Abstract
Human immunodeficiency virus type 1 (HIV-1) CRF08_BC and CRF07_BC are two major recombinants descended from subtypes B′ and C. Despite their massive epidemic impact in China, their migration patterns and divergence times remain unknown. Phylogenetic and population genetic analyses were performed on 228 HIV-1 sequences representing CRF08_BC, CRF07_BC, and subtype C strains from different locations across China, India, and Myanmar. Genome-specific rates of evolution and divergence times were estimated using a Bayesian Markov chain Monte Carlo framework under various evolutionary models. CRF08_BC originated in 1990.3 (95% credible region [CR], 1988.6 to 1991.9) in Yunnan province before spreading to Guangxi (south) and Liaoning (northeast) around 1995. Inside Guangxi region, the eastward expansion of CRF08_BC continued from Baise city (west) to Binyang (central) between 1997 and 1998 and later spread into Pingxiang around 1999 in the south, mainly through injecting drug users. Additionally, CRF07_BC diverged from its common ancestor in 1993.3 (95% CR, 1991.2 to 1995.2) before crossing the border into southern Taiwan in late 1990s. Phylogenetic analysis indicates that both CRF08_BC and CRF07_BC can trace their origins to Yunnan. The parental Indian subtype C lineage likely entered China around 1981.2 (95% CR, 1976.7 to 1985.9). Using a multiple unlinked locus model, we also showed that the dates of divergence calculated in this study may not be significantly affected by intrasubtype recombination among different lineages. This is the first phylodynamic study depicting the spatiotemporal dynamics of HIV/AIDS in East Asia.
The human immunodeficiency virus type 1 (HIV-1) circulating recombinant forms (CRFs) 08_BC (CRF08_BC) and 07_BC (CRF07_BC) are two highly prevalent strains that are circulating in Asia and causing hundreds of thousands of infections in China (42). HIV-1 CRF08_BC, a descendant of the parental subtypes B′ and C, was first described in the Guangxi province in southern China around 1997 (26). CRF08_BC is thought to have originated in Yunnan province (20, 49) and has spread to other regions of China, particularly in the south (26, 51). HIV-1 CRF07_BC is a related but distinct B′/C recombinant that was first reported in 1997 in Xinjiang and is predominant in the north of China; its origin has also been traced to Yunnan province (38).
Yunnan is located in southwestern China, bordering the “Golden Triangle” region of Southeast Asia, one of the world's largest heroin-producing regions. Yunnan plays an important role as the entry point for heroin smuggling into, and possibly beyond, China (2). Yunnan province is considered an epicenter of HIV/AIDS in China, and an HIV-1 outbreak there was first detected among injecting drug users (IDU) in 1989 (53). The early phase of the epidemic was due to subtype B strains of both North American and Southeast Asian (B′ or Thai B) origin (47), with subtype B′ later becoming the dominant strain among IDU in the region (10, 45). The subtype distribution then shifted in the early 1990s, when a subtype C strain closely related to Indian isolates emerged and subsequently became the predominant circulating strain (19). CRF01_AE from Thailand may also have entered Yunnan as early as 1993, possibly through sexual transmission networks (3). Following cocirculation of subtypes B′ and C in the region, various phylogenetically distinct B′/C recombinants—including CRF08_BC and CRF07_BC—were formed and subsequently spread outside Yunnan province. Drug trafficking activities, compounded by local heroin use along the trafficking routes, have been implicated in the spread of CRF08_BC and CRF07_BC across China (2). Although molecular epidemiologic surveys have previously reported the identification and characterization in various parts of China of CRF08_BC, CRF07_BC and their putative subtype C parental lineages (19, 26, 32, 38), the specific times of emergence and divergence of these strains in the world's most populous nation remain uninvestigated and unknown. Genealogical analysis is needed to reconstruct the epidemiological history of viral populations (8, 9, 43), which can lead to better understanding of HIV transmission and improved prevention programs.
In this study, we investigated the spatial and temporal spread of HIV-1 CRF08_BC and CRF07_BC in China by reconstructing the evolutionary histories of these strains using recently developed Bayesian analysis methods. In addition, we also assessed the phylodynamics of the subtype C lineage of Indian origin in order to provide a more comprehensive picture of the movement of subtype C and its related recombinants in the region. Here, we demonstrate and provide a timescale for the eastwards spread of HIV/AIDS in East Asia, and we highlight the significance of HIV-1 evolutionary history upon viral epidemiology, migration and molecular taxonomy.
MATERIALS AND METHODS
HIV-1 CRF08_BC and CRF07_BC sequence information.
HIV-1 CRF08_BC nucleotide sequences with known sampling dates were retrieved from the Los Alamos HIV Sequence Database (www.hiv.lanl.gov). Eighty-one CRF08_BC sequences, including the reference strains previously reported by our laboratory and others, were isolated from the provinces of Yunnan (n = 31; southwestern China), Guangxi (n = 44; southern China), Gansu (n = 1; northwestern China), and Liaoning (n = 5; northeastern China) (11, 15, 17, 26, 32, 48, 49). Samples from Yunnan province were collected mainly from Kunming, Honghe, and Wenshan prefectures, while isolates from Baise city, Binyang, and Pingxiang counties represented the Guangxi autonomous region. Liaoning sequences were derived from the provincial capital Shenyang. The nonrecombinant gag-pol region of CRF08_BC was selected for codon-based alignment, phylogenetic reconstruction, and coalescent analysis (14). This spans the region from p2 in gag to the reverse transcriptase in pol (strain HXB2 nucleotides 1918 to 2852; 921 bp in length) and is of subtype C origin. Similarly, 22 CRF07_BC sequences with known sampling years were retrieved from the database. These CRF07_BC sequences were isolated from Yunnan (n = 10), Xinjiang (n = 6; northwestern China), and Liaoning (n = 6) provinces (11, 17, 22, 32, 38, 48, 49). Except for five sequences from Xinjiang, all of the CRF07_BC nucleotide sequences have been previously reported by our laboratory. Although the Liaoning patients were recruited in the provincial capital Shenyang, interviews with the patients revealed that only one was possibly infected in Liaoning, whereas four patients reported having acquired the infection in Guangdong (southeastern) province and one patient in Sichuan (central) province. The 420-bp nonrecombinant gag region of CRF07_BC, which is of subtype C origin and encompasses the p17 and partial p24 proteins (HXB2 nucleotides 790 to 1218) was aligned for phylogenetic and evolutionary analyses. Demographic information and risk category for the persons from whom studied isolates were obtained are summarized in Table 1.
TABLE 1.
Demographic information and risk category of persons infected with the CRF08_BC and CRF07_BC viruses
| Virus strain and location | No. of sequences | No. of patients at risk for:a
|
|
|---|---|---|---|
| IDU | Sexual behavior | ||
| CRF08_BC | |||
| Yunnan | 31 | 26 | 5 |
| Guangxi | 44 | 44 | |
| Liaoning | 5 | 2 | 3 |
| Gansu | 1 | UNK | UNK |
| CRF07_BC | |||
| Yunnan | 10 | 7 | 3 |
| Xinjiang | 6 | 5 | 1 |
| Liaoningb | 6 | 4 | 2 |
UNK, unknown.
Five patients recruited in Liaoning are reported as having acquired HIV infection outside Liaoning: four in Guangdong and one in Sichuan.
Bayesian MCMC evolutionary analyses.
Genome region-specific rates of evolution were obtained from a reference set of subtype C sequences using BEAST, version 1.4 (7), a program that employs a Markov chain Monte Carlo (MCMC) algorithm to estimate evolutionary parameters. To perform these analyses, subtype C regions of gag-pol and gag that correspond to the nonrecombinant subtype C segments of CRF08_BC and CRF07_BC, respectively, were retrieved from the HIV sequence database. The resulting heterochronous reference data set contains 41 subtype C sequences sampled between 1989 and 2005, a date range of about 17 years. Maximum-likelihood phylogenies were estimated for each subgenomic region of CRF08_BC and CRF07_BC and for the heterochronous subtype C reference data set using PAUP*, version 4.0 beta (40), and BEAST, version 1.4 (7). All nucleotide alignments are available from the authors upon request.
Strict and relaxed molecular clock (5) analyses were performed under the Hasegawa-Kishino-Yano (HKY) (12) and general time-reversible (33) substitution models, with a gamma-distributed model of among-site rate variation with four rate categories (Γ4) (50). Here, the demographic model component of the BEAST analysis was not of interest, and, hence, it was treated as a nuisance parameter. Each analysis was therefore repeated using constant size, exponential, and logistic growth models in order to investigate the degree to which dating estimates are affected by the demographic model chosen (6). Each MCMC analysis was run for 20 million steps and sampled every 10,000 states. The posterior densities were calculated with 10% burn-in and checked for convergence using Tracer, version 1.4 (28). The posterior distribution of evolutionary rate obtained from the heterochronous reference data set was subsequently incorporated as a prior distribution for the evolutionary rate in the HIV-1 CRF08_BC and CRF07_BC analyses, thereby placing a timescale on the histories of these strains and enabling an estimation of the time to the most recent common ancestor (tMRCA) for the strains (27).
In order to test the possible effects of intrasubtype recombination on the estimated dates, we applied in BEAST a multiple unlinked locus model that allows independent coalescent pathways for different gene loci (16). The “linked” gag-pol gene was partitioned into two unlinked loci of equivalent length, denoted locus 1 (HXB2 nucleotides 1918 to 2390) and 2 (HXB2 nucleotides 2391 to 2852), and their tMRCAs were estimated in the Bayesian coalescent framework assuming a shared demographic history among all loci. Genealogical correlation between the linked and unlinked loci was evaluated by assessing their divergence dates.
Evolutionary analysis of subtype C.
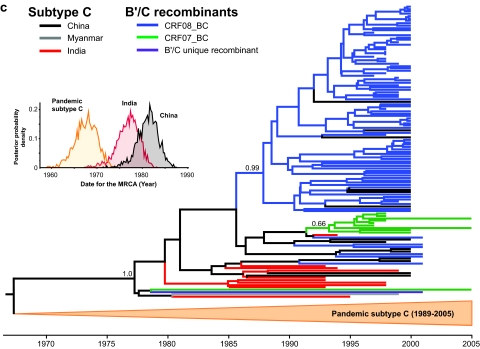
HIV-1 subtype C, probably of Indian origin, was identified as a major circulating strain in China in the early 1990s and is believed to be the putative parent for CRF08_BC and CRF07_BC (19, 20). To assess the divergence times of subtype C in both India and China, “pure” nonrecombinant subtype C genetic regions from 12 nearly full-length subtype C sequences of Indian origin (including a nearly full-length sequence from Myanmar [41]) were retrieved from the HIV Sequence Database. Additionally, 112 subtype C-related env sequences (HXB2 nucleotides 6984 to 7328; 336 bp) isolated from China were retrieved; these Chinese sequences were typed as subtype C (n = 14), CRF08_BC (n = 87), and CRF07_BC (n = 10) and one B′/C unique recombinant form. The methods described above were used to estimate phylogenies, genome region-specific evolutionary rates, and tMRCAs of the Indian and Chinese strains from the gag-pol and/or env genes. Furthermore, select subtype C sequences from other localities (31 full-length sequences from Africa and also Brazil) were also studied in order to assess the time of origin of the global HIV-1 subtype C.
RESULTS
A total of 103 HIV-1 B′/C circulating recombinant forms were retrieved from at least five provinces/autonomous regions representing a relatively wide range of locations across mainland China, three of which have the highest recorded HIV prevalences in the country (Yunnan, Guangxi, and Xinjiang) (23). Nucleotide sequences of the nonrecombinant regions of HIV-1 CRF08_BC (n = 81) and CRF07_BC (n = 22) from IDU (86.3%) and sexual risk patients (13.7%) were selected, codon aligned, and analyzed using likelihood and Bayesian approaches to determine their evolutionary relationships in various regions. Additionally, subtype C or subtype C-related sequences from India and China (n = 124) were also analyzed to determine the time of origin of these lineages.
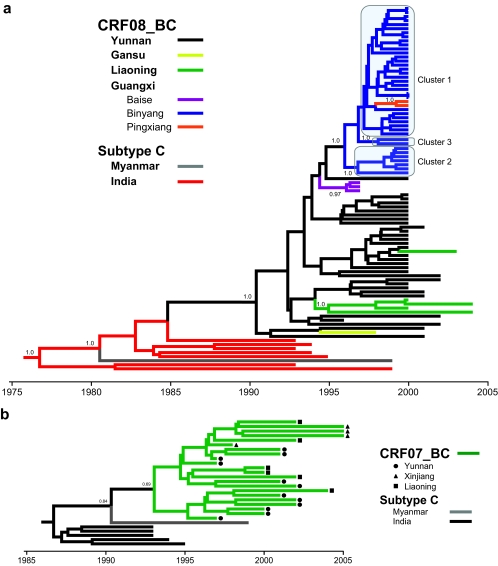
Phylogenetic reconstruction of the gag-pol gene (Fig. 1a) showed that all CRF08_BC sequences grouped in a single clade containing isolates from four Chinese regions: Yunnan, Gansu, Liaoning, and Guangxi (Baise city and Binyang and Pingxiang counties). These sequences were descended from subtype C isolates of Indian origin, confirming the parental relationship of Indian subtype C with respect to CRF08_BC. Yunnan sequences collected from different prefectures (Kunming, Honghe, and Wenshan) (49) were largely intermingled with sequences from Gansu, Liaoning, and Guangxi (Baise), suggesting close relationships among these sequences. However, other sequences from Binyang county (and also Pingxiang) in Guangxi formed a distinct monophyletic cluster that diverged from the Yunnan/Baise isolates and further bifurcated into three major clusters (indicated as clusters 1 to 3). Cluster 1 contained sequences from Binyang (n = 30) and Pingxiang (n = 2), while clusters 2 and 3 contained seven and two Binyang sequences, respectively. The robustness of each cluster was supported by high posterior clade probabilities (P > 0.95). Similarly, CRF08_BC sequences from Liaoning province (except that of one subject who acquired the infection in Yunnan) also formed a single cluster.
FIG. 1.
Maximum-likelihood phylogenetic analyses of HIV-1 CRF08_BC, CRF07_BC, and subtype C in China. (a) Phylogenetic reconstructions of CRF08_BC gag-pol genes (HXB2 nucleotides 1918 to 2852) isolated from HIV-1 patients in the Yunnan (Kunming, Honghe, and Wenshan), Gansu, Liaoning (Shenyang), and Guangxi (Baise, Binyang, and Pingxiang) regions of China. (b) Maximum clade credibility trees of the HIV-1 CRF07_BC sequences from Yunnan, Xinjiang, and Liaoning provinces, based on the gag gene (HXB2 nucleotides 790 to 1218). (c) The env (HXB2 nucleotides 6984 to 7328) phylogeny of subtype C, CRF08_BC, CRF07_BC, and a B′/C unique recombinant form isolated from China. Ancestral relationships are estimated using PAUP*, version 4.0 beta (40), and BEAST, version 1.4 (7). Subtype C of Indian origin, thought to be the putative parent of both CRF08_BC and CRF07_BC (19, 20), is also included. The CRF08_BC gag-pol and CRF07_BC gag tree branches are colored according to their respective geographical locations while the env tree branches show the respective HIV-1 subtype/CRF. Posterior probabilities greater than 0.5 are shown at their respective nodes.
Bayesian analyses under different evolutionary models estimated consistent mean rates of evolution (μ) for the CRF08_BC gag-pol gene, in the range of 1.7 × 10−3 to 1.8 × 10−3 substitutions/site/year. This range of rates was included as a prior probability distribution in subsequent analyses in order to calculate the tMRCA of the respective CRF08_BC clusters. When the constant size population model and HKY substitution model were used, the Yunnan (and Gansu) clusters were dated to 1990.3 (95% credible region [CR], 1988.6 to 1991.9) (Table 2). CRF08_BC was probably introduced later into Baise city in neighboring Guangxi around 1995.5 (95% CR, 1994.3 to 1996.5). This is in agreement with previous epidemiological investigations, which reported the first outbreaks of CRF08_BC (then classified as subtype C based on env gene sequencing) in this area (26, 51). Interestingly, the tMRCA of the Liaoning cluster is estimated at 1995.6 (95% CR, 1993.4 to 1997.5), comparable to that of Guangxi, and sequences from both these regions trace their origins to Yunnan. This suggests that the epidemics in Liaoning, an area with low HIV/AIDS prevalence, and Guangxi could have started at almost the same time. As shown in the gag-pol phylogeny (Fig. 1a), subsequent spread of CRF08_BC among the IDU populations in Binyang and Pingxiang involved at least three viral lineages (clusters 1 to 3) descended from the Yunnan or Baise sequences between 1997 and 1999 (Table 2). In addition, all Binyang and Pingxiang sequences contained signature amino acid deletions in gag (p7, p1, and p6) and in reverse transcriptase at position P25 (amino acid alignment not shown) that were not found in other CRF08_BC sequences, suggesting a shared common ancestor for these strains. Overall, the evolutionary and statistical assumptions applied in the coalescence analyses have almost no effect on the estimated dates (results generated under the general time-reversible nucleotide substitution model are shown in Table S1 in the supplemental material). In addition to strict molecular clock analyses, we also explored the relaxed phylogenetic method (5) to estimate the divergence times for each data set. The results, under selected evolutionary priors, were comparable to those described above (see Fig. S1 in the supplemental material for a summary). Our analysis reconstructs the chronological events of HIV-1 expansion by retracing the north-eastward and eastward migration of CRF08_BC to Liaoning and Guangxi, respectively, most likely from a common source in Yunnan province. However, we note that the estimated coalescence times, particularly for clusters with a small number of sequences, may be biased and hence more recent than the date of outbreak. This is because, among other factors, the extant virus diversity has been partially sampled; alternatively, the founding/parental lineages of the outbreak have since become extinct or are yet to be discovered (1, 43).
TABLE 2.
Evolutionary characteristics of CRF08_BC, CRF07_BC, and subtype C
| HIV-1 strain or subtype and location | Genetic region | Value (HPD) of the evolutionary parameter according to the indicated model:a
|
|||||
|---|---|---|---|---|---|---|---|
| HKY+Γ4 constant size
|
HKY+Γ4 exponential growth
|
HKY+Γ4 logistic growth
|
|||||
| Rate of evolution | tMRCA | Rate of evolution | tMRCA | Rate of evolution | tMRCA | ||
| CRF08_BC | |||||||
| Yunnan and Gansu | gag-pol | 1.8 (1.4, 2.3) | 1990.3 (1988.6, 1991.9) | 1.8 (1.4, 2.3) | 1989.8 (1988.0, 1991.6) | 1.7 (1.3, 2.1) | 1989.3 (1987.3, 1991.0) |
| Liaoning | 1995.6 (1993.4, 1997.5) | 1995.2 (1993.0, 1997.4) | 1995.1 (1992.6, 1997.3) | ||||
| Guangxi | |||||||
| Baise | 1995.5 (1994.3, 1996.5) | 1995.3 (1994.1, 1996.6) | 1995.3 (1993.8, 1996.4) | ||||
| Binyang | |||||||
| Cluster 1 | 1997.1 (1996.3, 1997.9) | 1996.9 (1996.0, 1997.7) | 1996.8 (1995.9, 1997.6) | ||||
| Cluster 2 | 1998.3 (1997.0, 1999.4) | 1998.0 (1996.6, 1999.3) | 1998.0 (1996.6, 1999.3) | ||||
| Cluster 3 | 1998.5 (1997.3, 1999.5) | 1998.4 (1997.1, 1999.5) | 1998.3 (1997.1, 1999.4) | ||||
| Pingxiang | 1999.3 (1998.5, 1999.9) | 1999.2 (1998.3, 1999.9) | 1999.2 (1998.3, 1999.9) | ||||
| CRF07_BC | |||||||
| Yunnan, Xinjiang, and Liaoningb | gag | 4.4 (3.1, 5.7) | 1993.3 (1991.2, 1995.2) | 3.5 (2.1, 4.7) | 1989.4 (1986.2, 1992.2) | 3.9 (2.6, 5.1) | 1992.0 (1989.5, 1994.3) |
| Taiwanc | |||||||
| Tainan (Southern) | env | 4.8 (3.0, 6.6) | 1999.7 (1998.2, 2001.1) | 4.9 (3.0, 6.6) | 1999.7 (1998.2, 2001.0) | 4.2 (2.6, 5.9) | 1999.1 (1997.5, 2000.6) |
| Taipei and Nantou (northern-central) | 2002.1 (2001.3, 2002.9) | 2002.1 (2001.1, 2002.8) | 2001.7 (2000.7, 2002.7) | ||||
| Subtype C | |||||||
| Global | gag-pol | 1.8 (1.4, 2.3) | 1967.6 (1962.5, 1972.0) | 1.8 (1.4, 2.3) | 1966.9 (1962.1, 1971.6) | 1.7 (1.3, 2.1) | 1970.3 (1967.0, 1973.7) |
| India | gag-pol | 1.8 (1.4, 2.3) | 1979.7 (1976.9, 1982.3) | 1.8 (1.4, 2.3) | 1979.5 (1976.3, 1982.6) | 1.7 (1.3, 2.1) | 1977.8 (1974.7, 1980.7) |
| env | 5.6 (3.1, 7.6) | 1976.9 (1972.1, 1981.5) | 5.2 (3.1, 7.0) | 1978.7 (1974.8, 1982.8) | 5.2 (3.3, 7.3) | 1978.2 (1973.9, 1982.1) | |
| China | env | 5.6 (3.1, 7.6) | 1981.2 (1976.7, 1985.9) | 5.2 (3.1, 7.0) | 1982.3 (1978.8, 1985.5) | 5.2 (3.3, 7.3) | 1982.0 (1978.1, 1985.2) |
Rates of evolution are expressed as 10−3 nucleotide substitutions per site per year. The 95% highest posterior density (HPD) confidence intervals are given in parentheses.
Five patients recruited in Liaoning are reported as having acquired HIV infection outside Liaoning (see Table 1).
Constant size and exponential growth analyses based on the hypervariable region-stripped env gene previously described (43).
Conversely, all gag sequences of CRF07_BC from Yunnan, Xinjiang, and Liaoning formed a single cluster, with subtype C isolates of Indian origin paraphyletic with respect to CRF07_BC (Fig. 1b). Coalescence analyses under a constant size population model (estimated evolutionary rate, μ = 4.4 × 10−3 substitutions/site/year) imply that the common ancestor of the Chinese CRF07_BC isolates existed around 1993.3 (95% CR, 1991.2 to 1995.2) (Table 2). This is in agreement with previous studies, based on the env gene, which dated the divergence time of CRF07_BC to about 1993 (43). The exponential and logistic demographic model gave slightly lower substitution rates and hence older MRCA estimates. Harmonic means of the coalescent likelihood estimated using marginal likelihood analysis (39), however, showed that other tree models do not significantly fit the gag data set better than the constant size population model (log10 Bayes factors of <3.0), suggesting that there could be insufficient information in the data to fit a more complex model. The logistic model meanwhile was strongly supported over the exponential model (log10 Bayes factors of 4.4). In addition, it is important to note that among the six Liaoning sequences, only one patient reported having acquired the infection in Liaoning while other patients referred to Guangdong or Sichuan provinces as the likely location of HIV infection, mainly through intravenous drug use. The evidence, therefore, suggests that CRF07_BC sequences from Xinjiang, Liaoning, Guangdong, and Sichuan can probably be traced to a common origin in Yunnan province, a similar pattern to that observed for the CRF08_BC sequences from different regions.
Maximum clade credibility trees revealed that the env sequences of subtype C (from India, China, and Myanmar) and of CRF08_BC, CRF07_BC, and other subtype C-related recombinants from China were intermingled in a single cluster, indicating a shared common ancestry, with subtype C strains of Indian origin as outgroup founder strains (Fig. 1c). Coalescence analyses of the gag-pol and env genes dated the tMRCA of subtype C strains from India and Myanmar to around the mid- to late 1970s (Table 2). Analysis of 14 env sequences suggests that subtype C spread into neighboring China later, in 1981.2 (95% CR, 1976.7 to 1985.9), most likely in Yunnan province. Together, the phylogenetic and coalescent results map the eastward movement of subtype C from India to China, probably through Myanmar (2), between the mid-1970s and early 1980s. Finally, the time of origin of the global HIV-1 subtype C was dated 1967.6 (95% CR, 1962.5 to 1972.0) and was highly consistent with previous estimates (44).
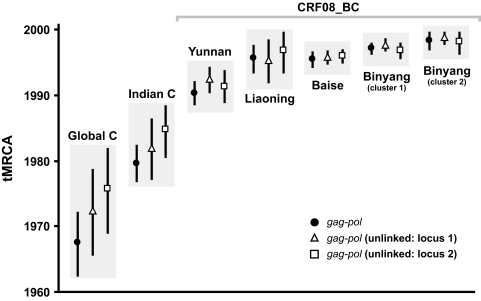
To evaluate the possible effects of intrasubtype recombination on the tMRCA estimates, we considered a multilocus model (16) that assumes different genealogies for each locus of the unlinked gag-pol data set. BEAST analyses revealed that the tree topologies (figures not shown) and the estimated tMRCAs for the unlinked loci were not significantly different from those of the linked gag-pol locus among the overlapping 95% CRs for each locus, especially in those states with younger tMRCAs (Fig. 2). The results suggest that intrasubtype recombination within HIV-1 subtype C and CRF08_BC may not have significant biases on the MRCA dates (16).
FIG. 2.
Dates of the MRCA of HIV-1 subtype C and CRF08_BC estimated in a multiple unlinked locus model. The gag-pol gene was partitioned (locus 1 and 2), and Bayesian estimation for each locus was performed in BEAST, assuming a shared demographic history among all loci. The mean coalescence time estimates with 95% highest posterior density for HIV subtype C and CRF08_BC from various geographical origins are illustrated.
DISCUSSION
To our knowledge, this is the first genealogy-based population genetic study of the divergence times of HIV-1 CRF08_BC and CRF07_BC across China. CRF08_BC and CRF07_BC are two subtype C-related recombinants with significant epidemic impact in China and beyond (42, 43). Prior to the emergence of CRF08_BC and CRF07_BC (26, 38), subtype C isolates closely related to Indian strains predominated, especially in the southern part of China (19). Later, subtype C recombined with subtype B′ (10, 45, 47) to form these recombinants. Both recombinants, long thought to have originated from a common birthplace in Yunnan province (20, 49), later dispersed to other areas primarily through drug use and trafficking activities (2). The present study provides new information on the evolutionary and epidemiological characteristics of HIV-1 by establishing the plausible dates of origin and geographic migration patterns of CRF08_BC and CRF07_BC across China. Although the early Indian subtype C strains play no major role in today's epidemic in China, the date of introduction of subtype C into China helps to provide a more complete picture of the epidemic history of subtype C and its related recombinants in East Asia (Fig. 3).
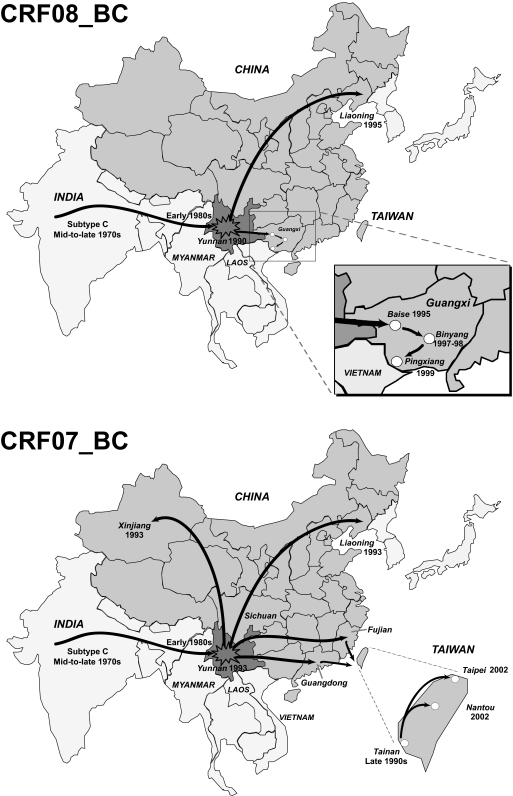
FIG. 3.
Plausible site of origin and migration routes of HIV-1 CRF08_BC and CRF07_BC. HIV-1 subtype C of Indian origin entered the Yunnan province (darker shade) in southern China in the early 1980s, possibly via Myanmar. Cocirculation of subtype C and the endemic subtype B′ (10, 45) led to genetic recombination and the generation of CRF08_BC (26) and CRF07_BC (38), two related but distinct B′/C recombinants in Yunnan. Further spread of CRF08_BC and CRF07_BC into other regions in the 1990s mainly through injection drug use (2) has been implicated as the major force spurring the HIV/AIDS epidemic in China. The tMRCAs of CRF08_BC, CRF07_BC, and subtype C strains representing different geographical locations are summarized in Table 2.
The Bayesian coalescence analyses show that CRF08_BC emerged in Yunnan province of China in the early 1990s. The virus then spread eastward to neighboring Guangxi (in Baise city near the Yunnan-Guangxi border) in the mid-1990s; this is consistent with historical accounts of CRF08_BC among IDU in this region (24). Inside Guangxi, highly homogeneous CRF08_BC strains from Baise (26, 52) were multiply introduced into Binyang county in 1996 to 1999 and are predominant among IDU (15). Moreover, the relatively recent introduction of CRF08_BC into Pingxiang in the late 1990s estimated here suggests a southward distribution of CRF08_BC near the China-Vietnam border, although CRF01_AE strains closely related to strains of northern Vietnam account for the majority of IDU infections in this region (13, 15). It is also interesting that the CRF08_BC epidemic in Liaoning province, a geographically remote area with low HIV prevalence located in northeastern China, started at almost the same time as that in Guangxi. Our results imply a rapid and simultaneous expansion of CRF08_BC from a common origin in Yunnan to Guangxi and as far as Liaoning province. Although a general trend of migration has been observed, it is essential to stress that the estimated dates—which merely imply the coalescence times of select viral strains—might not accurately reflect the actual time of CRF08_BC spread in a given region, especially when the sampled population clusters are small. Until more archival specimens are retrieved and thoroughly studied, the complex temporal movement of CRF08_BC lineages (and also other subtypes/CRFs) must be interpreted with discretion.
Phylogenetic reconstruction and coalescence inference show that the CRF07_BC isolates from different mainland Chinese regions (including Xinjiang, Liaoning, and probably Guangdong and Sichuan) most likely share a common ancestor that existed in 1993 in Yunnan province (20, 49). However, it is noteworthy that a single CRF07_BC sequence and a unique B′/C recombinant sequence were located at the base of the env maximum-likelihood tree, along with Indian subtype C sequences. This could be explained by two possibilities: (i) there is a recombination breakpoint within the region studied that rendered the phylogenetic inference inaccurate, or (ii) this is a new recombinant that has acquired an Indian subtype C env gene. Since no recombination breakpoint is identified within this region, it is likely that a recombination event in the env gene involving CRF07_BC and a “founder” Indian C lineage has occurred before the sequence was sampled (where the putative recombination breakpoints are probably located outside the studied region), a phenomenon which is not uncommon (49). Similar explanations also can be applied for the few CRF08_BC sequences that grouped outside the monophyletic cluster in the env region.
We previously investigated the spatiotemporal spread of CRF07_BC in Taiwan using env gene sequences (43). CRF07_BC was first introduced into southern Taiwan in the late 1990s, most probably from southwestern China (18), and later spread to the central-northern part of Taiwan in the early 2000s, resulting in the largest-ever HIV epidemic in Taiwan, mostly affecting IDU populations. Adding these results to our new analyses performed here, we have retraced the expansive migration of HIV-1 CRF07_BC from its origin in Yunnan to (a) Xinjiang by the north-westward drug trafficking paths, (b) to Liaoning either by direct introduction from Yunnan or through Guangdong/Sichuan, and (c) to Taiwan, possibly through southeastern China via drug trafficking routes.
Recombination is common in HIV infection (31) and serves as one of the intrinsic evolutionary mechanisms that shape the complex HIV diversity. Besides intersubtype recombination, recombination involving closely related lineages of the same subtype within a single individual has been reported (25, 34, 36), leading to possible quantitative discrepancies on linkage disequilibrium, including the loss of phylogenetic correlation between different loci and biases in the genealogical characteristics (i.e., overestimation of the tMRCA) along the HIV genome (35, 46). Our study, however, showed that intrasubtype recombination has no significant impact on the estimated coalescence dates (Fig. 2)—a feature previously observed in HIV-1 group O (16). This could possibly be explained by the high growth rates of HIV-1, as characterized by its star-like population phylogeny (29) where the terminal branches form a substantial proportion of a tree. In the rapidly growing HIV-1 population, recombination events are more likely to affect the terminal branches by increasing the variance in mutation rates rather than biasing the tree topology (16, 21). Consequently, intrasubtype recombination, despite occurring at high frequency, may not significantly influence the divergence times of HIV.
The currently adopted HIV-1 nomenclature guidelines classify HIV-1 CRFs sequentially, according to the first described full-length or nearly full-length genetic sequences obtained (30), without taking into account the evolutionary history of the viruses (1). Our studies of all available CRF08_BC and CRF07_BC sequences with known sampling dates have shown that CRF08_BC has an earlier evolutionary history than CRF07_BC, which is not readily indicated by the current naming system. We show that CRF08_BC was probably generated approximately 10 years after subtype C of Indian origin was first introduced to Yunnan province, around 1981. CRF07_BC was formed in the same province a few years later, around 1993. The findings highlight the feasibility and relevance of incorporating evolutionary population biology into HIV-1 classification to better reflect the evolutionary pathways of the viruses, particularly for those recombinants with close phylogenetic, genealogical and geographical links (4, 37).
Here, we reconstruct the epidemic histories of HIV-1 CRF08_BC, CRF07_BC and related sequences, inside and beyond China. Injection drug use remains a major transmission mode for HIV-1 CRF08_BC and CRF07_BC throughout China. Phylodynamic investigations are therefore important in unifying the epidemiological and evolutionary dynamics of the highly divergent and rapidly expanding HIV-1 subtypes and CRFs in Asia and elsewhere.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge Philippe Lemey and Chung-Chau Hon for assistance, Naoki Yamamoto for support, and Timothy D. Mastro for critical reading of the manuscript. We also thank the anonymous reviewers for their insightful comments.
This study was supported in part by grants from the Ministry of Health, Labour and Welfare (H18-AIDS-General-016); Ministry of Education, Culture, Sports, Science and Technology (Overseas Research Fund); the Japanese Foundation for AIDS Prevention (JFAP) to Y.T.; the Ministry of Science, Technology and Innovation (eScienceFund 02-01-03-SF0379) to A.K.; and the Royal Society University Research Fellowship to O.G.P. and Y.T. K.K.T. is a recipient of the JFAP research resident fellowship.
Footnotes
Published ahead of print on 2 July 2008.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Abecasis, A. B., P. Lemey, N. Vidal, T. de Oliveira, M. Peeters, R. Camacho, B. Shapiro, A. Rambaut, and A. M. Vandamme. 2007. Recombination confounds the early evolutionary history of human immunodeficiency virus type 1: subtype G is a circulating recombinant form. J. Virol. 818543-8551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beyrer, C., M. H. Razak, K. Lisam, J. Chen, W. Lui, and X. F. Yu. 2000. Overland heroin trafficking routes and HIV-1 spread in South and South-East Asia. AIDS 1475-83. [DOI] [PubMed] [Google Scholar]
- 3.Cheng, H., J. Zhang, J. Capizzi, N. L. Young, and T. D. Mastro. 1994. HIV-1 subtype E in Yunnan, China. Lancet 344953-954. [DOI] [PubMed] [Google Scholar]
- 4.De Sa Filho, D. J., M. C. Sucupira, M. M. Caseiro, E. C. Sabino, R. S. Diaz, and L. M. Janini. 2006. Identification of two HIV type 1 circulating recombinant forms in Brazil. AIDS Res. Hum. Retrovir. 221-13. [DOI] [PubMed] [Google Scholar]
- 5.Drummond, A. J., S. Y. Ho, M. J. Phillips, and A. Rambaut. 2006. Relaxed phylogenetics and dating with confidence. PLoS Biol. 4e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drummond, A. J., G. K. Nicholls, A. G. Rodrigo, and W. Solomon. 2002. Estimating mutation parameters, population history and genealogy simultaneously from temporally spaced sequence data. Genetics 1611307-1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drummond, A. J., and A. Rambaut. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gifford, R. J., T. de Oliveira, A. Rambaut, O. G. Pybus, D. Dunn, A. M. Vandamme, P. Kellam, and D. Pillay. 2007. Phylogenetic surveillance of viral genetic diversity and the evolving molecular epidemiology of human immunodeficiency virus type 1. J. Virol. 8113050-13056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gilbert, M. T., A. Rambaut, G. Wlasiuk, T. J. Spira, A. E. Pitchenik, and M. Worobey. 2007. The emergence of HIV/AIDS in the Americas and beyond. Proc. Natl. Acad. Sci. USA 10418566-18570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Graf, M., Y. Shao, Q. Zhao, T. Seidl, J. Kostler, H. Wolf, and R. Wagner. 1998. Cloning and characterization of a virtually full-length HIV type 1 genome from a subtype B′-Thai strain representing the most prevalent B-clade isolate in China. AIDS Res. Hum. Retrovir. 14285-288. [DOI] [PubMed] [Google Scholar]
- 11.Han, X., M. Zhang, D. Dai, Y. Wang, Z. Zhang, J. Liu, W. Geng, Y. Jiang, Y. Takebe, and H. Shang. 2007. Genotypic resistance mutations to antiretroviral drugs in treatment-naive HIV/AIDS patients living in Liaoning Province, China: baseline prevalence and subtype-specific difference. AIDS Res. Hum. Retrovir. 23357-364. [DOI] [PubMed] [Google Scholar]
- 12.Hasegawa, M., H. Kishino, and T. Yano. 1985. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 22160-174. [DOI] [PubMed] [Google Scholar]
- 13.Kato, K., T. Shiino, S. Kusagawa, H. Sato, K. Nohtomi, K. Shibamura, T. H. Nguyen, K. C. Pham, X. L. Truong, H. A. Mai, T. L. Hoang, G. Bunyaraksyotin, Y. Fukushima, M. Honda, C. Wasi, S. Yamazaki, Y. Nagai, and Y. Takebe. 1999. Genetic similarity of HIV type 1 subtype E in a recent outbreak among injecting drug users in northern Vietnam to strains in Guangxi Province of southern China. AIDS Res. Hum. Retrovir. 151157-1168. [DOI] [PubMed] [Google Scholar]
- 14.Kingman, J. F. C. 1982. The coalescent. Stochastic Processes Appl. 13235-248. [Google Scholar]
- 15.Laeyendecker, O., G. W. Zhang, T. C. Quinn, R. Garten, S. C. Ray, S. Lai, W. Liu, J. Chen, and X. F. Yu. 2005. Molecular epidemiology of HIV-1 subtypes in southern China. J. Acquir. Immune Defic. Syndr. 38356-362. [PubMed] [Google Scholar]
- 16.Lemey, P., O. G. Pybus, A. Rambaut, A. J. Drummond, D. L. Robertson, P. Roques, M. Worobey, and A. M. Vandamme. 2004. The molecular population genetics of HIV-1 group O. Genetics 1671059-1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li, X. J., S. Kusagawa, X. Xia, C. Yang, Q. Wang, Y. Yokota, Y. Hoshina, T. Onogi, K. Nohtomi, Y. Imamura, T. Shiino, R. Yang, N. Yamamoto, K. Ben, and Y. Takebe. 2005. Molecular epidemiology of the heterosexual HIV-1 transmission in Kunming, Yunnan Province of China suggests origin from the local IDU epidemic. AIDS Res. Hum. Retrovir. 21977-980. [DOI] [PubMed] [Google Scholar]
- 18.Lin, H. H., Y. L. Shih, Y. C. Liu, S. S. Lee, C. K. Huang, Y. L. Chen, C. Chin, C. H. Lai, H. C. Tsai, Y. C. Guo, and L. Zhang. 2006. An epidemic of HIV type I CRF07_BC infection among injection drug users in Taiwan. J. Acquir. Immune Defic. Syndr. 42248-255. [DOI] [PubMed] [Google Scholar]
- 19.Luo, C. C., C. Tian, D. J. Hu, M. Kai, T. Dondero, and X. Zheng. 1995. HIV-1 subtype C in China. Lancet 3451051-1052. [DOI] [PubMed] [Google Scholar]
- 20.McClutchan, F. E., J. K. Carr, D. Murphy, S. Piyasirisilp, F. Gao, B. Hahn, X. F. Yu, C. Beyrer, and D. L. Birx. 2002. Precise mapping of recombination breakpoints suggests a common parent of two BC recombinant HIV type 1 strains circulating in China. AIDS Res. Hum. Retrovir. 181135-1140. [DOI] [PubMed] [Google Scholar]
- 21.McVean, G. A. 2002. A genealogical interpretation of linkage disequilibrium. Genetics 162987-991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meng, Z., H. Xing, X. He, L. Ma, W. Xu, and Y. Shao. 2007. Genetic characterization of three newly isolated CRF07_BC near full-length genomes in China. AIDS Res. Hum. Retrovir. 231049-1054. [DOI] [PubMed] [Google Scholar]
- 23.Ministry of Health, People's Republic of China, Joint United Nations Programme on HIV/AIDs, and the World Health Organization. 2006. 2005 Update on the HIV/AIDS epidemic and response in China. National Center for AIDS/STD Prevention and Control, China CDC, Beijing, People's Republic of China.
- 24.Ministry of Health, People's Republic of China, and United Nations Theme Group on HIV/AIDS in China. 1997. China responds to AIDS: HIV/AIDS situation and needs assessment report. Chinese Ministry of Health, Beijing, People's Republic of China.
- 25.Philpott, S., H. Burger, C. Tsoukas, B. Foley, K. Anastos, C. Kitchen, and B. Weiser. 2005. Human immunodeficiency virus type 1 genomic RNA sequences in the female genital tract and blood: compartmentalization and intrapatient recombination. J. Virol. 79353-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Piyasirisilp, S., F. E. McCutchan, J. K. Carr, E. Sanders-Buell, W. Liu, J. Chen, R. Wagner, H. Wolf, Y. Shao, S. Lai, C. Beyrer, and X. F. Yu. 2000. A recent outbreak of human immunodeficiency virus type 1 infection in southern China was initiated by two highly homogeneous, geographically separated strains, circulating recombinant form AE and a novel BC recombinant. J. Virol. 7411286-11295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pybus, O. G., A. J. Drummond, T. Nakano, B. H. Robertson, and A. Rambaut. 2003. The epidemiology and iatrogenic transmission of hepatitis C virus in Egypt: a Bayesian coalescent approach. Mol. Biol. Evol. 20381-387. [DOI] [PubMed] [Google Scholar]
- 28.Rambaut, A., and A. J. Drummond. 2007. Tracer v1.4. Institute of Evolutionary Biology, University of Edinburgh, Edinburgh, Scotland. http://tree.bio.ed.ac.uk.
- 29.Robbins, K. E., P. Lemey, O. G. Pybus, H. W. Jaffe, A. S. Youngpairoj, T. M. Brown, M. Salemi, A. M. Vandamme, and M. L. Kalish. 2003. U.S. Human immunodeficiency virus type 1 epidemic: date of origin, population history, and characterization of early strains. J. Virol. 776359-6366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robertson, D. L., J. P. Anderson, J. A. Bradac, J. K. Carr, B. Foley, R. K. Funkhouser, F. Gao, B. H. Hahn, M. L. Kalish, C. Kuiken, G. H. Learn, T. Leitner, F. McCutchan, S. Osmanov, M. Peeters, D. Pieniazek, M. Salminen, P. M. Sharp, S. Wolinsky, and B. Korber. 2000. HIV-1 nomenclature proposal. Science 28855-56. [DOI] [PubMed] [Google Scholar]
- 31.Robertson, D. L., B. H. Hahn, and P. M. Sharp. 1995. Recombination in AIDS viruses. J. Mol. Evol. 40249-259. [DOI] [PubMed] [Google Scholar]
- 32.Rodenburg, C. M., Y. Li, S. A. Trask, Y. Chen, J. Decker, D. L. Robertson, M. L. Kalish, G. M. Shaw, S. Allen, B. H. Hahn, and F. Gao. 2001. Near full-length clones and reference sequences for subtype C isolates of HIV type 1 from three different continents. AIDS Res. Hum. Retrovir. 17161-168. [DOI] [PubMed] [Google Scholar]
- 33.Rodriguez, F., J. L. Oliver, A. Marin, and J. R. Medina. 1990. The general stochastic model of nucleotide substitution. J. Theor. Biol. 142485-501. [DOI] [PubMed] [Google Scholar]
- 34.Rousseau, C. M., G. H. Learn, T. Bhattacharya, D. C. Nickle, D. Heckerman, S. Chetty, C. Brander, P. J. Goulder, B. D. Walker, P. Kiepiela, B. T. Korber, and J. I. Mullins. 2007. Extensive intrasubtype recombination in South African human immunodeficiency virus type 1 subtype C infections. J. Virol. 814492-4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schierup, M. H., and J. Hein. 2000. Consequences of recombination on traditional phylogenetic analysis. Genetics 156879-891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shriner, D., A. G. Rodrigo, D. C. Nickle, and J. I. Mullins. 2004. Pervasive genomic recombination of HIV-1 in vivo. Genetics 1671573-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sierra, M., M. M. Thomson, D. Posada, L. Perez, C. Aragones, Z. Gonzalez, J. Perez, G. Casado, and R. Najera. 2007. Identification of 3 phylogenetically related HIV-1 BG intersubtype circulating recombinant forms in Cuba. J. Acquir. Immune Defic. Syndr. 45151-160. [DOI] [PubMed] [Google Scholar]
- 38.Su, L., M. Graf, Y. Zhang, H. von Briesen, H. Xing, J. Kostler, H. Melzl, H. Wolf, Y. Shao, and R. Wagner. 2000. Characterization of a virtually full-length human immunodeficiency virus type 1 genome of a prevalent intersubtype (C/B′) recombinant strain in China. J. Virol. 7411367-11376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suchard, M. A., R. E. Weiss, and J. S. Sinsheimer. 2001. Bayesian selection of continuous-time Markov chain evolutionary models. Mol. Biol. Evol. 181001-1013. [DOI] [PubMed] [Google Scholar]
- 40.Swofford, D. L. 2003. PAUP*, phylogenetic analysis using parsimony (*and other methods), 4.0 beta ed. Sinauer Associates, Sunderland, MA.
- 41.Takebe, Y., K. Motomura, M. Tatsumi, H. H. Lwin, M. Zaw, and S. Kusagawa. 2003. High prevalence of diverse forms of HIV-1 intersubtype recombinants in central Myanmar: geographical hot spot of extensive recombination. AIDS 172077-2087. [DOI] [PubMed] [Google Scholar]
- 42.Tebit, D. M., I. Nankya, E. J. Arts, and Y. Gao. 2007. HIV diversity, recombination and disease progression: how does fitness “fit” into the puzzle? AIDS Rev. 975-87. [PubMed] [Google Scholar]
- 43.Tee, K. K., O. G. Pybus, H. Liao, R. Uenishi, S. Hase, A. Kamarulzaman, X. J. Li, and Y. Takebe. 2008. Chronology of the HIV-1 CRF07_BC expansion in East Asia. AIDS 22156-158. [DOI] [PubMed] [Google Scholar]
- 44.Travers, S. A., J. P. Clewley, J. R. Glynn, P. E. Fine, A. C. Crampin, F. Sibande, D. Mulawa, J. O. McInerney, and G. P. McCormack. 2004. Timing and reconstruction of the most recent common ancestor of the subtype C clade of human immunodeficiency virus type 1. J. Virol. 7810501-10506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weniger, B. G., Y. Takebe, C. Y. Ou, and S. Yamazaki. 1994. The molecular epidemiology of HIV in Asia. AIDS 8(Suppl. 2)S13-S28. [PubMed] [Google Scholar]
- 46.Worobey, M. 2001. A novel approach to detecting and measuring recombination: new insights into evolution in viruses, bacteria, and mitochondria. Mol. Biol. Evol. 181425-1434. [DOI] [PubMed] [Google Scholar]
- 47.Xia, M., J. K. Kreiss, and K. K. Holmes. 1994. Risk factors for HIV infection among drug users in Yunnan province, China: association with intravenous drug use and protective effect of boiling reusable needles and syringes. AIDS 81701-1706. [PubMed] [Google Scholar]
- 48.Yang, R., S. Kusagawa, C. Zhang, X. Xia, K. Ben, and Y. Takebe. 2003. Identification and characterization of a new class of human immunodeficiency virus type 1 recombinants comprised of two circulating recombinant forms, CRF07_BC and CRF08_BC, in China. J. Virol. 77685-695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang, R., X. Xia, S. Kusagawa, C. Zhang, K. Ben, and Y. Takebe. 2002. On-going generation of multiple forms of HIV-1 intersubtype recombinants in the Yunnan Province of China. AIDS 161401-1407. [DOI] [PubMed] [Google Scholar]
- 50.Yang, Z. 1994. Maximum likelihood phylogenetic estimation from DNA sequences with variable rates over sites: approximate methods. J. Mol. Evol. 39306-314. [DOI] [PubMed] [Google Scholar]
- 51.Yu, X. F., J. Chen, Y. Shao, C. Beyrer, and S. Lai. 1998. Two subtypes of HIV-1 among injection-drug users in southern China. Lancet 3511250. [DOI] [PubMed] [Google Scholar]
- 52.Yu, X. F., W. Liu, J. Chen, W. Kong, B. Liu, Q. Zhu, F. Liang, F. McCutchan, S. Piyasirisilp, and S. Lai. 2002. Maintaining low HIV type 1 env genetic diversity among injection drug users infected with a B/C recombinant and CRF01_AE HIV type 1 in southern China. AIDS Res. Hum Retrovir. 18167-170. [DOI] [PubMed] [Google Scholar]
- 53.Zheng, X., C. Tian, K. H. Choi, J. Zhang, H. Cheng, X. Yang, D. Li, J. Lin, S. Qu, X. Sun, T. Hall, J. Mandel, and N. Hearst. 1994. Injecting drug use and HIV infection in southwest China. AIDS 81141-1147. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.