Abstract
The replication and transcription of double-stranded RNA (dsRNA) viruses occur within a polymerase complex particle in which the viral genome is enclosed throughout the entire life cycle of the virus. A single protein subunit in the polymerase complex is responsible for the template-dependent RNA polymerization activity. The isolated polymerase subunit of the dsRNA bacteriophage φ6 was previously shown to replicate and transcribe given RNA molecules. In this study, we show that this enzyme also catalyzes nontemplated nucleotide additions to single-stranded and double-stranded nucleic acid molecules. This terminal nucleotidyltransferase activity not only is a property of the isolated enzyme but also is detected to take place within the viral nucleocapsid. This is the first time terminal nucleotidyltransferase activity has been reported for a dsRNA virus as well as for a viral particle. The results obtained together with previous high-resolution structural data on the φ6 RNA-dependent RNA polymerase suggest a mechanism for terminal nucleotidyl addition. We propose that the activity is involved in the termination of the template-dependent RNA polymerization reaction on the linear φ6 genome.
Terminal nucleotidyltransferases (TNTases) are enzymes that catalyze the addition of nucleotides to the 3′ end of an RNA or DNA molecule. Such an activity has been described for several enzymes of eukaryotic, eubacterial, and archaeal origins as well as for several viral polymerases. TNTases are involved in numerous crucial cellular functions, such as maturation of tRNA and mRNA molecules, RNA degradation pathways, and generation of diversity in immunoglobulin- and T-cell receptor-encoding genes during oncogenesis (12, 20, 25, 64).
RNA polymerases of single-stranded RNA (ssRNA) viruses, such as hepatitis C virus (HCV), bovine viral diarrhea virus, GB virus G (family Flaviviridae) (3, 46, 50, 66) norovirus, sapovirus (family Caliciviridae) (16, 52), and poliovirus (family Picornaviridae) (33), possess TNTase activity on ssRNA substrates in addition to their template-dependent RNA polymerization activity. It has been suggested that the TNTase activity associated with some viral polymerases is designed to repair the 3′ ends of viral genomes that may have been subjected to degradation by cellular exonucleases (46). In addition, the reverse transcriptase of several retroviruses, including human immunodeficiency virus type 1 (Retroviridae), can add nontemplated nucleotides to the 3′ end of the newly synthesized DNA strand (10, 39). Similarly, the DNA-dependent RNA polymerase of coliphage T7 (Podoviridae) may incorporate one or two nucleotides beyond the end of the DNA template (30). However, such activity has been considered a malfunction that occurs on heterologous templates due to the lack of proper termination signals.
Interestingly, nontemplated nucleotides at the genome termini have been observed in the replicative intermediates of several ssRNA viruses, including bacteriophage Qβ (Leviviridae) (61), cucumber mosaic virus (Bromoviridae) (13), and Sindbis virus and Semliki Forest virus (Togaviridae) (62, 63). Also, double-stranded RNA (dsRNA) viruses replicating in yeast (Saccharomyces cerevisiae virus L-A and its satellite viruses [Totiviridae]) have a 3′ nontemplated adenosine in their dsRNA genomes (4, 7). Such observations suggest that the replication cycles of these viruses involve viral or cellular enzymes that possess TNTase activity.
Bacteriophage φ6 (Cystoviridae) is a dsRNA virus infecting gram-negative Pseudomonas syringae. The virion is a triple-layered structure including two protein layers: the innermost polymerase complex, the nucleocapsid surface shell, and the lipid envelope. The genome, enclosed within the polymerase complex, is trisegmented; the segments are designated S (2,948 nucleotides [nt]), M (4,063 nt), and L (6,374 nt) (44). The replication strategy of φ6 is similar to that of eukaryotic dsRNA viruses in the families Reoviridae, Birnaviridae, Totiviridae, and Partitiviridae (29).
A single polypeptide species is responsible for the RNA polymerization activity in the polymerase complex of dsRNA viruses (28). The polymerase protein, P2, of bacteriophage φ6 has been purified and possesses both RNA replication (synthesis of negative strand) and transcription (production of positive strand) activities in vitro, utilizing a de novo initiation (26, 27). Furthermore, the purified P2 self-assembles into viral polymerase complexes that become fully functional both in vitro and in vivo (42). Structural and biochemical studies of φ6 P2 have revealed an initiation mechanism for de novo RNA polymerization (8, 21), serving as a paradigm for the other structurally related polymerases, such as the HCV and bovine viral diarrhea virus polymerases (5, 11). In addition to the isolated P2, the transcription and replication activities of φ6 have been studied using either nucleocapsids or core particles isolated from infectious virions or recombinant empty polymerase complexes produced in Escherichia coli (34, 60). Presently, φ6 is one of the best characterized dsRNA viruses, especially with regard to viral RNA metabolism (28, 43, 44). Its RNA-dependent RNA polymerase is also a potential tool for efficient dsRNA production for RNA interference and other applications (1).
We describe here, for the first time, TNTase activity that is present in a dsRNA virus. We show that the TNTase activity not only is the property of the isolated viral polymerase but also is detected in purified viral particles. The nucleoside triphosphate (NTP) specificity is low, and both ssRNA and dsRNA molecules are accepted as substrates. Based on our biochemical results and the previously published φ6 polymerase structures (8, 53), we propose a mechanistic model for terminal nucleotide addition. Our model provides a working hypothesis for polymerases with both template-dependent and template-independent activities. Furthermore, we propose that the TNTase activity of φ6 P2 is a mechanism associated with the termination of nascent RNA strands.
MATERIALS AND METHODS
Plasmids.
To increase the production of the cloned φ6 P2 protein, the NdeI-HindIII restriction fragment (containing the P2 gene) from the P2 expression plasmid pEM2 (27) was transferred to the pMG60 expression plasmid (55) cut with the same restriction enzymes. The resultant plasmid, pEMG2, was used for the production of the wild-type P2 (P2WT) polymerase. In addition, an expression plasmid, pSVe14, was constructed for the production of a mutant polymerase (P2SAD) by introducing an amino acid change (D453A) into the P2WT gene in the pEMG2 plasmid, using oligonucleotides Phi6P2_SAD_EcoRV_down and Phi6P2_SAD_EcoRV_up according to Stratagene's QuikChange protocol (Table 1). P2E491Q was produced from plasmid pSVe4 (M. M. Poranen, P. S. Salgado, M. R. L. Koivunen, S. Wright, D. H. Bamford, D. I. Stuart, and J. M. Grimes, submitted for publication).
TABLE 1.
DNA oligonucleotides used in this study
| Name | Sequence |
|---|---|
| Phi6P2_SAD_EcoRV_down | 5′-GGTCCAGCCAAGCATAGCATCAGCAGATTTTGATATCTGACGGAT |
| Phi6P2_SAD_EcoRV_up | 5′-ATCCGTCAGATATCAAAATCTGCTGATGCTATGCTTGGCTGGACC |
| T7_1 | 5′-CGCGTAATACGACTCACTATAG |
| T7_2 | 5′-CGCGTAATACGACTCACTATAGGAAAAAAACTTTATATAAC |
| 3′end | 5′-AGAGAGAGAGCCCCCGA |
| 3′end_1 | 5′-AAGAGAGAGAGCCCCCGA |
| 3′end_2 | 5′-CAGAGAGAGAGCCCCCGA |
| 3′end_3 | 5′-GAGAGAGAGAGCCCCCGA |
| 3′end_4 | 5′-TAGAGAGAGAGCCCCCGA |
| 3′end_7 | 5′-AGAGAGAGCCCCCGAAG |
| 3′end_8 | 5′-GAGAGCCCCCGAAGGG |
| 3′end_10 | 5′-GCCCCCGAAGGGGCCGTCCTATTG |
Plasmids pLM659 (18), pLM656 (35), and pLM687 (31) were used to produce positive-sense ssRNA copies (s+, m+, and l+) of the small (S), medium (M), and large (L) φ6 genomic segments, respectively. Plasmid pEM15 contains the cDNA clone of the φ6 S segment with an internal deletion (26) and was used as a template to prepare sΔ+ ssRNAs with different 3′-end-labeled extensions and truncations.
Protein purification.
P2WT, P2SAD, and P2E491Q polymerases were expressed in E. coli BL21 (56) containing an appropriate expression plasmid at +20°C for 15 h (9) and purified to near homogeneity as described previously (27), except that a HiTrap Q column (Pharmacia) was used for anion-exchange chromatography and the entire purification was carried out at +4°C.
Nucleocapsid isolation.
For nucleocapsid production, bacteriophage φ6 was grown on P. syringae HB10Y and purified as described previously (2, 41). Nucleocapsid particles were isolated (2, 41) and collected by differential centrifugation through 20% (wt/vol) sucrose in 20 mM Tris, pH 7.4 (Sorvall T865; 40,000 rpm, 2.5 h, +10°C). The pellet was stored in 20 mM Tris, pH 7.4.
Preparation of RNA substrates.
Synthetic ssRNAs were produced by runoff in vitro transcription with T7 polymerase (15). Templates for T7 transcription were prepared from plasmid DNA either by cutting with suitable restriction endonucleases or by PCR amplification using a mixture of Taq (Promega) and Pfu Turbo (Stratagene) DNA polymerases. φ6-specific ssRNAs s+, m+, and l+ were produced from XbaI-cut (Roche), mung bean nuclease-treated (Promega) plasmids pLM659, pLM656, and pLM687, respectively, as described previously (14). RNA s+13+ was transcribed from SmaI-cut (Fermentas) plasmid pLM659 and contains 13 extra nucleotides at the 3′ end of the φ6 s+ segment that are derived from the plasmid (the plasmid-encoded sequence is CUAGAGGAUCCCC-3′). Similarly, sΔ+13+ ssRNA was prepared from SmaI-cut plasmid pEM15. The template for the synthesis of sΔ+ ssRNA (with a natural 3′-end-labeled sequence) was produced from plasmid pEM15 by PCR amplification using DNA oligonucleotides T7_1 and 3′end (Table 1) (65) as upstream and downstream primers, respectively. A set of sΔ+ ssRNA with different 3′-terminal sequences was produced using DNA oligonucleotides 3′end_1 to 3′end_8 (Table 1) as downstream primers for PCR amplification instead of the 3′end oligonucleotide. The template for sΔ+9− (sΔ+ with 9-nt-long 3′-terminal truncation) ssRNA was amplified using T7_2 and 3′end_10 oligonucleotides (Table 1). All ssRNA preparations were successively extracted with TRIzol (Invitrogen)-chloroform (5:1) and precipitated first with 4 M LiCl and then with 0.75 M ammonium acetate (NH4OAc) and ethanol. Viral dsRNA was isolated from polyethylene glycol-concentrated virus preparations with TRIzol-chloroform extraction, followed by isopropanol precipitation, stepwise precipitation with 2 M and 4 M LiCl, and NH4OAc-ethanol precipitation. The RNA concentrations were measured (A260), and the quality was checked by electrophoresis in a 1% agarose gel.
RNA oligonucleotides 26n_RNA (5′-AAUAAUAAUAAUAAGAUUUUUUCCCC-3′), RNA_anti_s117 (5′-CGACUCAUGGACCUUGGGAG-3′), and phi6spacRNA25-41 (5′-AUAUAAGUGCCCUUAGC-3′) were obtained from Dharmacon, and 6n_RNA (5′-UUUCCC-3′) was obtained from Eurogentec. Gel purification of ssRNA oligonucleotides in a 20% polyacrylamide gel containing 50% (wt/vol) urea and Tris-borate-EDTA was performed when required (15). The RNA oligonucleotides were 5′ end labeled using T4 polynucleotide kinase and [α-32P]ATP (>5,000 Ci/mmol; Amersham Biosciences) according to the manufacturer's instructions (Fermentas), after which the products were purified using Zeba micro desalt spin columns (Pierce).
φ6 polymerase assays.
The replication and TNTase activities of P2WT, P2SAD, and P2E491Q polymerases were assayed initially in 10-μl reaction mixtures containing 50 mM HEPES-KOH, pH 7.5, 20 mM NH4OAc, 6% (wt/vol) polyethylene glycol 4000, 5 mM MgCl2, 2 mM MnCl2, 0.1 mM EDTA, 0.1% Triton X-100, 0.8 unit/μl RNasin (Promega), and 0.1 mCi/ml [α-32P]UTP (3,000 Ci/mmol; Amersham Biosciences). The optimal TNTase buffer contained the following changes: 3 mM MgCl2, 0.5 mM MnCl2, and no NH4OAc. The final concentration of the RNA substrates was set equimolar to the final φ6 polymerase protein concentration (270 nM or 540 nM). For the replication reaction, the reaction mixture was supplemented with 0.2 mM UTP and CTP and 1 mM ATP and GTP. Unless indicated otherwise, the TNTase reaction mixture contained 30 nM unlabeled NTP in addition to 0.1 mCi/ml [α-32P]NTP. The mixtures were incubated at +30°C for 1 h and stopped with 2× U buffer (38). Reaction products were separated by standard agarose gel electrophoresis or strand-separating agarose gel electrophoresis (38). The gels were dried and analyzed using a phosphorimager (Fuji BAS1500 or FLA-5000). If reactions were carried out using short RNA oligonucleotides, the reaction products were purified through Zeba micro desalt spin columns (Pierce) and subjected to analysis in a denaturing 20% polyacrylamide gel in 50% (wt/vol) urea and Tris-borate-EDTA buffer.
RESULTS
φ6 RNA-dependent RNA polymerase catalyzes transfer of an α-32P label from an NTP to an ssRNA molecule.
φ6 RNA-dependent RNA polymerase (P2) catalyzes template-dependent de novo RNA synthesis, producing dsRNA molecules from single-stranded template RNAs (27). A standard assay mixture for RNA replication analysis contains the purified polymerase protein P2, the ssRNA template, four NTPs, buffer, and [α-32P]UTP for labeling (Fig. 1A, lane 1). When the unlabeled NTPs were omitted from the polymerase reaction mixture, a novel radioactively labeled reaction product was detected that migrated similarly to the ssRNA template during agarose gel electrophoresis (Fig. 1A, lane 3).
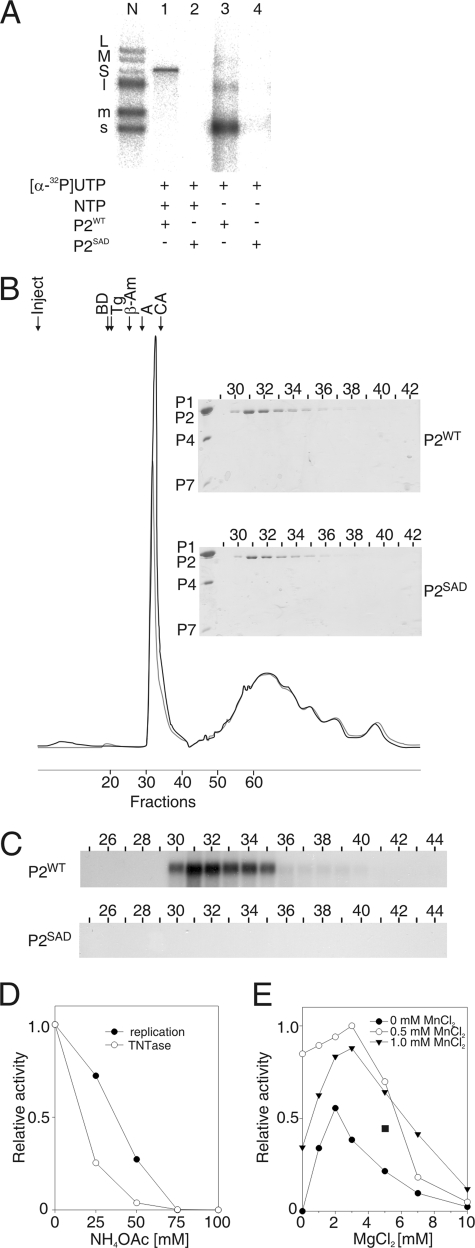
FIG. 1.
Purified φ6 P2 polymerase catalyzes the transfer of an α-phosphate from a donor UTP to an acceptor ssRNA molecule. (A) Native agarose gel electrophoresis of reactions with purified P2WT and P2SAD assayed under standard polymerization reaction conditions in the presence of [α-32P]UTP and a single-stranded φ6 s+ segment. The reactions were carried out with or without unlabeled NTPs as indicated. The positions of the labeled single-stranded (s+, m+, and l+) and double-stranded (S, M, and L) φ6 genome segments produced by nucleocapsid transcription (41) are shown on the left (lane N). (B) Absorbance (A280) elution profile of P2WT and P2SAD separated on a Superdex 200 gel filtration column (Amersham Biosciences). The arrows indicate the P2 injection time (Inject) and the position of selected gel filtration molecular mass markers (Sigma): BD, blue dextran (2,000 kDa); Tg, thyroglobulin (669 kDa); β-Am, β-amylase (200 kDa); A, albumin (66 kDa); and CA, carbonic anhydrase (29 kDa). The lower peak corresponds to nonionic detergents of the P2 storage buffer. (Insert) Sodium dodecyl sulfate-polyacrylamide gel analysis of the protein content in fractions 29 to 42 (upper panel, P2WT; lower panel, P2SAD). φ6 polymerase complex proteins (P1, P2, P4, and P7) are marked on the left. (C) Enzymatic activity in Superdex 200 fractions 25 to 44 of P2WT (upper panel) and P2SAD (lower panel) with s+13+ ssRNA template and [α-32P]UTP (in the absence of other NTPs). (D and E) Effect of the reaction conditions on the α-32P transfer activity of φ6 P2. The RNA products of the reaction mixtures containing sΔ+13+ ssRNA were separated by electrophoresis in a native agarose gel and analyzed by phosphorimager quantification. The graphs are normalized to the highest value (1) within each panel. The effect of the NH4OAc concentration on α-32P transfer activity is shown in panel D together with the replication activity under the same conditions for comparison. The MnCl2 and MgCl2 concentration effects are shown in panel E. The α-32P transfer activity under the optimal replication reaction conditions (2 mM MnCl2 and 5 mM MgCl2) is indicated by a filled square.
To confirm that the labeling of the ssRNA was catalyzed by the φ6 P2 polymerase and not by contaminating cellular proteins, we prepared a mutant polymerase in which one of the conserved amino acids in the SDD motif (6) was replaced with alanine (SDD→SAD) to destroy the catalytic center of P2. The mutant polymerase, P2SAD, was purified similarly to P2WT. However, no radioactively labeled products appeared in reactions in which P2WT was replaced with P2SAD (Fig. 1A, lanes 2 and 4), suggesting that no cellular proteins with ssRNA labeling activity copurified with P2. To verify that the loss of activity in P2SAD was due to the mutation in the catalytic site and not to differences in protein folding, both P2WT and P2SAD were subjected to analytical gel filtration under native conditions (Fig. 1B). The elution curves for P2WT and P2SAD were identical, indicating that P2SAD was properly folded. The position of the P2WT protein peak (Fig. 1B) coincided with that of the peak of the new enzymatic activity (Fig. 1C, upper panel). No activity was detected in the fractions containing P2SAD (Fig. 1C, lower panel). These results confirm that the activity was an inherent property of the P2 polymerase.
Several reaction condition parameters were tested to characterize the biochemical requirements of the P2-catalyzed transfer of an α-32P label from a donor NTP to an acceptor ssRNA molecule. The optimal conditions for this newly found activity differed slightly from those determined for the template-dependent polymerase activity of φ6 P2 (65). Notably, the new activity was more sensitive to increasing NH4OAc (Fig. 1D) or KCl (data not shown) concentration. Like the replication reaction, the new activity was dependent on divalent cations Mg2+ and/or Mn2+ (Fig. 1E). However, the optima for divalent cations were lower than in the replication reaction (0.5 mM MnCl2 and 3 mM MgCl2 versus 2 mM MnCl2 and 5 mM MgCl2 in replication), which is likely a reflection of the lower concentration of NTPs in the reaction mixture.
The reaction product is a 3′ extension of the acceptor RNA molecule.
To analyze the P2-catalyzed transfer reaction, we used short 5′-end-labeled ssRNA oligonucleotides with unlabeled nucleotides and visualized the reaction products via denaturing urea containing sodium dodecyl sulfate-polyacrylamide gel. It appeared that under the reaction conditions applied (equimolar concentrations of P2 and the acceptor RNA oligonucleotide and 50 M excess of UTP), the P2 polymerase typically added one or two extra nucleotides to a given acceptor ssRNA oligonucleotide (Fig. 2A and B, compare lanes 1 and 2). However, longer extensions were also detected but with lower frequency (Fig. 2A, lane 2). These longer products dominated if the donor UTP concentration was increased substantially (data not shown). More than half of the RNA oligonucleotide molecules were extended under the reaction conditions applied (Fig. 2A and B).
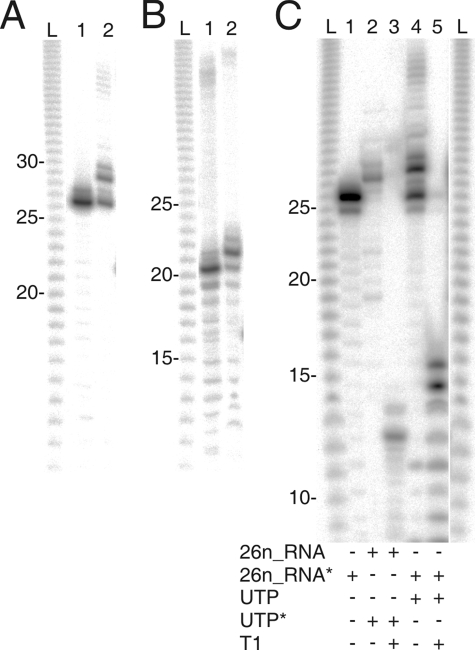
FIG. 2.
Characterization of the products of the φ6 P2-catalyzed α-32P transfer reaction. The reaction products were analyzed in a denaturing 20% polyacrylamide gel. Lane L contains a single nucleotide RNA ladder (under product development at Finnzymes). (A) 5′-end-labeled RNA oligonucleotide 26n_RNA (lane 1). Mobility shift of the same RNA after P2-catalyzed α-P transfer from an unlabeled donor UTP to the acceptor RNA (lane 2) is shown. (B) Same as panel A, but with RNA_anti_s117 (20 nt) as an acceptor. (C) End specificity of the P2-catalyzed α-P transfer reaction. RNA products of a reaction mixture containing an unlabeled 26n_RNA oligonucleotide acceptor and a [α-32P]UTP donor (lane 2) were digested with T1 nuclease (lane 3). As a control, the 5′-end-labeled RNA oligonucleotide was treated similarly (lanes 4 and 5), except that unlabeled UTP was used as a donor. The critical additives are indicated below the panel. The asterisk specifies a labeled substrate (RNA or UTP).
To characterize the end specificity of the reaction, the product RNAs of the P2-catalyzed TNTase reaction were digested with T1 RNase (Epicentre), which specifically cuts ssRNA molecules on the 3′ side of G nucleotides. A 26-nt-long RNA oligonucleotide, 26n_RNA (5′-AAUAAUAAUAAUAAGAUUUUUUCCCC-3′), was used for the analysis. This acceptor RNA was extended predominantly by 2 nt in the P2-catalyzed reaction (Fig. 2A, lane 2, and C, lane 4) and contained one G nucleotide that was 15 nt from the 5′ terminus. The analysis was accomplished with unlabeled oligonucleotide and [α-32P]UTP (Fig. 2C, lanes 2 and 3). If the transfer of an α-32P label from a donor UTP to the 26n_RNA oligonucleotide had occurred at the 3′ end of the acceptor RNA molecule, a radioactively labeled fragment of 13 nt (11 + 2 nt) would be expected after T1 digestion, whereas if the radioactivity were transferred to the 5′ end, a 17-nt-long (15 + 2 nt) T1 digestion product would be anticipated. A radioactively labeled reaction product of approximately 13 nt in length was detected after a P2-catalyzed TNTase reaction and T1 digestion (Fig. 2C, lane 3). As a control, the same experiment was repeated using a 5′-end-labeled oligonucleotide and unlabeled donor UTP (Fig. 2C, lanes 4 and 5). The T1 digestion of the control reaction produced an ∼15-nt-long digestion product (Fig. 2C, lane 5). This indicated that the UTP (as a UMP) was transferred to the 3′ end of the acceptor RNA molecule, proving that φ6 P2 possesses 3′-end-specific TNTase activity rather than 5′ ligase activity.
NTP specificity of φ6 P2-catalyzed TNTase activity.
The nucleotide specificity of the TNTase reaction was analyzed by adding different donor [α-32P] ribonucleotides to the standard TNTase reaction mixture containing s+ ssRNA (Fig. 3A), which is a natural template for φ6 P2 polymerase. All four [α-32P] NTPs could function as nucleotidyl donors in the TNTase reaction. The analysis was repeated using different acceptor ssRNA molecules with different terminal sequences (sΔ+13+, sΔ+A [sΔ+ with 3′-terminal A], sΔ+9−, and luciferase RNA). The efficiencies of the TNTase reactions with different acceptor RNA and donor NTP combinations were within the same magnitude (data not shown) although there were variations observed between the reactions.
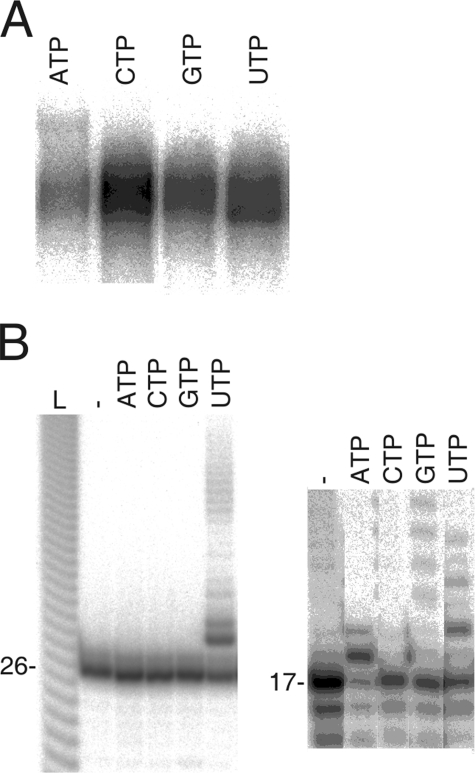
FIG. 3.
Donor nucleotide specificity of the φ6 P2 polymerase-catalyzed TNTase reaction. The nucleotides tested are indicated above the panels. (A) s+ ssRNA and each of the four [α-32P] NTPs (∼30 nM) were used as substrates in the TNTase reaction. The reaction products were analyzed by agarose gel electrophoresis and phosphorimaging. (B) Screening of donor nucleotide specificity by using unlabeled nucleotides and a radioactively labeled 26n_RNA (left panel) or phi6spacRNA25-41 oligonucleotide acceptor (17-nt-long RNA oligonucleotide) (right panel) with a gel shift assay (an autoradiogram of the reaction products analyzed on a denaturing 20% polyacrylamide gel). The lane containing the labeled oligonucleotide only is marked with a dash. Lane L is a single nucleotide ladder, as defined in the legend to Fig. 2.
Nucleotide specificity was also assayed with a gel shift assay using prelabeled short oligonucleotides (26n_RNA and phi6spacRNA25-41) and unlabeled donor NTPs. Elongated products were detected with 26n_RNA when UTP was used as a substrate and with ATP, GTP, and UTP when phi6spacRNA25-41 was used (Fig. 3B). Apparently the acceptor RNA molecules have influence on the NTP specificity of the reaction. This might reflect the competition between the formation of the replication initiation complex and the TNTase activity on the same ssRNA molecule. As UTP was the only NTP that was efficiently incorporated into all the tested ssRNA molecules, UTP was utilized as the donor NTP in all of the following experiments.
Specificity for the acceptor nucleic acid is low.
To evaluate the acceptor RNA specificity of the φ6 P2-catalyzed TNTase reaction, a set of φ6 s+-segment-specific ssRNA molecules was produced containing the natural φ6 terminus (viral s+ segment with internal deletion; sΔ+) and the φ6 terminus with a single nucleotide extension (3′ U, -G, -C, or -A; sΔ+U, sΔ+G, sΔ+C, or sΔ+A, respectively) and 2 (sΔ+2−)-, 5 (sΔ+5−)-, and 9 (sΔ9−)-nt-long 3′-end-labeled truncations (Fig. 4A). Although the RNA molecules were prepared using T7 polymerase, we assume that the majority of the RNA molecules had correct termini. It appeared that none of the modified ssRNA molecules was notably favored over the intact one; rather, sΔ+, with the natural 3′-end-labeled sequence, was slightly preferred over the truncated acceptor RNAs (sΔ+2−, sΔ+5−, and sΔ+9−) (Fig. 4B).
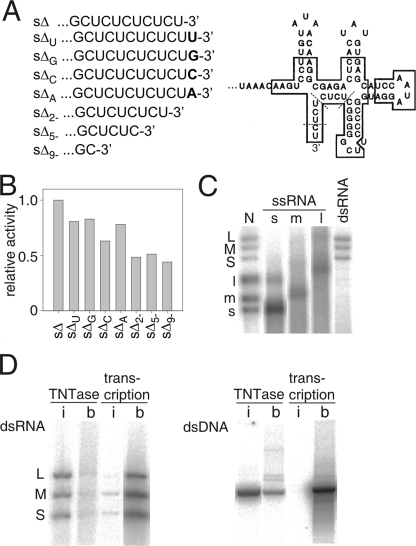
FIG. 4.
Acceptor RNA specificity of the φ6 P2-catalyzed TNTase reaction. (A) 3′-end-labeled sequences of φ6 s+-segment-specific, in vitro-produced ssRNAs. sΔ+ ssRNA contains wild-type 3′- and 5′-end-labeled sequences and an internal deletion (26). The 3′-terminal secondary structure of the s+ ssRNA is shown on the right, and the positions for 2-, 5-, and 9-nt truncations (sΔ+2−, sΔ+5−, and sΔ+9−) are indicated with dashed lines. The sequences in the boxes are conserved between the three φ6 genome segments (modified from reference 32). (B) Relative P2-catalyzed TNTase activity with different 3′-end-modified sΔ+ ssRNA acceptors. The reaction products were analyzed by agarose gel electrophoresis and phosphorimaging. The values obtained were normalized by setting the highest value to 1. (C and D) Autoradiograms of agarose gels of P2 reaction products. (C) Autologous ssRNAs and dsRNAs as substrates of the P2-catalyzed TNTase reaction. The acceptor ssRNAs are φ6 s+, m+, and l+, and genomic dsRNA (S, M, and L) isolated from virions. Each reaction mixture contained approximately equimolar amounts of 3′ ends of RNA. Lane N is as defined in the legend to Fig. 1A. (D) P2-catalyzed TNTase and transcription reactions with either intact (i) or boiled (b) dsRNA (left panel) or dsDNA (right panel). The nucleic acid molecules used were φ6 genomic dsRNA (S, M, and L segments) and SmaI-digested plasmid pEM15 (26).
The results obtained using modified φ6 s+-segment-specific ssRNA molecules suggested that φ6 P2 was a relatively nonspecific TNTase regarding the 3′-end-labeled sequence of the acceptor RNA. To evaluate this further, heterologous RNA molecules of different lengths (6 to 3,569 nts) and sequences were tested. The efficiencies of the TNTase reactions on the different ssRNA molecules differed significantly, and some of the shorter RNA molecules appeared to be totally inert acceptors. In addition, autologous ssRNA molecules, the full-length s+, m+, and l+ ssRNA segments of φ6, were labeled with different efficiencies (Fig. 4C) although their 3′-end-labeled sequences were similar for the 17 terminal nucleotides. These analyses suggested that conformational features in the acceptor ssRNA molecules may have a strong influence on the φ6 P2-catalyzed TNTase reaction.
Since the genome of φ6 is double stranded, we investigated whether dsRNA also could function as an acceptor in the φ6 P2-catalyzed TNTase reaction. The three φ6 genomic dsRNA segments (S, M, and L) isolated from the viral particles were all labeled with equal efficiencies (Fig. 4C). Furthermore, genomic dsRNA from rotavirus as well as different dsDNA molecules with blunt ends, 3′ or 5′ overhangs, were acceptors in the P2-catalyzed TNTase reaction (data not shown). The efficiencies of dsRNA and ssRNA labeling were within the same range (Fig. 4C and data not shown); more variation was detected in the utilization of different ssRNA molecules than between ssRNA and dsRNA acceptors.
Opening of the dsRNA substrate is not required for the TNTase reaction.
The φ6 P2-catalyzed transcription reaction on dsRNA templates is less efficient than the replication reaction using ssRNA templates (26). This phenomenon reflects the difficulty of P2 to open the dsRNA molecule in order to get the negative-strand RNA template into the template tunnel, which can accommodate only an ssRNA molecule (8, 53). If the dsRNA is heat denatured prior to the polymerization reaction (26), the efficiency of the reaction is increased considerably. Assuming that the same template tunnel of φ6 P2 is applied during the TNTase reaction, destabilization of the dsRNA duplex should also increase the TNTase activity of P2 on dsRNA substrates. Unexpectedly, denatured dsRNA was not preferred as an acceptor (Fig. 4D, left panel). Rather, denaturation of dsRNA decreased the φ6 P2-catalyzed TNTase reaction (to approximately one-sixth of that obtained using intact dsRNA). This decrease was not due to heat-induced degradation of the RNA, as the transcription reaction was substantially increased using the same RNA (100-fold increase in the activity) (Fig. 4D, left panel). Also, the denaturation of the dsDNA acceptor prior to the reaction reduced the TNTase activity (to approximately one-sixth of that obtained with intact dsDNA) (Fig. 4D, right panel). This indicates that double-stranded nucleic acid molecules are preferred acceptors in the φ6 P2-catalyzed TNTase reaction.
Reduction in the flexibility of φ6 P2 reduces its TNTase activity.
The structurally compact φ6 P2 apoenzyme does not contain any opening that would allow a double-stranded nucleic acid molecule to reach the catalytic site located in the interior of the polymerase, and thus, the high TNTase activity on double-stranded substrates indicates that the polymerase must at least transiently adopt a more open conformation. To test this hypothesis, we applied a mutant polymerase, P2E491Q, which is thermally more stable than P2WT (Poranen et al., submitted) and thus likely less prone to spontaneous conformational changes (opening). Replication and TNTase reactions were carried out with the mutant and wild-type polymerases by using single-stranded s+13+ RNA (data not shown). The TNTase activity of P2E491Q was only about 6% of that of P2WT, whereas the template-dependent replication activity of P2E491Q was close to that of the wild type (∼93%). This suggests that the thermal stability is associated with the TNTase activity.
Addition of nontemplated nucleotides during replication of φ6 P2.
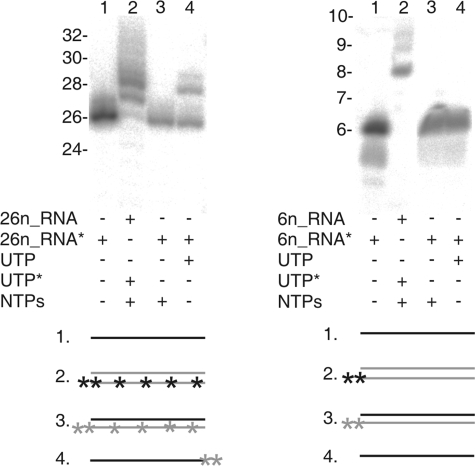
Since dsRNA molecules appeared to be substrates for the φ6 P2-catalyzed TNTase reaction, we investigated whether nascent dsRNA molecules synthesized by the P2 polymerase were extended. In this case, TNTase activity was analyzed under conditions in which replication would be active (in the presence of all of the NTPs, which would also be the condition within the infected cell). It appeared that the template RNA was not extended under those conditions (Fig. 5, lanes 3). However, the replication product (negative strand) was 1 or 2 nt longer than the template (Fig. 5, lanes 2), indicating that RNA synthesis could extend beyond the end of the template. The phenomenon was detected with the 26n_RNA oligonucleotide (the substrate for the P2 TNTase reaction in ssRNA form) (Fig. 5, left panel, lane 4) but also with the 6n_RNA oligonucleotide (5′-UUUCCC-3′), which was not used as an acceptor in the P2-catalyzed TNTase reaction in single-stranded form (Fig. 5, right panel, lane 4). Since the complementary strand of the oligonucleotide 6n_RNA does not contain U and thus the replication product as such should not be radioactively labeled (reactions with [α-32P]UTP and unlabeled NTPs), the detected extensions in the replication product must be UTP specific and nontemplated. Consequently, under experimental conditions containing only a single nucleotide substrate (UTP), one or few nucleotides may be added to the given ssRNA molecule, but when all NTPs were present, only the newly synthesized strand in the dsRNA product was extended. This suggests that the template-dependent replication reaction of φ6 P2 is favored over the TNTase reaction on ssRNA molecules and that the addition of nontemplated NTPs may occur as a termination step of the RNA-dependent RNA polymerization reaction.
FIG. 5.
TNTase activity under RNA replication conditions. 26n_RNA (left panel) and 6n_RNA (right panel) oligonucleotides were incubated with the P2 polymerase in the presence of different additives as indicated below the panel. Shown is an autoradiogram of the reaction products after separation on a denaturing 20% polyacrylamide gel. The RNA or the donor UTP was radioactively labeled (marked with an asterisk). The mobilities of RNA molecules of different sizes are indicated on the left sides of the panels (based on the single nucleotide ladder defined in the legend to Fig. 2). The reaction products within each lane (1 to 4) are schematically depicted below the panels. A black line represents the prelabeled RNA oligonucleotide, a gray line an unlabeled RNA, a black asterisk a labeled UMP, and a gray asterisk an unlabeled UMP incorporated into the RNA.
Virion-derived φ6 nucleocapsids catalyze the TNTase reaction by using genomic dsRNA segments as acceptors.
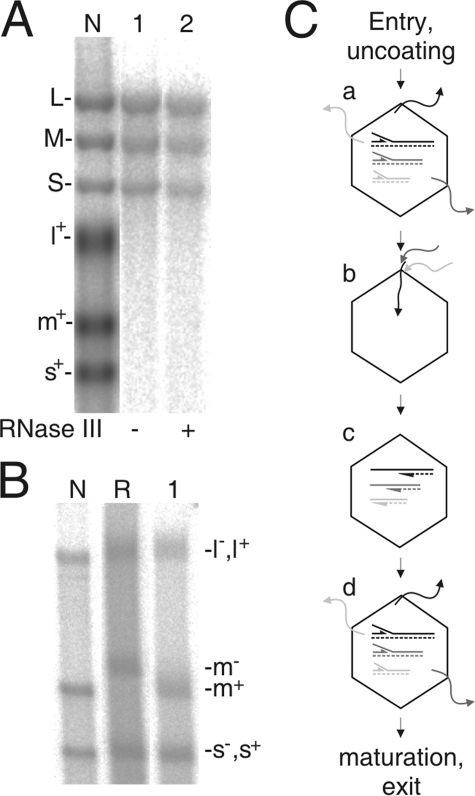
It was apparent that TNTase activity was associated with the purified φ6 P2 protein. It is intriguing to consider whether such activity is also an intrinsic property of the P2 protein within the viral particle. Therefore, φ6 nucleocapsids were isolated and the incorporation of label from the [α-32P]UTP donor to the viral genomic dsRNA, encapsidated within the viral particle, was analyzed (Fig. 6). It appeared that the P2 polymerase protein produced in P. syringae during φ6 infection and located within φ6 virions could catalyze label incorporation into the viral genome. All three genomic segments were equally labeled (Fig. 6A, lane 1), and the labeled products were resistant to RNase III treatment, indicating that the reaction products reside protected within the viral particles and do not originate from reactions carried out by disrupted particles (Fig. 6A, lane 2).
FIG. 6.
TNTase activity within φ6 nucleocapsids. Nucleocapsids were incubated with a [α-32P]UTP donor under standard TNTase reaction conditions, and the reaction products (lanes 1 and 2 in panel A and lane 1 in panel B) were separated by either native agarose gel electrophoresis (A) or strand-separating gel electrophoresis (B). (A) The reaction products were treated with RNase III prior to gel analysis as indicated below the panel. Lane N is as defined in the legend to Fig. 1A (control for labeled positive strands). Lane R is a product of the replication reaction using purified P2 and positive strands (s+, m+, and l+) of the φ6 genome (control for labeled negative strands). The mobilities of the dsRNA segments (S, M, and L in panel A), the positive strands (s+, m+, and l+ in panels A and B), and the negative strands (s−, m−, and l− in panel B) are indicated. (C) Schematic presentation of the φ6 replication cycle. (a) The viral particles partly uncoated upon entry initiate the production of positive-sense RNA segments within the cell cytoplasm. Transcription occurs via a semiconservative strand displacement mechanism (59); the newly produced positive strand stays connected with the negative strand in the dsRNA, while the positive strand produced within the previous host exits the viral particle. The viral mRNA molecules extruded from the polymerase complexes direct viral protein synthesis. (b) The newly produced proteins (including P2) assemble to form empty polymerase complexes which subsequently package one copy of each of the positive-sense RNA segments (s+, m+, and l+) (solid lines). (c) The packaged segments direct the synthesis of complementary negative strands (dotted lines). (d) To induce the production of additional viral proteins, the dsRNA-filled particles initiate a new cycle of transcription. Subsequently, the particles mature and the progeny virions exit the cell (44).
The above observations raise a question of strand specificity in the φ6 P2-catalyzed TNTase reaction within the viral particles. The TNTase reaction products were analyzed in an agarose gel (38) in which the positive (m+) and negative (m−) strands of the φ6 M segment could be separated (Fig. 6B). The nucleocapsids originate from viral particles that have been transcribing prior to their maturation into virions and are also programmed to initiate transcription upon entering into a new host cell (Fig. 6C). Consequently, labeling of the positive strand would result from the termination of transcription (that took place prior to maturation) (Fig. 6C), while labeling of the negative strand would suggest that the template strand is extended prior to the initiation of a new round of transcription. It was observed that within φ6 nucleocapsids, the genomic segment M was labeled solely at its positive strand (Fig. 6B, lane 1). The positive-strand specificity most likely also applies for the other two segments, although it was not possible to separate positive and negative strands of S and L in the gel system applied (Fig. 6B, compare lanes N and R). These results propose that, within virions, the P2-catalyzed TNTase reaction is a termination step for the P2-catalyzed transcription reaction.
DISCUSSION
dsRNA virus replication and transcription are carried out by the RNA-dependent RNA polymerase residing inside the polymerase complex particle (28). We report here the first evidence that the polymerase complex of a dsRNA virus as well as the isolated polymerase protein is capable of template-independent terminal nucleotidyl addition. This activity is an inherent property of the purified φ6 protein P2 (Fig. 1A to C) and also the φ6 nucleocapsid carrying the P2 protein (Fig. 6A). P2 was shown to catalyze a typical TNTase reaction specific for the 3′ terminus of the nucleic acid molecule (Fig. 2C). The Mg2+ ion dependence (Fig. 1E) and the sensitivity to mutation at the active center of the polymerase (Fig. 1A to C) suggest that the terminal nucleotidyl addition is catalyzed using the two-metal ion mechanism common to all polymerases (54). The new activity described here extends the potential applications of φ6 RNA-dependent RNA polymerase.
The φ6 P2-catalyzed TNTase reaction readily accepts different acceptor nucleic acids (Fig. 4). This observation correlates with the low template specificity of the P2-catalyzed replication reaction (27). Both observations reflect the fact that P2 resides within the viral polymerase complex that selectively packages only virus-specific ssRNA molecules (s+, m+, and l+) for replication (for a review, see reference 43) Initially, the TNTase activity of P2 was discovered using single-stranded RNA substrates, but double-stranded molecules seem to be the preferred acceptors (Fig. 4D). The narrow template tunnel utilized during the P2-catalyzed template-dependent RNA polymerization reaction can accommodate ssRNA but not dsRNA (Fig. 7A) (8, 26), and consequently, isolated P2 strongly favors single-stranded templates (26) for template-dependent reactions. The high TNTase activity on both ssRNA and dsRNA molecules (Fig. 4) indicates that a path other than the template tunnel is utilized to access the catalytic site for terminal nucleotidyl addition. In contrast to the template-dependent RNA transcription activity which requires opening of the double-stranded template RNA, the TNTase reaction apparently depends on opening of the compact apoenzyme structure but not the dsRNA. The ability to accept and replicate artificial circular templates also indicates that the apoenzyme may adopt different conformations which allow the polymerase to enclose circular templates (49). Thus, structural flexibility seems to be an intrinsic property of the apoenzyme.
FIG. 7.
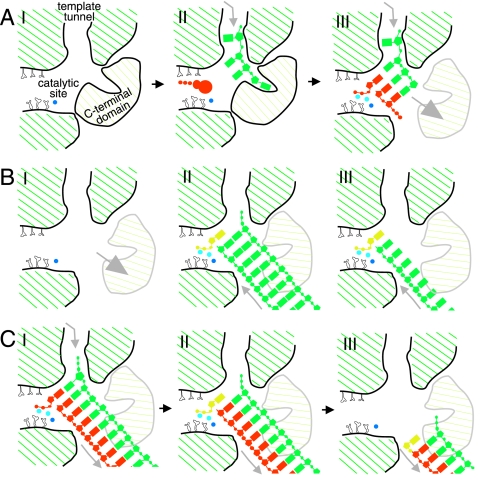
Model for the φ6 P2-catalyzed TNTase reaction. The template/acceptor RNA is depicted in bluish green and the product strand in vermillion. The Mn2+ ion is in blue and the Mg2+ ion in sky blue. (A) Cartoon presentation of the apoenzyme (I), an incoming nucleotide (vermillion), and a template ssRNA having a 3′ end positioned in the specificity pocket, well past the catalytic site (II). There is an opening of the exit path (C-terminal subdomain moves) after catalysis of the dinucleotide product to allow egress of the duplex (III) (modified from reference 8). (B) Model for the TNTase reaction with isolated P2 and added single- or double-stranded RNA. (I) Apoenzyme with the exit path open. dsRNA (II) and ssRNA (III) acceptor molecules reaching the catalytic site via the exit route of the polymerase for the terminal nucleotidyl addition are shown. The donor NTP molecule to be added at the 3′ end of the acceptor RNA is in yellow. (C) Termination of the RNA-dependent RNA polymerization reaction by nontemplated terminal nucleotidyl addition. (I) The last complementary nucleotide is added to the growing duplex RNA. (II) A donor NTP is assembled in the catalytic site, and a nucleotidyl (yellow) is transferred to the 3′ end of the newly synthesized strand of the duplex RNA. (III) The double-stranded product is released from the polymerase.
The mutant polymerase P2E491Q has almost normal replication activity, while its TNTase activity is less than 10% of that of the wild-type polymerase. Interestingly, when this mutation is introduced into the cDNA clone of the viral genome segment L, which is then transformed into competent host cells together with the cDNA clones of the S and M segments (57), the formation of live viruses is reduced to approximately 1/10 of that obtained with wild-type cDNA clones (M. M. Poranen and D. H. Bamford, unpublished data). This reduction could imply that the TNTase activity has biological relevance, although we cannot exclude other defects in the P2E491Q polymerase.
The P2-catalyzed TNTase activity shares several features with the TNTase reaction catalyzed by HCV NS5B polymerase. Both enzymes have an ability to transfer one or a few nucleotides to a given RNA molecule, and the reaction is dependent on divalent cations (Fig. 1D and 2A and B) (3, 46). In addition, the HCV polymerase shows low specificity for acceptor RNA and donor NTP (46). Apparently, the φ6 and HCV polymerases not only are structurally alike but also share very similar biochemical properties (8, 21-24, 47, 48). Therefore, the results obtained here for the φ6 P2 polymerase could also apply to other viral RNA-dependent RNA polymerases.
Although TNTase activity has been reported for several viral RNA-dependent RNA polymerases, there has been a lack of mechanistic insight into how the viral polymerases, directed to perform template-dependent RNA synthesis, may catalyze the template-independent nucleotidyltransfer reaction. The dual function of these polymerases makes the situation unique in comparison to that with the cellular TNTases, which are dedicated to template-independent nucleotidyltransfer only. Based on the biochemical data presented here and the previously described φ6 P2 structure (8), we propose a model for the mechanism of template-independent nucleotide addition (Fig. 7).
The high TNTase activity on double-stranded RNA and DNA substrates (Fig. 4C) as well as the reduced TNTase activity on denatured double-stranded molecules (Fig. 4D) suggests that the acceptor nucleic acid molecules do not enter through the narrow template tunnel utilized for replication (Fig. 7) (8). During the replication reaction, the double-stranded product is translocated via an exit path that is covered by the C-terminal subdomain during initiation (Fig. 7A) (8). It appears that the C terminus of the polymerase is a dynamic domain that could adopt a (transient) conformation in which a tunnel wide enough for dsRNA egress would appear (Fig. 7B, panel I). This repositioning of the C-terminal subdomain would allow the acceptor nucleic acid (both single and double stranded) to dock near the catalytic site of P2 via the exit path for terminal nucleotidyl addition (Fig. 7B, panels II and III). This model is supported by several observations based on data obtained here as well as previously: (i) double-stranded nucleic acid molecules are favored acceptors in the TNTase reaction (Fig. 4D); (ii) double-stranded molecules cannot enter via the template tunnel due to volume constraints (Fig. 7A) (8); (iii) reduction in the structural flexibility of the polymerase reduces its TNTase activity (results obtained with P2E491Q); and (iv) the catalytic site of the enzyme is not readily accessible for the 3′ end of the acceptor nucleic acid entering via the template tunnel, as observed from the φ6 P2 crystal structure (Fig. 7A, panel II) (8). Furthermore, the 3′ end of a nucleic acid molecule entering via the exit path would be in the correct position with respect to the catalytic site to allow the nucleotidyl transfer (Fig. 7B, panels II and III). After the catalysis of nontemplated nucleotide addition, the acceptor nucleic acid could leave the catalytic site via the same exit path.
Although TNTase activities have been reported for several viral polymerases of different origins, it is not clear whether these activities have biological relevance. The labeling of the ssRNA or dsRNA in a buffer containing only a single NTP (Fig. 1 to 4) is most likely an in vitro activity of the isolated enzyme which is suppressed in the presence of all four NTPs (Fig. 5) due to the assembly of the replication/transcription initiation complex. The optimal, low-salt reaction conditions for the TNTase reaction possibly allow RNA molecules to slip into the exit channel (Fig. 7B) in a manner not readily reached under higher, more-physiological salt conditions. However, the addition of nontemplated nucleotides to the 3′ end of the newly synthesized strand (Fig. 5) and the positive strand-specific TNTase activity of the viral nucleocapsids (Fig. 6) might reflect biological functions. Interestingly, both of these observations position the TNTase reaction at the termination phase of template-dependent RNA synthesis (Fig. 7C). The addition of nontemplated nucleotides at the termination phase of the RNA-dependent RNA polymerization reaction would be mechanistically the same as that proposed above for the TNTase reaction on double-stranded acceptor molecules (Fig. 7B, panel II), with the exception that the double-stranded substrate is produced by the enzyme, and thus, the 3′ end of the product strand is inevitably in the correct position with respect to the catalytic site for nucleotidyl addition. The results obtained with φ6 nucleocapsids also imply that, within the virions, the polymerase stays in contact with the 3′ end of the previously synthesized positive strand and that the initiation complex for the next round of transcription will assemble upon infection of a new host.
The potential of adding nontemplated nucleotides by the φ6 P2 polymerase as a termination step of template-dependent RNA polymerization predicts that the φ6 genome should contain single nucleotide overhangs on one or both ends of the genome depending on whether the TNTase activity was operating in the termination of both transcription and replication. No such overhangs have been reported; the direct RNA sequencing of the genome ends has implied that each φ6 genome segment has flush ends at both termini (19, 58). In addition, nontemplated residues were not detected in a more recent rapid amplification of cDNA ends of the φ6 genome termini (Poranen and Bamford, unpublished). However, the addition of a terminal nucleotide might occur as the polymerase disengages from the 3′ end of the most recently synthesized positive strand to initiate a new round of transcription within a new host (this would be congruent with the observed TNTase activity in nucleocapsids) (Fig. 6). Such addition of a terminal nucleotide would not be detected in the mature virions.
Single, nontemplated nucleotides at the genome termini would have consequences for the replication mechanism of the virus. To maintain the integrity of the genome from generation to generation, the replication should initiate not from the very end of the template strand but from the penultimate nucleotide. In fact, such a mechanism has been proposed for the replication complex of bacteriophage Qβ (51). Interestingly, structural studies of the φ6 polymerase have depicted that the first complementary nucleotide assembled in the initiation complex is actually complementary to the penultimate nucleotide of the template. Later, the template strand ratchets back and the nucleotide complementary to the very 3′ end of the template is assembled (8, 53). This type of initiation mechanism applies to a template mimicking the 3′ end of the φ6 negative strands (…UUUUUCC-3′), used in the initiation of transcription. Whether similar template sliding occurs during the initiation of replication is not known. This would require structural analysis using template RNAs mimicking the conserved 3′ end of the plus strands (…CUCUCUCUCU-3′). The specific contacts with the 3′-terminal cytidine in the initiation of transcription (8, 53) indicate that the polymerase may actually distinguish the termini of the negative and positive strands (C-3′ and U-3′, respectively) and thus potentially may apply different initiation pathways.
The nontemplated nucleotide addition by the reverse transcriptase of human immunodeficiency virus promotes strand transfer and is potentially an important source of new mutations (17, 39). The φ6 P2-catalyzed TNTase activity could also operate in the recombination process (31, 36, 37, 45). In fact, the crossover sites within φ6 genome segments as well as the rearrangement sites within the rotavirus genome occasionally contain nucleotides that do not originate from the donor or from the receptor strand (40, 45). These extra nucleotides may present an addition of nontemplated nucleotides by the viral polymerases. Together, our findings demonstrate that the template-dependent addition of nucleotides not only is a property of positive-sense RNA viruses or retroviruses but is also found in the polymerase of dsRNA bacteriophage φ6. This finding highlights the flexibility of viral RNA polymerases.
Acknowledgments
Sampo Vehma, Riitta Tarkiainen, Anna Latva-Käyrä, and Antti Aalto are acknowledged for their excellent technical assistance, and Eugene Makeyev is acknowledged for constructing plasmid pEMG2. We also greatly appreciate discussions with David Stuart and Jonathan Grimes (Oxford University).
This investigation was supported by grants from Finnish Centre of Excellence Program 2006-2011 (grant 1213467) from the Academy of Finland (to D.H.B.), Helsinki Graduate School in Biotechnology and Molecular Biology (to M.R.L.K.), and Kuopio Naturalists' Society (to M.M.P.).
Footnotes
Published ahead of print on 9 July 2008.
REFERENCES
- 1.Aalto, A. P., L. P. Sarin, A. A. van Dijk, M. Saarma, M. M. Poranen, U. Arumae, and D. H. Bamford. 2007. Large-scale production of dsRNA and siRNA pools for RNA interference utilizing bacteriophage φ6 RNA-dependent RNA polymerase. RNA 13422-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bamford, D. H., P. M. Ojala, M. Frilander, L. Walin, and J. K. H. Bamford. 1995. Isolation, purification, and function of assembly intermediates and subviral particles of bacteriophages PRD1 and φ6, p. 455-474. In K. W. Adolph (ed.), Methods in molecular genetics, vol. 6. Academic Press, San Diego, CA.
- 3.Behrens, S. E., L. Tomei, and R. De Francesco. 1996. Identification and properties of the RNA-dependent RNA polymerase of hepatitis C virus. EMBO J. 1512-22. [PMC free article] [PubMed] [Google Scholar]
- 4.Brennan, V. E., L. Field, P. Cizdziel, and J. A. Bruenn. 1981. Sequences at the 3′ ends of yeast viral dsRNAs: proposed transcriptase and replicase initiation sites. Nucleic Acids Res. 94007-4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bressanelli, S., L. Tomei, F. A. Rey, and R. De Francesco. 2002. Structural analysis of the hepatitis C virus RNA polymerase in complex with ribonucleotides. J. Virol. 763482-3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bruenn, J. A. 2003. A structural and primary sequence comparison of the viral RNA-dependent RNA polymerases. Nucleic Acids Res. 311821-1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bruenn, J. A., and V. E. Brennan. 1980. Yeast viral double-stranded RNAs have heterogeneous 3′ termini. Cell 19923-933. [DOI] [PubMed] [Google Scholar]
- 8.Butcher, S. J., J. M. Grimes, E. V. Makeyev, D. H. Bamford, and D. I. Stuart. 2001. A mechanism for initiating RNA-dependent RNA polymerization. Nature 410235-240. [DOI] [PubMed] [Google Scholar]
- 9.Butcher, S. J., E. V. Makeyev, J. M. Grimes, D. I. Stuart, and D. H. Bamford. 2000. Crystallization and preliminary X-ray crystallographic studies on the bacteriophage φ6 RNA-dependent RNA polymerase. Acta Crystallogr. D 561473-1475. [DOI] [PubMed] [Google Scholar]
- 10.Chen, D., and J. T. Patton. 2001. Reverse transcriptase adds nontemplated nucleotides to cDNAs during 5′-RACE and primer extension. BioTechniques 30574-582. [DOI] [PubMed] [Google Scholar]
- 11.Choi, K. H., J. M. Groarke, D. C. Young, R. J. Kuhn, J. L. Smith, D. C. Pevear, and M. G. Rossmann. 2004. The structure of the RNA-dependent RNA polymerase from bovine viral diarrhea virus establishes the role of GTP in de novo initiation. Proc. Natl. Acad. Sci. USA 1014425-4430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cohen, S. N. 1995. Surprises at the 3′ end of prokaryotic RNA. Cell 80829-832. [DOI] [PubMed] [Google Scholar]
- 13.Collmer, C. W., and J. M. Kaper. 1985. Double-stranded RNAs of cucumber mosaic virus and its satellite contain an unpaired terminal guanosine: implications for replication. Virology 145249-259. [DOI] [PubMed] [Google Scholar]
- 14.Frilander, M., and D. H. Bamford. 1995. In vitro packaging of the single-stranded RNA genomic precursors of the segmented double-stranded RNA bacteriophage φ6: the three segments modulate each other's packaging efficiency. J. Mol. Biol. 246418-428. [DOI] [PubMed] [Google Scholar]
- 15.Frilander, M. J., and J. J. Turunen. 2005. RNA ligation by T4 DNA ligase, p. 36-52. In R. K. Hartmann, A. Bindereif, A. Schön, and E. Westhof (ed.), Handbook of RNA biochemistry. Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim, Germany.
- 16.Fullerton, S. W., M. Blaschke, B. Coutard, J. Gebhardt, A. Gorbalenya, B. Canard, P. A. Tucker, and J. Rohayem. 2007. Structural and functional characterization of sapovirus RNA-dependent RNA polymerase. J. Virol. 811858-1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Golinelli, M. P., and S. H. Hughes. 2002. Nontemplated base addition by HIV-1 RT can induce nonspecific strand transfer in vitro. Virology 294122-134. [DOI] [PubMed] [Google Scholar]
- 18.Gottlieb, P., J. Strassman, X. Qiao, M. Frilander, A. Frucht, and L. Mindich. 1992. In vitro packaging and replication of individual genomic segments of bacteriophage φ6 RNA. J. Virol. 662611-2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iba, H., T. Watanabe, Y. Emori, and Y. Okada. 1982. Three double-stranded RNA genome segments of bacteriophage φ6 have homologous terminal sequences. FEBS Lett. 141111-115. [DOI] [PubMed] [Google Scholar]
- 20.Kudla, J., R. Hayes, and W. Gruissem. 1996. Polyadenylation accelerates degradation of chloroplast mRNA. EMBO J. 157137-7146. [PMC free article] [PubMed] [Google Scholar]
- 21.Laurila, M. R., E. V. Makeyev, and D. H. Bamford. 2002. Bacteriophage φ6 RNA-dependent RNA polymerase: molecular details of initiating nucleic acid synthesis without primer. J. Biol. Chem. 27717117-17124. [DOI] [PubMed] [Google Scholar]
- 22.Laurila, M. R., P. S. Salgado, D. I. Stuart, J. M. Grimes, and D. H. Bamford. 2005. Back-priming mode of φ6 RNA-dependent RNA polymerase. J. Gen. Virol. 86521-526. [DOI] [PubMed] [Google Scholar]
- 23.Lesburg, C. A., M. B. Cable, E. Ferrari, Z. Hong, A. F. Mannarino, and P. C. Weber. 1999. Crystal structure of the RNA-dependent RNA polymerase from hepatitis C virus reveals a fully encircled active site. Nat. Struct. Biol. 6937-943. [DOI] [PubMed] [Google Scholar]
- 24.Lévêque, V. J., R. B. Johnson, S. Parsons, J. Ren, C. Xie, F. Zhang, and Q. M. Wang. 2003. Identification of a C-terminal regulatory motif in hepatitis C virus RNA-dependent RNA polymerase: structural and biochemical analysis. J. Virol. 779020-9028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lupold, D. S., A. G. Caoile, and D. B. Stern. 1999. Polyadenylation occurs at multiple sites in maize mitochondrial cox2 mRNA and is independent of editing status. Plant Cell 111565-1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Makeyev, E. V., and D. H. Bamford. 2000. The polymerase subunit of a dsRNA virus plays a central role in the regulation of viral RNA metabolism. EMBO J. 196275-6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Makeyev, E. V., and D. H. Bamford. 2000. Replicase activity of purified recombinant protein P2 of double-stranded RNA bacteriophage φ6. EMBO J. 19124-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Makeyev, E. V., and J. M. Grimes. 2004. RNA-dependent RNA polymerases of dsRNA bacteriophages. Virus Res. 10145-55. [DOI] [PubMed] [Google Scholar]
- 29.Mertens, P. 2004. The dsRNA viruses. Virus Res. 1013-13. [DOI] [PubMed] [Google Scholar]
- 30.Milligan, J. F., D. R. Groebe, G. W. Witherell, and O. C. Uhlenbeck. 1987. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 158783-8798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mindich, L., X. Qiao, S. Onodera, P. Gottlieb, and M. Frilander. 1994. RNA structural requirements for stability and minus-strand synthesis in the dsRNA bacteriophage φ6. Virology 202258-263. [DOI] [PubMed] [Google Scholar]
- 32.Mindich, L., X. Qiao, S. Onodera, P. Gottlieb, and J. Strassman. 1992. Heterologous recombination in the double-stranded RNA bacteriophage φ6. J. Virol. 662605-2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neufeld, K. L., J. M. Galarza, O. C. Richards, D. F. Summers, and E. Ehrenfeld. 1994. Identification of terminal adenylyl transferase activity of the poliovirus polymerase 3Dpol. J. Virol. 685811-5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ojala, P. M., and D. H. Bamford. 1995. In vitro transcription of the double-stranded RNA bacteriophage φ6 is influenced by purine NTPs and calcium. Virology 207400-408. [DOI] [PubMed] [Google Scholar]
- 35.Olkkonen, V. M., P. Gottlieb, J. Strassman, X. Y. Qiao, D. H. Bamford, and L. Mindich. 1990. In vitro assembly of infectious nucleocapsids of bacteriophage φ6: formation of a recombinant double-stranded RNA virus. Proc. Natl. Acad. Sci. USA 879173-9177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Onodera, S., X. Qiao, P. Gottlieb, J. Strassman, M. Frilander, and L. Mindich. 1993. RNA structure and heterologous recombination in the double-stranded RNA bacteriophage φ6. J. Virol. 674914-4922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Onodera, S., Y. Sun, and L. Mindich. 2001. Reverse genetics and recombination in φ8, a dsRNA bacteriophage. Virology 286113-118. [DOI] [PubMed] [Google Scholar]
- 38.Pagratis, N., and H. R. Revel. 1990. Detection of bacteriophage φ6 minus-strand RNA and novel mRNA isoconformers synthesized in vivo and in vitro, by strand-separating agarose gels. Virology 177273-280. [DOI] [PubMed] [Google Scholar]
- 39.Patel, P. H., and B. D. Preston. 1994. Marked infidelity of human immunodeficiency virus type 1 reverse transcriptase at RNA and DNA template ends. Proc. Natl. Acad. Sci. USA 91549-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Patton, J. T., Z. Taraporewala, D. Chen, V. Chizhikov, M. Jones, A. Elhelu, M. Collins, K. Kearney, M. Wagner, Y. Hoshino, and V. Gouvea. 2001. Effect of intragenic rearrangement and changes in the 3′ consensus sequence on NSP1 expression and rotavirus replication. J. Virol. 752076-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pirttimaa, M. J., and D. H. Bamford. 2000. RNA secondary structures of the bacteriophage φ6 packaging regions. RNA 6880-889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Poranen, M. M., A. O. Paatero, R. Tuma, and D. H. Bamford. 2001. Self-assembly of a viral molecular machine from purified protein and RNA constituents. Mol. Cell 7845-854. [DOI] [PubMed] [Google Scholar]
- 43.Poranen, M. M., M. J. Pirttimaa, and D. H. Bamford. 2005. Encapsidation of the segmented double-stranded RNA genome of bacteiophage φ6. In C. Catalano (ed.), Viral genome packaging machines: genetics, structure, and mechanism. Landes Bioscience, Georgetown, TX.
- 44.Poranen, M. M., R. Tuma, and D. H. Bamford. 2005. Assembly of double-stranded RNA bacteriophages. Adv. Virus Res. 6415-43. [DOI] [PubMed] [Google Scholar]
- 45.Qiao, X., J. Qiao, and L. Mindich. 1997. An in vitro system for the investigation of heterologous RNA recombination. Virology 227103-110. [DOI] [PubMed] [Google Scholar]
- 46.Ranjith-Kumar, C. T., J. Gajewski, L. Gutshall, D. Maley, R. T. Sarisky, and C. C. Kao. 2001. Terminal nucleotidyl transferase activity of recombinant Flaviviridae RNA-dependent RNA polymerases: implication for viral RNA synthesis. J. Virol. 758615-8623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ranjith-Kumar, C. T., L. Gutshall, M. J. Kim, R. T. Sarisky, and C. C. Kao. 2002. Requirements for de novo initiation of RNA synthesis by recombinant flaviviral RNA-dependent RNA polymerases. J. Virol. 7612526-12536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ranjith-Kumar, C. T., L. Gutshall, R. T. Sarisky, and C. C. Kao. 2003. Multiple interactions within the hepatitis C virus RNA polymerase repress primer-dependent RNA synthesis. J. Mol. Biol. 330675-685. [DOI] [PubMed] [Google Scholar]
- 49.Ranjith-Kumar, C. T., and C. C. Kao. 2006. Recombinant viral RdRps can initiate RNA synthesis from circular templates. RNA 12303-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ranjith-Kumar, C. T., J. L. Santos, L. L. Gutshall, V. K. Johnston, J. Lin-Goerke, M. J. Kim, D. J. Porter, D. Maley, C. Greenwood, D. L. Earnshaw, A. Baker, B. Gu, C. Silverman, R. T. Sarisky, and C. Kao. 2003. Enzymatic activities of the GB virus-B RNA-dependent RNA polymerase. Virology 312270-280. [DOI] [PubMed] [Google Scholar]
- 51.Rensing, U., and J. T. August. 1969. The 3′-terminus and the replication of phage RNA. Nature 224853-856. [DOI] [PubMed] [Google Scholar]
- 52.Rohayem, J., K. Jager, I. Robel, U. Scheffler, A. Temme, and W. Rudolph. 2006. Characterization of norovirus 3Dpol RNA-dependent RNA polymerase activity and initiation of RNA synthesis. J. Gen. Virol. 872621-2630. [DOI] [PubMed] [Google Scholar]
- 53.Salgado, P. S., E. V. Makeyev, S. J. Butcher, D. H. Bamford, D. I. Stuart, and J. M. Grimes. 2004. The structural basis for RNA specificity and Ca2+ inhibition of an RNA-dependent RNA polymerase. Structure 12307-316. [DOI] [PubMed] [Google Scholar]
- 54.Steitz, T. A. 1998. A mechanism for all polymerases. Nature 391231-232. [DOI] [PubMed] [Google Scholar]
- 55.Strömsten, N. J., D. H. Bamford, and J. K. Bamford. 2005. In vitro DNA packaging of PRD1: a common mechanism for internal-membrane viruses. J. Mol. Biol. 348617-629. [DOI] [PubMed] [Google Scholar]
- 56.Studier, F. W., and B. A. Moffatt. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189113-130. [DOI] [PubMed] [Google Scholar]
- 57.Sun, Y., X. Qiao, and L. Mindich. 2004. Construction of carrier state viruses with partial genomes of the segmented dsRNA bacteriophages. Virology 319274-279. [DOI] [PubMed] [Google Scholar]
- 58.Szekeres, M., B. H. Brownstein, H. R. Revel, and R. Haselkorn. 1985. Terminal sequences of the bacteriophage φ6 segmented dsRNA genome and its messenger RNAs. Virology 1421-11. [DOI] [PubMed] [Google Scholar]
- 59.Usala, S. J., B. H. Brownstein, and R. Haselkorn. 1980. Displacement of parental RNA strands during in vitro transcription by bacteriophage φ6 nucleocapsids. Cell 19855-862. [DOI] [PubMed] [Google Scholar]
- 60.van Dijk, A. A., M. Frilander, and D. H. Bamford. 1995. Differentiation between minus- and plus-strand synthesis: polymerase activity of dsRNA bacteriophage φ6 in an in vitro packaging and replication system. Virology 211320-323. [DOI] [PubMed] [Google Scholar]
- 61.Weber, H., and C. Weissmann. 1970. The 3′-termini of bacteriophage Qβ plus and minus strands. J. Mol. Biol. 51215-224. [DOI] [PubMed] [Google Scholar]
- 62.Wengler, G., and H. J. Gross. 1982. Terminal sequences of Sindbis virus-specific nucleic acids: identity in molecules synthesized in vertebrate and insect cells and characteristic properties of the replicative form RNA. Virology 123273-283. [DOI] [PubMed] [Google Scholar]
- 63.Wengler, G., and H. S. Gross. 1979. Replicative form of Semliki Forest virus RNA contains an unpaired guanosine. Nature 282754-756. [DOI] [PubMed] [Google Scholar]
- 64.Xu, F., and S. N. Cohen. 1995. RNA degradation in Escherichia coli regulated by 3′ adenylation and 5′ phosphorylation. Nature 374180-183. [DOI] [PubMed] [Google Scholar]
- 65.Yang, H., E. V. Makeyev, and D. H. Bamford. 2001. Comparison of polymerase subunits from double-stranded RNA bacteriophages. J. Virol. 7511088-11095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhong, W., L. L. Gutshall, and A. M. Del Vecchio. 1998. Identification and characterization of an RNA-dependent RNA polymerase activity within the nonstructural protein 5B region of bovine viral diarrhea virus. J. Virol. 729365-9369. [DOI] [PMC free article] [PubMed] [Google Scholar]