Abstract
During acute human immunodeficiency virus type 1 (HIV-1) infection, early host cellular immune responses drive viral evolution. The rates and extent of these mutations, however, remain incompletely characterized. In a cohort of 98 individuals newly infected with HIV-1 subtype B, we longitudinally characterized the rates and extent of HLA-mediated escape and reversion in Gag, Pol, and Nef using a rational definition of HLA-attributable mutation based on the analysis of a large independent subtype B data set. We demonstrate rapid and dramatic HIV evolution in response to immune pressures that in general reflect established cytotoxic T-lymphocyte (CTL) response hierarchies in early infection. On a population level, HLA-driven evolution was observed in ∼80% of published CTL epitopes. Five of the 10 most rapidly evolving epitopes were restricted by protective HLA alleles (HLA-B*13/B*51/B*57/B*5801; P = 0.01), supporting the importance of a strong early CTL response in HIV control. Consistent with known fitness costs of escape, B*57-associated mutations in Gag were among the most rapidly reverting positions upon transmission to non-B*57-expressing individuals, whereas many other HLA-associated polymorphisms displayed slow or negligible reversion. Overall, an estimated minimum of 30% of observed substitutions in Gag/Pol and 60% in Nef were attributable to HLA-associated escape and reversion events. Results underscore the dominant role of immune pressures in driving early within-host HIV evolution. Dramatic differences in escape and reversion rates across codons, genes, and HLA restrictions are observed, highlighting the complexity of viral adaptation to the host immune response.
Cytotoxic T lymphocytes (CTL) recognizing HLA class-I-restricted viral epitopes presented on the infected cell surface are critical for the resolution of acute-phase plasma viremia (13, 39, 53). However, durable human immunodeficiency virus (HIV) immune control rarely is achieved, due in part to rapid viral evolution within the new host. Indeed, the course of HIV disease is influenced by the strength and specificity of the early CTL response (7, 76), combined with the virus' ability to adapt to changing immune pressures through the selection of HLA-restricted CTL escape mutations and the reversion of transmitted escape mutations from the previous host (14, 22, 36, 38, 39, 52, 58, 59, 67, 68). Given the extent of CD4 T-cell destruction that occurs during the acute phase (24, 25), it is important to achieve a deeper understanding of the interplay between immune response and viral adaptation in early infection. Recently, immunodominance hierarchies of CTL epitope targeting in early HIV infection have been characterized (7, 76); however, a comprehensive population-based assessment of the rates and extent of HLA-associated immune adaptation in early HIV infection remains to be undertaken.
It is now understood that CTL escape occurs along generally predictable pathways based on the host HLA profile (2, 17, 27), and that escape and reversion represent major forces driving viral evolution and diversity at both the individual and population levels (2, 11, 14, 34, 39, 52, 57-59, 65, 67, 68, 70). However, due in part to the lack of a consistent definition of HLA-associated mutation, as well as the lack of large longitudinal HIV sequence datasets (2, 9, 46, 61), HLA-driven viral adaptation in early HIV infection remains incompletely characterized. In this study, we employ a strict definition of HLA-attributable substitution based on a comprehensive, predefined list of HLA-associated polymorphisms in HIV type 1 (HIV-1) subtype B in order to estimate the proportion of viral evolution attributable to HLA-associated selection pressures in Gag, Pol, and Nef in the first year of HIV infection in a cohort of 98 untreated, subtype B-infected seroconverters. In addition, we systematically compute the rates of escape and estimate the rates of the reversion of these HLA-associated polymorphisms in early HIV infection, revealing marked differences in the rates of escape and reversion across codons, genes, and HLA restrictions.
MATERIALS AND METHODS
HIV seroconverter cohort.
The HIV seroconverter cohort consisted of 98 untreated, HIV subtype B-infected individuals enrolled through a private medical clinic (Jessen-Praxis) in Berlin, Germany (n = 38), and three sites within the Acute Infection and Early Disease Research Program (AIEDRP): Massachusetts General Hospital, Boston (n = 25), Aaron Diamond AIDS Research Center, New York, NY (n = 24), and the National Centre in HIV Epidemiology and Clinical Research, University of New South Wales, Sydney, Australia (n = 11). Of these, 61 (62%) individuals were identified during acute infection as defined by either documented positive HIV RNA (>5,000 copies/ml) and either (i) a negative HIV-1 enzyme immunoassay (EIA) or (ii) a positive EIA but a negative or indeterminate Western blotting result (AIEDRP stage 1; n = 53) or by a detectable serum p24 antigen and either (i) a negative EIA or (ii) a positive EIA but a negative or indeterminate Western blotting result (AIEDRP stage 2; n = 8). The time frame for acute infection as defined here ranges from 2 to 6 weeks following infection (31). The remaining 37 (38%) individuals were identified during early HIV infection, as defined by (i) a negative EIA during the previous 6 months or (ii) a negative detuned HIV-1 EIA (Vironostika-LS EIA; BioMerieux, Raleigh, NC) (44) at enrolment (AIEDRP stages 3a and 3b, respectively).
The date of HIV infection was estimated using clinical history (where available) by subtracting 4 weeks from the baseline (for stages 1a or 2a), by subtracting 6 weeks from the baseline (for stages 1b or 2b), by calculating the midpoint between the last negative and the first positive EIA (for stage 3a), or by subtracting 4 months from the baseline (for stage 3b). Subjects were monitored for a median of 425 days after the estimated infection date. The study was approved by the respective institutional review boards and was conducted in accordance with human experimentation guidelines of Massachusetts General Hospital. All subjects provided written informed consent.
HIV RNA genotyping and HLA typing.
HIV-1 RNA genotyping of gag, pol, and nef was performed on serial plasma samples (median of 5 samples/patient). In general, patients were monitored at baseline, 1 month, and every 2 or 3 months thereafter, although in some cases more frequent sampling was performed. HIV-1 gag (codons 1 to 500; HXB2 nucleotides [nt] 790 to 2289), a portion of pol (protease codons 1 to 99 and reverse transcriptase [RT] codons 1 to 400; nt 2253 to 3749), and nef (codons 1 to 206; nt 8797 to 9414) were amplified by nested reverse transcription-PCR from extracted plasma HIV RNA using gene-specific primers. Population (bulk) sequencing was performed on an Applied Biosystems 3730 automated DNA sequencer. Data were analyzed using Sequencher (Genecodes). Nucleotide mixtures were assigned if the secondary peak height exceeded 25% of the dominant peak height. Data were aligned to the HIV-1 subtype B reference HXB2 sequence (GenBank accession no. K03455) using a modified NAP algorithm (42). All subjects harbored subtype B infections, as confirmed by comparing gag/pol/nef sequences to all HIV subtype reference sequences (http://hiv-web.lanl.gov/content/hiv-db/mainpage.html). HLA class I typing was performed by standard sequence-specific PCR or sequence-based typing.
Predefined list of HLA-associated polymorphisms.
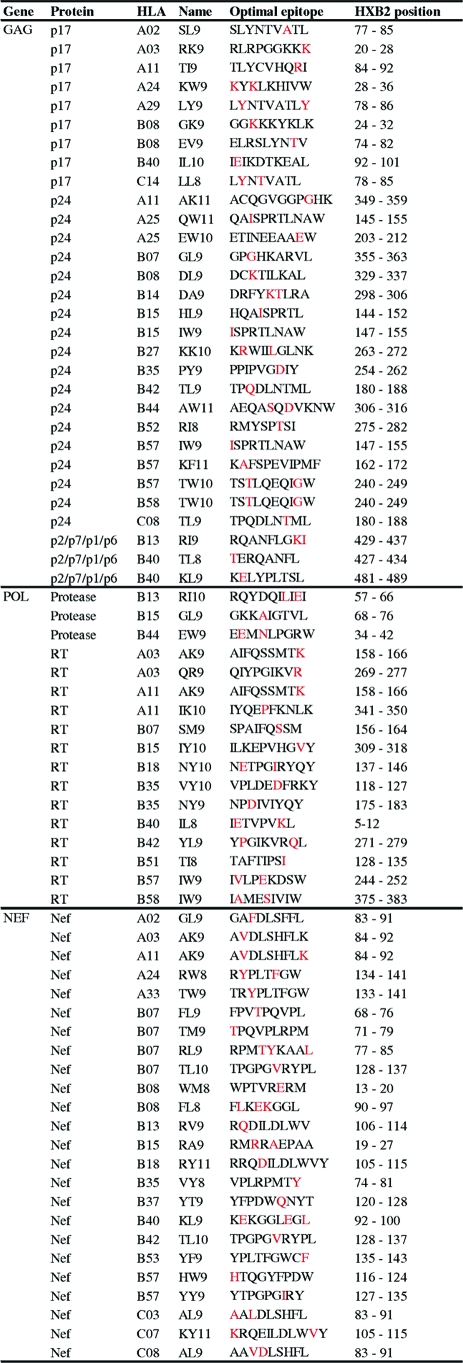
Previous studies of immune-driven HIV evolution have lacked a consistent definition of HLA-attributable mutation. We address this by defining HLA-associated escape and reversion based on a predefined, comprehensive list of HLA-associated polymorphisms identified in a cross-sectional analysis of >1,200 chronically infected, antiretroviral-naïve individuals from three published cohort studies in Canada (17, 18), the United States (45), and western Australia (11) using phylogenetically corrected methods and a q-value correction for multiple tests (q < 0.2, which translates to a 20% false discovery rate) (60, 75). As described previously (17; D. Heckerman, C. J. Brumme, M. John, J. Carlson, R. Haubrich, S. Riddler, L. Swenson, I. Tao, S. Szeto, D. Chan, C. Kadie, N. Frahm, C. Brander, B. D. Walker, Z. L. Brumme, P. R. Harrigan, and S. Mallal, presented at the 15th International Workshop on HIV Dynamics and Evolution, Santa Fe, NM, 27 to 30 April 2008), HLA-associated polymorphisms are dichotomized based on the presence or absence of selection pressure: escape amino acids represent the residue most likely to emerge under HLA-restricted CTL selection pressure, while reversion amino acids represent the immunologically susceptible residues most likely to reemerge following transmission to an HLA-unmatched individual. In general, the reversion amino acids at any particular HLA-associated position tend to be consensus, while the escaped forms tend to be nonconsensus, but this is not always the case. The list contains over 1,000 unique HLA-associated polymorphisms occurring at ∼20% of codons in Gag/Pol and ∼50% of codons in Nef (Heckerman et al., 15th International Workshop on HIV Dynamics and Evolution). Of these, ∼30% map within optimally defined CTL epitopes (http://www.hiv.lanl.gov/content/immunology/tables/optimal_ctl_summary.html) (Fig. 1).
FIG. 1.
Locations of HLA-associated substitutions within optimally defined CTL epitopes. Published sequences of the 71 optimally defined CTL epitopes harboring at least one HLA-associated polymorphic site, based on analysis of over 1,200 persons with chronic infection, are listed. HLA-associated polymorphic residues are indicated in red. Overlapping and/or variant epitopes restricted by the same HLA allele were removed to avoid double counting of escape mutations. A complete list of all optimally defined epitopes is available at http://www.hiv.lanl.gov/content/immunology/tables/optimal_ctl_summary.html.
Of the ∼180 optimally defined epitopes in Gag, Pol, and Nef, specific sites and pathways of HLA-driven substitutions were defined for 74 epitopes. Of these 74, 3 were restricted by HLA alleles not observed in the seroconverter cohort and were excluded from analysis; the remaining 71 epitopes, along with their escape sites, are listed in Fig. 1.
Data analysis and definitions. (i) Rates of escape and frequencies of epitope recognition in early infection.
The time to escape for each CTL epitope was defined as the number of days that elapsed between the estimated infection date and the first detection of a full or partial amino acid change consistent (for HLA, codon, and the direction of amino acid selection) with the predefined list of HLA-associated polymorphisms (Fig. 1). Note that this definition is independent of whether an amino acid is consensus or nonconsensus: in the presence of the restricting HLA, any observed full or partial amino acid change away from the reversion form and/or toward the escaped form was considered escape. Note that some epitopes (e.g., B*57 TW10) have more than one escapable site; in these cases, we computed the time to the first escape event at any of these sites. In addition, the baseline sequence was analyzed for the presence of HLA-associated polymorphisms to identify cases where escape may have occurred very early (or, indistinguishably, rare cases where an escaped variant may have been acquired at transmission). When such a polymorphism was present at baseline in a person with the restricting allele, it was counted as an escape mutation.
Corresponding epitope recognition frequencies were derived from an independent, partially published, cross-sectional data set of 289 individuals enrolled through the AIEDRP network who were screened for HIV-1-specific CTL responses against optimally defined epitopes using a gamma interferon (IFN-γ) enzyme-linked immunospot (ELISpot) assay at a single time point ∼8 weeks following the initial presentation with primary HIV infection, as described previously (7, 76); this effectively means that individuals were tested anywhere between ∼3 and 8 months after the estimated infection date. In addition, a subset of seroconverters for whom peripheral blood mononuclear cell samples were available during study follow-up (n = 8) were longitudinally investigated for HLA-restricted CTL responses to optimally described epitopes using IFN-γ ELISpot assays.
(ii) Estimating rates and frequencies of reversion, and the total proportion of amino acid substitutions attributable to HLA-associated selection pressures.
Rates and frequencies of reversion were conservatively estimated using methods similar to those used for escape, with one important difference. Since the vast majority of reversions are toward the consensus, it was not possible to infer reversion events occurring prior to the baseline. In this analysis, therefore, time zero was defined as the estimated infection date, and the baseline sequence was treated as the transmitted sequence. Reversion was defined as the presence of a specific known HLA-associated polymorphism at the baseline time point in an individual not bearing this HLA allele, followed by full or partial reversion toward the predefined immunologically susceptible (usually the consensus) amino acid during follow-up.
Similarly, the proportion of overall amino acid substitutions attributable to HLA-associated selection pressures (escape and/or reversion) on a gene-wide basis was conservatively calculated as the fraction of the total observed substitutions that achieved an exact match to an HLA-associated polymorphism in the predefined list.
RESULTS
Rates of recognition and escape in CTL epitopes.
CTL escape mutations were defined according to a comprehensive list of known HLA-associated polymorphisms derived from combined cohorts numbering >1,200 (Heckerman et al., 15th International Workshop on HIV Dynamics and Evolution). Using this list, specific sites and pathways of HLA-driven substitutions were defined for 74 of the ∼180 published optimally defined epitopes in Gag, Pol, and Nef (Heckerman et al., 15th International Workshop on HIV Dynamics and Evolution). Of these, 71 were restricted by HLA alleles observed in the presently described seroconverter cohort (Fig. 1) and, thus, were evaluated for rates of escape in the first year of infection.
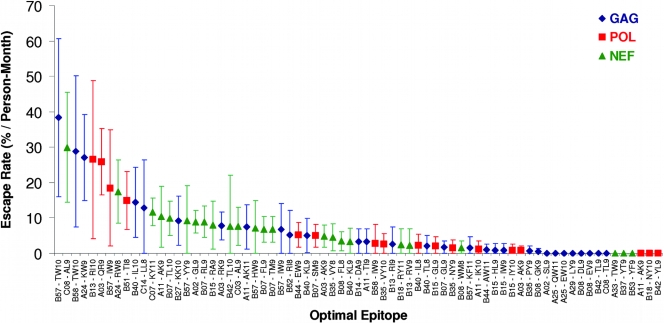
Epitope escape rates were characterized using Kaplan-Meier methods, where the time to escape was defined as the time elapsed between the estimated infection date and the first observation of a full or partial substitution that was consistent with the predefined list of HLA-associated polymorphisms. Evidence for escape was observed in 57 of 71 (80%) of these epitopes at the 1-year time point (Fig. 2). Escaping epitopes were relatively equally distributed across Gag, Pol, and Nef. Among evolving epitopes, escape was generally rapid: 42 (74%) of evolving epitopes escaped within the first 6 months of infection (an estimate that included cases where the specific HLA-associated polymorphism was already present at baseline in the context of the restricting allele).
FIG. 2.
First-year rates of escape among HLA-restricted, optimally defined CTL epitopes. The rates of escape in optimally defined CTL epitopes in Gag (blue diamonds), Pol (red squares), and Nef (green triangles) in the first year of infection are shown. Vertical bars indicate 95% confidence intervals. Epitopes were restricted to those containing a predefined HLA-associated polymorphism (Fig. 1). Note that this analysis incorporates an estimate of cases where the epitope may have escaped prior to baseline sampling, performed by analyzing each individual's baseline HIV sequence for known HLA-restricted escape mutations. Note that in doing so, we cannot discriminate cases of the very early selection of escape mutations from rare cases where an escaped variant may have been acquired at transmission.
The most rapidly escaping epitope was HLA-B*57 TW10 in p24 Gag (Fig. 2) (58). All evaluable B*57-expressing individuals in the cohort either already harbored the T242N and/or G248A escape mutations (18, 58) at baseline (n = 6) or selected one or both within the first 6 months (n = 1). Notably, T242 is a highly conserved residue in clade B, with over 90% of sequences in our clinically derived data set (n > 1,200) exhibiting the consensus T, arguing against the likelihood of this mutation having been transmitted. The single B*57-expressing individual in whom escape was not documented was lost to follow-up less than 2 months after infection. Thus, the crude first-year epitope escape rate for TW10 was calculated as 38%/person/month. Overall, first-year escape rates for all B*57-restricted epitopes were relatively high and could be ranked in the following order from most rapidly to slowest escaping: TW10-Gag (escape rate, 38%/month) > IW9-RT (18.5%/month) > YY9-Nef (9.2%/month) > HW9-Nef (7.2%/month) > IW9-Gag (6.7%/month) > KF11-Gag (1.6%/month) (Fig. 2 and 3). Note that QW9 (Gag) was not assigned an escape rate, as no B*57-associated polymorphisms within QW9 were predefined (Fig. 1). With the exception of KF11, all B*57-restricted epitopes examined ranked within the top half of the distribution of evolving CTL epitopes, consistently with the observation that the B*57-restricted CTL response features the broad, robust targeting of multiple epitopes in early infection (7, 76).
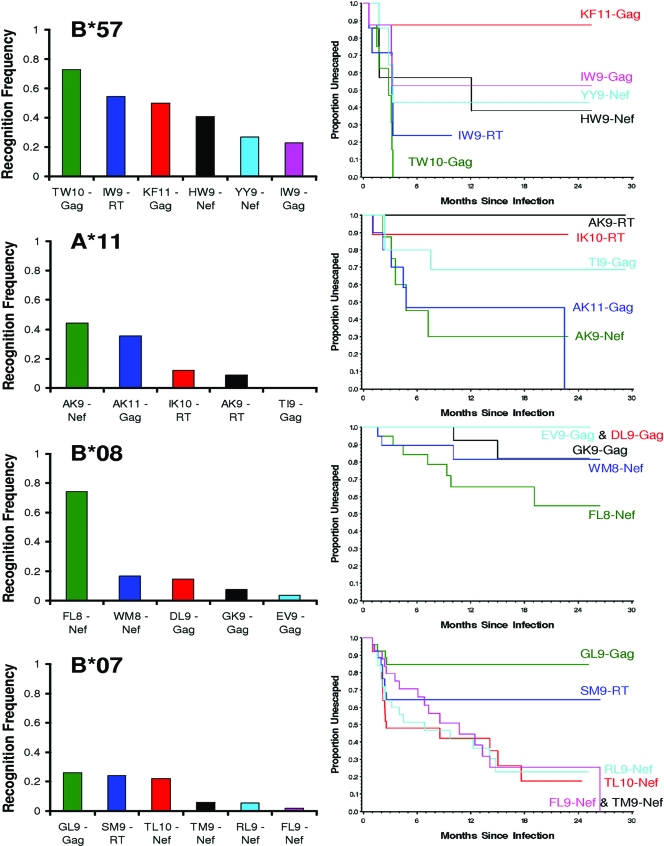
FIG. 3.
Rates of CTL escape generally reflect known immunodominance hierarchies of CTL epitope recognition in primary infection. The recognition frequencies of specific HLA-restricted epitopes (assessed by IFN-γ ELISpot assays measuring CTL responses to optimally described HIV peptides in a cohort of 289 individuals in primary HIV infection) (7, 76) are indicated in the left panels. Kaplan-Meier curves representing the corresponding rates of escape are indicated in the right panels in matching colors. Note that there are two cases where Kaplan-Meier curves for a pair of epitopes are superimposed so that only one curve is visible; these are B*08 EV9 and DL9 (which do not escape) and B*07 FL9 and TM9 (which overlap and share a single escaping site; Fig. 1).
Five of the 10 most rapidly evolving epitopes were restricted by protective HLA-B alleles, including B*57, B*58(01), B*13, and B*51 (21, 41) (P = 0.01 by Fisher's exact test). TW10 (Gag) also represented the most rapidly escaping epitope in B*58-expressing individuals (and the third most rapidly escaping epitope overall), while B*13-RI10 (Pol), B*57-IW9 (RT), and B*51-TI8 (RT) represented the fifth, seventh, and ninth most rapidly escaping epitopes, respectively (Fig. 2).
In order to characterize the relationship between the frequency of CTL responses and the corresponding frequencies of escape, we analyzed a data set of 289 individuals in primary infection screened for CTL responses against optimally defined epitopes using an IFN-γ ELISpot assay (7, 76). As expected, the rates of escape generally mirrored epitope-targeting frequencies (Fig. 3). For example, with the exception of KF11, the escape order for B*57-restricted epitopes generally reflected the established immunodominance hierarchies in primary infection (7), with TW10 representing the most frequently targeted (70%) and most rapidly escaping B*57-restricted epitope. HLA-A*11 and HLA-B*08 alleles also illustrate this relationship (Fig. 3). However, for some alleles, notably B*07, we observed a relatively poor relationship between the frequency of recognition and escape (Fig. 3).
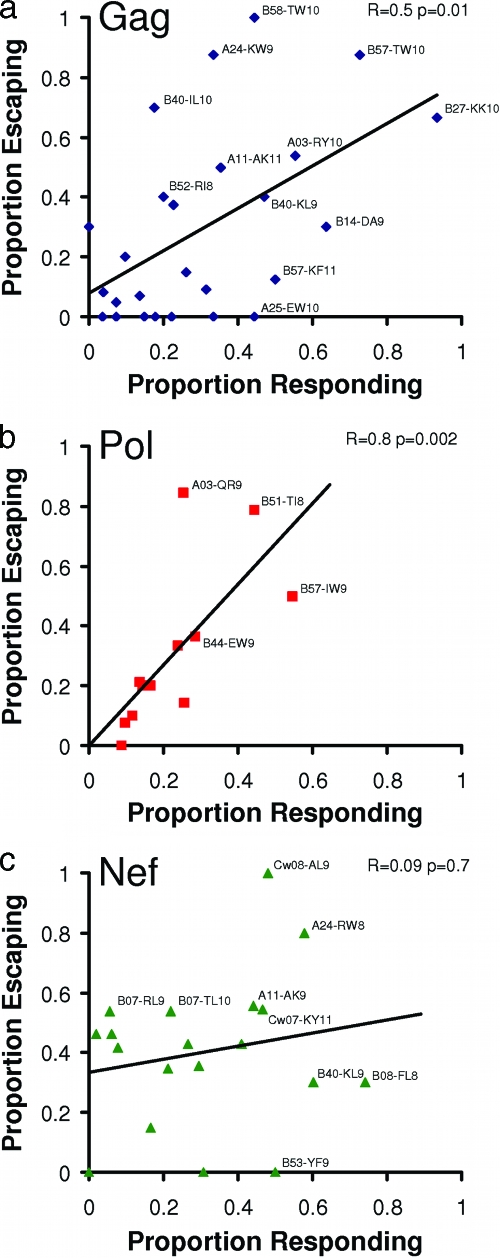
Overall, we observed a robust positive correlation between frequencies of epitope recognition and escape in Gag (Spearman's r = 0.5; P = 0.01) and Pol (r = 0.8; P = 0.002) epitopes, highlighting the ability of HIV to rapidly adapt to CTL-induced selection pressures, even in more conserved regions of the viral proteome (Fig. 4a, b). Of interest, B*27-KK10, the most frequently (90%) targeted epitope in early infection, escaped at a slower rate relative to other frequently targeted epitopes, consistently with previous reports (35, 39, 49). B*27-KK10 ranked as the 20th most rapidly evolving epitope overall, with 5 of 11 B*27-positive individuals selecting the L268M mutation during follow-up and an additional two exhibiting both R264K and L268M at the earliest time point (indicating either the transmission of these mutations or very early escape within this epitope [10]).
FIG. 4.
Correlation between frequencies of recognition and escape by protein. Spearman's rank correlation was used to characterize the relationship between the frequencies of recognition of CTL epitopes in Gag, Pol, and Nef with their corresponding frequencies of escape in the first year of infection. A regression line was drawn to highlight trends. All tested CTL epitopes are represented; those recognized and/or evolving at frequencies of >∼40% are labeled with the epitope name and HLA restriction.
Notably, we identified a small number of epitopes that exhibited little or no HLA-driven sequence evolution despite relatively high (>40%) frequencies of recognition during acute infection. These included the well-characterized B*57-KF11 epitope (23, 63), A*25-EW10 (Gag) (51), and others (Fig. 4). Of interest, the frequency of escape in A*02-SL9 (Gag) was zero, despite it representing one of the most immunodominant A*02-restricted responses in acute infection (with an 18% response frequency) (7) and large numbers of A*02-expressing persons to evaluate. In contrast to previous reports identifying escape mutations at codons 3,6, and/or 8 of this epitope (29, 37, 43), in our predefined list of HLA-associated polymorphisms, mutations at these codons were attributed to other HLA-restricted epitopes that overlapped SL9 (A*29 and Cw*14) (Fig. 1). The only A*02-associated polymorphism preidentified in SL9 occurred at position 7 (Gag codon 83) (Fig. 1). No substitutions at this position were observed in any A*02-expressing seroconverter during study follow-up, resulting in an escape rate of zero.
We observed a number of cases where epitope escape frequencies exceeded recognition frequencies in Gag and Pol (for example, A*24-KW9 [Gag] and A*03-QR9 [RT], the second and sixth most rapidly evolving epitopes overall, as well as a number of B*07-restricted epitopes; Fig. 4). This type of discordance, however, was observed most frequently in Nef and contributed to the poor correlation between recognition and escape frequencies in this protein (r = 0.09; P = 0.7) (Fig. 4c).
Rates of reversion in CTL epitopes.
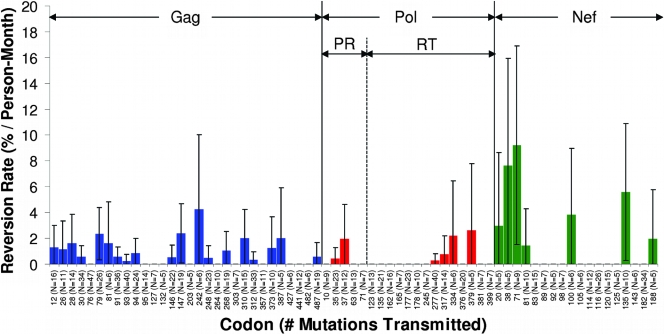
Reversion was conservatively defined as the presence of a known HLA-associated polymorphism at the baseline time point in an individual not bearing this HLA allele, followed by full or partial reversion toward the immunologically susceptible (usually the consensus) amino acid during follow-up. Within the first year of infection, reversions were observed at 29 (6%), 14 (3%), and 26 (13%) Gag, Pol and Nef codons, respectively, while an additional 9, 7, and 10 sites reverted after the first year. The majority of reverting codons (65%) were within published HLA-restricted CTL epitopes. On average, reversions within Gag epitopes restricted by protective HLA alleles (B*13, B*51, B*57, and B*58) occurred more rapidly than reversions outside these regions (P = 0.02); however, no such differences were observed in Pol or Nef. The rates of reversion at known escape sites within optimally described epitopes in the first year of infection are summarized in Fig. 5. The most rapidly reverting mutations within Gag CTL epitopes were B*57-associated escape mutations at codons 147 and 242 in the IW9 and TW10 epitopes, respectively, while the most rapidly reverting epitope-associated mutation within Nef was at codon 135 (residue 2 of A*24-RW8). Although it is important to emphasize that the rates of escape and reversion are not directly comparable due to the differences in calculations (see Materials and Methods and Discussion), it is nevertheless interesting that these three mutations were relatively rapidly escaping as well, implying a substantial fitness cost. However, the factors that shape HIV evolution are complex and, thus, one would not expect rates of escape to correlate with rates of reversion in all cases. RT codon 135 (residue 8 of B*51-TI8), for example, was not observed to revert despite the relatively frequent transmission of the escaped variant (n = 21), implying a negligible fitness cost to this mutation.
FIG. 5.
Locations and first-year rates of commonly observed reversions within published HLA-restricted epitopes in Gag, Pol, and Nef. Conservative estimates of the rates of reversion of known HLA-associated escape mutations in the absence of the restricting HLA (expressed as percent reversions/person-month) at Gag, Pol, and Nef codons within published HLA-restricted CTL epitopes are shown, along with the total number of observations (transmitted mutations) at each codon. A minimum of five observed cases of the transmission of the escaped variant was required for display. Note that in contrast to the escape analysis, this analysis does not take into consideration reversions that may have occurred prior to the baseline sampling. Thus, estimated reversion rates represent considerable underestimates of the true rate and are not directly comparable to epitope escape rates. PR, protease.
Proportion of HIV evolution attributable to HLA-mediated selection pressures.
The proportion of overall evolution in Gag, Pol, and Nef, within and outside published epitopes, was calculated by computing the proportion of the total observed amino acid changes between the baseline sequence and the final sequence matching an HLA-associated polymorphism on the predefined list. Means of six, four, and seven nonsynonymous substitutions per person were observed in Gag, protease/RT, and Nef, respectively, corresponding to overall nonsynonymous substitution rates of 0.01, 0.008, and 0.03 substitutions/subject/codon. A total of 36, 32, and 58% of observed substitutions in Gag, Pol, and Nef were attributable to HLA-associated selection pressures (P < 0.0001); overall, reversions accounted for ∼70% of these observations.
DISCUSSION
The resolution of acute-phase viremia is influenced by the strength and repertoire of the cellular immune response and the speed and efficiency by which the virus is able to adapt to these responses. Previous studies have demonstrated substantial viral evolution in acute infection (2, 9, 59); however, in general, most longitudinal studies of HLA-mediated viral evolution have been limited to small numbers of patients, have been largely biased toward protective HLA alleles associated with long-term viremic control, and have lacked a standardized classification scheme for what constitutes an HLA-attributable mutation.
The current study overcomes many of the limitations of previous investigations in this area, as it represents the first large-scale study of HLA-driven HIV evolution in acute clade B infection that employs a rational list of HLA-associated polymorphisms (defined through the analysis of a large [n > 1,200] independent data set) to systematically characterize the rates of immune-driven evolution on a protein-wide basis in the context of all HLA restrictions. The study focused on Gag, Pol, and Nef, because these proteins have been featured in candidate CTL vaccine design strategies, including the recent failed STEP vaccine trial (74); thus, developing a more in-depth understanding of CTL-driven evolution in these proteins is of key importance. We estimate that a minimum of 30 to 35% of nonsynonymous substitutions in Gag/Pol and a minimum of 60% of substitutions in Nef are attributable to HLA-restricted immune pressures in the first year of infection (of which the majority represent reversions), confirming dramatically different levels of HLA-mediated adaptation in HIV proteins (17).
Using Kaplan-Meier analysis, we systematically computed the rates of escape in published CTL epitopes, revealing markedly different kinetics of escape among them. Although we were not able to evaluate CTL responses in this cohort due to a lack of cryopreserved cells, we were able to integrate the sequence data derived here with epitope recognition frequencies characterized in a cohort of 289 persons with primary HIV infection (7, 76). A robust correlation between the epitope response frequencies and the rates of escape in Gag and Pol was observed, illustrating that in general, if the immune response mounts pressure against an epitope, in most cases HLA-driven mutations will appear shortly thereafter. There were relatively few examples of frequently targeted epitopes that maintained their sequences for extended periods of time without evidence of escape.
Of interest, protective HLA alleles (21, 41, 47) were overrepresented among rapidly escaping epitopes, consistent with the observation that these alleles impose stronger in vivo immune selection pressures than others (33, 72). In particular, B*57, the strongest host genetic factor associated with protection against HIV progression (21), restricted the most rapidly escaping epitope overall (TW10), and all but one of the remaining B*57-restricted epitopes displayed rapid escape as well. The somewhat paradoxical observation that certain alleles remain protective despite rapid and frequent escape may be explained by the fact that a key correlate of protection is the ability to mount a broad and robust CTL response very early in infection (4, 64), which indirectly manifests itself as CTL-driven sequence changes at these key sites. On average, B*57-expressing individuals respond to a mean of 4.7 B*57-restricted epitopes (genome-wide) in early infection, the highest number for all HLA restrictions, compared to a mean of two or three epitopes for the vast majority of other alleles (7, 76). From the perspective of HLA imprinting on HIV sequences at the population level, HLA-B*57 represents one of the alleles with the largest total number of identified HLA-associated polymorphic sites in Gag/Pol/Nef, with associations identified at 29 unique residues in these proteins (compared to 17 and 18 for B*07 and B*08, respectively) (Heckerman et al., 15th International Workshop on HIV Dynamics and Evolution). Furthermore, the earliest CTL responses likely are the most potent and possess the greatest antiviral potential; in no other stage of disease does one observe such a dramatic decline in viremia from the acute peak to set-point levels (19).
A number of additional reasons may explain why certain alleles remain protective despite the rapid selection of HLA-driven substitutions. First, not all of the identified HLA-associated polymorphisms confer an absolute escape phenotype: often, the variant will maintain at least some capacity to bind HLA and/or be cross-recognized by CTL (77), and there is evidence that B*57-restricted CTL have a superior ability to cross-recognize peptide variants compared to that of less protective alleles (77). Furthermore, the ability to mount a de novo CTL response against a selected variant (3, 30, 78) may be greater in earlier disease while immune function is relatively intact. Finally, recent data suggest that the breadth of CTL responses to Gag contributes strongly to HIV control (1, 28, 50, 79), and that the fitness costs of escape mutations in Gag are substantial (16, 23, 62, 73), suggesting that long-term protective effects are due to strong immune selective pressures driving viral evolution toward less-fit forms (6). Indeed, the observed rapid and frequent reversion of escape mutations within B*57-TW10 and IW9 in Gag supports this hypothesis (Fig. 4). Taken together, these results support the importance of a strong early CTL response in the control of HIV viremia, even if a consequence of this is the rapid selection of mutations.
The observation of discordant cases where in vivo escape frequencies exceeded in vitro recognition frequencies merits discussion. The fact that epitope recognition (7, 76) and escape were evaluated in independent cohorts accounts for a portion of these discordances (escape frequencies in the seroconverter cohort represent cumulative longitudinal frequencies calculated at 1 year postinfection, whereas recognition frequencies represent measurements taken at a single time point between 3 and 8 months following the estimated date of infection). The remainder may be attributed to an underestimation of CTL responses and/or an overestimation of escape frequencies, as follows. First, as CTL responses to wild-type peptides decline following escape in the autologous viral sequence (2), the recognition frequencies of rapidly escaping epitopes could have been underestimated if subjects were screened after escape had occurred. Indeed, for the eight seroconverters for whom longitudinal peripheral blood mononuclear cells were available, we observed a decline in responses to B*51 TI8-RT following escape in both B*51-expressing individuals in this subgroup (not shown). Similarly, CTL responses may be underestimated in cases where the autologous viral sequence differs from the tested epitope sequence (5, 62), a fact that would more substantially affect variable proteins such as Nef.
Another contributing factor may be the overestimation of escape. This is particularly an issue in cases where the identified escape variant for a given epitope represents a commonly occurring (or in some cases, the consensus) residue. Both A*24 KW9-Gag and A*03 QR9-RT are examples of such cases: for both epitopes, the immunologically susceptible form (position 1 of KW9 and position 9 of QR9, corresponding to Gag and RT codons 28 and 277, respectively) represents a nonconsensus amino acid, meaning that the subtype B consensus residue is considered the escaped form. The fact that our analysis (Fig. 2 to 4) included subjects exhibiting an escaped residue at baseline could have resulted in an overestimation of escape for these epitopes if these mutations were present at transmission. Finally, it is worth noting that in both cases (as well as for all B*07-restricted epitopes examined here), the published epitope sequence features at least one residue in its HLA-associated escaped form. Work is ongoing to assess whether these cases represent examples where the population HIV consensus has adapted to frequently observed HLA alleles (57); in any event, this observation merits consideration, as it could result in the systematic underestimation of CTL response frequencies to such epitopes.
There are a number of additional limitations that are important to discuss. Most importantly, in the absence of knowing the transmitted sequence, we cannot definitively identify sequence changes occurring prior to baseline sampling. Although the rates of escape analysis take into consideration likely escape events occurring prior to baseline sampling, it is not possible to infer reversion events (or to calculate the total amount of evolution) occurring prior to baseline sampling. Thus, not only does this limitation render the reversion rates to be substantial underestimates of the true rates, it also renders them not directly comparable to the escape rates. Nevertheless, the analysis still yields informative data regarding the locations, frequencies, and estimated rates of reversion, which illuminate potential positions at which mutations likely occur at substantial costs to fitness. For example, it was interesting that two of the most rapidly reverting codons were Gag 147 and 242 within B*57-TW10 and IW9 epitopes, respectively (39, 58), consistent with measurable costs to viral fitness associated with escape in B*57-restricted Gag epitopes (16, 62). Conversely, the rapidly selected B*51-associated escape mutation at RT codon 135 (residue 8 of B*51-TI8) is not observed to revert despite relatively frequent transmission, suggesting a relatively minor fitness cost.
If the rates of reversion and/or escape differ substantially before and after baseline sampling, this difference could contribute to errors in our estimates. Note, however, that no significant difference in the proportion of HLA-attributable evolution was observed among individuals captured within <3 (n = 61) or >3 to 6 months (n = 37) after infection. The inconsistent detection of minority variants below a threshold of ∼10 to 20% of the circulating species is a known limitation of the bulk PCR and sequencing techniques employed (55, 56); however, comprehensive clonal sequencing was not feasible in a cohort of this size and length of follow-up.
In addition, there are some limitations associated with defining HLA-associated substitutions through the analysis of an independent large clade B data set. As with any method, there will be both false-positive as well as false-negative results within this list (20) (J. Carlson, submitted for publication); thus, this list will not necessarily contain all escape mutations previously reported in the literature. Potential differences in the ethnic composition in the two cohorts also should be acknowledged. Information on ethnicity was unavailable; however, the HLA composition of both cohorts reflected expected allele frequencies among North American populations (not shown). Finally, although many mutational pathways are broadly predictable at the population level (17, 18, 65, 70), unique HLA-driven escape, reversion, or secondary/compensatory changes also will occur in individual patients; thus, employing a population-level definition of HLA-associated substitution at the individual level may underestimate the amount of evolution attributable to HLA. Nevertheless, the use of a predefined list of HLA-associated substitutions allows the comprehensive, systematic classification of amino acid substitutions both within and outside CTL epitopes, as well as the ability to investigate escape and reversion across entire proteins and across all HLA restrictions. Indeed, a similar classification technique was employed in a recent study of transmitted CTL escape mutations in HIV clade C (36). The current study investigated HLA-associated polymorphisms in Gag, Pol, and Nef only; however, as comprehensive lists of HLA-associated polymorphisms become available for additional genes, the analysis of escape and reversion rates in other HIV proteins will become possible. Finally, it is important to acknowledge the incompleteness as well as potential bias toward common and/or protective HLA alleles in the current published CTL epitope lists. As new epitopes are discovered, however, the rates of escape and reversion can easily be calculated using the current data set.
The relatively short follow-up period limits our ability to assess the long-term impact of early escape on HIV disease outcomes. In addition, we were unable to confirm the results of recent studies that demonstrate reduced viral loads in individuals transmitting HLA-associated escape mutations (22, 36); however, the lack of statistical power to detect these effects in the current study must be noted.
Given these limitations, our estimates that ∼30 to 35% and ∼60% of overall substitutions in Gag/Pol and Nef, respectively, are attributable to CTL escape and reversion confirm a substantial role of HLA-associated immune pressures in driving early within-host HIV evolution (2, 9, 39, 59). Selective forces responsible for the non-HLA-attributable fraction may include CD4+ T-cell responses (48, 71), adaptations to other host factors (12), reversions of such mutations selected in previous hosts, or random drift. A small number of substitutions in protease/RT (and possibly Gag (26) could be due to the reversion of transmitted resistance mutations. Indeed, a small number of resistance-associated polymorphisms and surveillance mutations (54) were observed in this cohort; note, however, that there was no overlap between major resistance-associated and HLA-associated polymorphic sites in protease/RT.
Substantial variability in the degree to which different viral proteins adapt to HLA-associated immune pressures during early HIV infection underscores the importance of selecting appropriate immunogens and determining how to best incorporate sequence diversity in vaccine design (5, 15, 32, 66, 69). In particular, strongly targeted yet slowly escaping epitopes (such as B*57-KF11 and A*25-EW10) may be of particular interest, as may be strongly targeted epitopes that can escape only at substantial fitness costs to the virus (such as B*57-TW10) (16). Specifically, immunogenic viral regions exhibiting high mutational barriers to escape may be good vaccine candidates, as they may mediate the effective long-term control of HIV CTL (39). On the other hand, epitopes that escape at considerable costs to viral replicative capacity also may be relevant to vaccine design: a vaccine capable of inducing immune responses that drive HIV toward crippled forms also may be effective in reducing viral replication to levels that slow disease progression (6). Indeed, the observation that HLA-B*57-restricted TW10 and IW9 are among the fastest escaping and the fastest reverting epitopes in Gag strongly supports the fitness costs of CTL escape as a key mediator of long-term viremia control in B*57-expressing individuals.
Taken together, our results confirm a substantial role of HLA-associated selection pressures on early within-host HIV evolution and support further research into CTL-based vaccine strategies incorporating information on common escape pathways, despite recent setbacks in the HIV vaccine field (8, 74).
Acknowledgments
We thank the patients for their participation.
We thank Toshiyuki Miura, Mark Brockman, Nicole Frahm, and Christian Brander for helpful discussions and Philip Goulder for the critical reading of the manuscript.
Z.L.B. is supported by a postdoctoral fellowship from the Canadian Institutes for Health Research (CIHR). H.S. is supported by a fellowship from the Deutscher Akademischer Austauschdienst (DAAD). D.H., J.C., and C.K. are funded by Microsoft Research. This study was supported in part by the Howard Hughes Medical Institute, the Mark and Lisa Schwartz Foundation, the Harvard University Center for AIDS Research (CFAR), National Institutes of Health grant R01AI50429, and Acute Infection Early Disease Research Network (AIEDRP) grant U01AI052403. This project has been funded in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract N01-CO-12400. This research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Sources of support were not involved in the design and conduct of the study, nor were they involved in the collection, analysis, and interpretation of the data or in the preparation, review, or approval of the manuscript.
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does the mention of trade names, commercial products, or organizations imply endorsement by the U.S. government.
Footnotes
Published ahead of print on 9 July 2008.
REFERENCES
- 1.Addo, M. M., X. G. Yu, A. Rathod, D. Cohen, R. L. Eldridge, D. Strick, M. N. Johnston, C. Corcoran, A. G. Wurcel, C. A. Fitzpatrick, M. E. Feeney, W. R. Rodriguez, N. Basgoz, R. Draenert, D. R. Stone, C. Brander, P. J. Goulder, E. S. Rosenberg, M. Altfeld, and B. D. Walker. 2003. Comprehensive epitope analysis of human immunodeficiency virus type 1 (HIV-1)-specific T-cell responses directed against the entire expressed HIV-1 genome demonstrate broadly directed responses, but no correlation to viral load. J. Virol. 772081-2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allen, T. M., M. Altfeld, S. C. Geer, E. T. Kalife, C. Moore, M. O'Sullivan, K. I. Desouza, M. E. Feeney, R. L. Eldridge, E. L. Maier, D. E. Kaufmann, M. P. Lahaie, L. Reyor, G. Tanzi, M. N. Johnston, C. Brander, R. Draenert, J. K. Rockstroh, H. Jessen, E. S. Rosenberg, S. A. Mallal, and B. D. Walker. 2005. Selective escape from CD8+ T-cell responses represents a major driving force of human immunodeficiency virus type 1 (HIV-1) sequence diversity and reveals constraints on HIV-1 evolution. J. Virol. 7913239-13249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allen, T. M., X. G. Yu, E. T. Kalife, L. L. Reyor, M. Lichterfeld, M. John, M. Cheng, R. L. Allgaier, S. Mui, N. Frahm, G. Alter, N. V. Brown, M. N. Johnston, E. S. Rosenberg, S. A. Mallal, C. Brander, B. D. Walker, and M. Altfeld. 2005. De novo generation of escape variant-specific CD8+ T-cell responses following cytotoxic T-lymphocyte escape in chronic human immunodeficiency virus type 1 infection. J. Virol. 7912952-12960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Altfeld, M., M. M. Addo, E. S. Rosenberg, F. M. Hecht, P. K. Lee, M. Vogel, X. G. Yu, R. Draenert, M. N. Johnston, D. Strick, T. M. Allen, M. E. Feeney, J. O. Kahn, R. P. Sekaly, J. A. Levy, J. K. Rockstroh, P. J. Goulder, and B. D. Walker. 2003. Influence of HLA-B57 on clinical presentation and viral control during acute HIV-1 infection. AIDS 172581-2591. [DOI] [PubMed] [Google Scholar]
- 5.Altfeld, M., M. M. Addo, R. Shankarappa, P. K. Lee, T. M. Allen, X. G. Yu, A. Rathod, J. Harlow, K. O'Sullivan, M. N. Johnston, P. J. Goulder, J. I. Mullins, E. S. Rosenberg, C. Brander, B. Korber, and B. D. Walker. 2003. Enhanced detection of human immunodeficiency virus type 1-specific T-cell responses to highly variable regions by using peptides based on autologous virus sequences. J. Virol. 777330-7340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Altfeld, M., and T. M. Allen. 2006. Hitting HIV where it hurts: an alternative approach to HIV vaccine design. Trends Immunol. 27504-510. [DOI] [PubMed] [Google Scholar]
- 7.Altfeld, M., E. T. Kalife, Y. Qi, H. Streeck, M. Lichterfeld, M. N. Johnston, N. Burgett, M. E. Swartz, A. Yang, G. Alter, X. G. Yu, A. Meier, J. K. Rockstroh, T. M. Allen, H. Jessen, E. S. Rosenberg, M. Carrington, and B. D. Walker. 2006. HLA alleles associated with delayed progression to AIDS contribute strongly to the initial CD8+ T cell response against HIV-1. PLoS Med. 3e403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anonymous. 2007. HIV vaccine failure prompts Merck to halt trial. Nature 449390. [DOI] [PubMed] [Google Scholar]
- 9.Bernardin, F., D. Kong, L. Peddada, L. A. Baxter-Lowe, and E. Delwart. 2005. Human immunodeficiency virus mutations during the first month of infection are preferentially found in known cytotoxic T-lymphocyte epitopes. J. Virol. 7911523-11528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Betts, M. R., B. Exley, D. A. Price, A. Bansal, Z. T. Camacho, V. Teaberry, S. M. West, D. R. Ambrozak, G. Tomaras, M. Roederer, J. M. Kilby, J. Tartaglia, R. Belshe, F. Gao, D. C. Douek, K. J. Weinhold, R. A. Koup, P. Goepfert, and G. Ferrari. 2005. Characterization of functional and phenotypic changes in anti-Gag vaccine-induced T cell responses and their role in protection after HIV-1 infection. Proc. Natl. Acad. Sci. USA 1024512-4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bhattacharya, T., M. Daniels, D. Heckerman, B. Foley, N. Frahm, C. Kadie, J. Carlson, K. Yusim, B. McMahon, B. Gaschen, S. Mallal, J. I. Mullins, D. C. Nickle, J. Herbeck, C. Rousseau, G. H. Learn, T. Miura, C. Brander, B. Walker, and B. Korber. 2007. Founder effects in the assessment of HIV polymorphisms and HLA allele associations. Science 3151583-1586. [DOI] [PubMed] [Google Scholar]
- 12.Bishop, K. N., R. K. Holmes, A. M. Sheehy, and M. H. Malim. 2004. APOBEC-mediated editing of viral RNA. Science 305645. [DOI] [PubMed] [Google Scholar]
- 13.Borrow, P., H. Lewicki, B. H. Hahn, G. M. Shaw, and M. B. Oldstone. 1994. Virus-specific CD8+ cytotoxic T-lymphocyte activity associated with control of viremia in primary human immunodeficiency virus type 1 infection. J. Virol. 686103-6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Borrow, P., H. Lewicki, X. Wei, M. S. Horwitz, N. Peffer, H. Meyers, J. A. Nelson, J. E. Gairin, B. H. Hahn, M. B. Oldstone, and G. M. Shaw. 1997. Antiviral pressure exerted by HIV-1-specific cytotoxic T lymphocytes (CTLs) during primary infection demonstrated by rapid selection of CTL escape virus. Nat. Med. 3205-211. [DOI] [PubMed] [Google Scholar]
- 15.Brander, C., N. Frahm, and B. D. Walker. 2006. The challenges of host and viral diversity in HIV vaccine design. Curr. Opin. Immunol. 18430-437. [DOI] [PubMed] [Google Scholar]
- 16.Brockman, M. A., A. Schneidewind, M. Lahaie, A. Schmidt, T. Miura, I. Desouza, F. Ryvkin, C. A. Derdeyn, S. Allen, E. Hunter, J. Mulenga, P. A. Goepfert, B. D. Walker, and T. M. Allen. 2007. Escape and compensation from early HLA-B57-mediated cytotoxic T-lymphocyte pressure on human immunodeficiency virus type 1 Gag alter capsid interactions with cyclophilin A. J. Virol. 8112608-12618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brumme, Z. L., C. J. Brumme, D. Heckerman, B. T. Korber, M. Daniels, J. Carlson, C. Kadie, T. Bhattacharya, C. Chui, J. Szinger, T. Mo, R. S. Hogg, J. S. Montaner, N. Frahm, C. Brander, B. D. Walker, and P. R. Harrigan. 2007. Evidence of differential HLA class I-mediated viral evolution in functional and accessory/regulatory genes of HIV-1. PLoS Pathog. 3e94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brumme, Z. L., I. Tao, S. Szeto, C. J. Brumme, J. M. Carlson, D. Chan, C. Kadie, N. Frahm, C. Brander, B. D. Walker, D. Heckerman, and P. R. Harrigan. 2008. HLA-specific polymorphisms in HIV-1 Gag and their association with viral load in chronic untreated infection. AIDS 221277-1286. [DOI] [PubMed] [Google Scholar]
- 19.Cao, J., J. McNevin, U. Malhotra, and M. J. McElrath. 2003. Evolution of CD8+ T cell immunity and viral escape following acute HIV-1 infection. J. Immunol. 1713837-3846. [DOI] [PubMed] [Google Scholar]
- 20.Carlson, J., C. Kadie, S. Mallal, and D. Heckerman. 2007. Leveraging hierarchical population structure in discrete association studies. PLoS ONE 2e591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carrington, M., and S. J. O'Brien. 2003. The influence of HLA genotype on AIDS. Annu. Rev. Med. 54535-551. [DOI] [PubMed] [Google Scholar]
- 22.Chopera, D. R., Z. Woodman, K. Mlisana, M. Mlotshwa, D. P. Martin, C. Seoighe, F. Treurnicht, D. A. de Rosa, W. Hide, S. A. Karim, C. M. Gray, and C. Williamson. 2008. Transmission of HIV-1 CTL escape variants provides HLA-mismatched recipients with a survival advantage. PLoS Pathog. 4e1000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crawford, H., J. G. Prado, A. Leslie, S. Hue, I. Honeyborne, S. Reddy, M. van der Stok, Z. Mncube, C. Brander, C. Rousseau, J. I. Mullins, R. Kaslow, P. Goepfert, S. Allen, E. Hunter, J. Mulenga, P. Kiepiela, B. D. Walker, and P. J. Goulder. 2007. Compensatory mutation partially restores fitness and delays reversion of escape mutation within the immunodominant HLA-B*5703-restricted Gag epitope in chronic human immunodeficiency virus type 1 infection. J. Virol. 818346-8351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Derdeyn, C. A., and G. Silvestri. 2005. Viral and host factors in the pathogenesis of HIV infection. Curr. Opin. Immunol. 17366-373. [DOI] [PubMed] [Google Scholar]
- 25.Douek, D. C., L. J. Picker, and R. A. Koup. 2003. T cell dynamics in HIV-1 infection. Annu. Rev. Immunol. 21265-304. [DOI] [PubMed] [Google Scholar]
- 26.Doyon, L., G. Croteau, D. Thibeault, F. Poulin, L. Pilote, and D. Lamarre. 1996. Second locus involved in human immunodeficiency virus type 1 resistance to protease inhibitors. J. Virol. 703763-3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Draenert, R., T. M. Allen, Y. Liu, T. Wrin, C. Chappey, C. L. Verrill, G. Sirera, R. L. Eldridge, M. P. Lahaie, L. Ruiz, B. Clotet, C. J. Petropoulos, B. D. Walker, and J. Martinez-Picado. 2006. Constraints on HIV-1 evolution and immunodominance revealed in monozygotic adult twins infected with the same virus. J. Exp. Med. 203529-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Edwards, B. H., A. Bansal, S. Sabbaj, J. Bakari, M. J. Mulligan, and P. A. Goepfert. 2002. Magnitude of functional CD8+ T-cell responses to the gag protein of human immunodeficiency virus type 1 correlates inversely with viral load in plasma. J. Virol. 762298-2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Edwards, C. T., K. J. Pfafferott, P. J. Goulder, R. E. Phillips, and E. C. Holmes. 2005. Intrapatient escape in the A*0201-restricted epitope SLYNTVATL drives evolution of human immunodeficiency virus type 1 at the population level. J. Virol. 799363-9366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feeney, M. E., Y. Tang, K. Pfafferott, K. A. Roosevelt, R. Draenert, A. Trocha, X. G. Yu, C. Verrill, T. Allen, C. Moore, S. Mallal, S. Burchett, K. McIntosh, S. I. Pelton, M. A. St. John, R. Hazra, P. Klenerman, M. Altfeld, B. D. Walker, and P. J. Goulder. 2005. HIV-1 viral escape in infancy followed by emergence of a variant-specific CTL response. J. Immunol. 1747524-7530. [DOI] [PubMed] [Google Scholar]
- 31.Fiebig, E. W., D. J. Wright, B. D. Rawal, P. E. Garrett, R. T. Schumacher, L. Peddada, C. Heldebrant, R. Smith, A. Conrad, S. H. Kleinman, and M. P. Busch. 2003. Dynamics of HIV viremia and antibody seroconversion in plasma donors: implications for diagnosis and staging of primary HIV infection. AIDS 171871-1879. [DOI] [PubMed] [Google Scholar]
- 32.Fischer, W., S. Perkins, J. Theiler, T. Bhattacharya, K. Yusim, R. Funkhouser, C. Kuiken, B. Haynes, N. L. Letvin, B. D. Walker, B. H. Hahn, and B. T. Korber. 2007. Polyvalent vaccines for optimal coverage of potential T-cell epitopes in global HIV-1 variants. Nat. Med. 13100-106. [DOI] [PubMed] [Google Scholar]
- 33.Frater, A. J., H. Brown, A. Oxenius, H. F. Gunthard, B. Hirschel, N. Robinson, A. J. Leslie, R. Payne, H. Crawford, A. Prendergast, C. Brander, P. Kiepiela, B. D. Walker, P. J. Goulder, A. McLean, and R. E. Phillips. 2007. Effective T-cell responses select human immunodeficiency virus mutants and slow disease progression. J. Virol. 816742-6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Friedrich, T. C., E. J. Dodds, L. J. Yant, L. Vojnov, R. Rudersdorf, C. Cullen, D. T. Evans, R. C. Desrosiers, B. R. Mothe, J. Sidney, A. Sette, K. Kunstman, S. Wolinsky, M. Piatak, J. Lifson, A. L. Hughes, N. Wilson, D. H. O'Connor, and D. I. Watkins. 2004. Reversion of CTL escape-variant immunodeficiency viruses in vivo. Nat. Med. 10275-281. [DOI] [PubMed] [Google Scholar]
- 35.Gao, X., A. Bashirova, A. K. Iversen, J. Phair, J. J. Goedert, S. Buchbinder, K. Hoots, D. Vlahov, M. Altfeld, S. J. O'Brien, and M. Carrington. 2005. AIDS restriction HLA allotypes target distinct intervals of HIV-1 pathogenesis. Nat. Med. 111290-1292. [DOI] [PubMed] [Google Scholar]
- 36.Goepfert, P. A., W. Lumm, P. Farmer, P. Matthews, A. Prendergast, J. M. Carlson, C. A. Derdeyn, J. Tang, R. A. Kaslow, A. Bansal, K. Yusim, D. Heckerman, J. Mulenga, S. Allen, P. J. Goulder, and E. Hunter. 2008. Transmission of HIV-1 Gag immune escape mutations is associated with reduced viral load in linked recipients. J. Exp. Med. 2051009-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goulder, P. J., M. A. Altfeld, E. S. Rosenberg, T. Nguyen, Y. Tang, R. L. Eldridge, M. M. Addo, S. He, J. S. Mukherjee, M. N. Phillips, M. Bunce, S. A. Kalams, R. P. Sekaly, B. D. Walker, and C. Brander. 2001. Substantial differences in specificity of HIV-specific cytotoxic T cells in acute and chronic HIV infection. J. Exp. Med. 193181-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goulder, P. J., R. E. Phillips, R. A. Colbert, S. McAdam, G. Ogg, M. A. Nowak, P. Giangrande, G. Luzzi, B. Morgan, A. Edwards, A. J. McMichael, and S. Rowland-Jones. 1997. Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nat. Med. 3212-217. [DOI] [PubMed] [Google Scholar]
- 39.Goulder, P. J., and D. I. Watkins. 2004. HIV and SIV CTL escape: implications for vaccine design. Nat. Rev. Immunol. 4630-640. [DOI] [PubMed] [Google Scholar]
- 40.Reference deleted.
- 41.Honeyborne, I., A. Prendergast, F. Pereyra, A. Leslie, H. Crawford, R. Payne, S. Reddy, K. Bishop, E. Moodley, K. Nair, M. van der Stok, N. McCarthy, C. M. Rousseau, M. Addo, J. I. Mullins, C. Brander, P. Kiepiela, B. D. Walker, and P. J. Goulder. 2007. Control of human immunodeficiency virus type 1 is associated with HLA-B*13 and targeting of multiple gag-specific CD8+ T-cell epitopes. J. Virol. 813667-3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang, X., and J. Zhang. 1996. Methods for comparing a DNA sequence with a protein sequence. Comput. Appl. Biosci. 12497-506. [DOI] [PubMed] [Google Scholar]
- 43.Iversen, A. K., G. Stewart-Jones, G. H. Learn, N. Christie, C. Sylvester-Hviid, A. E. Armitage, R. Kaul, T. Beattie, J. K. Lee, Y. Li, P. Chotiyarnwong, T. Dong, X. Xu, M. A. Luscher, K. MacDonald, H. Ullum, B. Klarlund-Pedersen, P. Skinhoj, L. Fugger, S. Buus, J. I. Mullins, E. Y. Jones, P. A. van der Merwe, and A. J. McMichael. 2006. Conflicting selective forces affect T cell receptor contacts in an immunodominant human immunodeficiency virus epitope. Nat. Immunol. 7179-189. [DOI] [PubMed] [Google Scholar]
- 44.Janssen, R. S., G. A. Satten, S. L. Stramer, B. D. Rawal, T. R. O'Brien, B. J. Weiblen, F. M. Hecht, N. Jack, F. R. Cleghorn, J. O. Kahn, M. A. Chesney, and M. P. Busch. 1998. New testing strategy to detect early HIV-1 infection for use in incidence estimates and for clinical and prevention purposes. JAMA 28042-48. [DOI] [PubMed] [Google Scholar]
- 45.John, M., D. Heckerman, L. Park, S. Gaudieri, A. Chopra, J. Carlson, I. James, D. Nolan, D. Haubrich, S. Mallal, and A. S. Team. 2008. Abstr. 15th Conf. Retrovir. Opportun. Infect., abstr. 312.
- 46.Jones, N. A., X. Wei, D. R. Flower, M. Wong, F. Michor, M. S. Saag, B. H. Hahn, M. A. Nowak, G. M. Shaw, and P. Borrow. 2004. Determinants of human immunodeficiency virus type 1 escape from the primary CD8+ cytotoxic T lymphocyte response. J. Exp. Med. 2001243-1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kaslow, R. A., M. Carrington, R. Apple, L. Park, A. Munoz, A. J. Saah, J. J. Goedert, C. Winkler, S. J. O'Brien, C. Rinaldo, R. Detels, W. Blattner, J. Phair, H. Erlich, and D. L. Mann. 1996. Influence of combinations of human major histocompatibility complex genes on the course of HIV-1 infection. Nat. Med. 2405-411. [DOI] [PubMed] [Google Scholar]
- 48.Kaufmann, D. E., P. M. Bailey, J. Sidney, B. Wagner, P. J. Norris, M. N. Johnston, L. A. Cosimi, M. M. Addo, M. Lichterfeld, M. Altfeld, N. Frahm, C. Brander, A. Sette, B. D. Walker, and E. S. Rosenberg. 2004. Comprehensive analysis of human immunodeficiency virus type 1-specific CD4 responses reveals marked immunodominance of gag and nef and the presence of broadly recognized peptides. J. Virol. 784463-4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kelleher, A. D., C. Long, E. C. Holmes, R. L. Allen, J. Wilson, C. Conlon, C. Workman, S. Shaunak, K. Olson, P. Goulder, C. Brander, G. Ogg, J. S. Sullivan, W. Dyer, I. Jones, A. J. McMichael, S. Rowland-Jones, and R. E. Phillips. 2001. Clustered mutations in HIV-1 gag are consistently required for escape from HLA-B27-restricted cytotoxic T lymphocyte responses. J. Exp. Med. 193375-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kiepiela, P., K. Ngumbela, C. Thobakgale, D. Ramduth, I. Honeyborne, E. Moodley, S. Reddy, C. de Pierres, Z. Mncube, N. Mkhwanazi, K. Bishop, M. van der Stok, K. Nair, N. Khan, H. Crawford, R. Payne, A. Leslie, J. Prado, A. Prendergast, J. Frater, N. McCarthy, C. Brander, G. H. Learn, D. Nickle, C. Rousseau, H. Coovadia, J. I. Mullins, D. Heckerman, B. D. Walker, and P. Goulder. 2007. CD8+ T-cell responses to different HIV proteins have discordant associations with viral load. Nat. Med. 1346-53. [DOI] [PubMed] [Google Scholar]
- 51.Klenerman, P., G. Luzzi, K. McIntyre, R. Phillips, and A. McMichael. 1996. Identification of a novel HLA-A25-restricted epitope in a conserved region of p24 gag (positions 71-80). AIDS 10348-350. [DOI] [PubMed] [Google Scholar]
- 52.Koenig, S., A. J. Conley, Y. A. Brewah, G. M. Jones, S. Leath, L. J. Boots, V. Davey, G. Pantaleo, J. F. Demarest, C. Carter, et al. 1995. Transfer of HIV-1-specific cytotoxic T lymphocytes to an AIDS patient leads to selection for mutant HIV variants and subsequent disease progression. Nat. Med. 1330-336. [DOI] [PubMed] [Google Scholar]
- 53.Koup, R. A., J. T. Safrit, Y. Cao, C. A. Andrews, G. McLeod, W. Borkowsky, C. Farthing, and D. D. Ho. 1994. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J. Virol. 684650-4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kuritzkes, D. R. 2001. A fossil record of zidovudine resistance in transmitted isolates of HIV-1. Proc. Natl. Acad. Sci. USA 9813485-13487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Larder, B. A., A. Kohli, P. Kellam, S. D. Kemp, M. Kronick, and R. D. Henfrey. 1993. Quantitative detection of HIV-1 drug resistance mutations by automated DNA sequencing. Nature 365671-673. [DOI] [PubMed] [Google Scholar]
- 56.Leitner, T., E. Halapi, G. Scarlatti, P. Rossi, J. Albert, E. M. Fenyo, and M. Uhlen. 1993. Analysis of heterogeneous viral populations by direct DNA sequencing. BioTechniques 15120-127. [PubMed] [Google Scholar]
- 57.Leslie, A., D. Kavanagh, I. Honeyborne, K. Pfafferott, C. Edwards, T. Pillay, L. Hilton, C. Thobakgale, D. Ramduth, R. Draenert, S. Le Gall, G. Luzzi, A. Edwards, C. Brander, A. K. Sewell, S. Moore, J. Mullins, C. Moore, S. Mallal, N. Bhardwaj, K. Yusim, R. Phillips, P. Klenerman, B. Korber, P. Kiepiela, B. Walker, and P. Goulder. 2005. Transmission and accumulation of CTL escape variants drive negative associations between HIV polymorphisms and HLA. J. Exp. Med. 201891-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Leslie, A. J., K. J. Pfafferott, P. Chetty, R. Draenert, M. M. Addo, M. Feeney, Y. Tang, E. C. Holmes, T. Allen, J. G. Prado, M. Altfeld, C. Brander, C. Dixon, D. Ramduth, P. Jeena, S. A. Thomas, A. St. John, T. A. Roach, B. Kupfer, G. Luzzi, A. Edwards, G. Taylor, H. Lyall, G. Tudor-Williams, V. Novelli, J. Martinez-Picado, P. Kiepiela, B. D. Walker, and P. J. Goulder. 2004. HIV evolution: CTL escape mutation and reversion after transmission. Nat. Med. 10282-289. [DOI] [PubMed] [Google Scholar]
- 59.Li, B., A. D. Gladden, M. Altfeld, J. M. Kaldor, D. A. Cooper, A. D. Kelleher, and T. M. Allen. 2007. Rapid reversion of sequence polymorphisms dominates early human immunodeficiency virus type 1 evolution. J. Virol. 81193-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Listgarten, J., and D. Heckerman. 2007. Determining the number of non-spurious arcs in a learned DAG model: investigation of a Bayesian and a frequentist approach. In Proceedings of the Twenty-Third Conference on Uncertainty in Artificial Intelligence. UAI Press, Vancouver, Canada.
- 61.Liu, Y., J. McNevin, J. Cao, H. Zhao, I. Genowati, K. Wong, S. McLaughlin, M. D. McSweyn, K. Diem, C. E. Stevens, J. Maenza, H. He, D. C. Nickle, D. Shriner, S. E. Holte, A. C. Collier, L. Corey, M. J. McElrath, and J. I. Mullins. 2006. Selection on the human immunodeficiency virus type 1 proteome following primary infection. J. Virol. 809519-9529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Martinez-Picado, J., J. G. Prado, E. E. Fry, K. Pfafferott, A. Leslie, S. Chetty, C. Thobakgale, I. Honeyborne, H. Crawford, P. Matthews, T. Pillay, C. Rousseau, J. I. Mullins, C. Brander, B. D. Walker, D. I. Stuart, P. Kiepiela, and P. Goulder. 2006. Fitness cost of escape mutations in p24 Gag in association with control of human immunodeficiency virus type 1. J. Virol. 803617-3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Migueles, S. A., A. C. Laborico, H. Imamichi, W. L. Shupert, C. Royce, M. McLaughlin, L. Ehler, J. Metcalf, S. Liu, C. W. Hallahan, and M. Connors. 2003. The differential ability of HLA B*5701+ long-term nonprogressors and progressors to restrict human immunodeficiency virus replication is not caused by loss of recognition of autologous viral gag sequences. J. Virol. 776889-6898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Migueles, S. A., M. S. Sabbaghian, W. L. Shupert, M. P. Bettinotti, F. M. Marincola, L. Martino, C. W. Hallahan, S. M. Selig, D. Schwartz, J. Sullivan, and M. Connors. 2000. HLA B*5701 is highly associated with restriction of virus replication in a subgroup of HIV-infected long term nonprogressors. Proc. Natl. Acad. Sci. USA 972709-2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Moore, C. B., M. John, I. R. James, F. T. Christiansen, C. S. Witt, and S. A. Mallal. 2002. Evidence of HIV-1 adaptation to HLA-restricted immune responses at a population level. Science 2961439-1443. [DOI] [PubMed] [Google Scholar]
- 66.Nickle, D. C., M. Rolland, M. A. Jensen, S. L. Pond, W. Deng, M. Seligman, D. Heckerman, J. I. Mullins, and N. Jojic. 2007. Coping with viral diversity in HIV vaccine design. PLoS Comput. Biol. 3e75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Phillips, R. E., S. Rowland-Jones, D. F. Nixon, F. M. Gotch, J. P. Edwards, A. O. Ogunlesi, J. G. Elvin, J. A. Rothbard, C. R. Bangham, C. R. Rizza, et al. 1991. Human immunodeficiency virus genetic variation that can escape cytotoxic T cell recognition. Nature 354453-459. [DOI] [PubMed] [Google Scholar]
- 68.Price, D. A., P. J. Goulder, P. Klenerman, A. K. Sewell, P. J. Easterbrook, M. Troop, C. R. Bangham, and R. E. Phillips. 1997. Positive selection of HIV-1 cytotoxic T lymphocyte escape variants during primary infection. Proc. Natl. Acad. Sci. USA 941890-1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rolland, M., M. A. Jensen, D. C. Nickle, J. Yan, G. H. Learn, L. Heath, D. Weiner, and J. I. Mullins. 2007. Reconstruction and function of ancestral center-of-tree human immunodeficiency virus type 1 proteins. J. Virol. 818507-8514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rousseau, C. M., M. G. Daniels, J. M. Carlson, C. Kadie, H. Crawford, A. Prendergast, P. Matthews, R. Payne, M. Rolland, D. N. Raugi, B. S. Maust, G. H. Learn, D. C. Nickle, H. Coovadia, T. Ndung'u, N. Frahm, C. Brander, B. D. Walker, P. J. Goulder, T. Bhattacharya, D. E. Heckerman, B. T. Korber, and J. I. Mullins. 2008. HLA class-I driven evolution of human immunodeficiency virus type 1 subtype C proteome: immune escape and viral load. J. Virol. 826434-6446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rychert, J., S. Saindon, S. Placek, D. Daskalakis, and E. Rosenberg. 2007. Sequence variation occurs in CD4 epitopes during early HIV infection. J. Acquir. Immun. Defic. Syndr. 46261-267. [DOI] [PubMed] [Google Scholar]
- 72.Scherer, A., J. Frater, A. Oxenius, J. Agudelo, D. A. Price, H. F. Gunthard, M. Barnardo, L. Perrin, B. Hirschel, R. E. Phillips, and A. R. McLean. 2004. Quantifiable cytotoxic T lymphocyte responses and HLA-related risk of progression to AIDS. Proc. Natl. Acad. Sci. USA 10112266-12270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schneidewind, A., M. A. Brockman, R. Yang, R. I. Adam, B. Li, S. Le Gall, C. R. Rinaldo, S. L. Craggs, R. L. Allgaier, K. A. Power, T. Kuntzen, C. S. Tung, M. X. LaBute, S. M. Mueller, T. Harrer, A. J. McMichael, P. J. Goulder, C. Aiken, C. Brander, A. D. Kelleher, and T. M. Allen. 2007. Escape from the dominant HLA-B27-restricted cytotoxic T-lymphocyte response in Gag is associated with a dramatic reduction in human immunodeficiency virus type 1 replication. J. Virol. 8112382-12393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sekaly, R. P. 2008. The failed HIV Merck vaccine study: a step back or a launching point for future vaccine development? J. Exp. Med. 2057-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Storey, J. D., and R. Tibshirani. 2003. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 1009440-9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Streeck, H., M. Lichterfeld, G. Alter, A. Meier, N. Teigen, B. Yassine-Diab, H. K. Sidhu, S. Little, A. Kelleher, J. P. Routy, E. S. Rosenberg, R. P. Sekaly, B. D. Walker, and M. Altfeld. 2007. Recognition of a defined region within p24 gag by CD8+ T cells during primary human immunodeficiency virus type 1 infection in individuals expressing protective HLA class I alleles. J. Virol. 817725-7731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Turnbull, E. L., A. R. Lopes, N. A. Jones, D. Cornforth, P. Newton, D. Aldam, P. Pellegrino, J. Turner, I. Williams, C. M. Wilson, P. A. Goepfert, M. K. Maini, and P. Borrow. 2006. HIV-1 epitope-specific CD8+ T cell responses strongly associated with delayed disease progression cross-recognize epitope variants efficiently. J. Immunol. 1766130-6146. [DOI] [PubMed] [Google Scholar]
- 78.Ueno, T., Y. Idegami, C. Motozono, S. Oka, and M. Takiguchi. 2007. Altering effects of antigenic variations in HIV-1 on antiviral effectiveness of HIV-specific CTLs. J. Immunol. 1785513-5523. [DOI] [PubMed] [Google Scholar]
- 79.Zuñiga, R., A. Lucchetti, P. Galvan, S. Sanchez, C. Sanchez, A. Hernandez, H. Sanchez, N. Frahm, C. H. Linde, H. S. Hewitt, W. Hildebrand, M. Altfeld, T. M. Allen, B. D. Walker, B. T. Korber, T. Leitner, J. Sanchez, and C. Brander. 2006. Relative dominance of Gag p24-specific cytotoxic T lymphocytes is associated with human immunodeficiency virus control. J. Virol. 803122-3125. [DOI] [PMC free article] [PubMed] [Google Scholar]