Abstract
Bats are increasingly recognized to harbor a wide range of viruses, and in most instances these viruses appear to establish long-term persistence in these animals. They are the reservoir of a number of human zoonotic diseases including Nipah, Ebola, and severe acute respiratory syndrome. We report the identification of novel groups of astroviruses in apparently healthy insectivorous bats found in Hong Kong, in particular, bats belonging to the genera Miniopterus and Myotis. Astroviruses are important causes of diarrhea in many animal species, including humans. Many of the bat astroviruses form distinct phylogenetic clusters in the genus Mamastrovirus within the family Astroviridae. Virus detection rates of 36% to 100% and 50% to 70% were found in Miniopterus magnater and Miniopterus pusillus bats, respectively, captured within a single bat habitat during four consecutive visits spanning 1 year. There was high genetic diversity of viruses in bats found within this single habitat. Some bat astroviruses may be phylogenetically related to human astroviruses, and further studies with a wider range of bat species in different geographic locations are warranted. These findings are likely to provide new insights into the ecology and evolution of astroviruses and reinforce the role of bats as a reservoir of viruses with potential to pose a zoonotic threat to human health.
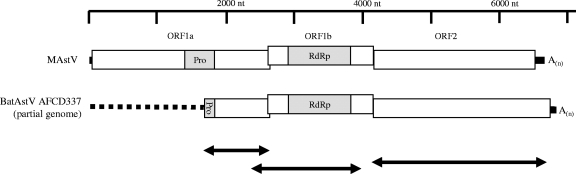
The Astroviridae are a family of nonenveloped, positive-sense single-stranded RNA viruses approximately 28 to 30 nm in size with a characteristic star-like surface structure. The genomes of these viruses range in size from 6.4 to 7.3 kb and are polyadenlyated at their 3′ ends (19). The viral RNA is synthesized by its RNA-dependent RNA polymerase (RdRp). The genome consists of three open reading frames (ORFs) designated ORF1a, ORF1b, and ORF2 (Fig. 1). ORF1a encodes nonstructural polyprotein 1a while the entire ORF1 encodes polyprotein 1ab with a ribosomal frameshift at the ORF1a/1b junction (9). Efficiency of ORF1b translation is estimated as only 25 to 28% of ORF1a (18). ORF2 encodes the viral structural polyprotein that is required for virion formation. Studies with human astroviruses showed that the structural polyprotein is intracellularly cleaved into functional units including a capsid-forming unit and a host binding motif unit (7, 12, 20).
FIG. 1.
Schematic diagrams of the mink astrovirus (MAstV) genome and bat astrovirus (BatAstV) AFCD337 partial genome. ORFs, protease motif (Pro), and RdRp motif are shown in the diagram. The unsequenced putative 5′ end of the genome region of BatAstV AFCD337 is represented by a dotted line. The 2.5-kb ORF1a (partial) region and the ORF1b and ORF2 regions used for phylogenetic analysis in Fig. 2 are indicated by arrows. An, poly(A) tail.
Astroviruses have been identified from a variety of mammals (genus Mamastrovirus) and birds (genus Avastrovirus) including humans, bovine, pigs, ovine, mink, dogs, cats, mice, chickens, and turkeys. In most species, these viruses are associated with gastroenteritis but some avian astroviruses have been associated with both intestinal and extraintestinal manifestations (reviewed in reference 19). Human astroviruses appear to cause milder disease than rotaviruses but are the second or third most common viral agent found in children with diarrhea (5, 8). Astroviruses can also cause significant disease in the elderly (16) and in immunocompromised patients (6).
Most of the surveillance studies of astroviruses focused on humans and domesticated animals, and relatively little is known about the prevalence of astroviruses in wildlife. The role of bats as the reservoirs for zoonotic diseases including rabies, Hendra, Nipah, and Ebola viruses has been highlighted in recent years (reviewed in reference2). Insectivorous bats have also been shown to harbor a range of novel coronaviruses including the precursor of severe acute respiratory syndrome coronavirus (15, 25). Some species of bats live in close proximity to human habitation, and thus it is important to have a better understanding of the virus ecology found in bats. The range and diversity of coronaviruses found in bats have led to the hypothesis that bat coronaviruses may be the precursors of most other mammalian group 1 and 2 coronaviruses (30). These findings highlight the importance of identifying novel viruses in wildlife in general and bats in particular. We used random primers to detect novel viruses in bat fecal specimens. Here, we report the discovery of novel astroviruses in bats in Hong Kong. The remarkably high prevalence and genetic diversity of astroviruses in various bat species found within a relatively small geographic area highlight the need for study in other species of bats and in other geographic locations.
MATERIALS AND METHODS
Sample collection.
The sampling of bats was carried out in two phases, and the sampling methods have been described previously (4, 25). Phase 1 was carried out in 2004 and 2005, and during this phase, swab samples were collected from different species of bats captured in the wild in different habitats in Hong Kong. Phase 2 was carried out in 2005 and 2006, and swab samples were collected from bats of the genus Miniopterus captured on four sampling occasions (June, August, and December 2005 and March 2006) in a single habitat, an abandoned mine cave in Lin Ma Hang, Hong Kong, near the border with mainland China (4). In both phases, species of bats captured for sampling were healthy and identified by a bat taxonomist. Rectal and throat swabs, together with fresh fecal samples if available, were collected. Swabs were placed in viral transport medium (Earle's balanced salt solution consisting of 0.2% sodium bicarbonate, 0.5% bovine serum albumin, 200 μg of vancomycin per liter, 18 μg of amikacin per liter, and 160 U of nystatin per liter) in screw-cap tubes and transported in a cool box to the laboratory for processing. Bats were released at the site after sampling.
Viral nucleic acid extraction and reverse transcription-PCR (RT-PCR).
RNA from 140 μl of sample in transport medium was extracted by a QIAamp virus RNA mini kit (Qiagen) following the protocol provided by the manufacturer. Purified RNA was eluted in 60 μl of elution buffer provided in the extraction kit. cDNA was generated from RNA using Superscript III reverse transcriptase (Invitrogen) in a 20-μl reaction mixture containing 150 ng of random hexamers or a 0.5 μM concentration of a gene-specific reverse primer (5′-TTTGGTCCNCCNCTCCAAA-3′) targeting the 3′ end of ORF1b, 10 mM dithiothreitol, 0.5 mM deoxynucleoside triphosphate mix, 1× First-Strand buffer (Invitrogen), and 200 U of reverse transcriptase. Reaction mixtures were incubated at 25°C for 5 min, followed by 50°C for 60 min, and then the enzyme was inactivated by heating at 70°C for 15 min.
Random hexamer-generated cDNA was screened for the presence of astrovirus using heminested PCR targeting the RdRp gene. A 50-μl PCR mixture was set up containing 1 U of Accuprime Taq DNA polymerase in 1× reaction buffer (Invitrogen), a 2 μM concentration (each) of forward and reverse primers, and 2 μl of cDNA or 1 μl of the first PCR product as a template. First-round PCR was carried out with a mixture of two forward primers, 5′-GARTTYGATTGGRCKCGKTAYGA-3′ and 5′-GARTTYGATTGGRCKAGGTAYGA-3′, and reverse primer 5′-GGYTTKACCCACATNCCRAA-3′. After an initial incubation at 94°C for 1 min, 30 cycles of amplification were carried out consisting of denaturation at 94°C for 30 s, annealing at 50°C for 30 s, and extension at 68°C for 30 s. Heminested PCR was carried out with a mixture of two forward primers, 5′-CGKTAYGATGGKACKATHCC-3′ and 5′-AGGTAYGATGGKACKATHCC-3′, and the same reverse primers used in the first-round PCR; the thermocycling conditions were the same as those used for the first-round PCR, except that 40 cycles of amplification were performed. Water controls were included in each run of the RT-PCR assay. PCR products were analyzed by standard agarose gel electrophoresis. The expected product size of the second PCR was 422 bp. All positive results were verified by direct DNA sequencing of the PCR amplicons.
Cloning and sequencing of PCR products.
PCR products were purified by a QIAquick PCR purification kit (Qiagen) following the manufacturer's instructions. Long PCR products (product sizes of >1,000 bp) were gel purified with a QIAquick gel extraction kit (Qiagen) and then cloned into pCR2.1-TOPO plasmids (Invitrogen) for DNA sequencing. Multiple clones of a PCR product were picked and sequenced by using a BigDye Terminator, version 3.1, Cycle Sequencing kit (Applied Biosystems). Sequencing products were analyzed by a PRISM 3700 DNA analyzer (Applied Biosystems).
One astrovirus-positive bat specimen, bat astrovirus AFCD337, was chosen for sequencing of the viral genome from PCR products derived from random or sequence-specific primed cDNA. The ORF2 region and the 3′ end of the virus sample were amplified using a 3′ rapid amplification of cDNA ends system (Invitrogen) following the protocol provided by the manufacturer (primers and PCR conditions available on request). ORF1a (partial) and ORF1b sequences were assembled from multiple overlapping sequences derived from PCR amplicons. Nine additional ORF1a (partial)/ORF1b astrovirus sequences from other samples were obtained using a similar method. The deduced sequences of these samples have at least threefold sequence coverage.
Phylogenetic analysis.
Sequence editing and sequence identity calculations were done using BioEdit, version 7.0.4 (http://www.mbio.ncsu.edu/BioEdit/bioedit.html). Alignments of nucleotide sequences and amino acid sequences were done using Clustal W (29) with default parameters. Phylogenetic trees were constructed using Clustal X, version 2.0 (14), and Mega 4 (28) by the neighbor-joining method with the nucleotide substitution model of maximum composite likelihood and default parameters. Bootstrap values of the phylogenetic tree constructed were generated by doing 1,000 replicates.
Nucleotide sequence accession numbers.
The sequence of bat astrovirus AFCD337 reported in this paper was deposited in the GenBank database under accession number EU847155 and the RdRp and the partial ORF1b sequences of other strains of bat astroviruses were deposited under accession numbers EU847144 through EU847154 and EU847156 through EU847220. For the genetic analysis in this paper, other astrovirus genomes were retrieved from the GenBank, including human astrovirus type 1 strain Oxford (L23513), human astrovirus type 1 strain Dresden (AY720892), human astrovirus type 4 strain Dresden (AY720891), human astrovirus type 4 isolate Goiania/GO/12/95/Brazil (DQ070852), human astrovirus type 4 isolate Guangzhou (DQ344027), human astrovirus type 5 isolate Goiania/GO/12/94/Brazil (DQ028633), human astrovirus type 8 (AF260508), mink astrovirus (NC_004579), ovine astrovirus (NC_002469) and turkey astrovirus type 1 (NC_002470).
RESULTS
Detection of astroviruses in bats.
A total of 262 bats were captured, and rectal and throat swabs were sampled in two phases of sample collection in Hong Kong in 2004 to 2006. Bats sampled included nine species: Cynopterus sphinx, Hipposideros armiger, Miniopterus magnater, Miniopterus pusillus, Miniopterus schreibersii, Myotis chinensis, Myotis ricketti, Pipistrellus abramus, and Rhinolophus rouxi. A total of 116 positives were detected from 250 available rectal samples tested, representing a positive rate of 46% (Table 1). On the other hand, only 19 (8%) positive throat swabs were found in 246 available samples tested. With the exception of two throat swabs collected from M. pusillus and R. rouxi bats in 2004, all of the positive throat swab results came from bats with astrovirus detected in rectal samples. All of the positive PCR products were sequenced to confirm the identity of the amplicon, and since the sequences of the different PCR amplicons were largely nonidentical, PCR cross-contamination could be excluded.
TABLE 1.
Detection of astrovirus in bats by RT-PCR
| Bat species | Astrovirus detection rate in:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Rectal samples
|
Throat samples
|
Tested bats
|
|||||||
| No. tested | No. positive | % Positive | No. tested | No. positive | % Positive | No. tested | No. positivea | % Positive | |
| Insectivorous bats | |||||||||
| H. armiger | 10 | 0 | 0 | 10 | 0 | 0 | 10 | 0 | 0 |
| M. magnater | 122 | 67 | 55 | 123 | 4 | 3 | 132 | 67 | 51 |
| M. pusillus | 73 | 31 | 42 | 71 | 6 | 8 | 74 | 32 | 43 |
| M. schreibersii | 3 | 3 | 100 | 2 | 1 | 50 | 3 | 3 | 100 |
| M. chinensis | 9 | 3 | 33 | 9 | 1 | 11 | 9 | 3 | 33 |
| M. ricketti | 12 | 10 | 83 | 12 | 5 | 42 | 12 | 10 | 83 |
| P. abramus | 2 | 1 | 50 | 1 | 0 | 0 | 3 | 1 | 33 |
| R. rouxi | 8 | 1 | 13 | 8 | 2 | 25 | 8 | 2 | 25 |
| Fruit bat, C. sphinx | 11 | 0 | 0 | 10 | 0 | 0 | 11 | 0 | 0 |
| Total | 250 | 116 | 46 | 246 | 19 | 8 | 262 | 118 | 45 |
Rectal and/or throat swabs tested positive.
Astroviruses were detected in seven out of the nine species of bats screened, i.e., M. magnater, M. pusillus, M. schreibersii, M. chinensis, M. ricketti, P. abramus, and R. rouxi (Table 1). The detection rates of astroviruses in these species of bats were remarkably high and ranged from 25% to 100%. However, astrovirus was not detectable in our samples collected from C. sphinx (n = 11) and H. armiger (n = 10).
These same specimens had previously been tested for bat coronaviruses (3, 4, 25). While 6% of bats were coinfected with both a bat astrovirus and a bat coronavirus, such coinfection appears to be randomly distributed, and there was no positive or negative statistical association between the presence of these two viruses (chi-square test with Yates correction, P = 0.82).
Longitudinal study of astrovirus in bats at a single habitat.
The bats listed in Table 1 include 157 M. magnater and M. pusillus bats that were captured at four separate visits over a 2-year period at one habitat, an abandoned mine cave in Hong Kong. M. magnater bats were found throughout the period while M. pusillus bats were mainly found in two visits carried out in December 2005 and March 2006; only one was found in a visit in August 2005. Sixty-two (54%) out of 115 rectal swabs and 2 (2%) out of 116 throat swabs collected from M. magnater bats and 18 (55%) out of 33 rectal swabs and 5 (15%) out of 33 throat swabs collected from M. pusillus bats were positive for astrovirus. The overall positive rate in individual bats (either rectal or throat swab or both positive) for M. magnater ranged from 36 to 100% at each of the four visits which spanned the winter, spring, and summer seasons; for M. pusillus the positive rates were 50% and 70% in the two instances when adequate bats were sampled.
Genetic and phylogenetic analysis of a novel astrovirus from M. pusillus.
The bat astrovirus AFCD337 detected in a rectal specimen from a M. pusillus bat collected in March 2006 was chosen as a representative virus for more extensive genome sequencing. Approximately 74% of the genome of this novel astrovirus was obtained by direct RT-PCR amplification from a rectal swab sample. The partial [excluding the poly(A) tail] 5,067-nucleotide (nt) genome was constructed by aligning sequences from multiple overlapping regions. Amino acid sequences deduced from the viral genome include part of ORF1a and the complete ORF1b, ORF2, and 3′ untranslated region (UTR) followed by the poly(A) tail at the 3′ end (Fig. 1).
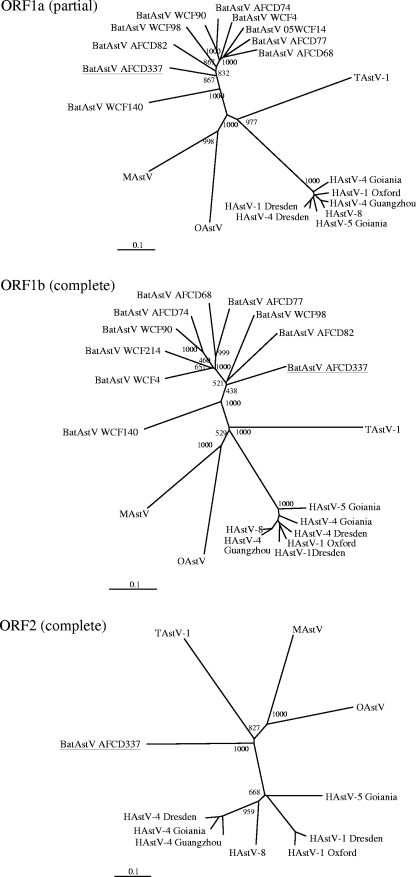
Amino acid sequences encoded by ORFs of the novel bat astrovirus were compared with sequences of other astrovirus genomes including human astrovirus types 1, 4, 5, and 8 (17, 26) as well as mink astrovirus (22), ovine astrovirus, and turkey astrovirus type 1 (11) (Table 2). The findings show that the identified bat astrovirus AFCD337 is a novel mamastrovirus clearly distinct from other known astroviruses. It has <53% and <27% genetic similarity to other known astroviruses in the ORF1b and ORF2 regions, respectively.
TABLE 2.
Amino acid sequence similarities between prototype bat astrovirus AFCD337 and subgroup A (AFCD68) and B (WCF140) bat astroviruses and other astroviruses
| Straina | % Amino acid identity with BatAstV AFCD337
|
||
|---|---|---|---|
| ORF1a (partial) | ORF1b | ORF2 | |
| BatAstV AFCD68 | 79.9 | 74.2 | |
| BatAstV WCF140 | 54.0 | 68.5 | |
| HAstV-1 Dresden | 17.2 | 48.4 | 22.3 |
| HAstV-1 Oxford | 17.9 | 49.2 | 22.1 |
| HAstV-4 Guangzhou | 17.5 | 49.0 | 22.3 |
| HAstV-4 Goiania/95/Brazil | 17.9 | 49.2 | 22.2 |
| HAstV-4 Dresden | 17.9 | 48.8 | 21.8 |
| HAstV-5 Goiania/94/Brazil | 17.5 | 49.2 | 20.9 |
| HAstV-8 | 17.2 | 48.8 | 20.5 |
| MAstV | 31.2 | 52.7 | 24.1 |
| OAstV | 28.9 | 52.6 | 26.2 |
| TAstV-1 | 13.9 | 40.2 | 14.1 |
Astroviruses are indicated as follows: BatAstV, bat; HAstV, human (types 1, 4, 5, and 8); MAstV, mink; OAstV, ovine; TAstV-1, turkey type 1.
The putative ORF1a (partial) and ORF1b of the bat astrovirus sequenced have sizes of 909 nt and 1,572 nt, respectively. A region at the 5′ end of ORF1a remains unsequenced so far. ORF1a and ORF1b encode nonstructural proteins that are essential for virus replication. Characteristic features of bat astrovirus AFCD337 ORF1a and ORF1b include the protease motif in ORF1a, an astrovirus “slippery sequence” (AAAAAC) at the junction between ORF1a and ORF1b that is required for inducing a ribosomal shift event, an RdRp motif in ORF1b, and a conserved sequence at the end of ORF1b of astroviruses (11). A conserved stem-loop structure that is predicted at the 3′ end of the genomic RNA of human, ovine, and porcine astroviruses and turkey astrovirus type 1 was not found in bat astrovirus AFCD337 (10).
The putative ORF2 of the virus has a size of 2,553 nt, which is the largest astrovirus capsid gene known. The N-terminal half of the ORF2 protein, which was previously shown to be more conserved among astroviruses and proposed to be the core assembly domain of the viral capsid (12), was also found to be relatively conserved in this bat astrovirus. The amino acid sequence similarities of this N-terminal half of the bat astrovirus capsid protein to human astrovirus type 1 Oxford, ovine astrovirus, and mink astrovirus are 36.3%, 45.0%, and 39.5%, respectively, compared with <27% similarity for the ORF2 region overall (Table 2). Thus, the C-terminal half of this bat astrovirus protein was highly divergent compared with other astroviruses. This observation supports the speculation that the C-terminal half of the protein is located on the surface of the viral particle and constitutes a region of the capsid that contributes to the species-specific tropism of the virus (12).
Phylogenetic analysis of astrovirus in bats.
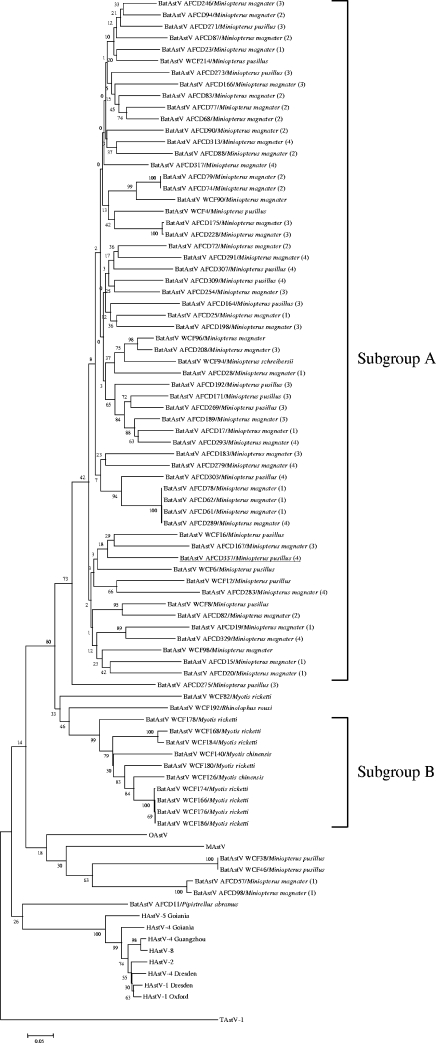
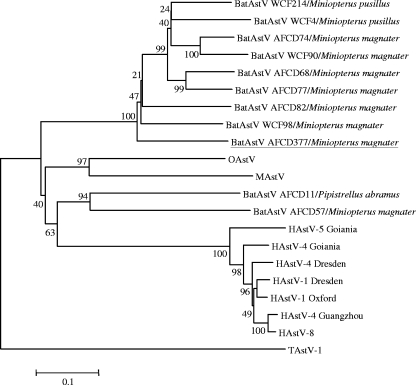
Phylogenetic analyses of the ORF1a (partial) and ORF1b regions and of ORF2 confirm that bat astrovirus AFCD337 is a novel, distinct astrovirus (Fig. 2). To better define the genetic diversity within the bat astroviruses by species, time, and geographic location, 77 PCR RdRp amplicons (422 nt) obtained from the rectal swab samples in the screening PCR assay were selected for genetic sequence analysis. Bat astrovirus gene sequences were aligned with sequences of other astroviruses including mink astrovirus, ovine astrovirus, human astrovirus type 1 Oxford, human astrovirus type 1 Dresden, human astrovirus type 2, human astrovirus type 4 Dresden, human astrovirus type 4 Goiania, human astrovirus type 4 Guangzhou, human astrovirus type 5 Goiania, and human astrovirus type 8. An avian astrovirus, turkey astrovirus, was included as an outgroup. A phylogenetic tree was constructed from the sequence alignment (Fig. 3). The 72 astroviruses detected in bats cluster together to form a novel group of viruses within the cluster of mamastroviruses (Fig. 3). Within this group are found two subgroups of viruses, one that includes the majority of astroviruses detected from M. magnater, M. pusillus, and M. schreibersii (including bat astrovirus AFCD337) bats and another subgroup that includes most of the viruses detected in M. chinensis and M. ricketti bats. Other than these two major groups of bat astroviruses, a few astroviruses detected in M. magnater and M. pusillus bats and a virus detected in a P. abramus bat appear to have an outgroup relationship to the others, albeit with weak levels of statistical confidence in this phylogenetic topology (Fig. 3). To further investigate this, a 750-nt region of the RdRp and ORF1b (3′ end) of these and other representative bat astroviruses were sequenced. A phylogenetic tree based on the aligned protein-encoding sequences was produced (Fig. 4). The phylogeny of the major group of viruses related to bat astrovirus AFCD337 is confirmed. Interestingly, however, AFCD11 from P. abramus and AFCD57 from M. magnater appear phylogenetically related to the human astroviruses although with modest bootstrap support. Their close relationship has also been confirmed by phylogenetic analysis using an alternative method, MrBayes (data not shown).
FIG. 2.
Phylogenetic analysis of the partial ORF1a (∼800 nt), ORF1b, and ORF2 nucleotide sequences comparing bat astrovirus AFCD337 (underlined) with astroviruses of other species. Nine other bat astrovirus sequences are included in the ORF1a (partial) and ORF1b phylogenetic trees. Alignment was based on the encoded amino acid sequences. Astroviruses are indicated as follows: BatAstV, bat; MAstV, mink; OAstV, ovine; HAstV-1, human type 1 (also types 4, 5, and 8); and TAstV-1, turkey type 1.
FIG. 3.
Phylogenetic tree constructed with RdRp gene sequences (422 nt) amplified by an RT-PCR screening assay. Sequences of 77 bat astroviruses (BatAstVs) and other respective sequences of different astroviruses isolated from human (HAstV types 1, 2, 4, 5, and 8), mink (MAstV), ovine (OAstV), and turkey type 1 (TAstV-1) viruses were included and aligned based on the nucleotide sequences. For the bat specimens collected during the phase 2 longitudinal study, the sampling dates are indicated in parentheses on the right according to the following code: 1, June 2005; 2, August 2005; 3, December 2005; 4, March 2006.
FIG. 4.
Phylogenetic tree constructed with 750-nt sequences of the RdRp gene and ORF1b (3′ end) of representative astroviruses isolated from bat (BatAstV), human (HAstV types 1, 4, 5, and 8), mink (MAstV), ovine (OAstV), and turkey type 1 (TAstV-1) viruses. These sequences were aligned based on the encoded amino acid sequences and were reverse-translated back to nucleotides for the phylogenetic analysis.
Occasionally the same virus strain can be found in multiple bats sampled at the same habitat in a single sampling trip (i.e., strains AFCD74 and AFCD79 and strains AFCD175 and AFCD228). However, most viruses detected even at the same sampling occasion at a single habitat were genetically diverse, and no dominant strain could be discerned even though the detection rates of the viruses were remarkably high.
DISCUSSION
We report the discovery of novel astroviruses in seven out of nine species of apparently healthy bats captured in Hong Kong. These astrovirus-positive species include M. magnater, M. pusillus, M. schreibersii, M. chinensis, M. ricketti, P. abramus, and R. rouxi. Attempts at viral culture thus far had been unsuccessful (unpublished data). Phylogenetic analysis revealed that 72 out of 77 bat astrovirus RdRp genes analyzed clustered together to form a novel group of astroviruses. This virus group can be divided into two subgroups, subgroup A detected from Miniopterus species and subgroup B detected from Myotis species. In the longitudinal study carried out in the abandoned mine cave habitat, the subgroup A viruses appeared to be circulating between M. magnater and M. pusillus bats without any evidence of species restriction. This is in marked contrast to the bat coronaviruses within the same habitat, where bat coronaviruses 1A and 1B appeared to have a marked host restriction to M. magnater and M. pusillus, respectively (4). Multiple clones of a partial ORF1 sequence (approximately 1,000 nt) for 10 representative samples were analyzed, and no evidence of multiple infection was found although more systematic studies on this aspect are needed.
The diversity of astroviruses in bats is remarkable. There is no significant phylogenetic clustering of viruses found within a single sampling occasion. The values of astrovirus RdRp amino acid pairwise similarity found within a single bat species, i.e., M. magnater, captured in a single habitat (mine cave) ranged between 51.1% and 100%, with 97.9% of these values lower than 90%. In contrast, the pairwise amino acid sequence similarities between the same gene region of human astroviruses from geographically diverse regions were estimated to range between 92.6% and 99.1%. It has been previously reported that the amino acid sequence identities of the RdRp gene (covering 80% of the RdRp gene regions analyzed in this report) between four groups of avastroviruses, i.e., turkey astrovirus type 1-like viruses, turkey astroviruses type 2-like viruses, avian nephritis virus-like viruses, and chicken-origin astroviruses, detected in different regions were also highly diverse, ranging from 50.1% to 73.8% identity (23). The high virus detection rates of bat astrovirus in our surveillance, taken together with the marked genetic diversity of viruses from bats within the same habitat, are reminiscent of our observations with bat coronaviruses (4). These findings may suggest that bats are persistently infected with astroviruses although mark-recapture studies are needed to confirm this contention. Other mammalian astrovirus infections tend to be short-lived in immunocompetent humans or other animals. However, type 3 human astrovirus has been associated with persistent gastroenteritis in immunocompetent children although the same virus serotype was not repeatedly demonstrated over the full period of clinical diarrhea (1).
Five bat astrovirus sequences from M. magnater, M. pusillus, and P. abramus failed to cluster with the subgroups A and B referred to above, and some of these (AFCD11 from P. abramus and AFCD57 from M. magnater) cluster, rather, with the human astroviruses although with weak statistical support (Fig. 4). Whether these bat viruses are related to the precursor of human astrovirus is yet to be further investigated. Further sequence data from these strains may help elucidate this phylogenetic association.
Evidence of recombination between astroviruses and also between coronaviruses is well documented (13, 24, 27). It has been reported that a stem-loop motif in the 3′ UTR was found conserved in mamastroviruses, turkey astrovirus type 1, and avian infectious bronchitis virus, which is a group 3 coronavirus (10). Interestingly, this stem-loop motif was not recognized in bat astrovirus AFCD337. This 3′ UTR stem-loop motif is also absent in turkey astrovirus type 2. However, a phylogenetic analysis of 3′ UTRs did not indicate a close phylogenetic relationship between the two sequences of bat astrovirus AFCD337 (81 nt) and turkey astrovirus type 2 (accession no. NC_005790) (196 nt), which lack the 3′ UTR stem-loop structure (data not shown). Recently, a report on the identification of a novel coronavirus from liver tissue of a whale with pulmonary disease and terminal acute liver failure showed that ORF6 of the novel coronavirus possessed significant amino acid similarity to human astrovirus capsid proteins (21). The high rates of infection of bats in the same mine cave habitat with coronaviruses and astroviruses imply frequent coinfection with both viruses. Therefore, we searched for sequences of bat astrovirus AFCD337 similar to bat coronaviruses using the BLAST program with algorithms allowing a word size down to seven bases. However, no sequences with significant similarity were detected between bat astroviruses and coronaviruses cocirculating within the same species within the same habitat.
The discovery of novel diverse astroviruses in bats and the genetic analysis of such viruses are likely to provide new insights into the ecology and evolution of astroviruses and reinforce the role of bats as a reservoir of viruses that sometimes pose a zoonotic threat to human health. More extensive surveillance for astroviruses in bats of different species and in different geographic areas is needed to further address these questions.
Acknowledgments
This project was supported by the National Institutes of Health (NIAID contract HHSN266200700005C) and by a Research Excellence Award to J.S.M.P. from The University of Hong Kong. The study was approved and supported by the Department of Agriculture, Fisheries and Conservation, Hong Kong, Special Administrative Region, People's Republic of China.
We thank K. S. Cheung, C. T. Shek, and C. S. M. Chan of the Department of Agriculture, Fisheries and Conservation, Hong Kong, for facilitating this study.
Footnotes
Published ahead of print on 11 June 2008.
REFERENCES
- 1.Caballero, S., S. Guix, W. M. El-Senousy, I. Calico, R. M. Pinto, and A. Bosch. 2003. Persistent gastroenteritis in children infected with astrovirus: association with serotype-3 strains. J. Med. Virol. 71245-250. [DOI] [PubMed] [Google Scholar]
- 2.Calisher, C. H., J. E. Childs, H. E. Field, K. V. Holmes, and T. Schountz. 2006. Bats: important reservoir hosts of emerging viruses. Clin. Microbiol. Rev. 19531-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chu, D. K., J. S. Peiris, H. Chen, Y. Guan, and L. L. Poon. 2008. Genomic characterizations of bat coronaviruses (1A, 1B and HKU8) and evidence for co-infections in Miniopterus bats. J. Gen. Virol. 891282-1287. [DOI] [PubMed] [Google Scholar]
- 4.Chu, D. K., L. L. Poon, K. H. Chan, H. Chen, Y. Guan, K. Y. Yuen, and J. S. Peiris. 2006. Coronaviruses in bent-winged bats (Miniopterus spp.). J. Gen. Virol. 872461-2466. [DOI] [PubMed] [Google Scholar]
- 5.Dalton, R. M., E. R. Roman, A. A. Negredo, I. D. Wilhelmi, R. I. Glass, and A. Sanchez-Fauquier. 2002. Astrovirus acute gastroenteritis among children in Madrid, Spain. Pediatr. Infect. Dis. J. 211038-1041. [DOI] [PubMed] [Google Scholar]
- 6.Gallimore, C. I., C. Taylor, A. R. Gennery, A. J. Cant, A. Galloway, D. Lewis, and J. J. Gray. 2005. Use of a heminested reverse transcriptase PCR assay for detection of astrovirus in environmental swabs from an outbreak of gastroenteritis in a pediatric primary immunodeficiency unit. J. Clin. Microbiol. 433890-3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geigenmuller, U., N. H. Ginzton, and S. M. Matsui. 2002. Studies on intracellular processing of the capsid protein of human astrovirus serotype 1 in infected cells. J. Gen. Virol. 831691-1695. [DOI] [PubMed] [Google Scholar]
- 8.Herrmann, J. E., D. N. Taylor, P. Echeverria, and N. R. Blacklow. 1991. Astroviruses as a cause of gastroenteritis in children. N. Engl. J. Med. 3241757-1760. [DOI] [PubMed] [Google Scholar]
- 9.Jiang, B., S. S. Monroe, E. V. Koonin, S. E. Stine, and R. I. Glass. 1993. RNA sequence of astrovirus: distinctive genomic organization and a putative retrovirus-like ribosomal frameshifting signal that directs the viral replicase synthesis. Proc. Natl. Acad. Sci. USA 9010539-10543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jonassen, C. M., T. O. Jonassen, and B. Grinde. 1998. A common RNA motif in the 3′ end of the genomes of astroviruses, avian infectious bronchitis virus and an equine rhinovirus. J. Gen. Virol. 79715-718. [DOI] [PubMed] [Google Scholar]
- 11.Jonassen, C. M., T. T. Jonassen, T. M. Sveen, and B. Grinde. 2003. Complete genomic sequences of astroviruses from sheep and turkey: comparison with related viruses. Virus Res. 91195-201. [DOI] [PubMed] [Google Scholar]
- 12.Krishna, N. K. 2005. Identification of structural domains involved in astrovirus capsid biology. Viral Immunol. 1817-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lai, M. M., and D. Cavanagh. 1997. The molecular biology of coronaviruses. Adv. Virus Res. 481-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Larkin, M. A., G. Blackshields, N. P. Brown, R. Chenna, P. A. McGettigan, H. McWilliam, F. Valentin, I. M. Wallace, A. Wilm, R. Lopez, J. D. Thompson, T. J. Gibson, and D. G. Higgins. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 232947-2948. [DOI] [PubMed] [Google Scholar]
- 15.Lau, S. K., P. C. Woo, K. S. Li, Y. Huang, H. W. Tsoi, B. H. Wong, S. S. Wong, S. Y. Leung, K. H. Chan, and K. Y. Yuen. 2005. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl. Acad. Sci. USA 10214040-14045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lewis, D. C., N. F. Lightfoot, W. D. Cubitt, and S. A. Wilson. 1989. Outbreaks of astrovirus type 1 and rotavirus gastroenteritis in a geriatric in-patient population. J. Hosp. Infect. 149-14. [DOI] [PubMed] [Google Scholar]
- 17.Lewis, T. L., H. B. Greenberg, J. E. Herrmann, L. S. Smith, and S. M. Matsui. 1994. Analysis of astrovirus serotype 1 RNA, identification of the viral RNA-dependent RNA polymerase motif, and expression of a viral structural protein. J. Virol. 6877-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lewis, T. L., and S. M. Matsui. 1996. Astrovirus ribosomal frameshifting in an infection-transfection transient expression system. J. Virol. 702869-2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mendez, E., and C. F. Arias. 2007. Astroviruses, p. 981-1000. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 20.Mendez, E., T. Fernandez-Luna, S. Lopez, M. Mendez-Toss, and C. F. Arias. 2002. Proteolytic processing of a serotype 8 human astrovirus ORF2 polyprotein. J. Virol. 767996-8002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mihindukulasuriya, K. A., G. Wu, J. St. Leger, R. W. Nordhausen, and D. Wang. 2008. Identification of a novel coronavirus from a beluga whale using a panviral microarray. J. Virol. 825084-5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mittelholzer, C., K. O. Hedlund, L. Englund, H. H. Dietz, and L. Svensson. 2003. Molecular characterization of a novel astrovirus associated with disease in mink. J. Gen. Virol. 843087-3094. [DOI] [PubMed] [Google Scholar]
- 23.Pantin-Jackwood, M. J., E. Spackman, and P. R. Woolcock. 2006. Molecular characterization and typing of chicken and turkey astroviruses circulating in the United States: implications for diagnostics. Avian Dis. 50397-404. [DOI] [PubMed] [Google Scholar]
- 24.Pantin-Jackwood, M. J., E. Spackman, and P. R. Woolcock. 2006. Phylogenetic analysis of turkey astroviruses reveals evidence of recombination. Virus Genes 32187-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poon, L. L., D. K. Chu, K. H. Chan, O. K. Wong, T. M. Ellis, Y. H. Leung, S. K. Lau, P. C. Woo, K. Y. Suen, K. Y. Yuen, Y. Guan, and J. S. Peiris. 2005. Identification of a novel coronavirus in bats. J. Virol. 792001-2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Silva, P. A., D. D. Cardoso, and E. Schreier. 2006. Molecular characterization of human astroviruses isolated in Brazil, including the complete sequences of astrovirus genotypes 4 and 5. Arch. Virol. 1511405-1417. [DOI] [PubMed] [Google Scholar]
- 27.Strain, E., L. A. Kelley, S. Schultz-Cherry, S. V. Muse, and M. D. Koci. 2008. Genomic analysis of closely related astroviruses. J. Virol. 825099-5103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 241596-1599. [DOI] [PubMed] [Google Scholar]
- 29.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 254876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vijaykrishna, D., G. J. Smith, J. X. Zhang, J. S. Peiris, H. Chen, and Y. Guan. 2007. Evolutionary insights into the ecology of coronaviruses. J. Virol. 814012-4020. [DOI] [PMC free article] [PubMed] [Google Scholar]