Abstract
Recent studies suggest a possible takeover of host antimicrobial autophagy machinery by positive-stranded RNA viruses to facilitate their own replication. In the present study, we investigated the role of autophagy in coxsackievirus replication. Coxsackievirus B3 (CVB3), a picornavirus associated with viral myocarditis, causes pronounced intracellular membrane reorganization after infection. We demonstrate that CVB3 infection induces an increased number of double-membrane vesicles, accompanied by an increase of the LC3-II/LC3-I ratio and an accumulation of punctate GFP-LC3-expressing cells, two hallmarks of cellular autophagosome formation. However, protein expression analysis of p62, a marker for autophagy-mediated protein degradation, showed no apparent changes after CVB3 infection. These results suggest that CVB3 infection triggers autophagosome formation without promoting protein degradation by the lysosome. We further examined the role of the autophagosome in CVB3 replication. We demonstrated that inhibition of autophagosome formation by 3-methyladenine or small interfering RNAs targeting the genes critical for autophagosome formation (ATG7, Beclin-1, and VPS34 genes) significantly reduced viral replication. Conversely, induction of autophagy by rapamycin or nutrient deprivation resulted in increased viral replication. Finally, we examined the role of autophagosome-lysosome fusion in viral replication. We showed that blockage of the fusion by gene silencing of the lysosomal protein LAMP2 significantly promoted viral replication. Taken together, our results suggest that the host's autophagy machinery is activated during CVB3 infection to enhance the efficiency of viral replication.
Cellular autophagy is a membrane trafficking process that leads to the degradation of long-lived cytoplasmic proteins and excess or damaged organelles by lysosomal hydrolyases (16, 19). The hallmark of autophagy is the emergence of double-membrane vesicles called autophagosomes, which are derived from the elongation of crescent-shaped double-membrane structures known as the isolation membranes, during which a portion of the cytoplasm, including organelles, is sequestrated. Matured autophagosomes eventually fuse with lysosomes to degrade and/or recycle their contents. Recent research efforts using Saccharomyces cerevisiae have greatly expanded our knowledge on the molecular aspects of autophagy, with the discovery of 31 autophagy-related genes (ATGs) (16). Further research with other organisms identified orthologs for most of the yeast ATG products, suggesting that autophagy is a highly conserved cellular mechanism in eukaryotes (25). The formation and maturation of the autophagosome require the activation of two ubiquitin-like molecules, Atg8p (also known as LC3 [microtubule-associated protein light chain 3 in mammals]) and Atg12p, by the activating enzyme Atg7p (28). This process is tightly regulated through the actions of various kinases, phosphatases, and GTPases. The autophagy regulatory mechanisms mediated by the mammalian target of rapamycin (mTOR), phosphatidylinositol 3-kinase (PI3K), and eukaryotic initiation factor 2α (eIF2α) signaling pathways are the best-characterized mechanisms of this process (5, 19).
Autophagy not only is important in cellular homeostasis but also participates in various physiological processes, such as responses to environmental stress, development, aging, and tumor suppression (22). Dysfunctional autophagy has been linked to diseases such as neurodegenerative diseases, cancer, and heart diseases (23). In addition, autophagy plays a role in both innate and adaptive immune responses and helps in the clearance of intracellular microorganisms (32). For example, induction of autophagy by rapamycin treatment has been shown to promote clearance of Mycobacterium tuberculosis in macrophages (9). Moreover, some viruses, such as herpes simplex virus type 1, evolved to encode autophagy-blocking viral proteins to promote their survival in host cells (36). Yet it has also been documented that certain positive-stranded RNA viruses, such as poliovirus and rhinovirus, are able to subvert cellular autophagy to facilitate their own replication (11). Thus, the role of autophagy in the host seems to be dependent on the viral strain, but its overall relevance remains controversial.
Coxsackievirus B3 (CVB3), an enterovirus in the Picornaviridae family, is one major human pathogen causing viral myocarditis (14). CVB3 has a positive-stranded RNA genome packaged in a nonenveloped icosahedral viral capsid. Like other small RNA viruses, CVB3 is believed to require host membrane structure for its replication. Here we explore the role of several constituents of autophagy machinery in CVB3-infected cells by monitoring the presence of double-membrane vesicles and the activation of LC3. Also, we demonstrate the effects of perturbing the autophagy pathway, using pharmacological inhibitors or RNA interference, on viral replication. We provide the first evidence that cellular autophagy is promoted in CVB3-infected cells and is involved in coxsackievirus replication.
MATERIALS AND METHODS
Cell culture and materials.
HeLa cells and HEK293A cells obtained from the American Type Culture Collection were used for this study. HeLa cells were grown in complete medium (Dulbecco's modified Eagle's medium [DMEM] supplemented with 10% heat-inactivated newborn calf serum). HEK293A cells were maintained in complete medium with the addition of 1% nonessential amino acids. The majority of the experiments were performed with HeK293A cells, and critical experiments were repeated using Hela cells.
Rapamycin, ammonium chloride (NH4Cl), bafilomycin A, 3-methyladenine (3-MA), and a monoclonal anti-β-actin antibody were purchased from Sigma. A monoclonal anti-VP1 antibody was obtained from DakoCytomation. Beclin-1 small interfering RNA (siRNA), lysosome-associated membrane protein 2 (LAMP2) siRNA, and horseradish peroxidase-conjugated secondary antibodies were purchased from Santa Cruz Biotechnology. A monoclonal anti-VPS34 antibody was obtained from Invitrogen. VPS34 siRNA, ATG7 siRNA, and scramble control siRNA were obtained from Qiagen. A monoclonal anti-Beclin-1 antibody was purchased from BD Biosciences. Polyclonal rabbit anti-p62 was purchased from Progen. A polyclonal rabbit anti-LC3 antibody was obtained from MBL International. A polyclonal rabbit anti-phospho-eIF2α antibody was obtained from Cell Signaling. Polyclonal rabbit anti-ATG7 and anti-LAMP2 antibodies were obtained from Cell Signaling and Abcam, respectively.
Virus infection.
HeLa cells were infected at a multiplicity of infection (MOI) of 10 with CVB3 (Kandolf strain) or sham infected with phosphate-buffered saline (PBS) for 1 h in serum-free DMEM. The cells were then washed with PBS and cultured in complete medium for various times until they were harvested. HEK cells were infected in a similar fashion, but with a higher MOI of 100. For autophagy induction, cells were either treated with rapamycin for 3 h or starved in Hank's balanced salt solution (137 mM NaCl, 5.4 mM KCl, 0.8 mM MgSO4, 0.8 mM Na2HPO4, 0.4 mM KH2PO4, 4.2 mM NaHCO3, 20 mM HEPES, and 1.5 mM CaCl2, pH 7.4) for 2 h prior to virus infection. For inhibition experiments, cells were infected with CVB3 in the presence or absence of various inhibitors or vehicle (dimethyl sulfoxide) for 1 h, washed with PBS, and then incubated with fresh DMEM containing different concentrations of inhibitors unless otherwise specified in the figure legends.
Plaque assay.
The virus titer in the cell supernatant was determined by an agar overlay plaque assay performed in triplicate, as described previously (34). In brief, the supernatant from CVB3-infected cells was serially diluted 10-fold and overlaid on a 90 to 95% confluent monolayer of HeLa cells. After 1 h of incubation, the HeLa cells were washed with PBS, followed by overlay with 2 ml of complete medium containing 0.75% agar in each well. The cells were incubated at 37°C for 72 h and then fixed with Carnoy's fixative (75% ethanol-25% acetic acid) and stained with 1% crystal violet. Plaques were counted, and the viral titer was calculated as PFU per milliliter.
Western blot analysis.
After treatment, the cells were incubated with lysis buffer (50 mM Tris-HCl, pH 7.2, 250 mM NaCl, 0.1% NP-40, 2 mM EDTA, 10% glycerol) containing protease inhibitor cocktail (Roche) on ice for 10 min. Cell lysates were then harvested by scraping, followed by brief sonication and centrifugation at 12,000 × g for 10 min at 4°C. The protein concentration was determined by the Bradford assay (Bio-Rad). Equal amounts of protein were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and then transferred to nitrocellulose membranes (Amersham). The membranes were blocked for 1 h with 5% nonfat dry milk solution containing 0.1% Tween 20. The blots were then incubated for 1 h with the primary antibody, followed by incubation for another hour with a secondary antibody. Immunoreactive bands were visualized by enhanced chemiluminescence (SuperSignal West Pico kit; Pierce).
siRNA transfection.
Cells were grown to 50 to 80% confluence and then transiently transfected with various siRNAs at a concentration of 50 to 80 nM, using Lipofectamine 2000 (Invitrogen) or HiPerFect (Qiagen) according to the manufacturer's instructions. A scrambled siRNA was used as a negative control. The silencing efficiency was detected by Western blot analyses using the respective antibodies. Forty-eight hours after transfection, cells were infected with CVB3 as described above.
Confocal microscopy.
For the detection of autophagosomes, cells were transiently transfected with GFP-LC3 plasmid (a generous gift from Sharon A. Tooze at the Cancer Research UK London Research Institute, United Kingdom), using Lipofectamine (Invitrogen). After 24 h of transfection, cells were infected with CVB3 for 5 h, and the fluorescence of GFP-LC3 was observed under a Leica SP2 AOBS confocal fluorescence microscope. The number of cells with punctate GFP-LC3 localization relative to all green fluorescent protein (GFP)-positive cells was counted (a minimum of 200 GFP-positive cells were counted in total for each condition) and presented as a percentage, as previously described (10).
Transmission electron microscopy.
For ultrastructural analysis, HeLa cells and HEK293A cells were sham infected or infected with CVB3 at MOIs of 10 and 100, respectively, for 5 and 7 h. Cells were fixed in 2.5% glutaraldehyde (Polysciences Inc.) in PBS, pH 7.2, for 10 min at room temperature. Following three washes in PBS, cells were postfixed in 1% osmium tetroxide (Polysciences Inc.), 1% potassium ferrocyanide, 100 mM sodium cacodylate buffer, pH 7.4, for 1 h at room temperature. After several rinses in distilled water, cells were dehydrated in a graded series of acetone washes and embedded in Eponate 12 resin (Ted Pella Inc.). Sections (70 to 80 nm) were cut and viewed on a Tecnai 12 transmission electron microscope (FEI Inc.). Approximately 15 cells were counted, and for each cell the number of double-membrane vesicles was examined, as previously described (1). Autophagosomes were defined as double-membrane vacuoles measuring 0.2 to 1.0 μm.
RESULTS
CVB3 infection increases autophagosome formation.
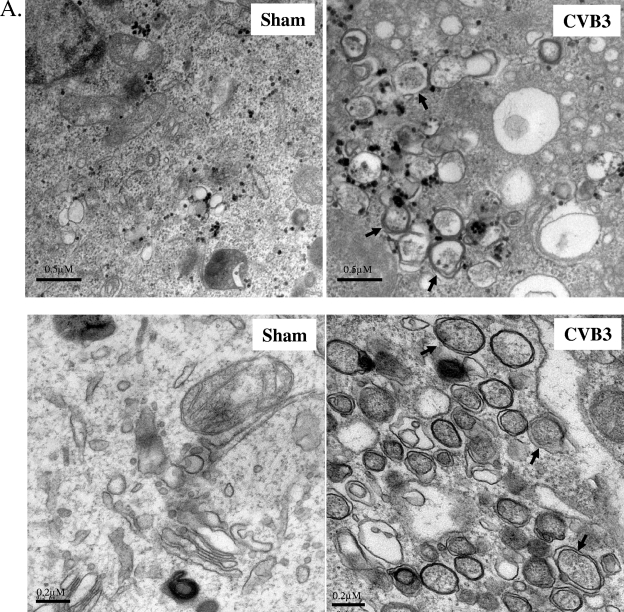
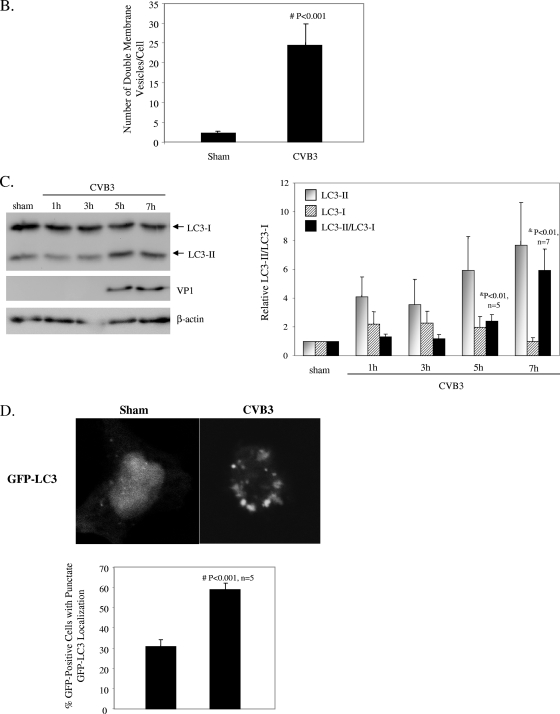
To determine whether CVB3 infection triggers cellular autophagy, ultrastructural analysis was performed with sham- or CVB3-infected HeLa and HEK293A cells. Electron micrographs of CVB3-infected cells revealed an increased prevalence of double-membrane vesicles in the perinuclear region compared to that for sham-infected cells (Fig. 1A). Further quantitative analysis (Fig. 1B) demonstrated a significant increase in autophagosome-like vesicles, which were characterized as double-membrane vesicles with a diameter of 0.2 to 1.0 μm present in the cytoplasm, in CVB3-infected cells.
FIG. 1.
CVB3 infection increases autophagosome formation. (A) Representative electron micrographs of sham- and CVB3-infected HeLa (top) and HEK293A (bottom) cells at 7 h postinfection. Arrows indicate representative autophagosomes that would be scored positive in panel B. (B) Quantitation of the numbers of autophagosomes in sham- and CVB3-infected HEK293A cells. Data shown represent the number of autophagosomes per cell under each condition (mean ± standard error [SE]). #, P < 0.001 compared to sham infection, by Student's t test. (C) The LC3-II/LC3-I ratio, which is a hallmark of autophagy, increases with the progression of CVB3 infection. (Left) Western blot analysis of LC3 and viral capsid protein VP1 expression in CVB3-infected cells at various times postinfection. β-Actin expression was examined as a protein loading control. (Right) Expression of LC3-II and LC3-I and ratio of LC3-II to LC3-I at the indicated times after CVB3 infection (means ± SE; n = 5 or 7 [as indicated]). LC3 expression was quantitated by densitometric analysis using NIH ImageJ v1.37 and normalized to sham infection, which was arbitrarily set to a value of 1.0. &, P < 0.01 compared to sham infection. (D) Representative confocal images and quantitation of autophagosomes in GFP-LC3-transfected HEK293A cells after CVB3 infection. HEK293A cells were transfected with GFP-LC3 plasmid for 24 h, followed by CVB3 infection. The GFP fluorescence was analyzed by confocal fluorescence microscopy. The percentage of cells with punctate GFP-LC3 localization relative to all GFP-positive cells was calculated and is presented as the mean ± SE (n = 5). #, P < 0.001 compared to sham infection.
To confirm that the observed double-membrane vesicles were indeed related to autophagy, we examined LC3 modification, another hallmark of autophagy (17, 24). Two forms of LC3 have been reported. In quiescent cells, LC3 resides in the cytoplasm in a precursor form, referred to as LC3-I. However, upon exposure to various autophagy stimuli, it is rapidly degraded and subsequently conjugated to phosphatidylethanolamine to form an LC3-phosphatidylethanolamine complex, commonly referred as LC3-II, which participates in the formation of autophagosomes and remains attached to matured autophagosomes (12). It has been shown that the LC3-II/LC3-I ratio correlates well with the number of autophagosomes (12). After CVB3 infection, Western blots were performed to detect the LC3-II/LC3-I ratio, using an anti-LC3 antibody which recognizes both forms of LC3. As shown in Fig. 1C, expression of LC3-II increased with the progression of CVB3 infection, which was accompanied by a decrease of LC3-I expression. The densitometry ratio of LC3-II to LC3-I demonstrated a cumulative increase in autophagosomes during CVB3 infection. Punctate accumulation of LC3, which represents the recruitment of LC3 to autophagic vacuoles, has also been suggested to be a characteristic of autophagy (12). After CVB3 infection, we showed that GFP-LC3-transfected cells with punctate GFP-LC3 distribution were markedly increased (Fig. 1D), further confirming the increased formation of autophagosomes after viral infection.
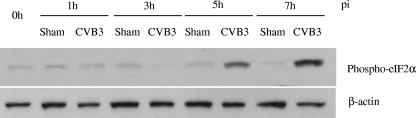
To determine the potential mechanism of induction of autophagosome formation, we examined the phosphorylation status of eIF2α and mTOR, two key regulators that control autophagosome formation and maturation. It has been shown that eIF2α phosphorylation positively regulates the autophagic pathway (36), whereas mTOR is a negative regulator of autophagosome formation (5). We found that eIF2α phosphorylation was increased at 5 and 7 h postinfection (Fig. 2), corresponding to an increased formation of autophagosomes in CVB3-infected cells. However, no significant changes in the levels of mTOR phosphorylation were observed after CVB3 infection (data not shown).
FIG. 2.
CVB3 infection induces eIF2α phosphorylation. Western blot analysis of phosphorylated eIF2α and β-actin (loading control) expression was performed at the indicated times postinfection. Similar results were obtained in two independent experiments.
Autophagy inhibitor 3-MA reduces CVB3 replication.
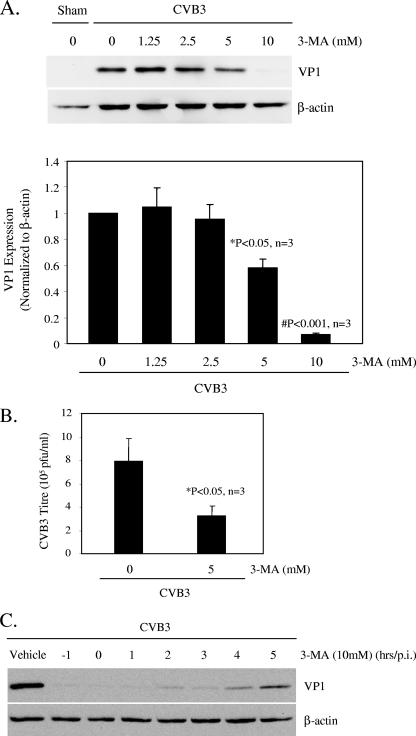
To determine whether the autophagosome induction during CVB3 infection was a host antiviral response or a viral replication mechanism, we tested the effect of 3-MA on CVB3 replication. 3-MA is a widely used selective inhibitor of autophagy which blocks the formation of autophagosomes and inhibits intracellular protein degradation without affecting protein synthesis (33). As shown in Fig. 3, treatment with 3-MA resulted in a dose-dependent reduction of viral capsid protein (VP1) expression (Fig. 3A) as well as in a significant reduction in CVB3 viral progeny titer (Fig. 3B). More importantly, we found that 3-MA treatment led to significant inhibition of viral protein synthesis, even when it was introduced during the later stages of the virus life cycle (Fig. 3C). This finding suggests that autophagic machinery may not have a destructive role during CVB3 infection and instead may play an important role in the replication of CVB3.
FIG. 3.
Autophagy inhibitor 3-MA reduces CVB3 replication. (A) (Top) Western blot analysis of viral protein VP1 and β-actin (loading control) expression 7 h following CVB3 infection, with or without 3-MA treatment, as indicated. (Bottom) VP1 expression was quantitated by densitometric analysis using NIH ImageJ v1.37 and normalized to β-actin expression. The VP1/β-actin ratio was further normalized to the CVB3-infected group without treatment, which was arbitrarily set to a value of 1.0. The data shown are means ± SE (n = 3), and significance was determined by Student's t test. *, P < 0.05; #, P < 0.001 (compared to nontreated cells). (B) Plaque assay measuring the CVB3 viral progeny titers in supernatants collected from infected cells treated with or without 3-MA. The data shown are means ± SE for three independent experiments. *, P < 0.05 compared to nontreated cells. (C) Western blot analysis of CVB3-infected cells treated with 10 mM 3-MA at different times, as indicated.
Induction of autophagy enhances viral growth.
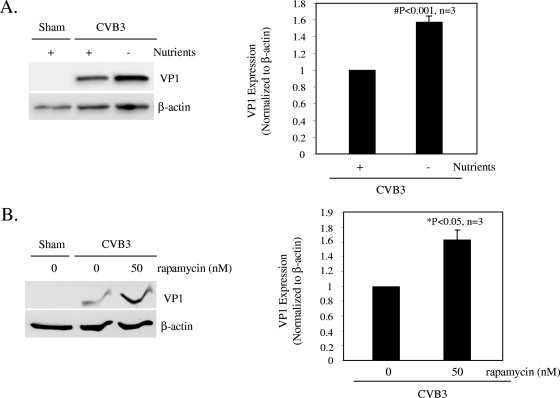
To further determine the role of autophagy, we investigated the effect of autophagosome induction on viral protein expression. Cells were deprived of amino acids (Fig. 4A), a condition that is known to be able to induce autophagy (5), or treated with the pharmacological reagent rapamycin, which has been shown to induce autophagy by inhibiting the mTOR pathway (3) (Fig. 4B). We showed that induction of autophagy through either nutrient deprivation or rapamycin treatment significantly increased viral protein expression (Fig. 4). These results further suggest a critical role of autophagy in the virus life cycle, although we cannot rule out other effects that these treatments might have on viral replication, independent of autophagy.
FIG. 4.
Induction of autophagy enhances viral replication. Western blot analysis of VP1 and β-actin expression was performed at 7 h postinfection. (A) Prior to CVB3 infection, cells were incubated in Hank's balanced salt solution for 2 h to induce cell starvation. (B) Cells were pretreated with rapamycin (50 nM) for 3 h. VP1 expression was quantitated, normalized, and analyzed as described in the legend to Fig. 3 (data are means ± SE; n = 3). *, P < 0.05; #, P < 0.001 (compared to nontreated cells).
Knockdown of the genes critical for autophagosome formation reduces CVB replication.
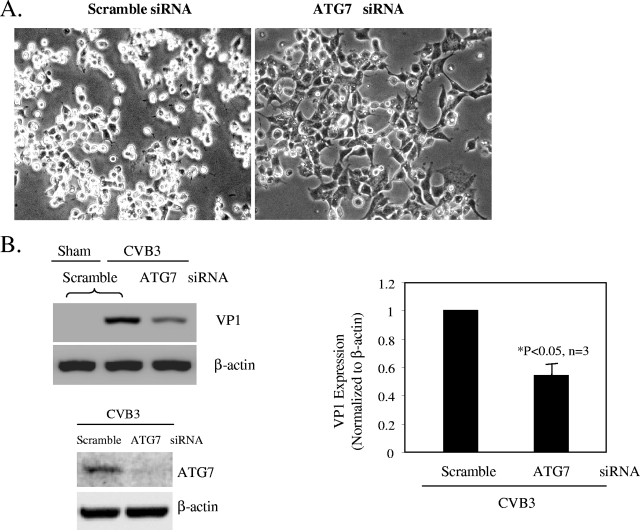
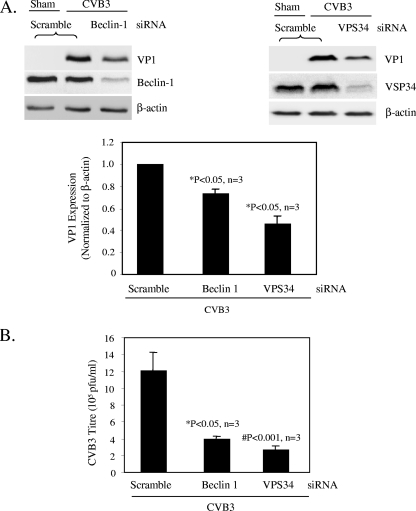
To extend the studies with pharmacological agents, we employed a target-specific RNA interference approach to assess the effect of autophagosome disruption on viral replication. Cells were transfected with siRNAs (for ATG7, Beclin-1, and VPS34) to knock down the genes required for different steps of autophagosome formation. The autophagy protein ATG7 is an enzyme essential for activation of autophagosome formation and maturation machinery (16, 19). Deletion of ATG7 in mice has been shown to result in reduced autophagosome formation and impaired degradation of proteins and organelles induced by starvation (20). As shown in the phase-contrast images in Fig. 5A, cells transfected with siRNA against ATG7 displayed greater resistance to CVB3 infection and maintained normal cell morphology, unlike the scramble siRNA-treated cells, which displayed severe virus-induced cytopathic effect. In addition, ATG7 siRNA treatment also blocked viral protein synthesis (Fig. 5B). Similarly, disruption of the class III PI3K signaling complex required for autophagosome formation by Beclin-1 and VPS34 siRNAs resulted in significant reductions in CVB3 replication (Fig. 6). Our results further demonstrate a requirement of autophagy for effective coxsackievirus infection.
FIG. 5.
Gene silencing of ATG7 by siRNA inhibits CVB3 replication. (A) Representative light microscopic images of CVB3-infected cells, with or without ATG7 siRNA transfection. (B) Western blot probing for VP1 and ATG7 expression, with or without ATG7 knockdown. VP1 expression was quantitated, normalized, and analyzed as described in the legend to Fig. 3 (data are means ± SE; n = 3). *, P < 0.05 compared to scramble siRNA-transfected cells.
FIG. 6.
Knockdown of class III PI3K components reduces CVB3 replication. (A) Western blot analysis of VP1 and Beclin-1 or VPS34 expression in CVB3-infected cells transfected with Beclin-1 (left) or VPS34 (right) siRNA prior to CVB3 infection. VP1 expression was quantitated, normalized, and analyzed as described in the legend to Fig. 3 (data are means ± SE; n = 3). *, P < 0.05 compared to scramble siRNA control. (B) Plaque assay results for scramble or Beclin-1 and VPS34 siRNA-transfected cells (means ± SE; n = 3). *, P < 0.05; #, P < 0.001 (compared to scramble siRNA-transfected cells).
Expression of p62 following CVB3 infection.
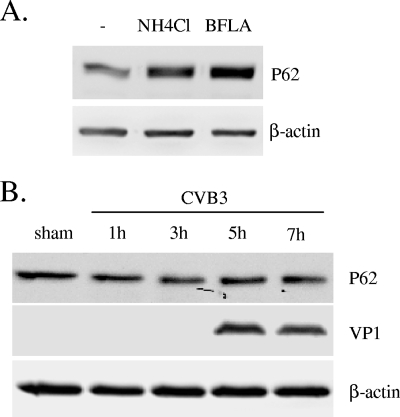
We next examined the effect of CVB3 infection on autophagy- or lysosome-mediated protein degradation. The protein p62 has been considered a marker for autophagy-mediated protein degradation or autophagic flux (18). As shown in Fig. 7, inhibition of lysosome function by bafilomycin A and NH4Cl, two pH-neutralizing agents, resulted in increased expression of p62. However, protein expression of p62 was unchanged during the course of CVB3 infection, suggesting that CVB3 infection triggers increased accumulation of autophagosomes without promoting protein degradation by lysosomes.
FIG. 7.
Expression of p62 is not affected by CVB3 infection. (A) Western blot analysis of p62, a marker of autophagic protein degradation activity, in cells treated with the lysosome inhibitors bafilomycin A (BFLA; 0.1 μM) and NH4Cl (2.5 mM) for 16 h. (B) Expression of p62 in CVB3-infected cells at the indicated times after virus infection. Similar results were obtained in two independent experiments.
Lysosomal inhibition enhances CVB3 replication.
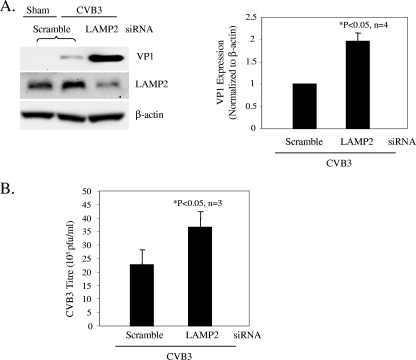
Finally, we investigated whether prevention of autophagosome-lysosome fusion promotes viral infectivity. Cells were transiently transfected with scramble or LAMP2 siRNA for 48 h and then infected with CVB3. We showed that knockdown of LAMP2 resulted in significant increases of viral protein expression and virus titer (Fig. 8). LAMP2 is a lysosomal membrane protein critical for autophagosome-lysosome fusion. Knockdown of LAMP2 has been shown to induce the cytoplasmic accumulation of LC3-associated vesicles (8). It was also reported that LAMP2-deficient mice exhibit a massive accumulation of autophagic vacuoles in various tissues (37). Thus, our results further suggest a key role of autophagosomes in supporting CVB3 replication.
FIG. 8.
Lysosomal inhibition enhances CVB3 replication. (A) Western blot analysis of VP1 and LAMP2 expression in CVB3-infected cells transiently transfected with scramble or LAMP2 siRNA. VP1 expression was quantitated, normalized, and analyzed as described in the legend to Fig. 3 (data are means ± SE; n = 3 or 4 [as indicated]). (B) Virus titer in scramble or LAMP2 siRNA-transfected cells, determined by plaque assay (mean ± SE; n = 3). *, P < 0.05 compared to scramble siRNA control.
DISCUSSION
Autophagy is an evolutionarily conserved cellular process that spans the entire spectrum of eukaryotes. It serves a housekeeping role in protein turnover and in the removal of damaged or unnecessary organelles. It also serves an important function in innate host defense by eliminating intracellular pathogens. For example, it has been shown that pharmacological induction of autophagy helps in the clearance of Mycobacterium tuberculosis (9) and that CD40 induction of autophagy promotes elimination of Toxoplasma gondii from macrophages (2). Furthermore, recent studies suggest a role for autophagy in host adaptive immune responses through the delivery of intracellular pathogenic antigens for major histocompatibility complex class II presentation to CD4 T cells (32).
Successful invading pathogens have evolved various strategies to evade host autophagy. For instance, some bacterial or viral proteins, such as IcsB secreted by Shigella flexneri (27) and ICP34.5 encoded by herpes simplex virus type 1 (36), have been documented to block or evade cellular autophagy. Other pathogens adopt different approaches in battling host autophagic defense by subverting it to facilitate their own procreation. In fact, several studies have shown that the autophagosome provides an intracellular compartment that benefits the survival and multiplication of bacteria, including Porphyromonas gingivalis, Brucella abortus, and Coxiella burnetii (32). In addition, some viruses have been reported to induce autophagosome formation to facilitate their own replication (15, 38).
Replication of positive-stranded RNA viruses requires intracellular membrane surfaces on which they assemble their replication complexes. Colocalization of viral RNA replication machineries with double-membrane vesicles in infected cells has been well documented for various positive-stranded RNA viruses, including poliovirus, rhinovirus, equine arterivirus, murine hepatitis virus, and severe acute respiratory syndrome virus (7, 38). Thus, it was speculated that the autophagosome, a double-membrane vesicle, may serve as a site for viral RNA replication. Recent studies on coronavirus and poliovirus seem to support this notion. It was reported that murine hepatitis virus, which belongs to the coronavirus family, colocalizes with the ubiquitin-like autophagy proteins (LC3 and Atg12p) and that viral replication is impaired in autophagy knockout (Atg5−/−) embryonic stem cell lines (31). Additionally, poliovirus proteins have been shown to colocalize with a cotransfected GFP-LC3 protein (11). Stimulation of autophagy increases the poliovirus yield, and inhibition of the autophagosomal pathway decreases the viral yield (11). Despite this exciting observation, recent data on the role of autophagy in positive-stranded RNA virus infection remain contradictory. A recent report showed that infection of rhinovirus type 2, a member of the picornavirus family, does not induce autophagosome formation (4). That paper further reported that neither induction nor inhibition of autophagy affects viral protein production. In addition, using ATG5-deficient cells, it was recently demonstrated that ATG5 and an intact autophagic pathway are not required for the replication of murine hepatitis virus (40). These studies challenge the view that the autophagosome is a common cellular component utilized by positive-stranded RNA viruses to facilitate their replication.
These discrepancies of the reported data prompted us to explore the role of autophagy in coxsackievirus infection in this study. We report here that CVB3 infection induces the formation of double-membrane vesicles with concurrent LC3 lipidation. We further showed that manipulation of the host autophagy pathway by pharmacological compounds or target-specific RNA interference resulted in significant alterations in the outcome of CVB3 replication. We demonstrated that inhibition of signaling pathways or autophagy genes critical for autophagosome formation reduces viral protein expression and the viral progeny titer. Conversely, increasing the cellular number of autophagosomes prior to viral infection by cellular starvation, rapamycin treatment, or lysosomal inhibition promotes viral replication. Taken together, our data suggest that the host double-membrane autophagosome is likely utilized by CVB3 to facilitate its own replication.
As alluded to earlier, the process of autophagosome formation is tightly controlled. The serine/threonine protein kinase mTOR is one of the key regulators and negatively controls autophagosome formation (5). Class I PI3K is the traditional upstream activator of mTOR. In addition, it has been shown that mTOR can be regulated by intracellular amino acids and ATP, independent of class I PI3K activation. ATP depletion or specific inhibition of mTOR by rapamycin has been shown to activate autophagy, whereas the addition of amino acids inhibits this process. In this study, we showed that levels of mTOR phosphorylation were unchanged after CVB3 infection, suggesting that CVB3-induced autophagy is not triggered by mTOR dephosphorylation.
In contrast, eIF2α kinase positively regulates autophagy in response to nutrient deprivation and viral infection. eIF2α kinase plays a key role in translation inhibition under various stress conditions, through phosphorylation of eIF2α. Restriction of herpes simplex virus growth has been linked to such a function of eIF2α phosphorylation (26). Recently, the eIF2α kinase signaling pathway was also shown to promote autophagy induced by nutrient deprivation or rapamycin treatment, likely through a mechanism that induces LC3 conversion (21, 36). Our present study demonstrates that eIF2α is phosphorylated after CVB3 infection, suggesting a potential signaling mechanism for CVB3-induced autophagosome formation. It is unclear how eIF2α phosphorylation occurs during CVB3 infection. One explanation could be that double-stranded RNA formed from a CVB3 intermediate activates double-stranded RNA-dependent protein kinase and subsequently leads to eIF2α phosphorylation.
Class III PI3K complexes also play a crucial role in the early steps of autophagosome formation. In mammalian cells, the autophagic protein Beclin-1 (a homologue of yeast ATG6) forms the class III PI3K complex with VPS34, a class III PI3K, at the trans-Golgi network (13, 39). This complex promotes autophagosome formation by supplying phosphatidylinositol 3-phosphates to the preautophagosome membrane (35). It has been shown that 3-MA blocks the formation of autophagosomes by inhibiting the activity of class III PI3K (3, 29). We demonstrates that 3-MA blocks CVB3 infection, which is in agreement with our previous report that inhibition of PI3K by LY294002 suppresses CVB3 replication (6). LY294002 is an inhibitor of both class I and class III PI3K and has also been shown to be a potent autophagy inhibitor (3).
Although the specific mechanism by which the autophagosome is utilized during coxsackievirus replication in host cells has not been elucidated, it is likely that, as demonstrated for poliovirus (11), the autophagosome provides a physical scaffold where the coxsackieviral complex may reside and viral RNA synthesis may occur. Furthermore, LC3 has been identified to be a binding protein of an AU-rich RNA element in the 3′-untranslated region of fibronectin mRNA, which facilitates the recruitment of RNA to polyribosomes for translation (41). Perhaps AU-rich element-containing viruses, such as CVB3, which carries an AU-rich element in the 3′-untranslated region (30), also adopt LC3-docking double-membrane vesicles as the sites of viral protein translation through the recruitment of polyribosomes. It is plausible to speculate that autophagosome double-membrane vesicles not only act as sites of viral RNA synthesis but also take an active role in CVB3 protein synthesis by bridging the nascent viral RNA with polyribosomes through LC3.
Acknowledgments
This work was supported by grants from the Heart and Stroke Foundation of British Columbia and Yukon (H.L.), the Canadian Institutes of Health Research (CIHR) (H.L.), and the Canada Foundation for Innovation (H.L.). J.W. is the recipient of master studentships from the Michael Smith Foundation for Health Research (MSFHR) and from the CIHR. X.S. is supported by a CIHR IMPACT postdoctoral fellowship and a CIHR Michael Smith postdoctoral fellowship. G.G. is supported by doctoral traineeships from the MSFHR and from the Heart and Stroke Foundation. H.L. is a new investigator under a CIHR/St. Paul's Hospital Foundation award and an MSFHR scholar.
Footnotes
Published ahead of print on 2 July 2008.
REFERENCES
- 1.Alexander, D. E., S. L. Ward, N. Mizushima, B. Levine, and D. A. Leib. 2007. Analysis of the role of autophagy in replication of herpes simplex virus in cell culture. J. Virol. 8112128-12134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrade, R. M., M. Wessendarp, M. J. Gubbels, B. Striepen, and C. S. Subauste. 2006. CD40 induces macrophage anti-Toxoplasma gondii activity by triggering autophagy-dependent fusion of pathogen-containing vacuoles and lysosomes. J. Clin. Investig. 1162366-2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blommaart, E. F., U. Krause, J. P. Schellens, H. Vreeling-Sindelarova, and A. J. Meijer. 1997. The phosphatidylinositol 3-kinase inhibitors wortmannin and LY294002 inhibit autophagy in isolated rat hepatocytes. Eur. J. Biochem. 243240-246. [DOI] [PubMed] [Google Scholar]
- 4.Brabec-Zaruba, M., U. Berka, D. Blaas, and R. Fuchs. 2007. Induction of autophagy does not affect human rhinovirus type 2 production. J. Virol. 8110815-10817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Codogno, P., and A. J. Meijer. 2005. Autophagy and signaling: their role in cell survival and cell death. Cell Death Differ. 12(Suppl. 2)1509-1518. [DOI] [PubMed] [Google Scholar]
- 6.Esfandiarei, M., H. Luo, B. Yanagawa, A. Suarez, D. Dabiri, J. Zhang, and B. M. McManus. 2004. Protein kinase B/Akt regulates coxsackievirus B3 replication through a mechanism which is not caspase dependent. J. Virol. 784289-4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Espert, L., P. Codogno, and M. Biard-Piechaczyk. 2007. Involvement of autophagy in viral infections: antiviral function and subversion by viruses. J. Mol. Med. 85811-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gonzalez-Polo, R. A., P. Boya, A. L. Pauleau, A. Jalil, N. Larochette, S. Souquere, E. L. Eskelinen, G. Pierron, P. Saftig, and G. Kroemer. 2005. The apoptosis/autophagy paradox: autophagic vacuolization before apoptotic death. J. Cell Sci. 1183091-3102. [DOI] [PubMed] [Google Scholar]
- 9.Gutierrez, M. G., S. S. Master, S. B. Singh, G. A. Taylor, M. I. Colombo, and V. Deretic. 2004. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell 119753-766. [DOI] [PubMed] [Google Scholar]
- 10.Hoyer-Hansen, M., L. Bastholm, P. Szyniarowski, M. Campanella, G. Szabadkai, T. Farkas, K. Bianchi, N. Fehrenbacher, F. Elling, R. Rizzuto, I. S. Mathiasen, and M. Jaattela. 2007. Control of macroautophagy by calcium, calmodulin-dependent kinase kinase-beta, and Bcl-2. Mol. Cell 25193-205. [DOI] [PubMed] [Google Scholar]
- 11.Jackson, W. T., T. H. Giddings, Jr., M. P. Taylor, S. Mulinyawe, M. Rabinovitch, R. R. Kopito, and K. Kirkegaard. 2005. Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol. 3e156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kabeya, Y., N. Mizushima, T. Ueno, A. Yamamoto, T. Kirisako, T. Noda, E. Kominami, Y. Ohsumi, and T. Yoshimori. 2000. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 195720-5728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kihara, A., Y. Kabeya, Y. Ohsumi, and T. Yoshimori. 2001. Beclin-phosphatidylinositol 3-kinase complex functions at the trans-Golgi network. EMBO Rep. 2330-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim, K. S., G. Hufnagel, N. M. Chapman, and S. Tracy. 2001. The group B coxsackieviruses and myocarditis. Rev. Med. Virol. 11355-368. [DOI] [PubMed] [Google Scholar]
- 15.Kirkegaard, K., M. P. Taylor, and W. T. Jackson. 2004. Cellular autophagy: surrender, avoidance and subversion by microorganisms. Nat. Rev. Microbiol. 2301-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klionsky, D. J. 2007. Autophagy: from phenomenology to molecular understanding in less than a decade. Nat. Rev. Mol. Cell Biol. 8931-937. [DOI] [PubMed] [Google Scholar]
- 17.Klionsky, D. J. 2005. The correct way to monitor autophagy in higher eukaryotes. Autophagy 165. [DOI] [PubMed] [Google Scholar]
- 18.Klionsky, D. J., H. Abeliovich, P. Agostinis, D. K. Agrawal, G. Aliev, D. S. Askew, M. Baba, E. H. Baehrecke, B. A. Bahr, A. Ballabio, B. A. Bamber, D. C. Bassham, E. Bergamini, X. Bi, M. Biard-Piechaczyk, J. S. Blum, D. E. Bredesen, J. L. Brodsky, J. H. Brumell, U. T. Brunk, W. Bursch, N. Camougrand, E. Cebollero, F. Cecconi, Y. Chen, L. S. Chin, A. Choi, C. T. Chu, J. Chung, P. G. Clarke, R. S. Clark, S. G. Clarke, C. Clave, J. L. Cleveland, P. Codogno, M. I. Colombo, A. Coto-Montes, J. M. Cregg, A. M. Cuervo, J. Debnath, F. Demarchi, P. B. Dennis, P. A. Dennis, V. Deretic, R. J. Devenish, F. Di Sano, J. F. Dice, M. Difiglia, S. Dinesh-Kumar, C. W. Distelhorst, M. Djavaheri-Mergny, F. C. Dorsey, W. Droge, M. Dron, W. A. Dunn, Jr., M. Duszenko, N. T. Eissa, Z. Elazar, A. Esclatine, E. L. Eskelinen, L. Fesus, K. D. Finley, J. M. Fuentes, J. Fueyo, K. Fujisaki, B. Galliot, F. B. Gao, D. A. Gewirtz, S. B. Gibson, A. Gohla, A. L. Goldberg, R. Gonzalez, C. Gonzalez-Estevez, S. Gorski, R. A. Gottlieb, D. Haussinger, Y. W. He, K. Heidenreich, J. A. Hill, M. Hoyer-Hansen, X. Hu, W. P. Huang, A. Iwasaki, M. Jaattela, W. T. Jackson, X. Jiang, S. Jin, T. Johansen, J. U. Jung, M. Kadowaki, C. Kang, A. Kelekar, D. H. Kessel, J. A. Kiel, H. P. Kim, A. Kimchi, T. J. Kinsella, K. Kiselyov, K. Kitamoto, E. Knecht, et al. 2008. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy 4151-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klionsky, D. J., and S. D. Emr. 2000. Autophagy as a regulated pathway of cellular degradation. Science 2901717-1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Komatsu, M., S. Waguri, T. Ueno, J. Iwata, S. Murata, I. Tanida, J. Ezaki, N. Mizushima, Y. Ohsumi, Y. Uchiyama, E. Kominami, K. Tanaka, and T. Chiba. 2005. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J. Cell Biol. 169425-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kouroku, Y., E. Fujita, I. Tanida, T. Ueno, A. Isoai, H. Kumagai, S. Ogawa, R. J. Kaufman, E. Kominami, and T. Momoi. 2007. ER stress (PERK/eIF2alpha phosphorylation) mediates the polyglutamine-induced LC3 conversion, an essential step for autophagy formation. Cell Death Differ. 14230-239. [DOI] [PubMed] [Google Scholar]
- 22.Levine, B. 2003. Autophagy in development, tumor suppression, and innate immunity. Harvey Lect. 9947-76. [PubMed] [Google Scholar]
- 23.Levine, B., and G. Kroemer. 2008. Autophagy in the pathogenesis of disease. Cell 13227-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mizushima, N. 2004. Methods for monitoring autophagy. Int. J. Biochem. Cell. Biol. 362491-2502. [DOI] [PubMed] [Google Scholar]
- 25.Mizushima, N., Y. Ohsumi, and T. Yoshimori. 2002. Autophagosome formation in mammalian cells. Cell Struct. Funct. 27421-429. [DOI] [PubMed] [Google Scholar]
- 26.Mohr, I. 2004. Neutralizing innate host defenses to control viral translation in HSV-1 infected cells. Int. Rev. Immunol. 23199-220. [DOI] [PubMed] [Google Scholar]
- 27.Ogawa, M., T. Yoshimori, T. Suzuki, H. Sagara, N. Mizushima, and C. Sasakawa. 2005. Escape of intracellular Shigella from autophagy. Science 307727-731. [DOI] [PubMed] [Google Scholar]
- 28.Ohsumi, Y. 2001. Molecular dissection of autophagy: two ubiquitin-like systems. Nat. Rev. Mol. Cell Biol. 2211-216. [DOI] [PubMed] [Google Scholar]
- 29.Petiot, A., E. Ogier-Denis, E. F. Blommaart, A. J. Meijer, and P. Codogno. 2000. Distinct classes of phosphatidylinositol 3′-kinases are involved in signaling pathways that control macroautophagy in HT-29 cells. J. Biol. Chem. 275992-998. [DOI] [PubMed] [Google Scholar]
- 30.Pilipenko, E. V., S. V. Maslova, A. N. Sinyakov, and V. I. Agol. 1992. Towards identification of cis-acting elements involved in the replication of enterovirus and rhinovirus RNAs: a proposal for the existence of tRNA-like terminal structures. Nucleic Acids Res. 201739-1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prentice, E., W. G. Jerome, T. Yoshimori, N. Mizushima, and M. R. Denison. 2004. Coronavirus replication complex formation utilizes components of cellular autophagy. J. Biol. Chem. 27910136-10141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmid, D., and C. Munz. 2007. Innate and adaptive immunity through autophagy. Immunity 2711-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seglen, P. O., and P. B. Gordon. 1982. 3-Methyladenine: specific inhibitor of autophagic/lysosomal protein degradation in isolated rat hepatocytes. Proc. Natl. Acad. Sci. USA 791889-1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Si, X., Y. Wang, J. Wong, J. Zhang, B. M. McManus, and H. Luo. 2007. Dysregulation of the ubiquitin-proteasome system by curcumin suppresses coxsackievirus B3 replication. J. Virol. 813142-3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suzuki, K., T. Kirisako, Y. Kamada, N. Mizushima, T. Noda, and Y. Ohsumi. 2001. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 205971-5981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Talloczy, Z., W. Jiang, H. W. T. Virgin, D. A. Leib, D. Scheuner, R. J. Kaufman, E. L. Eskelinen, and B. Levine. 2002. Regulation of starvation- and virus-induced autophagy by the eIF2alpha kinase signaling pathway. Proc. Natl. Acad. Sci. USA 99190-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tanaka, Y., G. Guhde, A. Suter, E. L. Eskelinen, D. Hartmann, R. Lullmann-Rauch, P. M. Janssen, J. Blanz, K. von Figura, and P. Saftig. 2000. Accumulation of autophagic vacuoles and cardiomyopathy in LAMP-2-deficient mice. Nature 406902-906. [DOI] [PubMed] [Google Scholar]
- 38.Wileman, T. 2006. Aggresomes and autophagy generate sites for virus replication. Science 312875-878. [DOI] [PubMed] [Google Scholar]
- 39.Zeng, X., J. H. Overmeyer, and W. A. Maltese. 2006. Functional specificity of the mammalian Beclin-Vps34 PI 3-kinase complex in macroautophagy versus endocytosis and lysosomal enzyme trafficking. J. Cell Sci. 119259-270. [DOI] [PubMed] [Google Scholar]
- 40.Zhao, Z., L. B. Thackray, B. C. Miller, T. M. Lynn, M. M. Becker, E. Ward, N. N. Mizushima, M. R. Denison, and H. W. T. Virgin. 2007. Coronavirus replication does not require the autophagy gene ATG5. Autophagy 3581-585. [DOI] [PubMed] [Google Scholar]
- 41.Zhou, B., N. Boudreau, C. Coulber, J. Hammarback, and M. Rabinovitch. 1997. Microtubule-associated protein 1 light chain 3 is a fibronectin mRNA-binding protein linked to mRNA translation in lamb vascular smooth muscle cells. J. Clin. Investig. 1003070-3082. [DOI] [PMC free article] [PubMed] [Google Scholar]