Western blot reagents are provided in Materials and Methods. (A) Expression of murine hemoglobin α1 protein; (B) expression of metallothionein protein; (C) expression of the p21 molecule.
Alterations in other stress response pathways and their potential role in countering oxidative stress.
In addition to the elevation of Hmox1 expression as a possible mechanism to cope with the toxicity of released heme, we observed the elevation of expression of various stress response genes that could potentially play a role in countering oxidative stress. These include uncoupling protein 2 (UCP2), a mitochondrial proton carrier, whose gene is upregulated more than twofold in the brain tissue of mice with ECM. Uncoupling proteins are a family of anion carrier transporters present in the inner mitochondrial membrane. Unlike other members of the mitochondrial carrier superfamily, UCP2 is expressed in neurons and microglial cells of the brain (21, 49) and has been demonstrated to play a major role in neuroprotection by reducing the mitochondrial membrane potential (64) and controlling the production of reactive oxygen species (4). Our finding that the expression of UCP2 is enhanced more than twofold in brain tissue samples during ECM suggests that this molecule may be part of a neuroprotective response to the oxidative stress arising from the pathogenesis of ECM. Other genes potentially related to this oxidative stress response or heme detoxification process that are upregulated as much as two- to threefold in ECM are Cyp4f18 (a cytochrome P450) and Cybb (a cytochrome b).
Additionally, in agreement with a recent report published by Wiese et al. (67), in our study, mice displaying symptoms of ECM had increased transcriptional and translational expression of the genes encoding metallothioneins I and II (MT-I and -II) (Table 2 and Fig. 1B). MT-I and MT-II belong to a superfamily of low-molecular-weight, cysteine-rich metalloproteins and are reported to participate in a wide array of activities, including immunoregulation, cell survival, and neuroprotection (8, 9, 26, 45).
Alterations in apoptotic and counterapoptotic pathways.
We observed that the expression of the gene encoding the kinase inhibitor of the Cip/Kip family, p21, is significantly upregulated during ECM (Table 2). p21 has been shown to be important in the regulation of apoptosis. Earlier studies have shown that expression of p21 increases when cells are damaged, and this increased expression might be a key element for cellular survival during oxidative stress (43). We examined the expression of p21 at the level of translation in order to further establish the role of p21 during ECM and to determine whether a correlation exists between the synthesis of mRNA and the synthesis of protein. Our microarray results show that the expression of p21 in moribund mice is 5.5-fold greater than its expression in nonmoribund mice at the level of transcription. Quantification of p21 protein in brain tissue by Western blotting demonstrated a 14.5-fold increase in expression at the level of translation (Fig. 1C). Thus, the expression pattern of the p21 gene reflects its elevated production, which potentially represents a counterapoptotic response in face of strong oxidative stress.
At least 10 other genes with potential roles in the regulation of apoptosis also showed altered expression in mice with ECM. In particular, we found that the levels of transcripts of two members of the TNF family, namely, TRAIL/CD253, were lower in mice with ECM. Furthermore, in the experiments with CD8-KO mice, we found that the expression of Fas (a receptor of ligands of the TNF family) was upregulated in moribund mice relative to that in resistant mutant mice. This finding suggests that multiple signals involving the TNF pathway may be involved in regulating cell death during ECM. Interestingly, we also observed that levels of two lipoxygenases, Aloxe3 and Alox12b, involved in leukotriene production (18) were elevated in mice with ECM. Given the observed role of leukotrienes in neural stem cell survival (66), it is conceivable that the implied increase in leukotriene production might have an antiapoptotic role. However, the leukotrienes could also mediate inflammatory responses in the brain that account in part for the pathogenesis of ECM (18). Other molecules with antiapoptotic roles that were uncovered in the comparisons between WT mice with ECM and resistant CD8-KO mice were Bcl2a1a and Bcl2a1b. Both of these could potentially act in synergy with p21 in inhibiting apoptosis. In terms of neural surface molecules, of particular interest was the downregulation in ECM of Unc5a, a netrin receptor that has previously been shown to be downregulated in response to neural injury (1). This, again, might represent a neural regenerative mechanism responding to the tissue damage triggered by the pathogenesis.
Expression changes suggestive of repair responses to cell damage in ECM.
A total of 17 genes involved in cytoskeleton organization and remodeling showed altered expression in ECM; the majority of them showed at least a twofold upregulation. These include β-actin and associated microfilament binding proteins such as coronin, cofilin-1, kelch-like-15, and transgelin, microtubular components such as tubulin-δ, intermediate filament proteins such as nestin and lamin-A, and the tight-junction component claudin-11. These hint at a repair response to the tissue damage triggered by ECM pathogenesis. Importantly, the repair process mediated by these cytoskeleton genes might also be central to the observed rapid clinical reversibility of, and recovery from, CM. At least two molecules, β-actin and claudin-11, are components of intercellular tight junctions. Claudin-11, also known as oligodendrocyte-specific protein (OSP), contributes 7% of total protein in purified myelin (6). Studies with OSP-null mice suggest that this gene plays a pivotal role in the formation of intramyelinic tight junctions in the central nervous system (CNS). These mice also exhibit hind limb weakness and slowed conduction velocities in the CNS (19), characteristics that appear to be consistent with the neurological effects observed in CM.
Inference of regulatory pathways and cell interactions associated with ECM.
Our systematic analysis of the genes with altered expression in the course of ECM in the brain suggested that several regulatory pathways were being activated or modulated. These included transcription factors (TFs) suggestive of secondary-transcription-level regulatory cascades, as well as signaling proteins from diverse signaling cascades. Additionally, we also found numerous secreted and cell surface proteins involved in cell-cell and cell-matrix interactions whose genes showed altered expression. Indeed, in terms of the number of genes with altered expression patterns, the latter class was overrepresented among all functional classes (P < 0.001 by the χ2 test).
(i) TFs.
Overall, the expression of 30 TFs and 11 chromatin proteins was found to be altered in mice with ECM in our various experiments. In moribund mice with ECM, expression of 11 TFs was modulated, and 7 of these were significantly upregulated relative to their levels in nonmoribund and resistant BALB/c mice. Compared to resistant CD8-KO mice, WT mice with ECM showed upregulation of 12 TFs and downregulation of a single TF. The downregulation of Stat1, a proinflammatory and proapoptotic TF, in resistant CD8-KO mice suggests that this TF might be involved in the tissue damage observed in ECM. Interestingly, several of the other TFs with altered mRNA levels, such as MafK, MafF, FosB, JunD1, Myc, Runx1, Rbpsuh, GATA2, and IκB-α (an NF-κB inhibitor), are related to hematopoiesis or lymphocyte-specific transcriptional responses. Specifically, Runx1 and Rbpsuh are critical for T-cell development and specialization, whereas FosB and JunD1 dimerize to constitute the AP1 transcription factor, required for downstream responses to T-cell-receptor-mediated signaling and interleukin-2 activation (24). Hence, the overexpression of the latter two genes might be related to the escalation of inflammation in ECM. The upregulation of GATA2 and MafK, both positive regulators of erythropoiesis, and the downregulation of Hox5a, an inhibitor of erythropoiesis (13), during ECM might be responses to the reduction in reticulocyte levels inferred above. The overexpression of MafK, a TF subunit important for normal platelet production (57), in ECM may also correlate with the previously characterized thrombocytopenia that occurs during ECM (2).
(ii) Signal transduction.
The expression of a number of signaling proteins, including polypeptide first messengers, diverse receptors, intracellular enzymes such as kinases, phosphatases, and ubiquitin E3 ligase subunits, and adaptor proteins with peptide-binding and protein-interacting domains such as SH2, SH3, and SAM domains, appears to be altered in our experiments. We systematically analyzed these to identify potential pathways activated during ECM. First, we observed that resistant CD8-KO mice exhibited downregulation of four chemokines, namely, Ccl3, Ccl9, Ccl5, and Ccl25, in contrast to moribund WT mice. This suggested that part of the resistance to ECM in CD8-KO mice may have emerged from the inability to activate these chemokines, which probably trigger a neural inflammatory response (23). Our hypothesis is supported by an earlier study that showed elevated expression of Ccl5 (RANTES) in postmortem brain tissue samples from CM patients (53). Similarly, the overexpression in ECM of the adipokine Angptl4 is consistent with recent reports that it is induced upon brain injury, leading to hypoxia/ischemia (68). This first messenger might represent an as yet unexplored pathway involved in the response to oxygen deprivation in the morbid brain.
At the level of the cell membrane, P2Y12, one of two ADP receptors expressed on platelets, was significantly downregulated (−2.6- to −3-fold) during ECM. We were particularly interested in this molecule due to its role in platelet activation. In vessel injury, platelets are activated and play an important role in the arrest of bleeding (20). Grau and colleagues have conducted a series of experiments suggesting an important role for platelets and microparticles in the pathogenesis of ECM. In these studies, expression of P-selectin on endothelial cells (but not platelets) (10) and microparticle formation (11) are both associated with the development of ECM. Downregulation of P2Y12 could disrupt the formation of microparticles, and future studies exploring the effect of P2Y12 expression on microparticle formation during ECM may throw further light on the role of microparticles in the pathogenesis of ECM. In contrast, five other G protein-coupled seven-transmembrane-domain receptors, namely, Gprc5a, Olfr705, Bdkrb1, Npy6r, and V1rf3, are upregulated in mice with ECM. Of these, the action of the bradykinin receptor Bdkrb1 and the neuropeptide Y receptor Npy6r might be of particular interest in terms of understanding the action of neuropeptides on leukocyte and thrombocyte function, especially in relation to inflammatory responses (12). Strikingly, we observed that Toll-like receptor 11 (Tlr11) has lowered expression in mice with ECM relative to that in nonmoribund C57BL/6 and resistant BALB/c mice. It has recently been proposed that Tlr11 is a key mediator of immunity against apicomplexan parasites (70); hence, its lowered expression might have significance for the pathogenesis of ECM.
Genes of nine kinases and one phosphatase, as well as two regulatory phosphatase subunits, show altered expression in our data sets. The majority of kinases are upregulated in ECM, suggesting the activation of diverse phosphorylation responses. Among these, of interest is casein kinase 2 (CK2), a ubiquitous and conserved nuclear and cytoplasmic serine/threonine protein kinase involved in cell growth and proliferation and in the suppression of apoptosis. Upregulation of CK2 is consistent with activation of other antiapoptotic factors (see above) and points to a concerted host response via multiple independent pathways to minimize additional damage to brain tissue during ECM. The GTPase GBP2 is upregulated in mice with ECM. This molecule belongs to the family of IFN-inducible GTPases and is induced in macrophages by IFN-γ (32, 37), which is consistent with the upregulation of the IFN-γ gene observed in moribund mice compared to resistant CD8-KO mice. The strong overexpression of the candidate tumor suppressor gene SASH1, which encodes an adaptor molecule with SAM and SH3 domains, during ECM suggests that this molecule might be an uncharacterized component of a lymphocyte signaling pathway, consistent with previous observations on Btk− B cells (28).
DISCUSSION
It is widely reported that approximately 1 million individuals die each year globally due to the effect of malaria, although the precise numbers of attributed deaths are known to vary. In two independent studies, P. falciparum malaria was reported to be the direct cause of 803,620 and 545,000 deaths of children below the age of 5 years in sub-Saharan Africa in the year 2000, respectively (50, 51). Although the clinical syndromes associated with the pathogenesis of CM are considered a major contributor to malaria deaths, the precise estimates of CM as a direct cause of death differ greatly and depend on the children's age and the rate, seasonality, and setting of transmission. For example, in an urban setting of mesoendemicity in a hospital in Bamako, Mali, 94% of children who died due to malaria had CM (48), while in the Kassena-Nankana District of Ghana, only 5.4% of children who had severe malaria were diagnosed with CM (41). The only method available for predicting the outcome of P. falciparum infection for an African child who is brought to the hospital with severe malaria is the clinical indicators that are found to be associated with CM or severe malaria anemia (48). It is a well-accepted fact that an early diagnosis of the clinical symptoms of CM allows accelerated care and significantly improves the prospects of survival. Therefore, validated prognostic tests that could reliably predict CM before the onset of fatal pathogenic symptoms might help save thousands of lives. This argument is supported by a report that even blood film microscopy for the clinical diagnosis of P. falciparum infections reduced the fatality rate by more than half (7.5% versus 3.2%; P < 0.001) (42). We believe that a prognostic test based on the host molecular factors that are associated with susceptibility or resistance to CM will be a great aid in clinical management and will reduce mortality.
Recently, several laboratories have utilized microarray technology to identify the host molecules associated with the pathogenesis of ECM in mice (15, 31, 35, 55). The major biomarkers of ECM identified by transcriptional analysis included immune response genes and molecules associated with apoptosis pathways (15, 31, 35). In one study that measured transcriptional alterations based on changes during the early stages of P. berghei infection in CM-susceptible and CM-resistant mice, detection of potent inflammatory responses, expression of molecules associated with brain injury repair, and early disturbances of brain metabolic energy metabolism were reported (14). In spite of these efforts, our knowledge regarding the molecules and molecular pathways that are linked with the pathology of CM remains incomplete, and further studies are needed to fill the critical lacuna. In this communication, we report the identification of several previously unknown host molecules and associated biochemical pathways that are potential mediators of the pathogenesis of ECM in the murine P. berghei ANKA model. These molecules were identified by using a combination of approaches, including comparative global gene expression profiling, bioinformatic analysis of protein function, and pathway reconstruction; for a few of the molecules considered biologically relevant, we determined a possible association between protein expression (by Western blotting) in brain tissue and real-time expression of ECM (for moribund versus nonmoribund mice) in susceptible WT C57BL/6 and resistant CD8-KO C57BL/6 mice.
The unique strength of our approach relied on determining the differential expression of ECM-associated genes in brain samples from moribund and nonmoribund C57BL/6 mice and from BALB/c mice, which are inherently resistant to ECM. In addition, to define the molecular mechanism underlying the pathogenic role of CD8 T cells and the perforin molecule during ECM, we compared gene expression profiles for ECM-susceptible C57BL/6 mice with ECM to those for resistant CD8-KO C57BL/6 mice and PFP-KO C57BL/6 mice during the cerebral phase of P. berghei ANKA infection.
Our comparative microarray analyses revealed novel and highly specific host responses; we found that 210 genes (66% upregulated, 34% downregulated) showed significantly altered expression in moribund WT C57BL/6 mice relative to that in both nonmoribund WT C57BL/6 mice and resistant BALB/c mice. In comparison, the resistance phenotypes of CD8-KO and PFP-KO mice were associated with alterations in only 166 and 9 genes, respectively. Our comprehensive computational sequence analysis of the genes with altered expression, followed by functional classification and identification of cellular interactions based on reconstruction of biological pathways, revealed that several critical biological processes were affected during the period when mice were experiencing the pathology of ECM. Notable among the host genes with ECM-associated responses are several molecules of the immune system, a wide variety of antiapoptotic factors, TFs, receptors, and proteins involved in signal transduction and cell-cell interaction. Several of the ECM-associated alterations in molecular pathways are reminiscent of septic-shock-like immunological responses and of cellular responses to severe stress and trauma-related injury (25, 29, 36). To offset these sudden and dramatic events, the host cells appear to mount vigorous cellular responses, particularly in the form of antiapoptotic and neuroprotectant molecules. Some of these important cellular responses related to ECM are discussed below.
A major finding of our study is the observation that there is a significant downregulation in the transcription of the hemoglobin α1 gene (Table 2) but a significant overexpression of the hemoglobin α1 protein in moribund compared to nonmoribund mice (Table 2; Fig. 1). We posit that a marked reduction in the reticulocyte counts along with a simultaneous influx of mature erythrocytes, possibly infected with Plasmodium parasites, may account for the increased levels of hemoglobin α1 protein in the brain during ECM. Importantly, this increase in hemoglobin α1 protein levels observed in our study may be a consequence of an increase in brain hemorrhaging and/or increased blood deposition in the brain resulting from parasite sequestration and an inflammation-induced decrease in blood flow. Hemoglobin α1 is further cannibalized by malaria parasites to produce heme, a molecule recently implicated as a major “host toxin” that triggers the inflammatory responses leading to the clinical syndromes of ECM (44). We also find that a significant portion of the host response is directed toward countering the damaging cellular events triggered due to the pathology of ECM. This contention is supported by results demonstrating that mice undergoing the clinical symptoms of ECM had increased transcriptional and translational expression of the MT-I and -II molecules (Table 2 and Fig. 1). In the brain, astrocytes are the main source of MT-I and -II (67), which might play an active role in reducing damage to the CNS by at least two mechanisms. First, MT-I and -II reduce the activation and recruitment of macrophages and T cells and also inhibit the production and secretion of inflammatory cytokines, including TNF-α and lymphotoxin (58). Second, MT-I and -II are powerful scavengers of free radicals (63) and therefore may moderate oxidative stress during CM. Importantly, exogenous treatment with MT-I and -II is also neuroprotective, is well tolerated, and has been shown to inhibit experimental autoimmune encephalomyelitis (46), the murine model of multiple sclerosis. These results suggest that the CNS response to damage inflicted by CM may bear similarities to neuroprotection induced by external stimuli or chronic illness.
Further evidence of vigorous host responses mounted in response to the deleterious effects of ECM was provided by significant alterations in the expression of the host genes involved in the regulation of both proapoptotic and antiapoptotic pathways (Table 2; see also Results). In our data set, levels of both proapoptotic and antiapoptotic genes were altered during ECM. Two recent studies also reported alterations in the expression of proapoptotic pathway genes during the cerebral phase in mice (31, 35). We focused our attention on p21 (the kinase inhibitor of the Cip/Kip family), a molecule important in the regulation of apoptosis. Cytoplasmic p21 has been shown to prevent apoptosis during neuronal differentiation (62) by inhibiting the catalytic activity of stress-activated protein kinase (56), and mitochondrial p21 prevents Fas-mediated apoptosis by inhibiting the activation of caspase-3 through direct interactions with its precursor, procaspase-3 (59, 60).
Compared to that in nonmoribund mice, expression of p21 in moribund mice is 5.5-fold increased at the level of transcription and 14.5-fold increased at the level of translation (Fig. 1). We think that the greatly enhanced expression of the p21 molecule during ECM represents a counterapoptotic response in the face of strong oxidative stress. Furthermore, differential expression of p21 in mice with ECM indicates that this molecule might serve as an important host biomarker of ECM. While it remains to be determined whether p21 expression is elevated in patients exhibiting the symptoms of CM, this molecule has been used to prognosticate certain cancers (52).
In summary, we believe that directed exploration of the data set generated for the ECM-susceptible and ECM-resistant mice and in-depth experimental analysis of predicted candidate molecules has improved our understanding of the repertoire of host molecules that mediate the pathology of CM. Further validation studies using brain biopsy specimens from patients who have succumbed to CM will be needed in order to determine the relevance of the identified molecules as predictors of CM in humans. Nonetheless, the presentation of this data set along with the accompanying “proof of concept” by Western blot studies will provide the research community with novel candidates for the development of diagnostics and therapeutics against CM, especially for young children in the areas of high P. falciparum transmission in Africa.
Supplementary Material
Acknowledgments
We thank Tim Myers and Guojian Jiang at the NIAID microarray research facility for assistance with the microarray experiments. We also thank the veterinary staff at the Twinbrook III facility, NIAID, for the care and maintenance of mice.
The views and opinions expressed here are those of the authors and should not be construed as the official opinion of the Food and Drug Administration.
Editor: W. A. Petri, Jr.
Footnotes
Published ahead of print on 21 July 2008.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Ahn, K. J., I. A. Seo, H. K. Lee, E. J. Choi, E. H. Seo, H. J. Lee, and H. T. Park. 2007. Down-regulation of UNC5 homologue expression after the spinal cord injury in the adult rat. Neurosci. Lett. 41943-48. [DOI] [PubMed] [Google Scholar]
- 2.Akingbola, T. S., W. A. Shokunbi, and P. E. Olumese. 2006. Coagulation profile in Nigerian children with cerebral malaria. Niger. Postgrad. Med. J. 13195-199. [PubMed] [Google Scholar]
- 3.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 253389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arsenijevic, D., H. Onuma, C. Pecqueur, S. Raimbault, B. S. Manning, B. Miroux, E. Couplan, M. C. Alves-Guerra, M. Goubern, R. Surwit, F. Bouillaud, D. Richard, S. Collins, and D. Ricquier. 2000. Disruption of the uncoupling protein-2 gene in mice reveals a role in immunity and reactive oxygen species production. Nat. Genet. 26435-439. [DOI] [PubMed] [Google Scholar]
- 5.Boubou, M. I., A. Collette, D. Voegtle, D. Mazier, P. A. Cazenave, and S. Pied. 1999. T cell response in malaria pathogenesis: selective increase in T cells carrying the TCR Vβ8 during experimental cerebral malaria. Int. Immunol. 111553-1562. [DOI] [PubMed] [Google Scholar]
- 6.Bronstein, J. M., P. E. Micevych, and K. Chen. 1997. Oligodendrocyte-specific protein (OSP) is a major component of CNS myelin. J. Neurosci. Res. 50713-720. [DOI] [PubMed] [Google Scholar]
- 7.Chang, K. H., M. Tam, and M. M. Stevenson. 2004. Inappropriately low reticulocytosis in severe malarial anemia correlates with suppression in the development of late erythroid precursors. Blood 1033727-3735. [DOI] [PubMed] [Google Scholar]
- 8.Chung, R. S., P. A. Adlard, J. Dittmann, J. C. Vickers, M. I. Chuah, and A. K. West. 2004. Neuron-glia communication: metallothionein expression is specifically up-regulated by astrocytes in response to neuronal injury. J. Neurochem. 88454-461. [DOI] [PubMed] [Google Scholar]
- 9.Chung, R. S., and A. K. West. 2004. A role for extracellular metallothioneins in CNS injury and repair. Neuroscience 123595-599. [DOI] [PubMed] [Google Scholar]
- 10.Combes, V., A. R. Rosenkranz, M. Redard, G. Pizzolato, H. Lepidi, D. Vestweber, T. N. Mayadas, and G. E. Grau. 2004. Pathogenic role of P-selectin in experimental cerebral malaria: importance of the endothelial compartment. Am. J. Pathol. 164781-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Combes, V., T. E. Taylor, I. Juhan-Vague, J. L. Mege, J. Mwenechanya, M. Tembo, G. E. Grau, and M. E. Molyneux. 2004. Circulating endothelial microparticles in Malawian children with severe falciparum malaria complicated with coma. JAMA 2912542-2544. [DOI] [PubMed] [Google Scholar]
- 12.Couture, R., M. Harrisson, R. M. Vianna, and F. Cloutier. 2001. Kinin receptors in pain and inflammation. Eur. J. Pharmacol. 429161-176. [DOI] [PubMed] [Google Scholar]
- 13.Crooks, G. M., J. Fuller, D. Petersen, P. Izadi, P. Malik, P. K. Pattengale, D. B. Kohn, and J. C. Gasson. 1999. Constitutive HOXA5 expression inhibits erythropoiesis and increases myelopoiesis from human hematopoietic progenitors. Blood 94519-528. [PubMed] [Google Scholar]
- 14.Delahaye, N. F., N. Coltel, D. Puthier, M. Barbier, P. Benech, F. Joly, F. A. Iraqi, G. E. Grau, C. Nguyen, and P. Rihet. 2007. Gene expression analysis reveals early changes in several molecular pathways in cerebral malaria-susceptible mice versus cerebral malaria-resistant mice. BMC Genomics 8452-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Delahaye, N. F., N. Coltel, D. Puthier, L. Flori, R. Houlgatte, F. A. Iraqi, C. Nguyen, G. E. Grau, and P. Rihet. 2006. Gene-expression profiling discriminates between cerebral malaria (CM)-susceptible mice and CM-resistant mice. J. Infect. Dis. 193312-321. [DOI] [PubMed] [Google Scholar]
- 16.de Souza, J. B., and E. M. Riley. 2002. Cerebral malaria: the contribution of studies in animal models to our understanding of immunopathogenesis. Microbes Infect. 4291-300. [DOI] [PubMed] [Google Scholar]
- 17.Engwerda, C., E. Belnoue, A. C. Gruner, and L. Renia. 2005. Experimental models of cerebral malaria. Curr. Top. Microbiol. Immunol. 297103-143. [PubMed] [Google Scholar]
- 18.Funk, C. D. 2001. Prostaglandins and leukotrienes: advances in eicosanoid biology. Science 2941871-1875. [DOI] [PubMed] [Google Scholar]
- 19.Gow, A., C. M. Southwood, J. S. Li, M. Pariali, G. P. Riordan, S. E. Brodie, J. Danias, J. M. Bronstein, B. Kachar, and R. A. Lazzarini. 1999. CNS myelin and Sertoli cell tight junction strands are absent in Osp/claudin-11 null mice. Cell 99649-659. [DOI] [PubMed] [Google Scholar]
- 20.Hechler, B., M. Cattaneo, and C. Gachet. 2005. The P2 receptors in platelet function. Semin. Thromb. Hemost. 31150-161. [DOI] [PubMed] [Google Scholar]
- 21.Horvath, T. L., C. H. Warden, M. Hajos, A. Lombardi, F. Goglia, and S. Diano. 1999. Brain uncoupling protein 2: uncoupled neuronal mitochondria predict thermal synapses in homeostatic centers. J. Neurosci. 1910417-10427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu, Z., D. M. Ng, T. Yamada, C. Chen, S. Kawashima, J. Mellor, B. Linghu, M. Kanehisa, J. M. Stuart, and C. DeLisi. 2007. VisANT 3.0: new modules for pathway visualization, editing, prediction and construction. Nucleic Acids Res. 35W625-W632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang, D., Y. Han, M. R. Rani, A. Glabinski, C. Trebst, T. Sorensen, M. Tani, J. Wang, P. Chien, S. O'Bryan, B. Bielecki, Z. L. Zhou, S. Majumder, and R. M. Ransohoff. 2000. Chemokines and chemokine receptors in inflammation of the nervous system: manifold roles and exquisite regulation. Immunol. Rev. 17752-67. [DOI] [PubMed] [Google Scholar]
- 24.Jain, J., V. E. Valge-Archer, and A. Rao. 1992. Analysis of the AP-1 sites in the IL-2 promoter. J. Immunol. 1481240-1250. [PubMed] [Google Scholar]
- 25.Kobori, N., G. L. Clifton, and P. Dash. 2002. Altered expression of novel genes in the cerebral cortex following experimental brain injury. Brain Res. Mol. Brain Res. 104148-158. [DOI] [PubMed] [Google Scholar]
- 26.Kohler, L. B., V. Berezin, E. Bock, and M. Penkowa. 2003. The role of metallothionein II in neuronal differentiation and survival. Brain Res. 992128-136. [DOI] [PubMed] [Google Scholar]
- 27.Lamikanra, A. A., D. Brown, A. Potocnik, C. Casals-Pascual, J. Langhorne, and D. J. Roberts. 2007. Malarial anemia: of mice and men. Blood 11018-28. [DOI] [PubMed] [Google Scholar]
- 28.Lindvall, J. M., K. E. Blomberg, A. Wennborg, and C. I. Smith. 2005. Differential expression and molecular characterisation of Lmo7, Myo1e, Sash1, and Mcoln2 genes in Btk-defective B-cells. Cell. Immunol. 23546-55. [DOI] [PubMed] [Google Scholar]
- 29.Lopez-Bojorquez, L. N., A. Z. Dehesa, and G. Reyes-Teran. 2004. Molecular mechanisms involved in the pathogenesis of septic shock. Arch. Med. Res. 35465-479. [DOI] [PubMed] [Google Scholar]
- 30.Lou, J., R. Lucas, and G. E. Grau. 2001. Pathogenesis of cerebral malaria: recent experimental data and possible applications for humans. Clin. Microbiol. Rev. 14810-820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lovegrove, F. E., S. A. Gharib, S. N. Patel, C. A. Hawkes, K. C. Kain, and W. C. Liles. 2007. Expression microarray analysis implicates apoptosis and interferon-responsive mechanisms in susceptibility to experimental cerebral malaria. Am. J. Pathol. 1711894-1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.MacMicking, J. D. 2004. IFN-inducible GTPases and immunity to intracellular pathogens. Trends Immunol. 25601-609. [DOI] [PubMed] [Google Scholar]
- 33.Marques, F., A. J. Rodrigues, J. C. Sousa, G. Coppola, D. H. Geschwind, N. Sousa, M. Correia-Neves, and J. A. Palha. 2008. Lipocalin 2 is a choroid plexus acute-phase protein. J. Cereb. Blood Flow Metab. 28450-455. [DOI] [PubMed] [Google Scholar]
- 34.Miller, L. H., D. I. Baruch, K. Marsh, and O. K. Doumbo. 2002. The pathogenic basis of malaria. Nature 415673-679. [DOI] [PubMed] [Google Scholar]
- 35.Miu, J., N. H. Hunt, and H. J. Ball. 2008. Predominance of interferon-related responses in the brain during murine malaria, as identified by microarray analysis. Infect. Immun. 761812-1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Natale, J. E., F. Ahmed, I. Cernak, B. Stoica, and A. I. Faden. 2003. Gene expression profile changes are commonly modulated across models and species after traumatic brain injury. J. Neurotrauma 20907-927. [DOI] [PubMed] [Google Scholar]
- 37.Nguyen, T. T., Y. Hu, D. P. Widney, R. A. Mar, and J. B. Smith. 2002. Murine GBP-5, a new member of the murine guanylate-binding protein family, is coordinately regulated with other GBPs in vivo and in vitro. J. Interferon Cytokine Res. 22899-909. [DOI] [PubMed] [Google Scholar]
- 38.Nitcheu, J., O. Bonduelle, C. Combadiere, M. Tefit, D. Seilhean, D. Mazier, and B. Combadiere. 2003. Perforin-dependent brain-infiltrating cytotoxic CD8+ T lymphocytes mediate experimental cerebral malaria pathogenesis. J. Immunol. 1702221-2228. [DOI] [PubMed] [Google Scholar]
- 39.Notredame, C., D. G. Higgins, and J. Heringa. 2000. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302205-217. [DOI] [PubMed] [Google Scholar]
- 40.Oakley, M. S., S. Kumar, V. Anantharaman, H. Zheng, B. Mahajan, J. D. Haynes, J. K. Moch, R. Fairhurst, T. F. McCutchan, and L. Aravind. 2007. Molecular factors and biochemical pathways induced by febrile temperature in intraerythrocytic Plasmodium falciparum parasites. Infect. Immun. 752012-2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oduro, A. R., K. A. Koram, W. Rogers, F. Atuguba, P. Ansah, T. Anyorigiya, A. Ansah, F. Anto, N. Mensah, A. Hodgson, and F. Nkrumah. 2007. Severe falciparum malaria in young children of the Kassena-Nankana district of northern Ghana. Malar. J. 696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Opoka, R. O., Z. Xia, P. Bangirana, and C. C. John. 2008. Inpatient mortality in children with clinically diagnosed malaria as compared with microscopically confirmed malaria. Pediatr. Infect. Dis. J. 27319-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O'Reilly, M. A. 2005. Redox activation of p21Cip1/WAF1/Sdi1: a multifunctional regulator of cell survival and death. Antioxid. Redox Signal. 7108-118. [DOI] [PubMed] [Google Scholar]
- 44.Pamplona, A., A. Ferreira, J. Balla, V. Jeney, G. Balla, S. Epiphanio, A. Chora, C. D. Rodrigues, I. P. Gregoire, M. Cunha-Rodrigues, S. Portugal, M. P. Soares, and M. M. Mota. 2007. Heme oxygenase-1 and carbon monoxide suppress the pathogenesis of experimental cerebral malaria. Nat. Med. 13703-710. [DOI] [PubMed] [Google Scholar]
- 45.Penkowa, M., and J. Hidalgo. 2001. Metallothionein treatment reduces proinflammatory cytokines IL-6 and TNF-α and apoptotic cell death during experimental autoimmune encephalomyelitis (EAE). Exp. Neurol. 1701-14. [DOI] [PubMed] [Google Scholar]
- 46.Penkowa, M., and J. Hidalgo. 2003. Treatment with metallothionein prevents demyelination and axonal damage and increases oligodendrocyte precursors and tissue repair during experimental autoimmune encephalomyelitis. J. Neurosci. Res. 72574-586. [DOI] [PubMed] [Google Scholar]
- 47.Potter, S., T. Chan-Ling, H. J. Ball, H. Mansour, A. Mitchell, L. Maluish, and N. H. Hunt. 2006. Perforin mediated apoptosis of cerebral microvascular endothelial cells during experimental cerebral malaria. Int. J. Parasitol. 36485-496. [DOI] [PubMed] [Google Scholar]
- 48.Ranque, S., B. Poudiougou, A. Traore, M. Keita, A. A. Oumar, I. Safeukui, S. Marquet, S. Cabantous, M. Diakite, D. Mintha, M. B. Cisse, M. M. Keita, A. J. Dessein, and O. K. Doumbo. 2008. Life-threatening malaria in African children: a prospective study in a mesoendemic urban setting. Pediatr. Infect. Dis. J. 27130-135. [DOI] [PubMed] [Google Scholar]
- 49.Richard, D., R. Rivest, Q. Huang, F. Bouillaud, D. Sanchis, O. Champigny, and D. Ricquier. 1998. Distribution of the uncoupling protein 2 mRNA in the mouse brain. J. Comp. Neurol. 397549-560. [PubMed] [Google Scholar]
- 50.Roca-Feltrer, A., I. Carneiro, and J. R. Armstrong Schellenberg. 2008. Estimates of the burden of malaria morbidity in Africa in children under the age of 5 years. Trop. Med. Int. Health 13771-783. [DOI] [PubMed] [Google Scholar]
- 51.Rowe, A. K., S. Y. Rowe, R. W. Snow, E. L. Korenromp, J. R. Schellenberg, C. Stein, B. L. Nahlen, J. Bryce, R. E. Black, and R. W. Steketee. 2006. The burden of malaria mortality among African children in the year 2000. Int. J. Epidemiol. 35691-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rundle, A., D. Tang, P. Brandt-Rauf, J. Zhou, A. Kelly, F. Schnabel, and F. P. Perera. 2002. Association between the ras p21 oncoprotein in blood samples and breast cancer. Cancer Lett. 18571-78. [DOI] [PubMed] [Google Scholar]
- 53.Sarfo, B. Y., S. Singh, J. W. Lillard, A. Quarshie, R. K. Gyasi, H. Armah, A. A. Adjei, P. Jolly, and J. K. Stiles. 2004. The cerebral-malaria-associated expression of RANTES, CCR3 and CCR5 in post-mortem tissue samples. Ann. Trop. Med. Parasitol. 98297-303. [DOI] [PubMed] [Google Scholar]
- 54.Schaecher, K., S. Kumar, A. Yadava, M. Vahey, and C. F. Ockenhouse. 2005. Genome-wide expression profiling in malaria infection reveals transcriptional changes associated with lethal and nonlethal outcomes. Infect. Immun. 736091-6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sexton, A. C., R. T. Good, D. S. Hansen, M. C. D'Ombrain, L. Buckingham, K. Simpson, and L. Schofield. 2004. Transcriptional profiling reveals suppressed erythropoiesis, up-regulated glycolysis, and interferon-associated responses in murine malaria. J. Infect. Dis. 1891245-1256. [DOI] [PubMed] [Google Scholar]
- 56.Shim, J., H. Lee, J. Park, H. Kim, and E. J. Choi. 1996. A non-enzymatic p21 protein inhibitor of stress-activated protein kinases. Nature 381804-806. [DOI] [PubMed] [Google Scholar]
- 57.Shivdasani, R. A., M. F. Rosenblatt, D. Zucker-Franklin, C. W. Jackson, P. Hunt, C. J. Saris, and S. H. Orkin. 1995. Transcription factor NF-E2 is required for platelet formation independent of the actions of thrombopoietin/MGDF in megakaryocyte development. Cell 81695-704. [DOI] [PubMed] [Google Scholar]
- 58.Stankovic, R. K., R. S. Chung, and M. Penkowa. 2007. Metallothioneins I and II: neuroprotective significance during CNS pathology. Int. J. Biochem. Cell Biol. 39484-489. [DOI] [PubMed] [Google Scholar]
- 59.Suzuki, A., Y. Tsutomi, K. Akahane, T. Araki, and M. Miura. 1998. Resistance to Fas-mediated apoptosis: activation of caspase 3 is regulated by cell cycle regulator p21WAF1 and IAP gene family ILP. Oncogene 17931-939. [DOI] [PubMed] [Google Scholar]
- 60.Suzuki, A., Y. Tsutomi, N. Yamamoto, T. Shibutani, and K. Akahane. 1999. Mitochondrial regulation of cell death: mitochondria are essential for procaspase 3-p21 complex formation to resist Fas-mediated cell death. Mol. Cell. Biol. 193842-3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Takeda, M., M. Kikuchi, R. Ubalee, K. Na-Bangchang, R. Ruangweerayut, S. Shibahara, S. Imai, and K. Hirayama. 2005. Microsatellite polymorphism in the heme oxygenase-1 gene promoter is associated with susceptibility to cerebral malaria in Myanmar. Jpn. J. Infect. Dis. 58268-271. [PubMed] [Google Scholar]
- 62.Tanaka, H., T. Yamashita, M. Asada, S. Mizutani, H. Yoshikawa, and M. Tohyama. 2002. Cytoplasmic p21Cip1/WAF1 regulates neurite remodeling by inhibiting Rho-kinase activity. J. Cell Biol. 158321-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Taylor, D. M., S. Minotti, J. N. Agar, and H. D. Durham. 2004. Overexpression of metallothionein protects cultured motor neurons against oxidative stress, but not mutant Cu/Zn-superoxide dismutase toxicity. Neurotoxicology 25779-792. [DOI] [PubMed] [Google Scholar]
- 64.Teshima, Y., M. Akao, S. P. Jones, and E. Marban. 2003. Uncoupling protein-2 overexpression inhibits mitochondrial death pathway in cardiomyocytes. Circ. Res. 93192-200. [DOI] [PubMed] [Google Scholar]
- 65.van der Heyde, H. C., J. Nolan, V. Combes, I. Gramaglia, and G. E. Grau. 2006. A unified hypothesis for the genesis of cerebral malaria: sequestration, inflammation and hemostasis leading to microcirculatory dysfunction. Trends Parasitol. 22503-508. [DOI] [PubMed] [Google Scholar]
- 66.Wada, K., M. Arita, A. Nakajima, K. Katayama, C. Kudo, Y. Kamisaki, and C. N. Serhan. 2006. Leukotriene B4 and lipoxin A4 are regulatory signals for neural stem cell proliferation and differentiation. FASEB J. 201785-1792. [DOI] [PubMed] [Google Scholar]
- 67.Wiese, L., J. A. Kurtzhals, and M. Penkowa. 2006. Neuronal apoptosis, metallothionein expression and proinflammatory responses during cerebral malaria in mice. Exp. Neurol. 200216-226. [DOI] [PubMed] [Google Scholar]
- 68.Wiesner, G., R. E. Brown, G. S. Robertson, S. A. Imran, E. Ur, and M. Wilkinson. 2006. Increased expression of the adipokine genes resistin and fasting-induced adipose factor in hypoxic/ischaemic mouse brain. Neuroreport 171195-1198. [DOI] [PubMed] [Google Scholar]
- 69.Yanez, D. M., D. D. Manning, A. J. Cooley, W. P. Weidanz, and H. C. van der Heyde. 1996. Participation of lymphocyte subpopulations in the pathogenesis of experimental murine cerebral malaria. J. Immunol. 1571620-1624. [PubMed] [Google Scholar]
- 70.Yarovinsky, F., and A. Sher. 2006. Toll-like receptor recognition of Toxoplasma gondii. Int. J. Parasitol. 36255-259. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.